Abstract
Background
Twitter is increasingly used to estimate disease prevalence, but such measurements can be biased, both due to biased sampling and to the inherent ambiguity of natural language.
Objective
We characterized the extent of these biases and how they vary with disease.
Methods
We correlated self-reported prevalence rates for 22 diseases from Experian's Simmons National Consumer Study (n=12,305) with the number of times these diseases were mentioned on Twitter during the same time period (2012). We also identified and corrected for two types of bias present in Twitter data: (1) demographic variance between US Twitter users and the general US population; and (2) natural language ambiguity, which creates the possibility that mention of a disease name may not actually refer to the disease (e.g., “heart attack” on Twitter often does not refer to myocardial infarction). We measured the correlation between disease prevalence and Twitter disease mentions both with and without bias correction. This allowed us to quantify each disease's overrepresentation or underrepresentation on Twitter, relative to its prevalence.
Results
Our sample included 80,680,449 tweets. Adjusting disease prevalence to correct for Twitter demographics more than doubles the correlation between Twitter disease mentions and disease prevalence in the general population (from 0.113 to 0.258, P < .001). Also, diseases varied widely in how often mentions of their names on Twitter actually referred to the diseases, from 14.9% of instances (for stroke) to 99.9% (for arthritis). Applying ambiguity correction to our Twitter corpus achieves a correlation between disease mentions and prevalence of 0.208 (P < .001). Simultaneously applying correction for both demographics and ambiguity more than triples the baseline correlation to 0.366 (P < .001). Compared to prevalence rates, cancer appeared most overrepresented in Twitter while high cholesterol appeared most underrepresented.
Conclusions
Twitter is a potentially useful tool to measure public interest in and concern about different diseases, but when comparing diseases, improvements can be made by adjusting for population demographics and word ambiguity.
Keywords: Social Media, Epidemiology, Public Health, Disease, Bias, Demographics, Prevalence, Data Mining
Introduction
Word use patterns in Twitter, Facebook, newsgroups, and Google queries have been used to investigate a wide array of health concerns. Twitter is perhaps the most popular online data source for such studies, due in part to its relative accessibility. It has been used to monitor health issues including influenza [1,2], cholera [3], H1N1 [4,5], swine flu [6], postpartum depression [7], concussion [8], epilepsy [9], migraine [10], cancer screening [11], antibiotic use [12], medical practitioner errors [13], dental pain [14], and attitudes about vaccination [15].
Such research has demonstrated the utility of mining social media for public health applications in spite of potential methodological challenges, including the facts that (a) Twitter users form a biased sample of the population [16-18], and (b) their word usage within tweets can be highly ambiguous. Focusing just on the medical domain, “stroke” has many non-medical uses (e.g. “stroke of genius”, “back stroke”), most mentions of “heart attack” are metaphorical, not literal, (“just had a heart attack and died the power went out while I was in the shower”) and although doctors associate “MI” with myocardial infarction, on Twitter it refers more often to the state of Michigan.
This paper quantifies, and provides a framework for partially correcting, the error arising when using sources such as Twitter as a proxy for measuring disease prevalence. We investigate the relationship between the frequency of disease mentions on Twitter in the US and the prevalence of the same diseases in the US population. Understanding this relationship could be useful for a variety of applications, including health care messaging and disease surveillance. We use Twitter as the venue for measuring discussion largely because it has already received substantial attention as an inexpensive proxy for tracking disease prevalence [19,20].
Our key contribution is to demonstrate that it is possible to better align Twitter disease mention statistics with actual disease prevalence statistics by correcting for ambiguous medical language on Twitter, and by correcting for the difference between the demographics of Twitter users and the general US population. We observe that a slight correlation exists between general population disease prevalence statistics (sourced from existing survey data) and the number of times each disease is mentioned on Twitter (according to our own counts). We find that we can significantly increase this correlation (a) by restricting the disease prevalence population specifically to Twitter users (i.e., by correlating with existing prevalence data focused specifically on that group), and (b) by adjusting our disease mention counts to correct for word sense ambiguity.
Methods
Overview
We first identified a list of diseases; then for each disease we constructed a list of terms that refer to it (i.e., a disease-specific lexicon). We also collected a large number of tweets and compiled them into a tweets corpus. Next we retrieved a random sample of tweets from our corpus that contained any of our disease-terms. We then determined the relative frequencies (percentage) of medical uses of the disease-terms (i.e., valid positives) versus non-medical uses (i.e., false positives due to ambiguity), using human annotation on the random sample. This allowed us to compute corrected counts of the number of tweets in the corpus that mention each disease (we call this a disease's validated tweet count, whereas an uncorrected count is termed a raw tweet count).
We correlated the corrected disease mention frequencies to US disease prevalence statistics from the Simmons National Consumer Study [21]. The resulting correlation serves as a measure of the relationship between the quantity of disease mentions in the corpus, and the quantity of disease cases in the US population (for either the general population or the Twitter-using population). Comparing the correlations with and without correction demonstrates the size of our corrections.
Data collection
Selection of diseases
We used two criteria for selecting diseases for this research: (1) diseases that could be paired with both US population prevalence data and Twitter-use data; and (2) diseases deemed by previous literature to be most impactful for the healthcare community. Each criterion is satisfied by a different dataset.
The first dataset comes from Experian, a global information services company known as a source of credit scores. Experian also conducts consumer surveys on a variety of topics, including health care. For this study, we used data from Experian's Simmons National Consumer Study and focused on survey questions pertaining to general demographics, health status, and social media use.
Results from the various Experian surveys are combined into a study universe database and released both quarterly and annually. Experian conducts post-stratification on its survey data to create demographically representative estimates for its measured variables. We queried this database to obtain a dataset for the year 2012 that cross-tabulates (a) disease prevalence for all available diseases (n=52) with (b) general demographics and with (c) Twitter use. For the estimated English-speaking or Spanish-speaking US adult population (n=230,124,220) we were able to find the estimated number who suffer from a disease (e.g., backache, n=42 million), and the subset of those disease sufferers who use Twitter (in the case of backache, n=2.6 million). This dataset, then, provides us with parallel sets of disease prevalence statistics for the general US population and for US Twitter users.
The second dataset is from a RAND study designed to broadly measure the quality of health care delivery in the US [22]. Through reviews of the literature and of national healthcare data, and through consultation with panels of medical experts, 46 “clinical areas” were identified in this report that represent the leading causes of illness, death, and healthcare utilization in the US.
