Abstract
Background & Aims
Intestinal metaplasia (Barrett's esophagus, BE) is the principal risk factor for esophageal adenocarcinoma (EAC). Study of the basis for BE has centered on intestinal factors, but loss of esophageal identity likely also reflects absence of key squamous-cell factors. As few determinants of stratified epithelial cell-specific gene expression are characterized, it is important to identify the necessary transcription factors.
Methods
We tested regional expression of mRNAs for all putative DNA-binding proteins in the mouse digestive tract and verified esophagus-specific factors in human tissues and cell lines. Integration of diverse data defined a human squamous esophagus-specific transcriptome. We used chromatin immunoprecipitation (ChIP-seq) to locate transcription factor binding sites, computational approaches to profile transcripts in cancer datasets, and immunohistochemistry to reveal protein expression.
Results
The transcription factor SOX15 is restricted to esophageal and other murine and human stratified epithelia. SOX15 mRNA levels are attenuated in BE and its depletion in human esophageal cells reduced esophageal transcripts significantly and specifically. SOX15 binding is highly enriched near esophagus-expressed genes, indicating direct transcriptional control. SOX15 and hundreds of genes co-expressed in squamous cells are reactivated in up to 30% of EAC specimens. Genes normally confined to the esophagus or intestine appear in different cells within the same malignant glands.
Conclusions
These data identify a novel transcriptional regulator of stratified epithelial cells and a subtype of EAC with bi-lineage gene expression. Broad activation of squamous-cell genes may shed light on whether EACs arise in the native stratified epithelium or in ectopic columnar cells.
Keywords: Esophageal gene regulation, Barrett's esophagus, esophageal transcriptome, SOX15 cistrome
Introduction
Intestinal metaplasia of the esophagus (Barrett's esophagus, BE) is a common, chronic condition in which an epithelium containing intestinal goblet and other columnar cells replaces the native stratified squamous mucosa (1). BE results from chronic acid and bile reflux. Over time, the metaplastic tissue may become dysplastic and it progresses to invasive cancer in 3 to 5 cases per 1,000 person-years (2). Esophageal adenocarcinoma (EAC) arises principally in the setting of BE and the incidence of this cancer in the West increased about 8-fold between 1970 and 2010, with about 18,000 new US cases and 15,000 deaths expected in 2015 (http://seer.cancer.gov).
Investigation into the mechanisms of BE has centered largely on determinants of intestinal identity (3), particularly the intestine-restricted transcription factors (TFs) CDX1 and CDX2, which specify the embryonic intestine (4). Forced expression of CDX2 or CDX1 in the mouse stomach induces ectopic intestinal differentiation (5, 6) and both factors are implicated in activating intestinal genes in BE (7, 8), though forced CDX2 expression in the mouse esophagus does not induce BE per se (9). Loss of esophagus-specific transcripts and of stratified squamous morphology probably reflects parallel loss of transcriptional determinants of the native epithelium, which are largely unknown. TP63 regulates differentiation of all stratified epithelia, such as those in the esophagus and skin (10, 11), acting in part through another transcription factor, BNC1 (12). SOX2 controls esophageal differentiation in embryos (13) and growth of adult progenitor cells (14, 15), an activity in which KLF4 and KLF5 also may participate (16). FOXA2 is expressed in embryonic, but not in adult, esophageal cells (17). We sought to identify other tissue-restricted TFs that might control the characteristic stratified epithelium.
Among all putative DNA-binding proteins, we searched first for those with esophagus-restricted expression among digestive epithelia and then for factors with attenuated expression in BE. We identified SOX15 as such a TF and show that it directly controls transcription of a large fraction of human esophagus-expressed genes. SOX15 is absent from most EACs, but up to 30% of cases retain expression of SOX15 and its target genes, co-expressing representative intestinal and squamous-specific genes within the same tissue. Together, these data identify a novel regulator of stratified epithelial genes and a sub-type of EAC with bi-lineage gene expression.
Results and Discussion
Identification of TFs that are specific to the esophageal epithelium and attenuated in BE
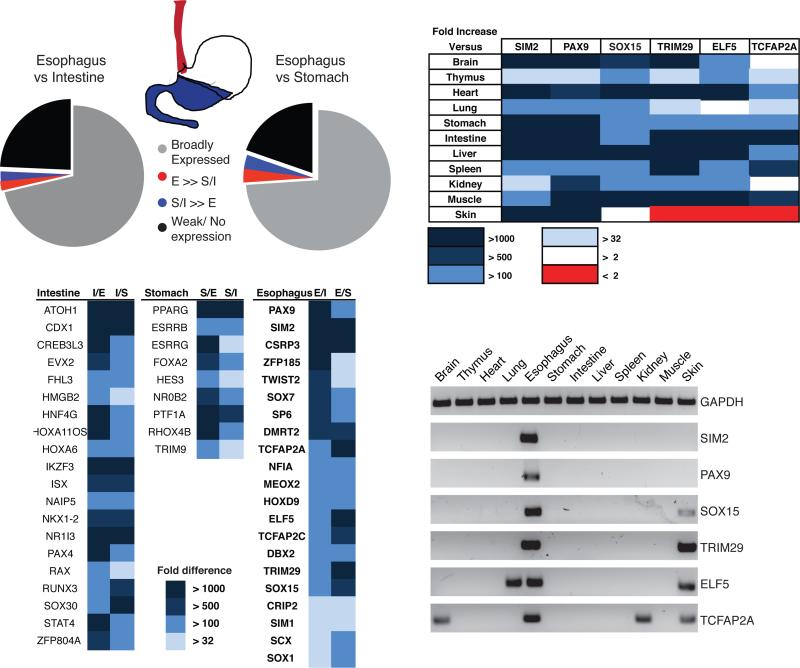
To identify candidate regulators of esophageal squamous identity, we first examined epithelia isolated from different regions of the mouse alimentary tract – esophagus, glandular stomach, and intestine (duodenum) – with a goal to identify TF mRNAs expressed selectively in the stratified esophageal epithelium (Fig. 1A). Among 1,880 known and putative DNA-binding proteins, those showing ≥32-fold higher expression in the intestinal mucosa included the known intestinal factors Atoh1, Cdx1, Creb3l3, Hnf4g and Isx (18), underscoring the fidelity of the experimental approach (Fig. 1B). Forty factors and 59 TF genes showed considerably higher expression in esophageal cells, compared to the gastric corpus and the intestine, respectively (Fig. 1A), and 21 TFs were common to the two esophagus-specific groups (Fig. 1B). To exclude variability among mouse strains and to assess specificity relative to non-digestive organs, we measured expression of these 21 mRNAs in 9 diverse tissues from C57BL/6 mice, including the skin. Six TFs gave consistent evidence of high tissue specificity (Fig.1C-D). Sim2 and Pax9 showed the greatest specificity, followed by Sox15 and Trim29, which showed some expression in murine skin. Additional data from 65 adult human tissues (19) revealed robust expression of each of these four TF mRNAs in the esophagus, with varying levels in other stratified squamous tissues, such as the tongue, mouth, pharynx, and skin derivatives (Fig. 2A).
