Abstract
Rotaviruses are a major cause of viral gastroenteritis in children. For accurate and sensitive detection of rotavirus RNA from stool samples by reverse transcription-polymerase chain reaction (RT-PCR), the extraction process must be robust. However, some extraction methods may not remove the strong RT-PCR inhibitors known to be present in stool samples. The objective of this study was to evaluate and compare the performance of six extraction methods used commonly for extraction of rotavirus RNA from stool, which have never been formally evaluated: the MagNA Pure Compact, KingFisher Flex and NucliSENS® easyMAG® instruments, the NucliSENS® miniMAG® semi-automated system, and two manual purification kits, the QIAamp Viral RNA kit and a modified RNaid® kit. Using each method, total nucleic acid or RNA was extracted from eight rotavirus-positive stool samples with enzyme immunoassay optical density (EIA OD) values ranging from 0.176 to 3.098. Extracts prepared using the MagNA Pure Compact instrument yielded the most consistent results by qRT-PCR and conventional RT-PCR. When extracts prepared from a dilution series were extracted by the 6 methods and tested, rotavirus RNA was detected in all samples by qRT-PCR but by conventional RT-PCR testing, only the MagNA Pure Compact and KingFisher Flex extracts were positive in all cases. RT-PCR inhibitors were detected in extracts produced with the QIAamp Viral RNA Mini kit. The findings of this study should prove useful for selection of extraction methods to be incorporated into future rotavirus detection and genotyping protocols.
Introduction
Group A rotaviruses are well established as the major cause of acute viral gastroenteritis in infants and young children worldwide. Rotaviruses belong to the Reoviridae family and possess a genome of 11 segments of double-stranded RNA (dsRNA). The binomial classification system for rotaviruses is based on the two outer capsid proteins, VP7 (G genotype) and VP4 (P genotype) (Estes and Kapikian, 2007). At least 27 G and 35 P genotypes have been designated for human and animal strains (Matthijnssens et al., 2011). Five strains, G1P1A[8], G2P1B [4] G3P1A[8], G4P1A[8], and G9P1A[8] are the globally predominant human pathogens (Gentsch et al., 2005) and have been targeted in vaccine development.
Two live-attenuated oral vaccines, RotaTeq® (Merck, Whitehouse Station, NJ, USA) and Rotarix® (GlaxoSmithKline, Research Triangle Park, NC, USA) have been introduced into childhood immunization programs in the United States and other countries (Glass et al., 2006). RotaTeq® is a pentavalent human-bovine reassortant rotavirus vaccine that includes genes of human rotavirus serotypes G1-G4 and P1A[8]. Rotarix® vaccine is a monovalent vaccine derived from a G1P1A[8] human rotavirus strain. Transmission and shedding of rotavirus vaccine strains has been reported (Donato et al., 2012; Payne et al., 2010; Rivera et al., 2011; Yen et al., 2011).
To monitor circulating rotavirus serotypes before and after vaccine introduction, including any possible emerging or novel strains post-vaccine introduction, many countries conduct regional rotavirus strain surveillance programs. In the United States, surveillance by the Centers for Disease Control and Prevention (CDC), in collaboration with laboratories of the National Rotavirus Strain Surveillance System (NRSSS) (Griffin et al., 2000; Ramachandran et al., 1998), and the New Vaccine Surveillance Network (Payne et al., 2008), has been ongoing since 1996 and 2006, respectively.
Rotavirus strain surveillance programs typically use reverse transcription-polymerase chain reaction (RT-PCR)-based methods to determine rotavirus genotypes directly from RNA extracted from stool specimens (Das et al., 1994; Gentsch et al., 1992; Gouvea et al., 1990) and rotavirus detection by real time RT-PCR (qRT-PCR) is increasing in use (Freeman et al., 2008). Fecal samples are among the most difficult clinical samples to process because of the presence of very potent inhibitors of nucleic acid amplification such as complex polysaccharides, bilirubin and bile salts (Chiu and Ou, 1996; Monteiro et al., 1997; Pandey et al., 1996). Difficulty in eliminating RT-PCR inhibitors from stool extracts has been reported extensively (Lantz et al., 1997; Makristathis et al., 1998; Monteiro et al., 1997; Petrich et al., 2006). Inhibitory effects can be reduced by adding amplification facilitators such as bovine serum albumin to the PCR reaction (Kreader, 1996), using thermostable polymerases that are more resistant to PCR inhibition (Abu Al-Soud and Radstrom, 1998), or using more efficient processes for extracting nucleic acid from stool samples. The efficiency of nucleic acid extraction and purification influences the sensitivity, reproducibility and the accuracy of RT-PCR target detection (Lim et al., 2005). During the last 10 years, several new manual, semi-automated and automated commercial nucleic acid or RNA extraction systems using magnetic beads or silica particles have been developed for DNA, RNA or total nucleic acid extraction which are attractive due to their flexibility, convenience, and ease of use (Tang et al., 2005a). A small number of studies have compared some of these novel extraction methods and reported that they differ in their ability to recover viral RNA, indicating that no single RNA extraction method is optimal for all viruses (Baert et al., 2007; Hale et al., 1996; Kok et al., 2000; Petrich et al., 2006).
The most recent study comparing nucleic acid extraction methods for rotavirus detection in stool was published in 2002 (Rasool et al., 2002). Since then, a number of new nucleic acid extraction systems using magnetic beads or silica particles have been developed, both in automated and manual formats (Dundas et al., 2008b; Perelle et al., 2009; Schuurman et al., 2007; Tang et al., 2005b). Although some of these extraction systems have been used extensively for the extraction of rotavirus RNA from stool and other clinical and environmental samples (Banyai et al., 2011; Doan et al., 2011; Esona et al., 2010a; Esona et al., 2010b; Freeman et al., 2008; Hull et al., 2011; Matthijnssens et al., 2006; Rahman et al., 2010; Rahman et al., 2007a; Rahman et al., 2007b; Zeller et al., 2010), they have not been evaluated formally to determine which method is most efficient for extraction of rotavirus RNA.