The list of 24 diseases (Multimedia Appendix 1) used in the present study is composed of the overlap between the diseases represented in the Experian dataset (n=52) and in the RAND study (n=46). This overlap may be explicit (e.g., asthma appears on both lists) or implicit (e.g., two separate Experian entries, stomach ulcers and acid reflux disease/GERD, are both suggested by the single RAND entry peptic ulcer disease and dyspepsia). The focus in this task was not pinpointing exact matches between the two lists, but rather finding areas of general agreement between them, in order to identify high impact diseases from the Experian dataset.
Compilation of disease-terms
For each disease on our list, we constructed a lexicon of disease-terms that are used to refer to that disease. For example, the lexicon for diabetes used in this study contains three disease-terms: “diabetes”, “diabete”, “niddm.” All lexica in this study are derived from terms found in the Consumer Health Vocabulary (CHV) [23], an online open source thesaurus that associates medical concepts (including diseases, medical procedures, drugs, anatomy, etc.) with a mix of colloquial and technical terms. At the time of this study the CHV contains 158,519 entries, covering 57,819 unique (but often closely related) concepts. Each entry collects (along with other data) at least three term elements: (a) a CHV term, (b) a descriptive phrase, and (c) a related term from a medical vocabulary called the Unified Medical Language System (UMLS). A CHV term can have multiple entries in the thesaurus, thereby associating the CHV term with any number of descriptive phrases or UMLS terms. Each CHV term, then, can be seen as a key-value pair, where the CHV term is the key and a network of associated terms (comprised of descriptive phrases and UMLS terms) is the value.
For each of the 24 diseases included in the study, we processed the CHV to retrieve an entire key-value network of associated terms if any one of the terms (in the key or the value) seemed to refer to the target disease. Multiple networks could be (and often were) collected for any disease. Together, these results constituted a disease's list of candidate disease-terms (these were then vetted, in a process described below). A term was judged to be a potential reference to a target disease (thereby triggering the retrieval of all associated terms) primarily if it contained a search string derived from the target disease's name (including both abbreviated and spelled-out forms). For example, “attention deficit” is a search string for ADD/ADHD; “heart disease” is a search string for heart disease; and “GERD” is a search string for acid reflux disease/GERD. Also included among the search strings were some common disease synonyms, such as “zit” for acne and “tumor” for cancer. The number of search strings per disease varied, ranging from one to seven.
Tweet text corpus
The tweets used in our analysis were taken from a random sample of 1% of all available tweets in 2012, as collected through the Twitter “1% random public stream” API [24]. In order to align our data more closely to the American and mostly English-speaking Experian Simmons sample, we filtered our Twitter corpus to keep only English language tweets originating in the US. To filter for English we only considered tweets with at least 50% of their words found in the HunSpell English dictionary [25]. Tweets were then further restricted to the US by finding tweets with “United States” or non-ambiguous US cities in their location field. (City names were taken from [26]). For example, “Chicago” would match the US while “London”, even though there is a London in Texas, would not. This resulted in a corpus of 80,680,449 tweets.
Vetting disease-term candidates
Grammatical
In this research we focused on finding tweets that specifically name our target diseases. Broadening this focus to include related concepts, such as symptoms and treatments, was desirable but was not possible for the scope of this paper. Because of our focus on terms that name diseases (as opposed to terms that describe or suggest them), we dropped all candidate disease-terms that were not nouns (e.g., adjectives such as “depressed” or “arthritic”).
We then manually expanded the list, adding plural forms where grammatically appropriate.
Medical
We mined the CHV using a keyword search strategy inclined toward inclusiveness. For example, a search on “acne” retrieved terms for medical concepts that might be at best tenuously related to acne. One of these concepts was acne rosacea, whose network of associated terms contains the terms “acne rosacea”, “disorders rosacea”, “rosacea”, and “rosacea acne”.
Because the concept acne rosacea incorporates at least one term containing the text string “acne”, its entire network of terms automatically became candidates for the acne lexicon. This inclusiveness raises the question of whether “rosacea”, “acne rosacea”, “disorders rosacea”, etc., denote acne. To solve this problem, a physician on the research team vetted the candidate terms. For each disease, she dropped candidates that did not denote the disease, ensuring that only medically appropriate terms were admitted into any disease lexicon.
Structural
We produced a list of text strings for each disease that we could use to search the Twitter corpus for mentions of that disease. To achieve this goal we took into account two realities. (1) Many CHV term elements use constructions that are uncommon in natural language (e.g., “fever hay” as in nasal allergies/hay fever), “attack heart”, “attacks hearts”, or “attacking heart” as in heart attack, and “pain, back, radiating” as in back pain). (2) During execution of searches within the Twitter corpus, only the shortest element of a search phrase is required; if a compound search phrase contains a shorter search phrase, the longer one is implied by the shorter (e.g., “asthma” retrieves asthma, allergic asthma, pollen asthma, etc.; “diabetes” retrieves diabetes mellitus, insulin dependent diabetes, diabetes screening, etc.).
Because of these two facts, we were able to significantly shorten the candidate disease-term lists that were produced by the semi-automated CHV search procedure. All reverse-order candidates (e.g., “fever hay”) and compound candidates (e.g., “allergic asthma”) were dropped.
After these three vetting procedures, the 24 disease-lexica contained a combined 488 disease-terms (Multimedia Appendix 1).
Manual Tweet appraisal
We determined how often each of the 488 disease-terms referred to its associated disease when included in a tweet. This began with a basic count for each disease-term of the number of tweets in the corpus in which the term appears (prior to any language ambiguity corrections being applied). This is a disease-term's raw tweet count. Note that we allow a single tweet to be counted twice if it contains multiple disease terms (regardless of whether the two terms refer to the same or a different disease). Throughout this study we consider random instances of disease-terms as they appear in tweets, without consideration for other terms that co-occur with them.
We then performed manual appraisal. For each disease-term we randomly selected 30 tweets containing the term from our tweet corpus for manual analysis. This number was chosen to balance research needs and time constraints. Some disease-terms occurred in 30 or fewer tweets in the tweet corpus. When this occurred, all available tweets were retrieved.