Figure 1. Differential TF expression in the normal mouse gut and other tissues.
(A) Distribution of all TFs in wild-type mouse digestive epithelia, as revealed in a qRT-PCR screen. Expression of 1,880 TF mRNAs was assessed in epithelial cell isolates from adult CD1 mouse esophagus (red), stomach and intestine (blue). (B) TFs restricted to intestinal (I), stomach (S) or esophageal (E) epithelium, with the fold-excess over other tissues represented in shades of blue. (C) Relative expression of Sim2, Pax9, Sox15, Trim29, Elf5 and Tcfap2a mRNAs in mouse tissues. The fold-excess values are represented in shades of color as indicated in the key. (D) Products of qRT-PCR for the 6 most highly esophagus-specific TF mRNAs in 12 adult mouse organs, showing selective expression in the esophagus and of some factors in the skin.
Figure 2. Differential TF expression in normal and metaplastic human esophagus.
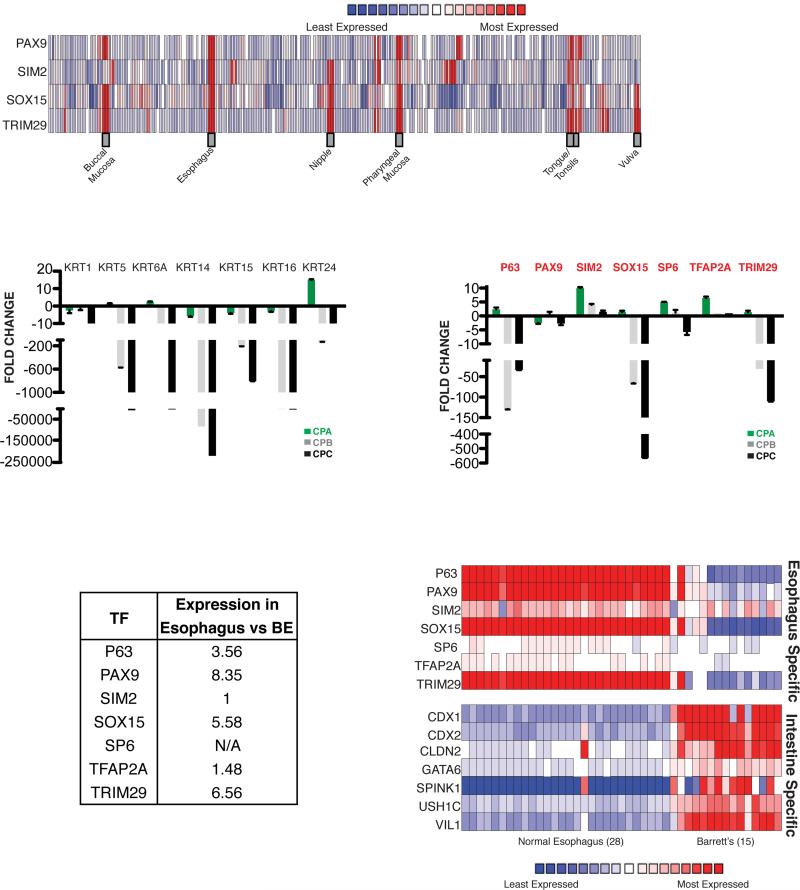
(A) Expression profiles of PAX9, SIM2, SOX15 and TRIM29 in 65 human organs. Data from necropsies (19), analyzed using Oncomine tools (22), show selective expression in the esophagus and other stratified epithelia, such as the oropharyngeal mucosa and skin derivatives. (B) Relative expression of esophagus-active keratin genes in the human BE cell line series (CPA, CPB and CPC) with increasing dysplasia. Results of qRT-PCR analysis are represented with respect to transcript levels in the immortalized human esophageal cell line EPC2-hTert (20). (C) Relative expression of esophagus-specific TF mRNAs in human BE cell lines CP-A, CP-B and CP-C, expressed in relation to levels in EPC2-hTert cells. (D) Fold-enriched expression of esophagus-specific TF mRNAs in fresh human esophageal epithelial biopsy samples (8), relative to areas of BE in the same patients. (E) Expression of esophagus-specific TFs and intestine-specific genes in normal human esophagus and BE resection specimens. Data from Ref. (23) were analyzed using Oncomine tools.
To determine if these TFs may function in the identity of stratified epithelia, we examined expression data from immortalized EPC2-hTert esophageal keratinocytes (20) and found high expression of each factor except ELF5 (data not shown). Next we tested a series of three cell lines: CP-A, which represents non-dysplastic BE; and CP-B and CP-C, which represent BE with high-grade dysplasia. This cell line series replicates disease progression (21), with reduced levels of multiple keratin mRNAs (Fig. 2B). We observed a concomitant decline in SOX15 and TRIM29 levels, matching or exceeding that of TP63 mRNA, with little variance in the other factors (Fig. 2C). Although these findings do not in isolation give robust information about a relation to mucosal dysplasia per se, they reveal the squamous cell specificity of SOX15 and TRIM29. Furthermore, gene expression data from a collection of human esophageal biopsy specimens (8) showed significantly less SOX15 and TRIM29 mRNAs in primary BE, compared to adjacent normal esophageal mucosa (Fig. 2D). Finally, we used Oncomine tools (22) to analyze mRNA data from an independent series of 28 frozen human normal esophagus and 15 frozen BE biopsy specimens (23). Levels of PAX9, SOX15 and TRIM29 were uniformly high in normal esophagus and attenuated in BE specimens (Fig. 2E).
Together, these data identify SOX15, PAX9 and TRIM29 as conserved candidate determinants of squamous cell identity. PAX9 levels were similar in CP-A, CP-B and CP-C cells (Fig. 2C), and although TRIM29 has a putative DNA-binding domain, its role in transcriptional regulation is poorly defined and uncertain (24). By contrast, SOX proteins control differentiation of diverse tissues, often in conjunction with other family members (25), and related factors such as SOX2 and SOX7 are known to regulate aspects of esophageal organogenesis and squamous cell cancer (13, 14). We therefore concentrated on human SOX15, which shares 85% homology (100% in the DNA-binding domain) with the mouse protein. SOX15 was previously noted as one among hundreds of genes in various expression profiling studies (26-28) and we proceeded to investigate its functions.