The objective of this study was to identify the best commercially-available nucleic acid extraction method for preparing stool samples for detection of rotaviruses by both conventional RT-PCR and qRT-PCR. This information will be very useful for rotavirus surveillance programs, diagnostics, and epidemiologic studies.
Materials and Methods
Stool samples
Eight rotavirus positive stool samples with optical density (OD) values ranging from 0.176 to 3.098 , as determined by enzyme immunoassay (EIA) (Premier™ RotaClone®, Meridian Bioscience, Cincinnati, OH, USA), were used to prepare 10% suspensions in phosphate-buffered saline solution. These samples (5627:EIA OD=0.854, genotype G2P[4]; 5628:OD=0.176, G2P[4]; 5629:OD=0.547, G2P[4]; 5630:OD=3.098, G4P[8]; 5631:OD=2.075, G4P[8]; 5633:OD=1.768, G2P[4]; 5634:OD=0.318, G1P[8]; and 5635:OD=0.315, G1P[8]) were collected in Nicaragua during the 2009 rotavirus season.
Extraction methods
Six commercial extraction methods for total nucleic acid or RNA were evaluated: the automated MagNA Pure Compact (Roche Applied Science, Indianapolis, IN, USA), KingFisher Flex-96 (ThermoFisher, Pittsburgh, PA, USA), and NucliSENS® easyMAG® (BioMerieux, Durham, NC, USA) methods; the semi-automated NucliSENS® miniMAG® (BioMerieux, Durham, NC, USA); and the manual QIAamp Viral RNA mini kit (Qiagen,Valencia, CA, USA) and RNaid® kit (Qbiogene, Carlsbad, CA, USA) with a modified protocol) (Table 1). Each extraction method was chosen because it had been used previously in studies for the extraction of nucleic acid from stool. The MagNA Pure LC instrument has been used to purify nucleic acid from many types of clinical specimens including stool samples for rotavirus (Dalesio et al., 2004; Petrich et al., 2006; Schmid et al., 2004; Wolk et al., 2002; Wu et al., 2011) but is no longer in production so the MagNA Pure Compact instrument, which uses the same paramagnetic bead technology, was evaluated instead . The MagNA Pure Compact was used with the MagNA Pure Compact RNA Isolation Kit (Roche Applied Science Indianapolis, IN, USA). The easyMAG and miniMAG extraction platforms utilize the same silica extraction technology and use the same reagents. Both methods are universal and can be applied to a broad range of different specimens such as blood, sputum, serum, throat swabs and stool (Dundas et al., 2008a; Freeman et al., 2008; Loens et al., 2008; Perelle et al., 2009; Tang et al., 2005b). Both methods have been used for extraction of rotavirus RNA from stool (Hull et al., 2011). The easyMAG and miniMAG are mainly used in laboratories that require moderate- and low-throughput, respectively (Yang et al., 2011). The KingFisher Flex (96 deep well magnet head) designed for high throughput laboratories, is an automated extractor also used for rotavirus RNA extraction (Freeman et al., 2008; Grant et al., 2011; Hull et al., 2011). In this study it was used with the MagMax™- 96 Total RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). The QIAamp Viral RNA Mini kit, which is based on silica membrane technology, has been used extensively for the extraction of rotavirus RNA from stool (Doan et al., 2011; Matthijnssens et al., 2006; Rahman et al., 2007a; Rahman et al., 2007b; Zeller et al., 2010).
Table 1.
Commercial extraction methods compared in this study.
| Instrument | Kit | Vendor | Core Technology | Mode of extraction |
Extracted product |
Sample vol. processed (μl) |
Elution vol. (μl) |
|---|---|---|---|---|---|---|---|
| Not applicable | RNaid® RNA extraction Kit |
Qbiogene Carlsbad, CA |
Glass powder | Manual | RNAa | 200 | 50 |
| Not applicable | QIAamp Viral RNA Mini Kit |
Qiagen Valencia, CA |
Silica membrane | Manual | RNAa | 140 | 60 |
| MagNa Pure Compact |
MagNA Pure Compact RNA Isolation Kit |
Roche Applied Science Indianapolis, IN |
Magnetic beads | Automated | RNA | 100 | 100 |
| KingFisher Flex 96 |
MagMax™-96 Total RNA Isolation Kit |
ThermoFisher Pittsburgh, PA |
Magnetic beads | Automated | RNAa | 50 | 75 |
| NucliSENS® easyMAG® |
NucliSENS® Nucleic Acid extraction Reagents for easyMAG® |
BioMerieux Durham, NC |
Magnetic beads | Automated | TNAb | 200 | 55 |
| NucliSENS® miniMAG® |
NucliSENS® Nucleic Acid extraction Reagents for miniMAG® |
BioMerieux Durham, NC |
Magnetic beads | Semi- automated |
TNAb | 200 | 50 |
These RNA extraction kits do not utilize a DNase treatment step and thus the extracted product may also contain DNA.
Total nucleic acid.
