Abstract
Short Abstract
To help assess the molecular mechanisms underlying zebrafish biliary-driven liver regeneration, we established a liver injury model in which the nitroreductase-expressing hepatocytes are genetically ablated upon metronidazole treatment. In this protocol, we describe how to adeptly manipulate, monitor and analyze hepatocyte ablation and biliary-driven liver regeneration.
Long abstract
The liver has a great capacity to regenerate. Hepatocytes, the parenchymal cells of the liver, can regenerate in one of two ways: hepatocyte- or biliary-driven liver regeneration. In hepatocyte-driven liver regeneration, regenerating hepatocytes are derived from preexisting hepatocytes, whereas, in biliary-driven regeneration, regenerating hepatocytes are derived from biliary epithelial cells (BECs). For hepatocyte-driven liver regeneration, there are excellent rodent models that have significantly contributed to the current understanding of liver regeneration. However, no such rodent model exists for biliary-driven liver regeneration. We recently reported on a zebrafish liver injury model in which BECs extensively give rise to hepatocytes upon severe hepatocyte loss. In this model, hepatocytes are specifically ablated by a pharmacogenetic means. Here we present in detail the methods to ablate hepatocytes and to analyze the BEC-driven liver regeneration process. This hepatocyte-specific ablation model can be further used to discover the underlying molecular and cellular mechanisms of biliary-driven liver regeneration. Moreover, these methods can be applied to chemical screens to identify small molecules that augment or suppress liver regeneration.
Keywords: nitroreductase, metronidazole, biliary epithelial cells, hepatocytes, zebrafish, liver regeneration, cell ablation, transgenic
Introduction
The liver is a highly regenerative organ. During liver regeneration, regenerating hepatocytes, the parenchymal cells of the liver, are derived from pre-existing hepatocytes (hepatocyte-driven liver regeneration) or BECs (biliary-driven liver regeneration)1,2. Liver injury usually elicits the proliferation of pre-existing hepatocytes; however, when hepatocyte proliferation is compromised, BECs can contribute to hepatocytes2–4. These two modes of liver regeneration are clinically significant. Upon surgical removal of a portion of the human liver (e.g., because of liver tumors or live liver donors), hepatocytes in the remaining liver proliferate to recover the lost liver mass. By contrast, in patients with severe liver diseases, hepatocyte proliferation is greatly compromised, so that BECs or liver progenitor cells (LPCs) appear to contribute to regenerating hepatocytes5,6. The rodent 2/3 partial hepatectomy model, in which hepatocytes proliferate to recover the lost liver mass, has significantly contributed to the current understanding of hepatocyte-driven liver regeneration7,8. However, there is no valid rodent model in which regenerating hepatocytes are mainly derived from BECs. Although several rodent liver toxin models led to the identification of biliary-driven liver regeneration2–4, recent lineage tracing studies in mice indicate a minimal contribution of BECs to regenerating hepatocytes in these models9,10. Some of the rodent liver injury models, including partial hepatectomy11–13 and acetaminophen-induced liver damage14,15, have been applied to zebrafish and led to the identification of novel genes or pathways implicated in liver regeneration. However, hepatocyte-driven, but not BEC-driven, liver regeneration occurs in these zebrafish liver injury models. Therefore, a novel liver injury model in which BECs extensively contribute to regenerating hepatocytes is needed for a better understanding of BEC-driven liver regeneration.
The overall goals of the hepatocyte ablation model described here are (1) to generate a liver injury model in which BECs extensively contribute to regenerating hepatocytes, and (2) elucidate the molecular and cellular mechanisms underlying BEC-driven liver regeneration. We hypothesized that severity of injury determines the mode of liver regeneration; thus, we predicted that biliary-driven liver regeneration would initiate upon severe hepatocyte injury. To test this hypothesis, we developed a zebrafish liver injury model by generating a transgenic line, Tg(fabp10a:CFP-NTR)s931, that highly expresses bacterial Nitroreductase (NTR) fused with cyan fluorescent protein (CFP) under the hepatocyte-specific fabp10a promoter. Since NTR converts the non-toxic prodrug, metronidazole (Mtz), into a cytotoxic drug, it ablates only the intended NTR-expressing cells16–18, in this case, the hepatocytes. By manipulating the duration of Mtz treatment, the extent of hepatocyte ablation can be controlled. Using this model, we recently reported that upon severe hepatocyte loss, BECs extensively give rise to regenerating hepatocytes19, which was further confirmed by two other independent studies20,21. Therefore, compared to the aforementioned rodent and zebrafish liver injury models, our hepatocyte ablation model is more advantageous for studying BEC-driven liver regeneration.