Two English-speaking research assistants independently read each tweet and made a simple appraisal, answering, “For each tweet, in your judgment does the disease-term that flagged the tweet's retrieval refer to a medical meaning of that term?” Each tweet required a Yes or No judgment, as shown in Table 1. The two raters each compiled a complete collection of Y/N judgments, held in secret from the other rater. After these tweet-level appraisals were completed, we aggregated the scores at the disease-term level (independently for each rater's collection of judgments). For each rater and each disease-term (n=488), we calculated the percentage of tweets from the sample that were appraised as referring to a medical meaning. The Cohen's kappa for inter-rater reliability was 0.77.
Table 1.
Example of rating whether each tweet does or does not refer to a medical meaning of the selected term. Here the term is “heart attacks”.
| Rater 1 | Rater 2 | Tweet |
|---|---|---|
| Y | Y | Visited a man who has had 2 heart attacks who feels privileged to be in circumstances that allow him to share his trust in God. #realdeal |
| Y | N | Got room for 1 more? RT @pjones59: Sausage balls, heart attacks on a stick, dip, chips, wings and cheese, cream cheese/pickle/ham wraps |
| N | N | I still can't believe I saw Kris at work the other day. Talk about mini heart attacks. U_U |
The disease-term percentages of the two raters were then averaged, resulting in a correction factor (cf) for each disease-term. We multiplied this coefficient by the disease-term's raw tweet count (rcount), to arrive at an estimated disease-term validated tweet count (vcount).
Once this estimate was completed for each disease-term in a disease-lexicon, the disease-term estimates were summed, producing our ultimate metric, a validated tweet count for each disease-lexicon. The validated tweet count for a disease-lexicon is the estimated number of tweets in our corpus that are a valid reference to the disease in question, i.e., correcting for the ambiguity error present in the disease-lexicon's raw tweet count.
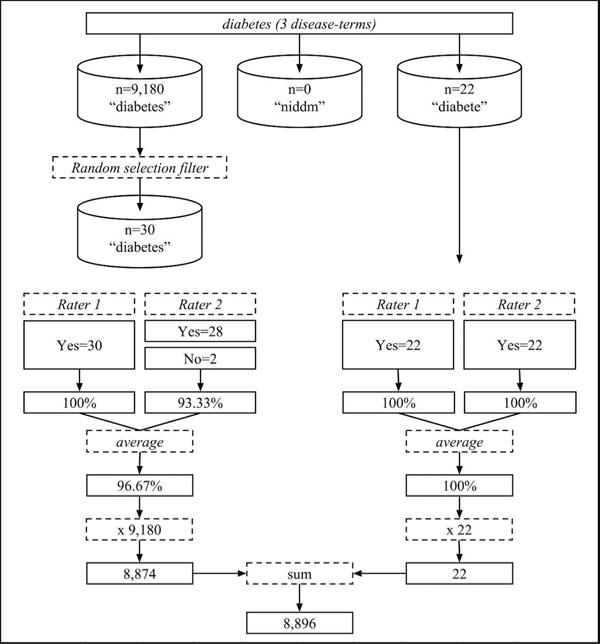
As an example, manual appraisal for the diabetes disease-lexicon (Figure 1) illustrates the evolution from a raw tweet count of 9,202 to a validated tweet count of 8,896.
Figure 1.
Disease-terms from the diabetes lexicon undergo manual appraisal. Each term receives appraisal on up to 30 instances. The term-level appraisals are then summed to reach the final lexicon-level diabetes validated tweet count (8,896).
Results
Of the 2,824 tweets containing disease-terms that we manually reviewed, the averaged judgments of our two human raters indicate that 2,276.5 (80.61%) actually referred to diseases, with validity rates that were highly variable across different diseases. For example, stroke terms rarely referred to the medical emergency (only 14.89% of the time), while diabetes terms almost always referred to the medical condition (96.67% of the time).
The raw tweet counts and validated tweet counts for the 24 diseases are compared in Table 2, along with a correction factor (an adjustment according to the percentage of evaluated tweets that were judged as valid). Table 2 also includes disease prevalence data (for both the general US population and among US Twitter users), which come directly from Experian's Simmons National Consumer Study. We see high levels of heterogeneity for all five measurements across diseases. This probably reflects the heterogeneity among the diseases themselves: included among them are acute viral infections (e.g., flu), general maladies (e.g., backache, nasal allergies/hay fever), chronic disorders (e.g., arthritis, osteoporosis), test measures (e.g., high cholesterol, hypertension/high blood pressure), medical emergencies (e.g., heart attack, stroke), and psychological disorders (e.g., depression, ADD/ADHD). Some of the diseases are transitory (e.g., urinary tract infection) and others are long-term (e.g., diabetes). Some are causes of mortality (e.g., cancer, congestive heart failure), while others are relatively superficial (e.g., acne). Given such variety, it is no surprise to see a wide range of values for tweet counts, correction factor, and prevalence across the list of diseases.
Table 2.
Raw and validated tweet counts, correction factor, and US and Twitter disease prevalence for each disease.a,b,c,d
| Disease | Raw tweet count | Validated tweet count | CF | Prev., US (millions) | Prev., US Twitter (millions) |
|---|---|---|---|---|---|
| acid reflux disease/GERD | 743 | 631 | 84.98 | 32.4 | 2.4 |
| acne | 6,936 | 6,027 | 86.89 | 11.2 | 2.0 |
| ADD/ADHD | 2,794 | 2,660 | 95.19 | 4.9 | 0.9 |
| arthritis | 2,524 | 2,522 | 99.92 | 34.4 | 1.3 |
| asthma | 3,952 | 3,754 | 95.00 | 12.4 | 1.0 |
| backache | 3,035 | 3,028 | 99.77 | 42.0 | 2.6 |
| cancer | 110,760 | 63,647 | 57.46 | 5.0 | 0.46 |
| congestive heart failure (CHF) | 928 | 313 | 33.76 | -- | -- |
| heart disease | 2,741 | 2,410 | 87.91 | -- | -- |
| CHF/heart disease | 3,669 | 2,723 | 74.21 | 5.9 | 0.46 |
| COPD | 226 | 188 | 83.37 | 5.5 | 0.86 |
| depression | 14,294 | 10,459 | 73.17 | 18.7 | 2.2 |
| diabetes | 9,202 | 8,896 | 96.67 | 20.8 | 1.2 |
| flu | 10,139 | 8,810 | 86.90 | 17.2 | 1.8 |
| genital herpes | 76 | 66 | 86.84 | 1.8 | 0.33 |
| heart attack | 15,027 | 2,311 | 15.38 | -- | -- |
| stroke | 12,852 | 1,914 | 14.89 | -- | -- |
| heart attack/stroke | 27,879 | 4,225 | 15.15 | 3.0 | 0.11 |
| high cholesterol | 225 | 218 | 96.67 | 37.9 | 1.7 |
| human papilloma virus | 636 | 545 | 85.73 | 1.5 | 0.12 |
| hypertension/high blood pressure | 1,630 | 1,491 | 91.49 | 43.5 | 1.5 |
| migraine headache | 5,958 | 5,615 | 94.24 | 16.4 | 1.8 |
| nasal allergies/hay fever | 481 | 473 | 98.27 | 18.2 | 1.3 |
| osteoporosis | 316 | 306 | 96.68 | 6.0 | 0.13 |
| stomach ulcers | 80 | 73 | 91.25 | 3.3 | 0.030 |
| urinary tract infection | 880 | 479 | 54.40 | 10.0 | 1.0 |
Header abbreviations: CF represents “correction factor” (i.e., the percentage of tweets that were appraised as valid); Prev., US (millions) represents a disease's prevalence in the US; Prev., US Twitter (millions) represents a disease's prevalence among US Twitter users.