SOX15 depletion affects genes specific to stratified epithelium
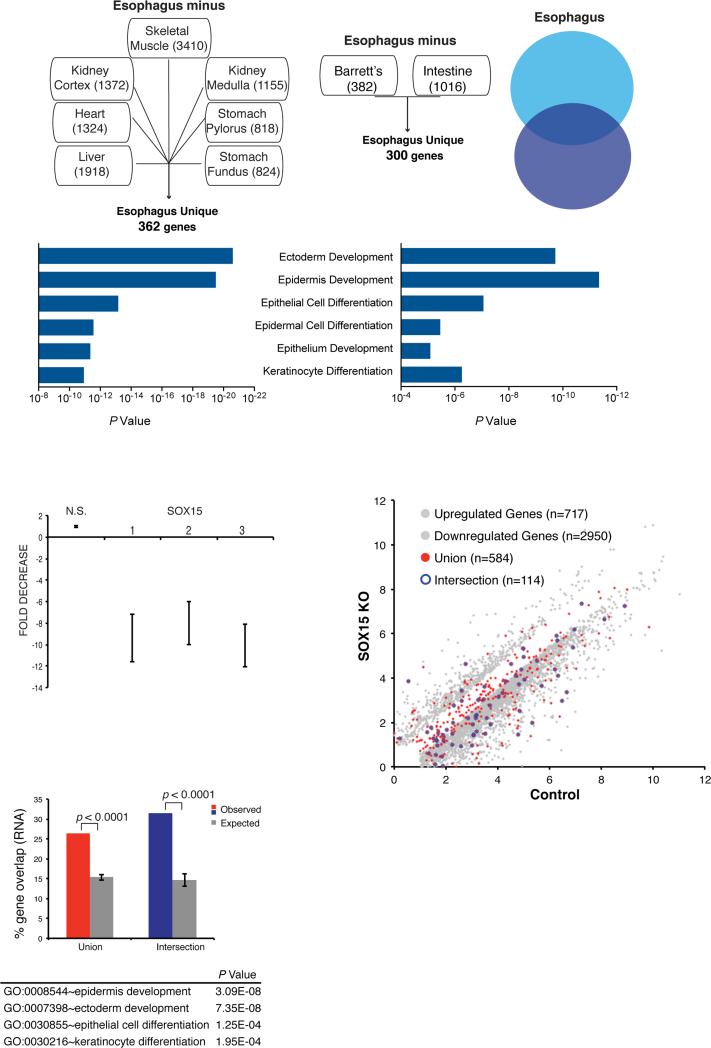
To test if SOX15 might regulate genes specific to the stratified human esophageal epithelium, we needed to delineate the corresponding transcriptome. To this end, first we considered public data from adult human postmortem tissues (19) (see Fig. 2A) and identified 362 genes that express at >3-fold higher levels (p<0.05) in the esophagus than in any of 7 diverse tissues, including glandular stomach, from the same collection of postmortem samples (Fig. 3A). Second, we identified 300 genes with >3-fold higher mRNA levels (p<0.05) in normal fresh human esophagus biopsy specimens than in adjacent areas of BE or in fresh intestinal biopsies from the same study (8). Consistent with specific roles in stratified epithelia, both gene sets were highly enriched for functions related to ectodermal, epidermal and keratinocyte differentiation, and they shared 114 genes (Fig. 3A). Accordingly, we regard the union set of 548 genes as a good representation of human esophagus-specific transcripts and the intersection set of 114 genes as an especially robust subset.
Figure 3. Impact of SOX15 depletion on esophageal gene expression.
(A) Delineation of the human esophageal transcriptome. mRNAs expressed in human esophageal necropsy specimens (left) were compared against transcripts from 7 other postmortem organs (19) and mRNAs present in fresh esophageal biopsy specimens (right) were compared against transcripts from fresh BE and intestinal biopsies (8). Numbers in each box represent squamous esophagus-specific genes relative to that tissue. We identified 362 and 300 esophagus-specific genes, respectively, with a significant 114-gene overlap (P <0.0001, χ2 test). The top Gene Ontology (GO) terms in each case are highly related to stratified epithelia. (B) SOX15 mRNA depletion in 3 representative experiments in which CPA cells were infected with lentiviruses carrying SOX15-specific or a non-specific (NS) 21-bp shRNAs. Knockdown efficiency, assessed by qRT-PCR 72 h after infection, was >8− to 10-fold in every experiment. (C) Results of duplicate RNA-seq analysis of genes differentially expressed in CP-A cells treated with SOX15-specific (y-axis) or control, non-specific (x-axis) shRNAs. Grey dots mark differential expression (log2 >1.5-fold, q <0.05) and genes present in the union (548 genes) or intersection (114) sets of esophagus-specific genes are represented by red and blue dots, respectively. (D) Fraction of esophagus-specific transcripts reduced upon SOX15 depletion (red, 548 union-set genes; blue, 114 intersection-set genes), compared to 5 random sets of equal numbers of genes expressed in CP-A cells (grey bars). The table lists GO terms enriched among SOX15-dependent genes.
To determine if SOX15 regulates any part of this transcriptome, we used lentiviral delivered shRNA to deplete the TF in CP-A cells. These cells express keratin and TF genes specific to stratified epithelia, including SOX15, at levels similar to immortalized EPC2-hTert esophageal epithelial cells (Fig. 2B-C) and they tolerate lentiviral infection and drug selection. Because SOX15 depletion retarded CP-A cell growth and survival, we harvested cells 72 h post-infection, when they appeared healthy but SOX15 mRNA levels were appreciably reduced (Fig. 3B). RNA-seq analysis showed reduced and increased levels of 2,950 and 717 transcripts, respectively, compared to cells treated with a non-specific shRNA (Fig. 3C). In agreement with the deficit in cell growth, these genes were enriched for Gene Ontology terms related to the cell cycle (Suppl. Table 1). More importantly, genes reduced in SOX15-depleted cells included 26.4% of the human esophagus-specific “union” transcriptome, compared to 15.34% overlap with multiple sets of 2,950 random genes expressed in CP-A cells (Fig. 3D, P <0.0001). Correspondence was even higher for the esophagus-specific “intersection” transcriptome, where 31.5% of genes were reduced in SOX15-depleted cells, compared to 14.68% of random genes (P <0.0001). SOX15-dependent genes were highly enriched for functions related to stratified epithelia (Fig. 3D). None of the 114 genes in the esophagus-specific “intersection set,” and only 23 genes in the “union set,” were increased in SOX15-depleted CP-A cells and we observed no increase in intestinal genes. Rather, the 717 increased transcripts were enriched for functions such as apoptosis and vesicular transport (Suppl. Table 1). Thus, beyond cell survival or proliferation, a substantial portion of the esophageal transcriptome depends on SOX15.