The RNaid kit extraction protocol, which uses glass powder to capture and purify rotavirus RNA, has been modified and used by the CDC Rotavirus Surveillance Laboratory routinely since the 1990’s along with other methods to extract rotavirus RNA from stool. (Gentsch et al., 1992; Jothikumar et al., 2009). In brief, 200 −L of clarified 10% stool suspension was mixed directly with 400 −L 6M guanidine thiocyanate (Ultrapure Grade; Boehringer Mannheim, Germany) and 10 −L of RNAID glass powder. The mixture was then vortexed and mixed on a Nutator rocker (Clay Adams Division, Becton-Dickinson, Parsippany, NJ, USA) for 15 min at room temperature. Each sample was then centrifuged for 60 s at 650 × g and the supernatant was removed by aspiration with a Pasteur pipette. The samples were washed once with 700 −L of guanidine thiocyanate wash solution (4 M guanidine thiocyanate, 16.7 mM Tris-HCl, pH 7.5) and centrifuged at 850 × g for 60 s. The supernatant was aspirated and the samples were then washed two times with 400 −L of the RNAID kit wash buffer and centrifuged at 850 × g for 60 s. The supernatant was aspirated, and the samples were washed once more with the same buffer and were then finally centrifuged at 10,000 × g for 120 s. After aspiration of the supernatant, the samples were dried at room temperature for 1 hour, resuspended in 35 −L of nuclease free water, and incubated for 10 min at 65°C. The samples were centrifuged at 10,000 × g for 120 s, and the supernatant was transferred to microcentrifuge tubes (Lube Tube; Marsh Co., Rochester NY, USA). The pellet was re-extracted with the same volume of water (35 −L), and the combined supernatants were stored at −80°C until they were used. Immediately before use for PCR, the supernatants were incubated at 56°C for 5 min and were then centrifuged at 10,000 × g for 60 s to pellet the residual RNAID from the sample.
With the exception of the RNaid kit as described, extractions were performed according to kit and instrument protocols. Single aliquots of each stool suspension and a negative control stool were extracted in triplicate by each method.
qRT-PCR
The total nucleic acid or RNA extracts were amplified in duplicate by rotavirus NSP3 gene qRT-PCR (Freeman et al., 2008) modified for use on ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with the 7500 Fast SDS software v1.4.0. No-template and positive controls were included in each run. All assays were run using the standard mode on the ABI 7500 Fast instrument. Before beginning the TaqMan cycling conditions, each RNA extract was first denatured in a thermocycler at 95°C for 5 min followed by a 1 min incubation on ice. The thermocycling program used was 30 min reverse transcription at 50°C (1 cycle), followed by a 15 min enzyme deactivation step at 95°C (1 cycle), and 45 cycles of amplification that consisted of denaturation at 95°C for 15 sec and annealing and extension at 60°C for 1 min.
To test for potential carryover of RT-PCR inhibitors from the stool samples into total nucleic acid or RNA extracts, extracts from three stool samples were diluted 1:10 in nuclease free water and re-tested in duplicate by qRT-PCR as previously described by Petrich et al (Petrich et al., 2006).
Comparison of dilution series extracts by conventional RT-PCR and qRT-PCR
A 10% suspension prepared from stool sample with a high viral concentration (OD value 3.098) was serially diluted 10−1 to 10−7 in a rotavirus-negative stool suspension and then re-tested by EIA. Total nucleic acid or RNA was extracted by each of the six methods and tested by VP6 RT-PCR and NSP3 qRT-PCR (Freeman et al., 2008; Iturriza Gomara et al., 2002). For the real-time assay, each dilution was tested in duplicate and the average threshold cycle (Ct) value for each reaction was calculated. VP6 RT-PCR amplicons were detected by electrophoresis of the amplified products through 1% agarose gels containing GelRed (Biotium, Heyward, CA, USA) and RT-PCR products were detected under UV transillumination.
Cost and time analysis
Extraction cost per sample was estimated using the 2012 manufacturer price of each extraction kit or reagents. Hands-on time for the MagNA Pure Compact system, miniMAG system, RNaid kit, and QIAamp Viral RNA Mini was measured as time per sample when eight extractions were performed in parallel. Also, the easyMAG and KingFisher Flex systems were timed using a run of eight samples. For all methods, operator’s hands-on time was measured as the total time to perform each step in the extraction process. Total time to completion (hands-on and hands-off) was determined by timing a complete run on each instrument, from the addition of the first reagent to the recovery of the RT-PCR-ready nucleic acid.
Statistical analysis
Prism Version 5.02 Software for Windows (GraphPad Software, La Jolla CA) was used to plot Ct values. To determine whether the differences in Ct values for nucleic acid extracts prepared using each extraction method were significant, Ct values were compared using the Friedman test. When significant differences were identified, Dunn’s Multiple Comparison Test was used to perform nonparametric pairwise analyses of Ct values.
Results
Comparison of extraction methods by qRT-PCR
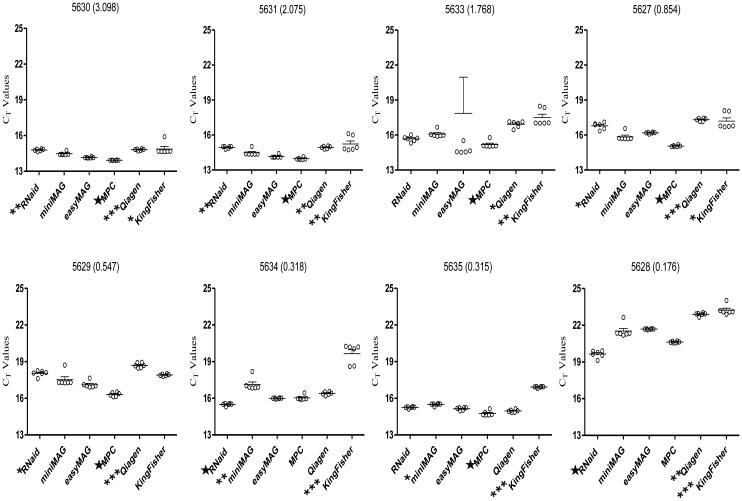
When qRT-PCR results were compared for all six extraction methods, samples extracted by using the MagNA Pure Compact system yielded the lowest average Ct values for 6 of 8 stools, including the 5 highest concentration samples as determined at EIA; the RNaid performed best with the other two samples which were among the 3 lowest concentration samples as determined by EIA (Fig. 1). Statistical analysis of method-to-method variability detected significant differences among the mean Ct values for all nucleic acid extracts obtained using the 6 methods (P < 0.001). When pairwise analyses of extraction method data were performed, the mean Ct values for KingFisher Flex extracts were significantly higher than those of the best performing method (MagNA Pure Compact or RNaid kit) for 7 of 8 samples as was the case for the QIAamp Viral RNA Mini kit with 6 samples (Fig. 1). In the case of 4 samples (5630, 5631, 5627, 5629) where the MagNA Pure Compact extracts yielded the lowest mean Ct values, the values for the RNaid kit were significantly higher (Fig. 1). In the two cases where the RNaid kit extracts yielded the lowest mean Ct values (samples 5634 and 5628), the mean Ct values for the MagNA Pure Compact extracts were not significantly different. With 2 samples (5634, 5635), the mean Ct values for miniMAG extracts were significantly higher than the best performing method. The mean Ct values for the easyMAG were never the lowest but never differed significantly from the best performing method.