The source for both Prev., US (millions) and Prev., US Twitter (millions) is the Experian Simmons National Consumer Study.
In the Experian dataset, congestive heart failure and heart disease are collapsed into a single data point. We mined Twitter for these diseases separately, and we applied our evaluation method to tweets containing disease-terms for each one separately. However, because Experian was our source for prevalence statistics, we can only report on the prevalence of these two diseases in a combined state.
The same is true for the diseases heart attack and stroke.
We determined the Spearman correlation coefficients (all P values < .001) between both raw and validated tweet counts and disease prevalence among both the general US population and among US Twitter users (Table 3). Correcting just for Twitter use more than doubles the correlation between tweet count and prevalence (from 0.113 to 0.258). Correcting only for word ambiguity has a similar but slightly smaller effect (0.208). Correcting for both more than triples the baseline correlation (0.366).
Table 3.
Spearman correlation coefficients among raw and validated tweet counts and US population and Twitter user disease prevalence (all P values <.001).
| Prevalence | Prevalence | |
|---|---|---|
| US pop. | US Twitter users | |
| Raw tweet count | 0.113 | 0.258 |
| Validated tweet count | 0.208 | 0.366 |
Discussion
Overview
The correlation improvements we found due to ambiguity correction may seem unsurprising. But the improvements due to demographic correction are less straightforward, particularly because no effort was made to restrict our tweet analysis to first-person self-report mentions of diseases. It is easy to assume that there must be a causal connection between disease prevalence and disease mentions. Indeed, we interpret an increased correlation due to demographic correction as supporting this assumption: it means the signal we measure (i.e., disease mentions on Twitter) demonstrates positive correspondence to a plausible source of that signal (i.e., disease sufferers who use Twitter). However, we find that for certain individual diseases, disease prevalence and disease mentions are wildly out of sync. For the time being just what causes someone to tweet (or not tweet) about a disease remains an open question, particularly since many people mentioning the disease are not suffering from it. In any case, methods utilizing social media to estimate disease prevalence do not need to explain a causal connection. They only demand that social media reliably captures the variance of disease prevalence. We have shown that such measurement can be improved by adjusting for demographic differences between disease sufferers and Twitter users.
Bias correction
We found that naively counting mentions of disease-terms in tweets produces results that are biased (in terms of correlation with known disease prevalence statistics) both due to the demographic pattern of Twitter users, and due to the ambiguity of natural language. These biases can be at least partially corrected, resulting in a three-fold increase in the correlation between counts of disease terms in tweets and known prevalence statistics for the 24 diseases we studied.
The observation that the Twitter population is a biased sample of the US is relatively easily corrected using standard stratified sampling methods, given the known demographics of the Twitter population. We identified this using data from an Experian survey, but other studies of Twitter demographics could be used. We demonstrated that the demographic corrections roughly doubled the correlation between disease mentions and disease prevalence.
Types of ambiguity
The intrinsic ambiguity of language requires more work to correct. We observed that language ambiguity varies significantly across diseases. The fraction of mentions of a disease term that actually refer to the disease ranged from highly specific terms like arthritis (99.92%) to less specific terms like stroke (14.89%). This language ambiguity takes two major forms.
The first is lexical ambiguity. Some diseases such as arthritis, diabetes, and high cholesterol are in practice referred to by terms that almost always refer to their associated disease concepts. In our analysis, tweeters rarely used words from the arthritis lexicon to refer to anything other than the disease arthritis. There are, however a number of disease-terms that are often used to refer to concepts that are not diseases (or not the intended diseases). Frequently occurring example words include “cancer” (the astrological sign), “depression,” “stroke” (non-medical usages and also heat stroke), and “flu” (as in stomach flu, versus influenza). Abbreviations are particularly ambiguous. For example, “copd” (i.e., chronic obstructive pulmonary disease) is a popular variant spelling of “copped” (as in the verb “took”); “uti” (urinary tract infection), “hpv” (human papillomavirus), and “zit” (acne) show up in Internet addresses (particularly in short links using URL redirection); and “CHF” (congestive heart failure) is an abbreviation for the Swiss Franc. Or conversely, “Gerd” is a masculine first name that coincides with an abbreviation for the disease gastroesophogeal reflux disease (part of the acid reflux disease/GERD lexicon). Lexical ambiguity also arises from metaphorical and slang usages of disease-terms. “Heart attack” and “heart failure” are used to mean surprise and “ADHD” to mean distracted.
The second type of ambiguity could be considered disease ambiguity. Some of the 24 diseases included in this study are less clearly delineated than others. One aspect of this problem is intensity. Is it medical depression if a Twitter user reports being depressed about her favorite sports team losing a game? What if she ends a seemingly grave tweet with “LOL”? A second aspect of disease ambiguity is specificity or accuracy. Some Twitter users may use the word migraine for other types of headache or say hay fever when actually they're allergic to cats. A third aspect of disease ambiguity is complexity. A prime example is the range of cardiovascular diseases in this study (i.e., congestive heart failure, heart disease, heart attack, high cholesterol, hypertension, possibly stroke), whose interrelations and exact boundaries are difficult or impossible to draw.