SOX15 directly regulates esophagus-specific genes
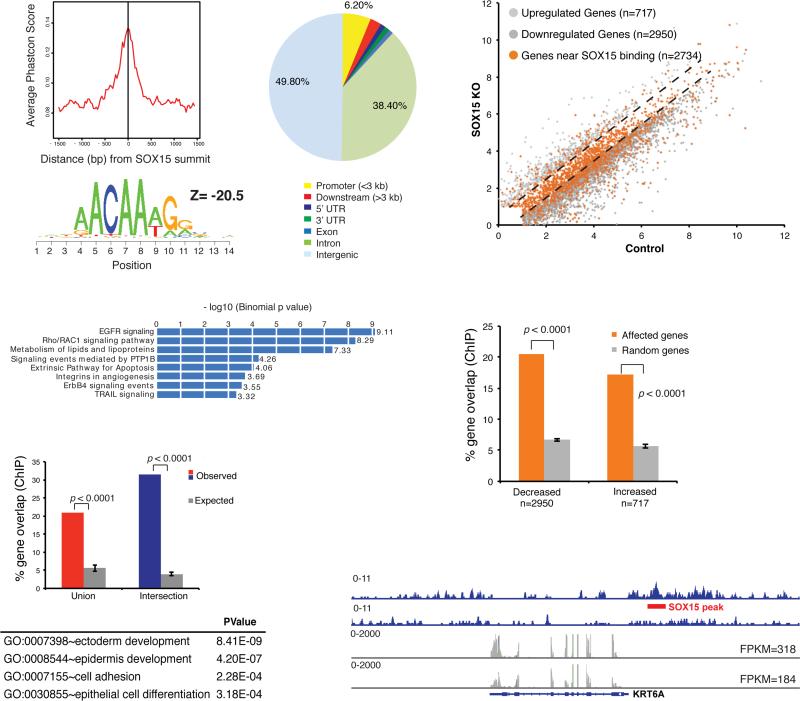
Depletion of SOX15 could affect transcript levels as a consequence of its cis-regulatory activity or indirectly. To determine if SOX15 might regulate dependent genes directly, we used chromatin immunoprecipitation with high-throughput sequencing (ChIP-seq) to map its cistrome. Because available antibodies performed poorly in ChIP assays, we expressed biotin-tagged SOX15 stably in CP-A cells and precipitated chromatin using streptavidin beads. The nearly 5,000 high-confidence binding sites we identified by this approach showed high sequence conservation and greatest enrichment for the SOX consensus motif, which was present in >97% of sites, implying direct TF occupancy (Fig. 4A). Similar to other tissue-specific TFs, SOX15 occupied few promoters (6.2% of all binding sites) and bound DNA predominantly in intergenic regions and introns (Fig. 4A). GREAT analysis (29) of the nearest flanking genes within 50 kb of SOX15 occupancy revealed enrichment of pathways known to be vital in stratified epithelia, such as Epidermal Growth Factor and Rho/Rac signaling, and in cell survival (Fig. 4B). Moreover, 20.9% of genes in the human esophagus-specific transcriptome and 31.5% of genes common to the two esophagus transcript sources showed at least one SOX15 binding site within 50 kb of the transcription start site, compared to about 5% of comparable numbers of random genes (P <0.0001, Fig. 4C). Gene Ontology terms related to stratified epithelia were further enriched among SOX15-bound genes (Fig. 4C). Most importantly, genes affected by SOX15 depletion in CP-A cells were highly enriched for nearby SOX15 binding, compared to random gene sets of equal size (P <0.0001) and SOX15-bound genes reduced in SOX15-depleted cells far outnumbered genes that were increased (Fig. 4D-E). Taken together, these data indicate direct SOX15 regulation of genes specific to the stratified squamous epithelium, with a strong bias toward gene activation. Canonical esophageal genes such as KRT6A illustrate SOX15 occupancy at putative cis-regulatory sites and reduced expression in SOX15-depleted cells (Fig. 4F).
Figure 4. Genome-wide SOX15 occupancy and gene dependence in human esophageal cells.
(A) Summary of ChIP analysis for biotinylated SOX15 in CP-A cells, showing high sequence conservation and significant enrichment of a canonical SOX recognition motif ACAA(A/T)G among 4,864 identified binding sites. SOX15 mainly binds DNA far from promoters. (B) GO terms enriched among the 2 nearest genes within 50 kb of SOX15 binding sites, determined using GREAT (29). (C) Percentages of esophagus-specific genes (as determined in Fig. 2A, red: union, blue: intersection set) that bind SOX15 within 50 kb of the transcription start site (TSS), compared to 5 random gene sets of equivalent size (grey bars, P <0.0001). The table lists GO terms enriched among genes from the esophagus transcriptome that lie within 50 kb of SOX15 binding sites. (D) SOX15 binding (orange dots) within 50 kb of genes expressed in SOX15-depleted and control CP-A cells (grey dots, q <0.05, as in Fig. 2B). Dashed lines demarcate genes unaffected by SOX15 loss. (E) Genes reduced or increased in SOX15-depleted cells are significantly enriched for nearby SOX15 binding. Together with the proportions of orange dots in D, the data imply direct SOX15 activation of many, and direct repression of fewer genes. (F) Integrated Genome Viewer representation of esophageal gene KRT6A, showing SOX15 binding at the locus (top rows, blue, ChIP-seq tags) and reduced expression in SOX15-depleted CP-A cells (bottom rows, grey, RNA-seq tags). Numbers represent the height of the y-axis.