Figure 1.
Scatter plots showing the distribution of Ct values obtained using RNA or total nucleic acid prepared from eight rotavirus samples using six different extraction protocols. The horizontal line in each group of observed values for a method corresponds to the mean value. Sample ID and OD values (in parentheses) are indicated above each scatter plot graph. T-bars indicate the standard deviation of the mean. The best performing method for each sample is labeled with a ★.
Methods with mean Ct values that were found to be significantly higher than the best performing method are labeled with * (P < 0.05), ** (P<0.01), or *** (P<0.001).
Inhibitors were detected in extracts prepared with the QIAamp Viral RNA Mini kit; the 1:10 dilutions of each QIAamp Viral RNA Mini kit extract exhibited lower Ct values than the undiluted extract (Table 2), with average Ct values decreasing 1.3 to 2.5 cycles. The 1:10 dilutions of the extracts prepared using the other extraction methods showed increases in Ct values by 2 to 4 cycles, consistent with a dilution effect of target RNA.
Table 2.
Mean and Standard Deviations (SD) of Ct values obtained by quantitative reverse transcriptase PCR using undiluted and 1:10 dilutions of rotavirus RNA/TNA extracts from 3 stool samples.
| RNaid | QIAamp Viral RNA | MagNA Pure Compact | KingFisher Flex 96 | NucliSENS easyMAG | NucliSENS miniMAG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Original | 1:10 | Original | 1:10 | Original | 1:10 | Original | 1:10 | Original | 1:10 | Original | 1:10 | ||
| Sample | OD Value | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD | Mean±SD |
| 5628 | 0.176 | 20.3±0.124 | 24.23±0.04 | 23.5±0.02 | 21.0±0.08 | 22.09±0.79 | 24.88±0.05 | 23.13±0.05 | 26.52±0.06 | 22.06±0.04 | 24.84±0.2 | 22.48±0.07 | 25.13±0.05 |
| 5627 | 0.854 | 17.21±0.06 | 19.48±0.04 | 16.0±0.02 | 14.7±0.03 | 16.3±0.66 | 19.15±0.06 | 16.97±0.07 | 20.62±0.03 | 16.93±0.06 | 19.08±0.03 | 17.24±0.03 | 19.61±0.04 |
| 5630 | 3.098 | 15.38±0.04 | 18.35±0.02 | 17.7±0.04 | 15.5±0.06 | 15.3±0.48 | 17.53±0.06 | 15.19±0.02 | 17.73±0.1 | 14.64±0.05 | 16.17±0.07 | 15.21±0.03 | 17.14±0.04 |
Comparison of dilution series extracts by conventional RT-PCR and qRT-PCR
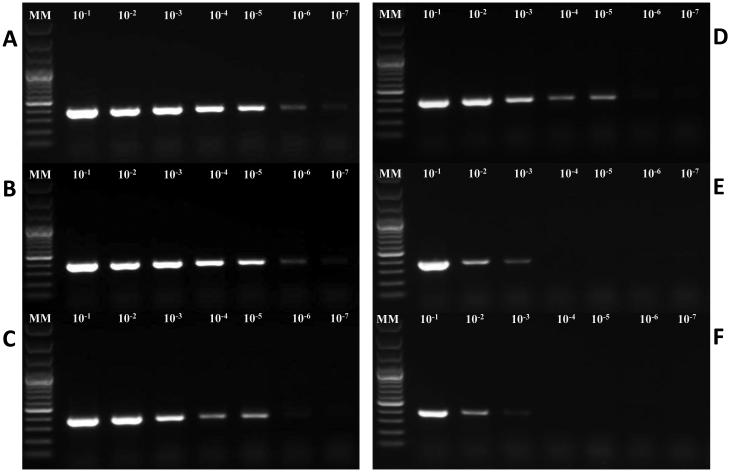
Extracts from serially diluted stool samples were analyzed for the presence of the template using VP6 RT-PCR (Iturriza Gomara et al., 2002) and the NSP3 qRT-PCR assay (Freeman et al., 2008). For the conventional RT-PCR assay, analysis by gel electrophoresis revealed a single band of the expected size for all rotavirus samples (Fig. 2). MagNA Pure Compact and KingFisher Flex were the only platforms capable of producing amplifiable template from all 7 dilutions (10−1 to 10−7). The QIAamp Viral RNA kit, and miniMAG systems yielded amplifiable template in 5 out of the 7 dilutions (10−1 to 10−5). The RNaid® kit and easyMAG® system yielded template with the poorest level of detection with template detected in the first 3 dilutions (10−1 to 10−3) only. With the qRT-PCR, the Ct values obtained with each method increased with each sample dilution and all rotavirus RNA was detected in all dilutions for all six extraction methods (Table 3) but there were significant differences in Ct values among the six methods (P<0.0001). Ct values obtained from extracts prepared using the RNaid® kit were significantly lower than those obtained for the easyMAG®, miniMAG®, and QIAamp Viral Mini RNA kit extracts (Table 3). Extracts prepared using the KingFisher Flex yielded Ct values that were significantly lower than those obtained from easyMAG® and miniMAG® extracts (Table 3). Ct values obtained from extracts prepared using the MagNA Pure Compact were significantly lower than those obtained for the miniMAG® (Table 3). Extracts prepared using the RNaid® kit, MagNA Pure Compact and KingFisher Flex did not differ significantly (Table 3).
Figure 2.