Both types of ambiguity can affect a disease's validation coefficient. The first type, lexical ambiguity (e.g., homographs or metaphorical word usage), is likely to affect the “back end” of the methodology, requiring corrections to the observed term counts (i.e., using the method described in this paper). The second type, disease boundary ambiguity, presents problems on the “front end” and it makes tailoring the disease lexica difficult. This type of ambiguity raises the question of whether the potentially hierarchical relationship between congestive heart failure and heart disease, or the potentially causal relationship between high cholesterol and either heart attack or stroke, could or should somehow be encoded in the disease-lexica.
In this research, we treated each disease-lexicon as a stand-alone entity, and the effects of that decision are necessarily written into the results we derived. We can expect that diseases of a more “stand-alone” quality (i.e., those that are relatively self-contained like osteoporosis, rather than part of a complex like heart disease) will naturally be better represented by their respective disease-lexica than are diseases that potentially harbor a complex relationship with other diseases. It is intuitive that mismatch between a disease's representation on Twitter and its representation within its disease-lexicon is essentially what causes the disease's validation coefficient to drop below 100%.
Correlation of validated tweet count with prevalence
Just as validity rates proved highly variable across diseases, the levels of Twitter discussion relative to disease prevalence also varied. Some diseases were discussed at levels outstripping their prevalence in the population, while others received little relative attention. The relationship between validated tweet count and US prevalence across diseases has a correlation of 0.208 (Table 3, P < .001). To provide a more detailed picture of this relationship, we calculated validated tweet count for each disease as a function of prevalence. The formula we used is, for each disease d: projected prevalence = (validated tweet count of d/sum of all validated tweet counts)*sum of all disease prevalences. This can be understood as the prevalence that validated tweet count (inaccurately) projects for each disease.
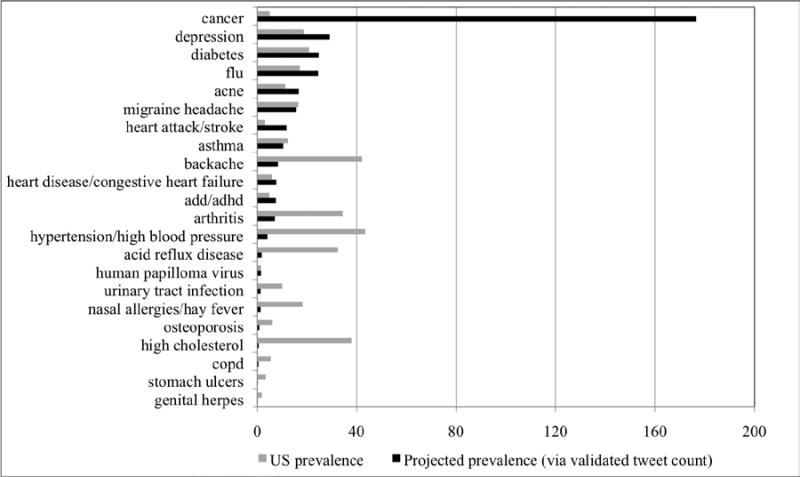
We compare this projected prevalence to actual prevalence in Figure 2. The sum of prevalence across all diseases (351,939,580) is identical for both the projected and the actual cases, but the distributions are quite different. We see that cancer is a major outlier, “taking up” over 50% of projected prevalence (176,605,210) while it accounts for less than 2% of actual prevalence (5,031,120). Clearly cancer receives far more attention than merely prevalence warrants. Projected prevalence is more than 35 times as great as, or 3,500% of, actual prevalence. Conversely, high cholesterol is on the extreme end of underrepresentation. Projected prevalence (604,898) is only 1.598% of actual prevalence (37,861,070). These figures demonstrate that other unknown factors besides prevalence influence the amount of discussion a disease receives on Twitter.
Figure 2.
Projected prevalence (as a function of validated tweet count) vs. actual US prevalence for 22 diseases, in millions (sorted by projected prevalence). Some diseases are “over-tweeted” (in particular, cancer) while others are “under-tweeted” (e.g., backache and arthritis).
One hypothesis is that Twitter demographics skew discussion levels upward for diseases that are of high concern to the population of users and downward for diseases that are of less concern. Given the generalization that Twitter users tend to be young, this could explain why arthritis seems to be drastically under-tweeted, and why acne and ADD/ADHD are over-tweeted. But demographics alone cannot explain the extraordinary projected prevalence of cancer. Nor are they likely to explain the over-tweeting of flu and diabetes. We assume that demographics do influence these results (see acne and hypertension/high blood pressure), but that multiple other factors also play roles. Likely candidates include the intensity and history of disease awareness and advocacy campaigns (see cancer, diabetes, and human papilloma virus); and disease stigma or body part stigma (see genital herpes and urinary tract infection). Investigation into these and other possible factors is an area for future research.
Limitations
It remains unclear precisely what is the nature of the relationship between disease discussion (on Twitter or even just in general) and disease prevalence. Twitter disease discussion is likely driven by many more factors than disease prevalence. People tweet about diseases for many reasons, and for the purposes of this paper, we do not attempt to disentangle such reasons. We do demonstrate, though, that Twitter disease mentions correlate with disease prevalence, and that this correlation improves after our demographic and word ambiguity corrections have been applied. This lesson can and should be incorporated into other research or tools that would seek to mine the language found on Twitter (or similar venues) for information about broader populations.
Despite our best efforts, the demographics of our Twitter corpus and of the Experian dataset do not entirely match. Most significantly, the Experian dataset includes disease prevalence estimates for both English-speaking and Spanish-speaking US residents whereas our tweet corpus was restricted to English language tweets. This research was conducted in English; future work should extend a similar analysis to other languages.
We did not account in this research for all possible variables that could influence the interplay of disease prevalence and tweets about diseases. Some of these missed variables are disease-centric. For example, some diseases may actually be more “tweet-able” than others due to any number of disease factors, including intensity, duration, stigma, social salience, and so on, or possibly even due to formal considerations (is the disease easy or quick to spell?). A less tweet-able disease might be expected to have fewer associated tweets, outside of any prevalence-based influence on tweet counts.
We only account for mentions of diseases that specifically name a (properly spelled) disease in a tweet. On the one hand, relying on correct noun-form disease names that are sourced from a recognized health vocabulary like the CHV helps push this study toward semi-automation, objectivity, and reproducibility. But on the other hand, this decision leaves an unknown, but possibly large, quantity of disease-relevant tweets unmined, and so unaccounted for in our analysis. We miss mentions that are slang terms (e.g., “diabeetus”) or are misspelled (e.g., “ashtma”, “hi colesterol”, etc.). On a strictly formal level, our current approach is tuned to precision at the expense of recall. Furthermore, people may discuss health concerns on Twitter by mentioning symptoms, sequelae, locations (such as a hospital), drugs, or treatments, etc. Our focus on disease names is unable to capture this broader domain of health-related tweets. Improving recall is left for future work.