SOX15 in human EAC
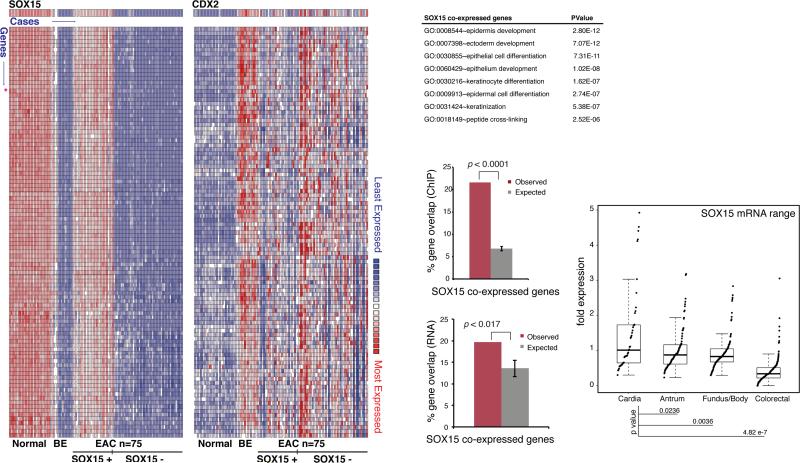
SOX15 is expressed highly in normal human esophagus, but not in the BE cell lines CP-B and CP-C (Fig. 2C) or in areas of intestinal metaplasia in vivo (Fig. 2D-E). To our surprise, RNA expression data from a large collection of frozen primary esophagus, BE and EAC biopsy specimens (23) revealed high SOX15 mRNA levels in up to 1/3 of EACs (Fig. 5A, left; note that all samples in this study, including EAC, were frozen biopsy specimens). Moreover, at least 317 transcripts that are strongly co-expressed with SOX15 in the normal esophagus (r>0.81, Suppl. Table 2) were also present in the same EAC specimens (Fig. 5A shows the 100 genes with highest correlation), suggesting broad activation of the squamous cell transcriptional program. Accordingly, functions related to stratified epithelia were significantly enriched among the genes co-expressed with SOX15, (Fig. 5B). The canonical intestinal marker CDX2 and its co-expressed genes (r>0.81) were expressed in many SOX15+ and also in SOX15− specimens (Fig. 5A, right), revealing co-expression of esophageal and intestinal genes in some cases. Moreover, 21.6% of genes co-expressed with SOX15 in this analysis showed SOX15 binding within 50 kb in CP-A cells, compared to ~6% of random genes (P <0.0001, Fig. 5C top), which implies that many of these genes are direct transcriptional targets. Indeed, the effects of SOX15 depletion were significantly greater on these genes than on random sets of genes expressed in CP-A cells (P <0.017, Fig. 5C bottom). These features collectively suggest direct SOX15 regulation of many esophagus-restricted genes that are silent in BE and reactivated in up to 1/3 of human EACs.
Figure 5. SOX15 expression in esophageal adenocarcinomas.
(A) Gene co-expression profiles for SOX15 (left) and CDX2 (right) in a large collection of normal, BE and EAC epithelium (23). 317 transcripts correlated strongly (r >0.81) with SOX15 mRNA levels in normal esophagus and in approximately 1/3 of 75 EACs in this series. The 100 most highly correlated genes are shown. (B) GO term enrichment among these 317 SOX15-coexpressed genes. (C) Top: Fraction of SOX15 co-expressed genes showing SOX15 occupancy (Observed) within 50 kb, compared to the fraction expected for appropriate random gene sets of equal size. Bottom: Fraction of SOX15 co-expressed genes affected by SOX15 depletion (Observed), compared to the fraction expected among random gene sets of equal size. (D) Ranges of SOX15 mRNA expression extracted from RNA-seq data on TCGA collections of cancers of the gastric cardia, fundus/body and antrum (30), or colon and rectum (42). Significance of differences was determined by the t-test.
To exclude the possibility that EACs expressing SOX15 were simply contaminated with normal SOX15+ esophageal cells, we studied cases from an independent collection, The Cancer Genome Atlas (TCGA), where non-malignant cells were meticulously minimized (30). Cancers of the gastro-esophageal (G-E) junction typically arise in a background of BE and, when associated with chromosomal instability (CIN), usually represent distal EACs. Among 30 cases of CIN+ tumors from the G-E junction or gastric cardia in the TCGA collection of gastric cancers, some samples showed robust levels of SOX15 and of genes co-expressed with SOX15 in normal esophageal epithelium (Suppl. Fig. S1A-B). Transcripts specific to the squamous esophageal epithelium were thus again evident in a fraction of EACs. To determine if this extent of SOX15 expression is specific to EACs, we evaluated other gastrointestinal cancers in the TCGA collection: gastric fundus, body and antrum, and colorectal tumors. Extreme outliers for SOX15 mRNA expression were present only among tumors of the gastric cardia (Fig. 5D).
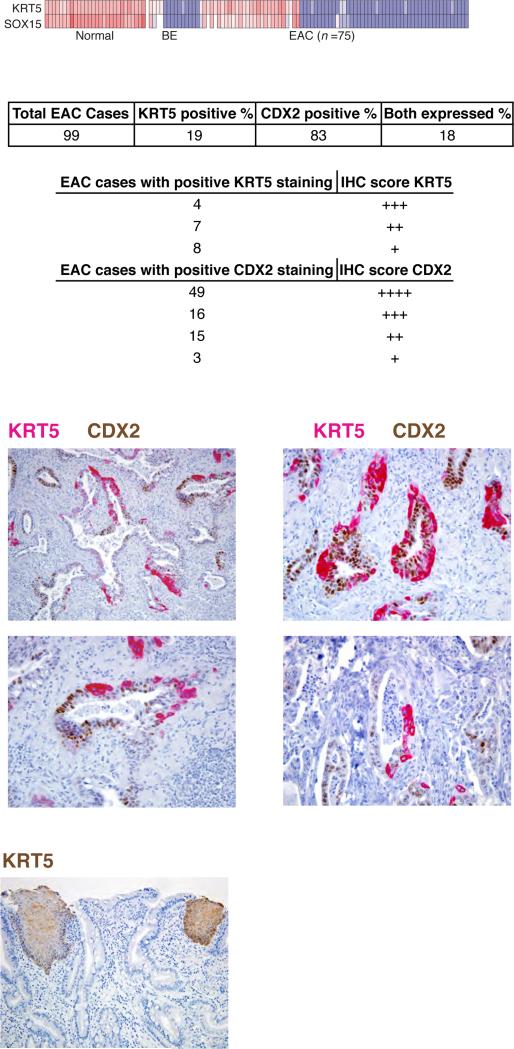
Finally, we examined 99 separate EACs by immunohistochemistry (IHC) on resection specimens. Because several antibodies failed to detect SOX15, we used KRT5 as a proxy for expression of SOX15 and other stratified cell-specific products, noting nearly total concordance of SOX15 and KRT5 mRNA expression in the large aforementioned tissue collection (23) (Fig. 6A, r=0.97); we also stained the same samples for the intestinal marker CDX2. Nineteen cases (19%) showed cytoplasmic KRT5 expression within malignant glands and most of these cases co-expressed nuclear CDX2 (Fig. 6B). Levels of KRT5 were variable (Fig. 6B) but did not correlate with tumor grade or other pathological features such as mucin production. Importantly, KRT5 was not expressed in rare pockets of squamous differentiation, but rather in bona fide glandular structures. In fact, and of particular note, cytoplasmic KRT5 and nuclear CDX2 were almost always present in different cells within the same glands (Figs. 6C-D show examples from 2 different cases). Co-expression of esophagus- and intestine-specific genes within individual glands reveals the malignant cells’ potential to express genes from distinct cell lineages.