Gels showing limit of detection of rotavirus VP6 gene fragment (379 bp) by conventional RT-PCR using RNA or total nucleic acid extracted from a stool sample containing a high concentration of rotavirus (EIA OD value = 3.0) diluted 10−1 to 10−7 in a negative stool suspension, by each extraction method. A= MagNA Pure Compact, B= KingFisher Flex, C= QIAamp Viral RNA, D= miniMAG®, E= easyMAG® and F= RNaid®. MM= 100 bp Plus DNA Ladder.
Table 3.
Mean and standard deviation (SD) of CT values obtained by qRT-PCR using TNA or RNA extracted from a stool sample with a high viral concentration diluted 10−1 to 10−7 in a negative stool sample by each extraction method.
| RNaida | MagNA Pureb
Compact |
NucliSENS miniMAG |
NucliSENS easyMAG |
KingFisherc
Flex |
QIAamp Viral RNA |
||
|---|---|---|---|---|---|---|---|
| Dilution | OD Value | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD |
| 10−1 | 2.81 | 13 ± 0.09 | 13.4 ± 0.23 | 17.3 ± 0.01 | 15.3 ± 0.18 | 12 ± 0.03 | 12.3 ± 0.11 |
| 10−2 | 1.51 | 17 ± 0.03 | 17.4 ± 0.34 | 19.5 ± 0.08 | 18.8 ± 0.01 | 17.3 ± 0.13 | 18 ± 0.04 |
| 10−3 | 0.32 | 20 ± 0.20 | 20.5 ± 0.07 | 22.3 ± 0.04 | 21.4 ± 0.22 | 20.7 ± 0.96 | 21.2 ± 0.13 |
| 10−4 | 0.10 | 23.1 ± 0.11 | 23.3 ± 0.06 | 25.8 ± 0.08 | 24.5 ± 0.23 | 23.6 ± 0.29 | 24 ± 0.01 |
| 10−5 | 0.08 | 24.9 ± 0.02 | 27± 0.21 | 27.4 ± 0.07 | 28.1 ± 0.22 | 25.6 ± 0.17 | 28.2 ± 0.09 |
| 10−6 | 0.08 | 28.4 ± 0.17 | 28.6 ± 0.07 | 31.6 ± 0.12 | 31.4 ± 0.12 | 30 ± 0.08 | 31.6 ± 0.51 |
| 10−7 | 0.07 | 29 ± 0.01 | 33.7 ± 0.19 | 33.2 ± 0.11 | 34.6 ± 0.40 | 33.3 ± 0.24 | 33.2 ± 0.24 |
CT values obtained for RNaid kit extracts were significantly lower than those obtained for the easyMAG (P< 0.001), miniMAG (P<0.001), and QIAamp (P<0.01) kits.
CT values obtained for MagNA Pure Compact extracts were significantly lower than those obtained for the miniMAG (P<0.05).
CT values obtained for KingFisher Flex extracts were significantly lower than those obtained for the easyMAG (P< 0.05) and miniMAG (P<0.01).
Cost and time analysis
The total extraction time for 8 samples ranged from approximately 40 min for QIAamp Viral RNA Mini kit and KingFisher Flex to approximately 110 min for the RNaid kit (Table 4). The RNaid kit had the lowest cost/per sample and MagNA Pure Compact was the most expensive. The KingFisher Flex had the highest sample throughput (up to 96 samples/run).
Table 4.
Total time and cost per extraction.
| Instrument/Kit | Total time (min) |
Hands-on time (min) |
Hands-off time (min) |
Cost per extraction (US $)a |
Extraction per run |
|---|---|---|---|---|---|
| RNaid® | ~110 | ~50 | ~60 | 0.87 | n/a |
| QIAamp Viral RNA Mini Kit |
~40 | ~30 | ~10 | 3.94 | n/a |
| MagNa Pure Compact |
~80 | ~20 | ~60 | 10.75 | 8 |
| KingFisher Flex |
~85 | ~55 | ~30 | 3.48 | 96 |
| easyMAG® | ~71 | ~25 | ~46 | 8.84 | 24 |
| miniMAG® | ~72 | ~57 | ~15 | 6.3 | 12 |
Cost calculated in 2012
Discussion
Evaluation of the six commercial extraction platforms using stool samples indicated that the MagNA Pure Compact performed best for isolating rotavirus RNA that could be amplified by both conventional RT-PCR and qRT-PCR. No PCR inhibitors were detected in the extracts, which most often yielded the lowest Ct values. Although it is the most costly, this method was the most consistent and had relatively low hands-on and hands-off time processing time. Without knowledge of the proprietary components of the kit used with MagNA Pure Compact RNA Isolation kit, the reason for its superior performance cannot be determined. However, a possible explanation is that the method uses silica-coated magnetic beads, and the extraction process includes a DNase treatment step. The easyMAG, miniMAG and KingFisher Flex methods also use magnetic beads but without a DNase treatment step. The Ct values for extracts obtained using these methods were always higher but the extracts were not found to contain inhibitors of RT-PCR. The easyMAG and miniMAG extraction platforms are total nucleic acid extraction methods, suggesting that DNA in the extracts may have interfered with the extraction process or RT-PCR. The KingFisher Flex is an RNA only extraction method, but without a DNase treatment step the extracted product is likely to be contaminated with DNA. Therefore, the sub-optimal performance of the KingFisher Flex may be a function of the small volume of sample processed (50 −L) as well as the presence of DNA in the extract.
The QIAamp Viral Mini RNA kit yielded extracts with mean Ct values which were significantly higher than the best performing method with 6 samples. This study showed that use of the QIAamp Viral RNA Mini kit can lead to carryover of RT-PCR inhibitors for rotavirus RNA detection, since the 1:10 dilution of extracts lowered the Ct values. Detection of partial amplification inhibition has been reported for stool samples extracted using the QIAamp Viral RNA kit when stool extracts were tested for SARS coronavirus RNA (Petrich et al., 2006). The rest of the extraction methods showed no notable inhibition, only an expected dilution effect which results in an increase in Ct values.