Other missed variables are Twitter-centric. It is well documented that Twitter does not reveal what sampling procedures are used in their APIs [27,28]. Therefore it is unclear how representative the tweeters (whose tweets were captured for this study) are of the US population of Twitter users. This is unavoidable, and it is a shortcoming common to all research using Twitter APIs.
We also did not discriminate in this research between tweeters. A Twitter “user” may not be an individual person. Many health-related or even disease-related organizations mention diseases on Twitter. Factors related to such organizations (their quantity, their social media strategies, etc.) may be relevant to counts of disease-naming tweets. Other researchers have addressed the problem of distinguishing tweets authored by health organizations [29], but this study did not make that distinction.
Comparison with Prior Work
In the normal course of life or business individuals and organizations generate vast amounts of text that can be mined. Much of it is shared or published online in one form or another, and this data is attractive to researchers, including those interested in epidemiology and public health.
Several “infodemiology” studies (e.g., many in the long list cited in the introduction) correlate word use in Twitter with prevalence of a disease or medical condition. Beyond Twitter, web search activity has also been used, for example to monitor Lyme disease [30] and dengue [31], as well as risk behaviors associated with dietary habits [32] and with suicide [33]. Blog posts have been used to predict influenza outbreaks [34]. Facebook has been used to predict “gross national happiness,” i.e., well-being across the US [35].
These studies primarily correlate word use in some medium (e.g., Twitter or Google search) over some time period (e.g., day or week) and in some region (e.g., US county or state) with a disease prevalence level. Such correlational approaches rely on certain assumptions about the homogeneity of the populations they study, which often go unstated and so presumably untested and uncorrected. It is not clear that demographic or word ambiguity biases are typically accounted for. We suppose that researchers implicitly assume that these factors will be handled automatically by the statistical regression methods they use. If demographic and ambiguity biases are constant over time and space, this will be true. However, if populations vary in their usage of the target medium (e.g., Twitter) prediction accuracy can vary, and this variation may be significant.
Google Flu Trends is perhaps the “poster child” for the correlational approach to prediction. It is a widely cited online tool that uses statistical correlations between a broad set of Google search terms and historical flu levels to predict regional changes in US flu levels [36]. Google Flu Trends was initially highly accurate. But it has also been used as a case study of how “big data” predictions can go awry when the statistical patterns upon which they are based are descriptively inaccurate (either from the start or due to drift over time) [37,38], with claims that at one point predictions became exaggerated by nearly a factor of two [39].
A limited number of studies have emphasized concerns about validity in social media analyses [40]. There has also been some work on selecting “high quality” disease-related tweets, mostly achieving high specificity at the cost of poor sensitivity. For example, [41] uses regular expressions and machine learning methods to filter out all but first person self-report tweets. We strive for higher coverage, including all “real” mentions of a disease, and then we seek out previously established data (i.e., disease prevalence) against which to validate our findings. In [42] researchers identify known sick persons, then study their Twitter data to characterize a kind identifying fingerprint for Twitter users who have the flu (utilizing their tweets and their Twitter profile metadata). They then use that model to “diagnose” individual Twitter users with influenza, an approach that the authors imply could be directed toward population level disease surveillance.
In the near term, we think the major use of social media for public health may be to understand attitudes toward health, disease, and treatment. Effective public policy depends on subjective inquiries into what people know and care about. Why do they seek or avoid treatment? How do they reveal disease status? What risk behaviors do they shrug off? Predictions about a phenomenon that one can measure, such as disease prevalence, may have limited utility, especially if the measurements are timely and accurate. Although traditional ground truth measurements have been questioned [43], Centers for Disease Control and Prevention flu estimates appear to be better than Google Flu Trends estimates [44]. Nevertheless, online disease detection and prediction is a rapidly growing research area, and as work continues in this field, our collective ability to make these types of estimates will likely increase.
Conclusions
Several types of research using social media to study public health will benefit from corrections for demographic variation and language ambiguity of the type that are outlined in this paper. Social media datasets are biased convenience samples, and word ambiguity is endemic. Nevertheless, social media provide a relatively cheap way to monitor countless domains, including public health and attitudes toward health and health care.
In this study we began with a large, “poor quality,” non-random dataset (i.e., Twitter), and compared it to a small, “high quality,” random (achieved via post-stratification) dataset, (i.e., the Simmons National Consumer Study from Experian). We filtered the Twitter dataset so that its demographics would match the Experian dataset's. Then we performed both naïve and ambiguity-corrected counts of Twitter disease mentions. Finally we compared both those counts to prevalence data found in the Experian survey. We found that the corrected Twitter counts correlated much more strongly than the naïve Twitter counts with the “high quality” Experian data. We think that this demonstrates both the need and the capacity for other studies using non-random convenience samples (e.g., social media data or Google queries) to take demographic and word ambiguity factors explicitly into account, for example by using our method or other related or novel methods.
Abbreviations
- ADD/ADHD
attention deficit disorder/attention deficit hyperactivity disorder
- API
Application Programming Interface
- CHF
congestive heart failure
- CHV
Consumer Health Vocabulary
- COPD
chronic obstructive pulmonary disease
- GERD
gastroesophageal reflux disease
- UMLS
Unified Medical Language System
Multimedia Appendix
Multimedia Appendix 1. This study focuses on a list of 24 diseases. Each of these diseases is represented by a disease lexicon composed of one or more disease-terms. There are 24 lexica, comprising 488 total disease terms. The first row in this appendix holds lexica names, subsequent rows hold disease-terms. Each column represents a different disease.
Footnotes
Conflicts of Interest
None declared.
Contributor Information
Christopher Weeg, Psychology, University of Pennsylvania, Philadelphia, PA, USA.
H Andrew Schwartz, Computer and Information Science, University of Pennsylvania, Philadelphia, PA, USA.
Shawndra Hill, Operations and Information Management, University of Pennsylvania, Philadelphia, PA, USA.
Raina M Merchant, Penn Medicine Social Media and Health Innovation Lab, Penn Medicine Center for Health Care Innovation, University of Pennsylvania, Philadelphia, PA, USA.