Figure 6. Bi-lineage gene expression in a subset of EACs.
(A) High correlation (r=0.97) of SOX15 and KRT5 mRNAs in normal esophagus, BE, and EAC, validating KRT5 as a proxy for SOX15 and other stratified epithelium-specific genes. (C-D) Representative immunohistochemistry for KRT5 (red, a surrogate marker for SOX15 and other squamous-specific gene products) and CDX2 (brown, a representative intestine-specific marker) in two separate cases (C and D) of human EAC. High KRT5 expression is evident, with almost mutually exclusive distribution of KRT5 (++ to +++) and CDX2 (+++ to ++++) in the same malignant glands. Original magnifications: Top, 200X; Bottom, 400X. (E) Absence of KRT5 immunostaining in areas of BE. Adjoining areas of normal stratified epithelium provide a positive control and contrast.
To corroborate the observation that mRNA levels of stratified cell genes are low in BE but elevated in many EACs (Figs. 5A and 6A), we used IHC to assess areas of BE that were present in 24 of the 99 resection specimens. KRT5 was uniformly absent from these areas, though the signal was clear in adjoining stratified epithelium (Fig. 6E). These findings extend previous reports of absent expression of stratified epithelium-specific keratins in BE (31-33) and low-level expression of squamous cell products in EACs (34). Our delineation of a squamous cell- restricted transcriptome (Fig. 3A), coupled with re-analysis of published RNA expression data (Fig. 5A-B) and investigation of additional cases by IHC (Fig. 6B-D), reveals for the first time the extent and breadth of an aberrant stratified-cell program in EACs.
Implications for esophageal squamous differentiation and EAC
Insights into transcriptional control of the esophageal squamous epithelium are largely limited to the broad functions of TP63 and SOX2 (10, 13). Our identification of SOX15 as a novel, conserved, and likely direct regulator of many human stratified epithelial genes extends understanding of esophageal differentiation and pathology. The lack of overt esophageal defects in Sox15 mutant mice (35, 36) is compatible with the considerable known redundancies among SOX-family TFs (25).
There is much debate whether BE, and particularly EAC, originates in the native esophageal epithelium through bona fide metaplasia or in ectopic cells that may colonize the esophagus from the gastric cardia, as in mice (11, 37). Clearly, the best way to answer the question is through lineage tracing studies, which are possible in animals, but not in humans. When lineage tracing is not feasible, cell-specific transcript patterns offer clues, and expression of SOX15 and other squamous epithelial genes may be informative. Consider, for example, the observation that the BE cell lines CP-A/B/C show reduced levels of SOX15, esophageal keratins, TP63 and other esophagus-specific TF genes, implying loss of an esophageal program, but some esophagus-restricted TF genes such as SIM2 and TFAP2A are highly expressed in these cells (Fig. 2C) and in BE biopsy specimens (Fig. 2D-E). Moreover, >300 esophageal genes, including SOX15, are active in up to 30% of human EACs (Fig. 5A), with intestinal genes such as CDX2 often co-expressed in the same glands as esophageal genes (Fig. 6C-D). These findings in BE and EAC could indicate residual squamous cell-specific transcription or fortuitous ectopic gene activity. If diseased cells are better equipped to express genes from their native transcriptional program than genes from a heterologous cell lineage, because native genes and their cis-regulatory elements are inherently primed and accessible, then the first possibility may be more likely. Our findings do not of course rule out the alternative model, which will require additional, independent lines of evidence.
EAC is a particularly recalcitrant disease, with poor 5-year survival rates. Surgery and empiric cytotoxic chemotherapy anchor current treatment approaches (38, 39), and although disease heterogeneity is apparent in the clinic, the underlying determinants are unclear. We show here that one-fifth to one-third of EACs simultaneously express products specific to the esophageal squamous epithelium and columnar intestinal cells. It will be important, in the future, to identify clinical and genetic correlates of these EACs showing bi-lineage gene expression and to determine if they reflect a distinctive pathophysiology or harbor unique therapeutic vulnerabilities.
Materials and Methods
All authors had access to the study and reviewed and approved the final manuscript.
Tissue preparation and TF expression screen
We isolated epithelial sheets from the esophagus, gastric corpus-antrum, and duodenum of 1-month old CD1 and C57BL/6 mice. Before peeling the mucosa using fine forceps, the esophagus was treated with 0.1% Collagenase-Dispase (Roche, catalog no. 11097113001) in phosphate-buffered saline (PBS) for 15 min at 37°C, whereas stomach and duodenum were incubated in 1 mM EDTA in PBS at 37°C. To determine relative transcript levels (Fig. 1A-C), we used qRT-PCR and a library containing oligonucleotide primers specific to 1,880 known and putative TFs (40). Tissue-specific TFs were identified using the comparative CT method (41). To further determine tissue specificity (Fig. 1D), other whole organs were harvested from adult C57BL/6 mice.
Cell lines
CP-A (KR-42421), CP-B (CP-52731) and CP-C (CP-94251) cells (American Type Culture Collection) were cultured in MCDB-153 medium (Sigma, catalog no. M7403) supplemented with 0.4 μg/ml hydrocortisone, 20 ng/ml recombinant human Epidermal Growth Factor (Sigma, catalog no. E9644), 8.4 μg/L cholera toxin (Sigma, catalog no. H0135), 20 mg/L adenine (Sigma, catalog no. A2786), 140 μg/ml bovine pituitary extract (Sigma, catalog no. P1476), ITS Supplement (Sigma, catalog no. I1884, final concentrations: 5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml sodium selenite), 4 mM glutamine and 5% fetal bovine serum. EPC2- hTert cells (20) were cultured in Keratinocyte-SFM medium (Gibco) supplemented with bovine pituitary extract and recombinant human epidermal growth factor (Gibco). Soybean trypsin inhibitor (Sigma) was used to quench trypsin activity during cell passage.