When a dilution series of rotavirus positive stool diluted in a negative stool was extracted by each of the six methods and then tested by qRT-PCR, rotavirus RNA was detected in all samples. When each set of extracts from the dilution series was tested by conventional RT-PCR assays, however, only the MagNA Pure Compact and KingFisher Flex extracts were able to yield a detectable product in all seven dilutions. This observation could be the result of differences in amplicon size between the two methods, 91 base-pairs for qRT-PCR versus 379 bp for conventional RT-PCR. Rotavirus RNA extracted by using the RNaid kit may be sheared or somehow damaged by the manual extraction procedure of this kit with reduced detection efficiency of larger RNA molecules. Automated methods such as the MagNA Pure Compact and KingFisher Flex may damage RNA less than manual methods. Although limitations for detection at low virus concentrations may not be of concern when testing samples from persons with acute rotavirus gastroenteritis, from whom rotavirus typically is shed at very high concentrations in stool (approximately 1010 to 1011 particles/gram of stool) (Ray et al., 2006), this limitation would be important when choosing a method to extract samples from convalescent cases or healthy controls in which the viral concentration would be expected to be much lower.
Two limitations of this study should be noted. Because of the differences in extraction products (total nucleic acid or RNA), as well the presence of carrier RNA in the QIAamp Viral Mini RNA kit extracts, no attempt was made to measure directly and compare the quantity of the nucleic acid in the sample extracts tested. Also, no internal positive control was included in the extractions. Another limitation of this study was the lack of an encapsulated RNA control that could be spiked into stool, without RNA degradation by nucleases present in feces, and detected post-extraction by qRT-PCR and RT-PCR. Efforts to develop such a control are currently underway.
In summary, this study reports the most extensive evaluation to date of extraction platforms for the purification of rotavirus RNA from stool samples. The findings of this study will be very useful for laboratories in the selection of extraction methods for detection of rotavirus RNA in stool and the development of future protocols for rotavirus testing and surveillance.
Acknowledgment
We wish to thank Jon Gentsch for helpful discussions and guidance and Mary McCauley for editorial assistance.
Footnotes
INSTITUTION WHERE WORK WAS COMPLETED: Centers for Disease Control and Prevention, Atlanta, Georgia.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
References
- Abu Al-Soud W, Radstrom P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-Inhibiting samples. Appl. Environ. Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert L, Uyttendaele M, Debevere J. Evaluation of two viral extraction methods for the detection of human noroviruses in shellfish with conventional and real-time reverse transcriptase PCR. Lett. Appl. Microbiol. 2007;44:106–111. doi: 10.1111/j.1472-765X.2006.02047.x. [DOI] [PubMed] [Google Scholar]
- Banyai K, Mijatovic-Rustempasic S, Hull JJ, Esona MD, Freeman MM, Frace AM, Bowen MD, Gentsch JR. Sequencing and phylogenetic analysis of the coding region of six common rotavirus strains: Evidence for intragenogroup reassortment among co-circulating G1P[8] and G2P[4] strains from the United States. J. Med. Virol. 2011;83:532–539. doi: 10.1002/jmv.21977. [DOI] [PubMed] [Google Scholar]
- Chiu CH, Ou JT. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 1996;34:2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalesio N, Marsiglia V, Quinn A, Quinn T. Performance of the MagNA pure LC robot for extraction of Chlamydia trachomatis and Neisseria gonorrhoeae DNA from urine and swab specimens. J. Clin. Microbiol. 2004;42:3300–3302. doi: 10.1128/JCM.42.7.3300-3302.2004. CA., G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BK, Gentsch JR, Cicirello HG, Woods PA, Gupta A, Ramachandran M, Kumar R, Bhan MK, Glass RI. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 1994;32:1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan YH, Nakagomi T, Cunliffe NA, Pandey BD, Sherchand JB, Nakagomi O. The occurrence of amino acid substitutions D96N and S242N in VP7 of emergent G2P[4] rotaviruses in Nepal in 2004-2005: a global and evolutionary perspective. Arch. Virol. 2011;156:1969–1978. doi: 10.1007/s00705-011-1083-z. [DOI] [PubMed] [Google Scholar]
- Donato CM, Ch'ng LS, Boniface KF, Crawford NW, Buttery JP, Lyon M, Bishop RF, Kirkwood CD. Identification of strains of RotaTeq rotavirus vaccine in infants with gastroenteritis following routine vaccination. J. Infect. Dis. 2012;206:377–383. doi: 10.1093/infdis/jis361. [DOI] [PubMed] [Google Scholar]
- Dundas N, Leos N, Mitui M, Revell P. Comparison of automated nucleic acid extraction methods with manual extraction. J. Mol. Diagn. 2008a;10:311–316. doi: 10.2353/jmoldx.2008.070149. BB., R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas N, Leos NK, Mitui M, Revell P, Rogers BB. Comparison of automated nucleic acid extraction methods with manual extraction. J. Mol. Diagn. 2008b;10:311–316. doi: 10.2353/jmoldx.2008.070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esona MD, Foytich K, Wang Y, Shin G, Wei G, Gentsch JR, Glass RI, Jiang B. Molecular characterization of human rotavirus vaccine strain CDC-9 during sequential passages in Vero cells. Hum. Vaccin. 2010a;6:247–253. doi: 10.4161/hv.6.3.10409. [DOI] [PubMed] [Google Scholar]