Catalina Arango, Wharton School, University of Pennsylvania, Philadelphia, PA, USA.
Lyle Ungar, Computer and Information Science, University of Pennsylvania, Philadelphia, PA, USA.
References
- 1.Collier N, Son NT, Nguyen NM. OMG U got flu? Analysis of shared health messages for bio-surveillance. Journal of Biomedical Semantics. 2011;2(Suppl 5):S9. doi: 10.1186/2041-1480-2-S5-S9. doi:10.1186/2041-1480-2-S5-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul MJ, Dredze M. Proceedings of the Fifth International AAAI Conference on Weblogs and Social Media (ICWSM) Barcelona, Spain: Jul 17-21, 2011. You Are What You Tweet: Analyzing Twitter for public health. pp. 265–272. ISBN: 781577355052. [Google Scholar]
- 3.Chunara R, Andrews JR, Brownstein JS. Social and news media enable estimation of epidemiological patterns early in the 2010 Haitian cholera outbreak. The American Journal of Tropical Medicine and Hygiene 2012. 86(1):39–45. doi: 10.4269/ajtmh.2012.11-0597. doi:10.4269/ajtmh.2012.11-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chew C, Eysenbach G. Pandemics in the age of Twitter: content analysis of Tweets during the 2009 H1N1 outbreak. PLoS One 2010. 5(11):e14118. doi: 10.1371/journal.pone.0014118. doi:10.1371/journal.pone.0014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Signorini A, Segre AM. Polgreen, PM. The use of Twitter to track levels of disease activity and public concern in the U.S. during the influenza A H1N1 pandemic. PLoS One. 2011;6(5):e19467. doi: 10.1371/journal.pone.0019467. doi:10.1371/journal.pone.0019467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szomszor M, Kostkova P, De Quincey E. # swineflu: Twitter predicts swine flu outbreak in.. 3rd International ICST Conference on Electronic Healthcare for the 21st Century (eHEALTH2010); Casablanca, Morocco. 2010 Dec 13-15; Berlin: Springer Berlin Heidelberg; 2009. 2012. pp. 18–26. ISBN:9783642236358. [Google Scholar]
- 7.De Choudhury M, Counts S, Horvitz E. Predicting Postpartum changes in emotion and behavior via social media.. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems; Paris, France. 2013 Apr 27-May 2; pp. 3267–3276. doi:10.1145/2470654.2466447. [Google Scholar]
- 8.Sullivan SJ, Schneiders AG, Cheang CW, Kitto E, Lee H, Redhead J, et al. “What”s happening?' A content analysis of concussion-related traffic on Twitter. Br J Sports Med. 2012;46(4):258–63. doi: 10.1136/bjsm.2010.080341. doi:10.1136/bjsm.2010.080341. [DOI] [PubMed] [Google Scholar]
- 9.McNeil K, Brna PM, Gordon KE. Epilepsy in the Twitter era: A need to re-tweet the way we think about seizures. Epilepsy & Behavior. 2012;23(2):127–130. doi: 10.1016/j.yebeh.2011.10.020. doi:10.1016/j.yebeh.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Nascimento TD, DosSantos MF, Danciu T, DeBoer M, van Holsbeeck H, Lucas SR, et al. Real-time sharing and expression of migraine headache suffering on Twitter: A cross-sectional infodemiology study. Journal of Medical Internet Research. 2014;16(4):e96. doi: 10.2196/jmir.3265. doi:10.2196/jmir.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyles CR, Lopez A, Pasick R, Sarkar U. “5 mins of uncomfyness is better than dealing with cancer 4 a lifetime”: an exploratory qualitative analysis of cervical and breast cancer screening dialogue on Twitter. J Cancer Educ. 2013;28(1):127–33. doi: 10.1007/s13187-012-0432-2. doi:10.1007/s13187-012-0432-2. [DOI] [PubMed] [Google Scholar]
- 12.Scanfeld D, Scanfeld V, Larson EL. Dissemination of health information through social networks: Twitter and antibiotics. American Journal of Infection Control. 2010;38(3):182–188. doi: 10.1016/j.ajic.2009.11.004. doi:10.1016/j.ajic.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakhasi A, Passarella R, Bell SG, Paul MJ, Dredze M, Pronovost P. Malpractice and malcontent: Analyzing medical complaints in Twitter.. 2012 AAAI Fall Symposium Series; Arlington, VA. 2012 Nov 2-4.pp. 84–85. [Google Scholar]
- 14.Heaivilin N, Gerbert B, Page JE, Gibbs JL. Public health surveillance of dental pain via Twitter. Journal of Dental Research. 2011;90(9):10471051. doi: 10.1177/0022034511415273. doi:10.1177/0022034511415273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salathé M, Khandelwal S. Assessing vaccination sentiments with online social media: Implications for infectious disease dynamics and control. PLoS Comput Biol. 2011;7(10):e1002199. doi: 10.1371/journal.pcbi.1002199. doi:10.1371/journal.pcbi.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner J, Smith A. 72% of online adults are social networking site users. Pew Research Center; Washington, DC: Aug 5, 2013. http://www.pewinternet.org/files/old- media/Files/Reports/2013/PIP Social networking sites update PDF.pdf Archived at: http://www.webcitation.org/6SGvEVQFj. [Google Scholar]
- 17.Duggan M, Smith A. Social media update 2013. Pew Research Center; Washington, DC: Jan. 2014. http://www.pewinternet.org/files/2013/12/PIP Social-Networking-2013.pdf. Archived at: http://www.webcitation.org/6SGvMaF2d. [Google Scholar]
- 18.Mislove A, Lehmann S, Ahn Y, Onnela J, Rosenquist JN. Understanding the demographics of Twitter users.. Proceedings of the Fifth International AAAI Conference on Weblogs and Social Media (ICWSM); Barcelona, Spain. 2011 July 1721; pp. 554–557. ISBN:781577355052. [Google Scholar]
- 19.Khan AS, Fleischauer A, Casani J, Groseclose SL. The Next Public Health Revolution: Public Health Information Fusion and Social Networks. American Journal of Public Health. 2010;100(7):1237–1242. doi: 10.2105/AJPH.2009.180489. doi:10.2105/AJPH.2009.180489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernardo TM, Rajic A, Young I, Robiadek K, Pham MT, Funk JA. Scoping review on search queries and social media for disease surveillance: a chronology of innovation. Journal of medical Internet research. 2013;15(7):e147. doi: 10.2196/jmir.2740. doi:10.2196/jmir.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Experian Marketing Services Simmons National Consumer Study. http://www.experian.com/simmons-research/consumer-study.html. Archived at: http://www.webcitation.org/6SGtWFNi5.