Gene analyses
Figs. 2A, 2D-E, 5A and 6A show analyses of relative mRNA expression levels from published studies of 65 adult human tissues (19), of human esophageal biopsy specimens (8), and of human normal esophagus, BE and EAC samples (23). The data were re-analyzed with respect to SOX15 using Oncomine tools (Compendia Bioscience, https://www.oncomine.com), considering all samples in each dataset. Genes significantly associated with SOX15 (Fig. 5A) were ranked on the basis of correlation values. Enriched Gene Ontology terms (Figs. 3A, 3D, 4B, 4C, 5B) were determined using DAVID tools (http://david.abcc.ncifcrf.gov/). We examined processed RNA-seq data from a TCGA study on stomach cancer (30), first isolating 30 CIN+ tumors arising at the G-E junction or gastric cardia for unsupervised clustering (Suppl. Fig. 1). To this group we applied hierarchical clustering (using hclust from the R package; http://cran.r-project.org) on the 1,000 most variable transcripts normalized according to expression z-scores, followed by a second hierarchical clustering on the set of 317 genes co-expressed with SOX15. To assess specificity of SOX15 overexpression in tumors of the gastric cardia (Fig. 5D), we compared with RNA-seq data from TCGA studies on colon (42) and distal gastric adenocarcinomas (30).
Experimental RNA analyses
Total RNA was isolated using Trizol (Invitrogen), treated with the RNeasy Mini Kit (Qiagen), and DNA was digested using Turbo DNA-Free (Ambion). For qRT-PCR analysis (Figs. 1A-C, 2B-C, 3B), 1 μg of total RNA was reverse transcribed with Superscript III First Strand Synthesis System (Invitrogen) and cDNA was amplified using SYBRGreen PCR Master Mix (Applied Biosystems). RNA-seq libraries (Fig. 3C, full dataset is deposited in the Gene Expression Omnibus with accession number GSE62909) were prepared from 300 ng of total RNA using TruSeq RNA Sample Preparation kits (Illumina) and 75-bp single-end sequences were obtained on a NextSeq 500 instrument (Illumina). Reads were aligned to human genome build Hg19 using TopHat v2.0.6. Expression levels of transcripts in duplicate samples were calculated as fragments per kb per 106 mapped reads (FPKM) using Cufflinks v2.0.2 and differential expression was determined using CuffDiff (43). Chi-square tests with 1 degree of freedom and two-tailed p values were used to assess significance. Log2 (FPKM+1) values for control and SOX15-depleted samples were plotted to display differential expression.
Depletion of SOX15 and expression of biotin-tagged SOX15
Cells were infected with lentiviruses generated from the pLKO.1 vector (Open Biosystems) carrying either a SOX15-targeting shRNA (TGCCTGGCAGCTATGGCTCTT) or a control, non-specific shRNA that does not complement any human gene and is not toxic to cultured human cells (CCTAAGGTTAAGTCGCCCTCG). Human SOX15 cDNA was cloned into the pUltra vector (Addgene, catalog no. 24129) together with cassettes for the T2A sequence, biotin, and BirA-V5 (gift of Ben Ebert, Brigham & Women's Hospital, Boston, MA).
Chromatin immunoprecipitation (ChIP) and ChIP-seq
Cells were cross-linked with 2 mM disuccinimidyl glutarate (DSG, Pierce, catalog no. 20593) in PBS for 45 min, followed by 10 min with 1% formaldehyde (Pierce) in PBS, at room temperature. ChIP and ChIP-seq were performed as described previously (44), using 30 μL slurry of streptavidin-conjugated magnetic beads (Invitrogen, catalog no. 65601). We used Cistrome tools (www.cistrome.org) to identify and annotate TF binding sites, generate wiggle files and conservation plots, identify enriched sequence motifs and linked genes, and compare data across ChIP-seq libraries. Wiggle traces were projected on the Integrative Genome Viewer (IGV) (45). Functions of genes within 50 kb of SOX15 occupancy were determined using GREAT (29). ChIP-seq data are deposited in the GEO database with accession number GSE62909.
Immunohistochemistry
4-μm-thick tissue paraffin sections were baked overnight at 37°C, deparaffinized in xylenes, rehydrated, and peroxidase activity was blocked with 1.5% H2O2 in methanol for 10 min. Slides were treated with 0.01 M citrate buffer, pH 6.0 in a pressure cooker at 120°C for 30 min for antigen retrieval, then transferred to Tris-buffered saline. Sections were first incubated with mouse CDX2 Ab (clone CDX2-88, Biogenex mu392A-uc, 1:200) for 40 min, followed by Dako Envision+ Mouse (Dako K4007) secondary Ab for 30 min, and developed with 3,3’-diaminobenzidine (Dako). Sections were then incubated with mouse KRT5 Ab (clone XM26, Leica NCL-L-CK, 1:500) for 40 min, followed by PowerVision AP mouse (Leica, catalog no. PV6110) secondary Ab for 30 min, developed with Permanent Red, and counterstained with Mayer's hematoxylin. To stain resection specimens that carried areas of BE, slides were treated with the same mouse KRT5 Ab, followed by Dako Envision+ Mouse (Dako K4007) secondary Ab for 30 min, and developed with 3,3’-diaminobenzidine (Dako).
Supplementary Material
Acknowledgments
Supported by the DFCI-Novartis Drug Discovery Program, generous gifts from the Lind family, and National Institutes of Health grant P50CA127003 (SPORE). A.J.B and A.K.R. were supported by grant P01CA098101 (National Cancer Institute). We thank Rishi Puram and Ben Ebert for BirA-V5 vectors and Austin Dulak and Hiroshi Nakagawa for assistance with esophageal cell culture.
Funding Information: Supported by the DFCI-Novartis Drug Discovery Program, generous gifts from the Lind family, and National Institutes of Health grants R01DK081113 (R.A.S), P01CA098101 (A.J.B. and A.K.R.), and P50CA127003 (Dana-Farber/Harvard).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Synopsis: This study identifies SOX15 as a direct transcriptional regulator of a substantial fraction of cell type-specific genes in stratified epithelial cells. SOX15 expression is attenuated in intestinal metaplasia (Barrett's esophagus) but is active in many esophageal adenocarcinomas.
Conflicts of interest: No conflicts of interest exist.