- Esona MD, Mijatovic-Rustempasic S, Conrardy C, Tong S, Kuzmin IV, Agwanda B, Breiman RF, Banyai K, Niezgoda M, Rupprecht CE, Gentsch JR, Bowen MD. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum) Emerg. Infect. Dis. 2010b;16:1844–1852. doi: 10.3201/eid1612.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes M, Kapikian A. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 5th Kluwer/Lippincott, Williams and Wilkins; Philadelphia, PA: 2007. pp. 1917–1974. [Google Scholar]
- Freeman MM, Kerin T, Hull J, McCaustland K, Gentsch J. Enhancement of detection and quantification of rotavirus in stool using a modified real-time RT-PCR assay. J. Med. Virol. 2008;80:1489–1496. doi: 10.1002/jmv.21228. [DOI] [PubMed] [Google Scholar]
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. Serotype diversity and reassortment between human and animal rotavirus strains: Implications for rotavirus vaccine programs. J. Infect. Dis. 2005;192:S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- Glass RI, Parashar UD, Bresee JS, Turcios R, Fischer TK, Widdowson MA, Jiang B, Gentsch JR. Rotavirus vaccines: current prospects and future challenges. Lancet. 2006;368:323–332. doi: 10.1016/S0140-6736(06)68815-6. [DOI] [PubMed] [Google Scholar]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant L, Esona M, Gentsch J, Watt J, Reid R, Weatherholtz R, Santosham M, Parashar U, O'Brien K. Detection of G3P[3] and G3P[9] rotavirus strains in American Indian children with evidence of gene reassortment between human and animal rotaviruses. J. Med. Virol. 2011;83:1288–1299. doi: 10.1002/jmv.22076. [DOI] [PubMed] [Google Scholar]
- Griffin DD, Kirkwood CD, Parashar UD, Woods PA, Bresee JS, Glass RI, Gentsch JR. Surveillance of rotavirus strains in the United States: identification of unusual strains. The National Rotavirus Strain Surveillance System collaborating laboratories. J. Clin. Microbiol. 2000;38:2784–2787. doi: 10.1128/jcm.38.7.2784-2787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale AD, Green J, Brown DWG. Comparison of four RNA extraction methods for the detection of small round structured viruses in faecal specimens. J. Virol. Methods. 1996;57:195–201. doi: 10.1016/0166-0934(95)01966-9. [DOI] [PubMed] [Google Scholar]
- Hull JJ, Teel EN, Kerin TK, Freeman MM, Esona MD, Gentsch JR, Cortese MM, Parashar UD, Glass RI, Bowen MD. United States rotavirus strain surveillance from 2005 to 2008: genotype prevalence before and after vaccine introduction. Pediatr. Infect. Dis. J. 2011;30:S42–47. doi: 10.1097/INF.0b013e3181fefd78. [DOI] [PubMed] [Google Scholar]
- Iturriza Gomara M, Wong C, Blome S, Desselberger U, Gray J. Rotavirus subgroup characterisation by restriction endonuclease digestion of a cDNA fragment of the VP6 gene. J. Virol. Methods. 2002;105:99–103. doi: 10.1016/s0166-0934(02)00087-3. [DOI] [PubMed] [Google Scholar]
- Jothikumar N, Kang G, Hill VR. Broadly reactive TaqMan assay for real-time RT-PCR detection of rotavirus in clinical and environmental samples. J. Virol. Methods. 2009;155:126–131. doi: 10.1016/j.jviromet.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Kok T, Wati S, Bayly B, Devonshire-Gill D, Higgins G. Comparison of six nucleic acid extraction methods for detection of viral DNA or RNA sequences in four different non-serum specimen types. J. Clin. Virol. 2000;16:59–63. doi: 10.1016/s1386-6532(99)00066-9. [DOI] [PubMed] [Google Scholar]
- Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz PG, Matsson M, Wadstrom T, Radstrom P. Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J. Microbiol. Meth. 1997;28:159–167. [Google Scholar]
- Lim DV, Simpson JM, Kearns EA, Kramer MF. Current and Developing Technologies for Monitoring Agents of Bioterrorism and Biowarfare. Clin. Microbiol. Rev. 2005;18:583–607. doi: 10.1128/CMR.18.4.583-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loens K, Ursi D, Goossens H, Ieven M. Evaluation of the NucliSens miniMAG RNA extraction and real-time NASBA applications for the detection of Mycoplasma pneumoniae and Chlamydophila pneumoniae in throat swabs. J. Microbiol. Methods. 2008;72:217–219. doi: 10.1016/j.mimet.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Makristathis A, Pasching E, Schutze K, Wimmer M, Rotter ML, Hirschl AM. Detection of Helicobacter pylori in stool specimens by PCR and antigen enzyme immunoassay. J. Clin. Microbiol. 1998;36:2772–2774. doi: 10.1128/jcm.36.9.2772-2774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch. Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Rahman M, Yang X, Delbeke T, Arijs I, Muyembe JJ. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. J. Clin. Microbiol. 2006;44:1801–1809. doi: 10.1128/JCM.44.5.1801-1809.2006. K.J. M., V.R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro L, Bonnemaison D, Vekris A, Petry KG, Bonnet J, Vidal R, Cabrita J, Megraud F. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 1997;35:995–998. doi: 10.1128/jcm.35.4.995-998.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RN, Adams RP, Flournoy LE. Inhibition of random amplified polymorphic DNAs (RAPDs) by plant polysaccharides, Plant Mol. Biol. Report. 1996;14:17–22. [Google Scholar]
- Payne DC, Edwards KM, Bowen MD, Keckley E, Peters J, Esona MD, Teel EN, Kent D, Parashar UD, Gentsch JR. Sibling transmission of vaccine-derived rotavirus (RotaTeq) associated with rotavirus gastroenteritis. Pediatrics. 2010;125:e438–441. doi: 10.1542/peds.2009-1901. [DOI] [PubMed] [Google Scholar]