- 22.Kerr EA, Asch SM, Hamilton EG, McGlynn EA, editors. Quality of care for general medical conditions: A review of the literature and quality indicators. RAND; Santa Monica, CA: 2000. ISBN:0833029169. [Google Scholar]
- 23.Consumer Health Vocabulary. http://consumerhealthvocab.org/. Archived at: http://www.webcitation.org/6SGtoLBel.
- 24.Twitter Public streams API documentation. https://dev.twitter.com/docs/streaming-apis/streams/public. Archived at: http://www.webcitation.org/6SGu2qB7Z.
- 25.Hunspell dictionary. http://hunspell.sourceforge.net/. Archived at: http://www.webcitation.org/6SGuCquNN.
- 26.US Cities List. http://www.uscitieslist.org/. Archived at: http://www.webcitation.org/6SGuPqocl.
- 27.González-Bailón S, Wang N, Rivero A, Borge-Holthoefer J, Moreno Y. Assessing the bias in samples of large online networks. Social Networks. 2014;38:16–27. doi:10.1016/j.socnet.2014.01.004. [Google Scholar]
- 28.Morstatter F, Pfeffer J, Liu H, Carley KM. Is the sample good enough? Comparing data from Twitter's streaming API with Twitter's firehose.. Proceedings of the Seventh International AAAI Conference on Weblogs and Social Media (ICWSM); Cambridge, MA. 2013 July 8-11; pp. 400–408. ISBN:9781577356103. [Google Scholar]
- 29.Dumbrell D, Steele R. Twitter and health in the Australian context: What types of information are health-related organizations tweeting?. Proceedings of the 46th Annual Hawaii International Conference on System Sciences (HICSS); Maui, HI. 2013 Jan 7-10; pp. 2666–2675. doi:10.1109/HICSS.2013.578. [Google Scholar]
- 30.Seifter A, Schwarzwalder A, Geis K, Aucott J. The utility of “Google Trends” for epidemiological research: Lyme disease as an example. Geospatial Health. 2010;4(2):135–137. doi: 10.4081/gh.2010.195. [DOI] [PubMed] [Google Scholar]
- 31.Chan EH, Sahai V, Conrad C, Brownstein JS. Using web search query data to monitor dengue epidemics: a new model for neglected tropical disease surveillance. PLoS Neglected Tropical Diseases. 2011;5(5):e1206. doi: 10.1371/journal.pntd.0001206. doi:10.1371/journal.pntd.0001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West R, White RW, Horvitz E. From cookies to cooks: Insights on dietary patterns via analysis of Web usage logs.. Proceedings of the 22nd International World Wide Web Conference; Rio de Janeiro, Brazil. 2013 May 13-17; pp. 1399–1410. ISBN:9781450320351. [Google Scholar]
- 33.McCarthy MJ. Internet monitoring of suicide risk in the population. Journal of Affective Disorders. 2010;122(3):277–279. doi: 10.1016/j.jad.2009.08.015. doi:10.1016/j.jad.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corley CD, Cook DJ, Mikler AR, Singh KP. Text and structural data mining of influenza mentions in Web and social media. International Journal of Environmental Research and Public Health. 2010;7(2):596–615. doi: 10.3390/ijerph7020596. doi:10.3390/ijerph7020596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer ADI. An unobtrusive behavioral model of gross national happiness.. Proceedings of the 28th International Conference on Human Factors in Computing Systems (CHI 2010); Atlanta, GA. 2010 Apr 10-15; 2010. pp. 287–290. doi:10.1145/1753326.1753369. [Google Scholar]
- 36.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457(7232):1012–1014. doi: 10.1038/nature07634. doi:10.1038/nature07634. [DOI] [PubMed] [Google Scholar]
- 37.Lazer D, Kennedy R, King G, Vespignani A. The parable of Google Flu: Traps in big data analysis. Science. 2014;343(6176):1203–1205. doi: 10.1126/science.1248506. doi:10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- 38.Olson DR, Konty KJ, Paladini M, Viboud C, Simonsen L. Reassessing Google Flu Trends data for detection of seasonal and pandemic influenza: A comparative epidemiological study at three geographic scales. PLoS Computational Biology. 2013;9(10):e1003256. doi: 10.1371/journal.pcbi.1003256. doi:10.1371/journal.pcbi.1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler D. When Google got flu wrong. Nature. 2013;494(7436):155–156. doi: 10.1038/494155a. doi:10.1038/494155a. [DOI] [PubMed] [Google Scholar]
- 40.Kass-Hout TA, Alhinnawi H. Social media in public health. British Medical Bulletin. 2013;108(1):5–24. doi: 10.1093/bmb/ldt028. doi:10.1093/bmb/ldt028. [DOI] [PubMed] [Google Scholar]
- 41.Prieto VM, Matos S, Alvarez M, Cacheda F, Oliveira JL. Twitter: A good place to detect health conditions. PLoS ONE. 2014;9(1):e86191. doi: 10.1371/journal.pone.0086191. doi:10.1371/journal.pone.0086191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodnar T, Barclay VC, Ram N, Tucker CS, Salathé M. On the ground validation of online diagnosis with Twitter and medical records.. Proceedings of the 23rd International Conference on World Wide Web companion volume; Seoul, Korea. 2014 Apr 7-11; pp. 651–656. ISBN:9781450327459. [Google Scholar]
- 43.Reed C, Angulo FJ, Swerdlow DL, Lipsitch M, Meltzer MI, Jernigan D, Finelli L. Estimates of the prevalence of pandemic (H1N1) 2009, United States, April-July 2009. Emerging Infectious Diseases 2009. 15(12):20042007. doi: 10.3201/eid1512.091413. doi:10.3201/eid1512.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ortiz JR, Zhou H, Shay DK, Neuzil KM, Fowlkes AL, Goss CH. Monitoring influenza activity in the United States: A comparison of traditional surveillance systems with Google Flu Trends. PLoS ONE. 2011;6(4):e18687. doi: 10.1371/journal.pone.0018687. doi:10.1371/journal.pone.0018687. [DOI] [PMC free article] [PubMed] [Google Scholar]