Author contributions: R.S. and R.A.S. conceived and designed the study. R.S. and J.C. performed experiments. Z.A. provided mouse TF libraries and A.K.R. provided cell lines. U.J. helped analyze RNA-seq data. S.P. and A.B. analyzed data from TCGA cancers. A.S. and J.L.H. provided EAC specimens and performed and scored immunohistochemistry. R.A.S. provided overall supervision. R.S. and R.A.S. drafted the manuscript, with contributions from all authors.
REFERENCES
- 1.Spechler SJ, Souza RF. Barrett's esophagus. N Engl J Med. 2014;371:836–845. doi: 10.1056/NEJMra1314704. [DOI] [PubMed] [Google Scholar]
- 2.Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med. 2011;365:1375–1383. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 3.Souza RF, Krishnan K, Spechler SJ. Acid, bile, and CDX: the ABCs of making Barrett's metaplasia. Am J Physiol Gastrointest Liver Physiol. 2008;295:G211–218. doi: 10.1152/ajpgi.90250.2008. [DOI] [PubMed] [Google Scholar]
- 4.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silberg DG, et al. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122:689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- 6.Mutoh H, et al. Cdx1 induced intestinal metaplasia in the transgenic mouse stomach: comparative study with Cdx2 transgenic mice. Gut. 2004;53:1416–1423. doi: 10.1136/gut.2003.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eda A, et al. Aberrant expression of CDX2 in Barrett's epithelium and inflammatory esophageal mucosa. Journal of gastroenterology. 2003;38:14–22. doi: 10.1007/s005350300001. [DOI] [PubMed] [Google Scholar]
- 8.Stairs DB, et al. Cdx1 and c-Myc foster the initiation of transdifferentiation of the normal esophageal squamous epithelium toward Barrett's esophagus. PLoS One. 2008;3:e3534. doi: 10.1371/journal.pone.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong J, Crissey MA, Funakoshi S, Kreindler JL, Lynch JP. Ectopic Cdx2 expression in murine esophagus models an intermediate stage in the emergence of Barrett's esophagus. PLoS One. 2011;6:e18280. doi: 10.1371/journal.pone.0018280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniely Y, et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287:C171–181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, et al. Residual embryonic cells as precursors of a Barrett's-like metaplasia. Cell. 2011;145:1023–1035. doi: 10.1016/j.cell.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boldrup L, Coates PJ, Laurell G, Nylander K. p63 Transcriptionally regulates BNC1, a Pol I and Pol II transcription factor that regulates ribosomal biogenesis and epithelial differentiation. Eur J Cancer. 2012;48:1401–1406. doi: 10.1016/j.ejca.2011.06.032. [DOI] [PubMed] [Google Scholar]
- 13.Que J, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass AJ, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41(11):1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu K, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tetreault MP, et al. Esophageal squamous cell dysplasia and delayed differentiation with deletion of kruppel-like factor 4 in murine esophagus. Gastroenterology. 2010;139:171–181. doi: 10.1053/j.gastro.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang DH, et al. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett's metaplasia. J Clin Invest. 2014;124:3767–3780. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi MY, et al. A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development. 2006;133:4119–4129. doi: 10.1242/dev.02537. [DOI] [PubMed] [Google Scholar]
- 19.Roth RB, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 20.Harada H, et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–738. [PubMed] [Google Scholar]
- 21.Palanca-Wessels MC, et al. Genetic analysis of long-term Barrett's esophagus epithelial cultures exhibiting cytogenetic and ploidy abnormalities. Gastroenterology. 1998;114:295–304. doi: 10.1016/s0016-5085(98)70480-9. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SM, et al. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS One. 2010;5(11):e15074. doi: 10.1371/journal.pone.0015074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 25.Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 26.Hackett NR, et al. The human airway epithelial basal cell transcriptome. PLoS One. 2011;6:e18378. doi: 10.1371/journal.pone.0018378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thu KL, et al. SOX15 and other SOX family members are important mediators of tumorigenesis in multiple cancer types. Oncoscience. 2014;1:326–335. doi: 10.18632/oncoscience.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyland PL, et al. Global changes in gene expression of Barrett's esophagus compared to normal squamous esophagus and gastric cardia tissues. PLoS One. 2014;9:e93219. doi: 10.1371/journal.pone.0093219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Network TCGAR Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014 doi: 10.1038/nature13480. 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Zimaity HM, Graham DY. Cytokeratin subsets for distinguishing Barrett's esophagus from intestinal metaplasia in the cardia using endoscopic biopsy specimens. Am J Gastroenterol. 2001;96:1378–1382. doi: 10.1111/j.1572-0241.2001.03792.x. [DOI] [PubMed] [Google Scholar]
- 32.Nurgalieva Z, Lowrey A, El-Serag HB. The use of cytokeratin stain to distinguish Barrett's esophagus from contiguous tissues: a systematic review. Dig Dis Sci. 2007;52:1345–1354. doi: 10.1007/s10620-006-9399-3. [DOI] [PubMed] [Google Scholar]
- 33.van Baal JW, et al. Cytokeratin and CDX-2 expression in Barrett's esophagus. Scand J Gastroenterol. 2008;43:132–140. doi: 10.1080/00365520701676575. [DOI] [PubMed] [Google Scholar]
- 34.DiMaio MA, Kwok S, Montgomery KD, Lowe AW, Pai RK. Immunohistochemical panel for distinguishing esophageal adenocarcinoma from squamous cell carcinoma: a combination of p63, cytokeratin 5/6, MUC5AC, and anterior gradient homolog 2 allows optimal subtyping. Human pathology. 2012;43:1799–1807. doi: 10.1016/j.humpath.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HJ, et al. Sox15 is required for skeletal muscle regeneration. Mol Cell Biol. 2004;24:8428–8436. doi: 10.1128/MCB.24.19.8428-8436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. J Biol Chem. 2005;280:24371–24379. doi: 10.1074/jbc.M501423200. [DOI] [PubMed] [Google Scholar]
- 37.Quante M, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21:36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ajani JA, et al. Multicenter phase III comparison of cisplatin/S-1 with cisplatin/infusional fluorouracil in advanced gastric or gastroesophageal adenocarcinoma study: the FLAGS trial. J Clin Oncol. 2010;28:1547–1553. doi: 10.1200/JCO.2009.25.4706. [DOI] [PubMed] [Google Scholar]
- 39.Cunningham D, et al. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N Engl J Med. 2008;358:36–46. doi: 10.1056/NEJMoa073149. [DOI] [PubMed] [Google Scholar]
- 40.Gupta RK, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 42.Network TCGA Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein- protein and protein-DNA interactions in mouse embryonic stem cells. Nature protocols. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- 45.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.