- Payne DC, Staat MA, Edwards KM, Szilagyi PG, Gentsch JR, Stockman LJ, Curns AT, Griffin M, Weinberg GA, Hall CB, Fairbrother G, Alexander J, Parashar UD. Active, population-based surveillance for severe rotavirus gastroenteritis in children in the United States. Pediatrics. 2008;122:1235–1243. doi: 10.1542/peds.2007-3378. [DOI] [PubMed] [Google Scholar]
- Perelle S, Cavellini L, Burger C, Blaise-Boisseau S, Hennechart-Collette C, Merle G, Fach P. Use of a robotic RNA purification protocol based on the NucliSens easyMAG for real-time RT-PCR detection of hepatitis A virus in bottled water. J. Virol. Methods. 2009;157:80–83. doi: 10.1016/j.jviromet.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Petrich A, Mahony J, Chong S, Broukhanski G, Gharabaghi F, Johnson G, Louie L, Luinstra K, Willey B, Akhaven P, Chui L, Jamieson F, Louie M, Mazzulli T, Tellier R, Smieja M, Cai W, Chernesky M, Richardson SE. Multicenter comparison of nucleic acid extraction methods for detection of severe acute respiratory syndrome coronavirus RNA in stool specimens. J. Clin. Microbiol. 2006;44:2681–2688. doi: 10.1128/JCM.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Matthijnssens J, Saiada F, Hassan Z, Heylen E, Azim T, Van Ranst M. Complete genomic analysis of a Bangladeshi G1P[8] rotavirus strain detected in 2003 reveals a close evolutionary relationship with contemporary human Wa-like strains. Infect. Genet. Evol. 2010;10:746–754. doi: 10.1016/j.meegid.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, Iturriza-Gomara M, Iftekharuddin N, Azim T, Van Ranst M. Evolutionary history and global spread of the emerging G12 human rotaviruses. J. Virol. 2007a;81:2382–2390. doi: 10.1128/JVI.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Sultana R, Ahmed G, Nahar S, Hassan ZM, Saiada F, Podder G, Faruque AS, Siddique AK, Sack DA, Matthijnssens J, Van Ranst M, Azim T. Prevalence of G2P[4] and G12P[6] rotavirus, Bangladesh. Emerg. Infect. Dis. 2007b;13:18–24. doi: 10.3201/eid1301.060910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran M, Gentsch JR, Parashar UD, Jin S, Woods PA, Holmes JL, Kirkwood CD, Bishop RF, Greenberg HB, Urasawa S, Gerna G, Coulson BS, Taniguchi K, Bresee JS, Glass RI. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasool NB, Monroe SS, Glass RI. Determination of a universal nucleic acid extraction procedure for PCR detection of gastroenteritis viruses in faecal specimens. J. Virol. Methods. 2002;100:1–16. doi: 10.1016/s0166-0934(01)00379-2. [DOI] [PubMed] [Google Scholar]
- Ray P, Fenaux M, Sharma S, Malik J, Subodh S, Bhatnagar S, Greenberg H, Glass RI, Gentsch J, Bhan MK. Quantitative evaluation of rotaviral antigenemia in children with acute rotaviral diarrhea. J. Infects. Dis. 2006;194:588–593. doi: 10.1086/505878. [DOI] [PubMed] [Google Scholar]
- Rivera L, Pena LM, Stainier I, Gillard P, Cheuvart B, Smolenov I, Ortega-Barria E, Han HH. Horizontal transmission of a human rotavirus vaccine strain--a randomized, placebo-controlled study in twins. Vaccine. 2011;29:9508–9513. doi: 10.1016/j.vaccine.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Schmid M, Oehme R, Schalasta G, Brockmann S, Kimmig P. Fast detection of Noroviruses using a real-time PCR assay and automated sample preparation. BMC Infect. Dis. 2004;4:1–8. doi: 10.1186/1471-2334-4-15. G., E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman T, de Boer R, Patty R, Kooistra-Smid M, van Zwet A. Comparative evaluation of in-house manual, and commercial semi-automated and automated DNA extraction platforms in the sample preparation of human stool specimens for a Salmonella enterica 5'-nuclease assay. J. Microbiol. Methods. 2007;71:238–245. doi: 10.1016/j.mimet.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Tang YW, Sefers SE, Li HJ, Kohn DJ, Procop GW. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J. Clin. Microbiol. 2005a;43:4830–4833. doi: 10.1128/JCM.43.9.4830-4833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YW, Sefers SE, Li HJ, Kohn DJ, Procop GW. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J. Clin. Microbiol. 2005b;43:5833–5833. doi: 10.1128/JCM.43.9.4830-4833.2005. (vol 43, pg 4830, 2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk D, Schneider SK, Wengenack NL, Sloan LM, JE R. Real-time PCR method for detection of Encephalitozoon intestinalis from stool specimens. J. Clin. Microbiol. 2002;40:3922–3928. doi: 10.1128/JCM.40.11.3922-3928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FT, Banyai K, Huang JC, Wu HS, Chang FY, Yang JY, Hsiung CA, Huang YC, Lin JS, Hwang KP, Jiang B, Gentsch JR. Diverse origin of P[19] rotaviruses in children with acute diarrhea in Taiwan: Detection of novel lineages of the G3, G5, and G9 VP7 genes. J. Med. Virol. 2011;83:1279–1287. doi: 10.1002/jmv.22052. [DOI] [PubMed] [Google Scholar]
- Yang G, Erdman DE, Kodani M, Kools J, Bowen MD, Fields BS. Comparison of commercial systems for extraction of nucleic acids from DNA/RNA respiratory pathogens. J. Virol. Methods. 2011;171:195–199. doi: 10.1016/j.jviromet.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C, Jakob K, Esona MD, Peckham X, Rausch J, Hull JJ, Whittier S, Gentsch JR, LaRussa P. Detection of fecal shedding of rotavirus vaccine in infants following their first dose of pentavalent rotavirus vaccine. Vaccine. 2011;29:4151–4155. doi: 10.1016/j.vaccine.2011.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller M, Rahman M, Heylen E, De Coster S, De Vos S, Arijs I, Novo L, Verstappen N, Van Ranst M, Matthijnssens J. Rotavirus incidence and genotype distribution before and after national rotavirus vaccine introduction in Belgium. Vaccine. 2010;28:7507–7513. doi: 10.1016/j.vaccine.2010.09.004. [DOI] [PubMed] [Google Scholar]