This protocol describes the procedure for performing liver regeneration experiments using the zebrafish hepatocyte ablation model. This model will be appropriate for determining the mechanisms underlying biliary-driven liver regeneration and for chemical screens to identify small molecules that can repress or augment liver regeneration.
Protocol
Zebrafish were raised and bred according to standard procedure; experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
1. Preparation of Embryos/Larvae
-
1.1
To conduct timed matings, set up adult male and female Tg(fabp10a:CFP-NTR)s931 hemizygous or homozygous fishes overnight and place a divider between them. Remove this divider the following morning when mating is desired. Collect embryos by straining the water using a fine plastic mesh strainer.
-
1.2
Turn the strainer upside down and rinse the mesh surface with a fine stream of egg water dispensed from a squeeze bottle. Transfer 100 embryos into a 100-mm petri dish with 25 ml of egg water. Keep no more than 100 embryos per petri dish and raise them at 28 °C.
-
1.3
To inhibit pigmentation, add phenylthiourea (PTU) into the petri dishes prior to 10 hours post-fertilization (hpf), and raise embryos/larvae until 80 hpf. The final concentration of PTU is 0.2 mM. CAUTION: PTU is toxic. Use in accordance with appropriate handling guidelines.
-
1.4
Anesthetize the larvae by adding 3-amino benzoic acid ethyl ester (Tricaine), at a final concentration of 0.016%, into the petri dishes. Then, using an epifluorescence microscope, sort CFP-positive larvae. Use larvae with a similar liver size and discard those with a smaller or larger liver. NOTE: any CFP-positive larvae, regardless of intensity, can be used because there is no difference in the extent of hepatocyte ablation between the hemizygous and homozygous larvae. CAUTION: tricaine is toxic. Use in accordance with appropriate handling guidelines.
-
1.5
Remove the egg water containing Tricaine and add egg water supplemented with 0.2 mM PTU. Keep the embryos/larvae at 28 °C.
2. Mtz treatment
-
2.1
Prepare fresh 10 mM Mtz solution by dissolving 0.171 g of Mtz powder in 100 ml egg water supplemented with 0.2% DMSO and 0.2 mM PTU. To completely dissolve the Mtz, mix the solution at room temperature (RT) with rapid stirring for 20–30 minutes. To help solubilize Mtz and to enhance Mtz permeation into the larvae, add DMSO when preparing Mtz. CAUTION: prolonged exposure or increased concentration of Mtz is toxic. Use in accordance with appropriate handling guidelines.
-
2.2
Separate the larvae into control and experimental groups by transferring desired number of larvae into two different 100-mm petri dish or multi-well plate. For the experimental group, keep the larvae in 10 mM Mtz solution; for the control group, keep them in egg water containing 0.2% DMSO.
-
2.3
Cover the plates or petri dishes containing the Mtz solution with aluminum foil to prevent the photo-inactivation of Mtz, and incubate at 28 °C. The duration of Mtz treatment depends on the larval stage and the transgenic line. To observe similar severe hepatocyte ablation in the Tg(fabp10a:CFP-NTR) line, treat the larvae with the Mtz solution from either 3.5 days post-fertilization (dpf) (for a duration of 36 hr) or from 5 dpf (for a duration of 24 hr).
3. Analysis of biliary-driven liver regeneration
-
3.1
At the end of the ablation period, remove the Mtz solution from the plates. Wash the Mtz-treated larvae by adding 25 ml of egg water. Swirl the plate 2–3 times to wash the larvae before discarding the egg water. Repeat three times and then immerse the larvae once more in egg water. To avoid heart edema and larval death in the Mtz-treated larvae, add a lower concentration of Tricaine, at a final concentration of 0.008%, to immobilize the larvae.
-
3.2
For liver analysis, examine the liver size of the Mtz-treated larvae under an epifluorescence microscope with the CFP filter. Assess hepatocyte ablation levels based on the relative liver size: large (non-ablated; 0–10% larvae), medium (partially ablated; 10–20%), and very small (fully ablated; 80–90%). Remove the larvae with non-ablated or partially ablated livers. Only keep larvae with a tiny liver, which is indicative of severe hepatocyte ablation. For liver analysis at the regeneration 0 hr time point (R0h), harvest the larvae after Mtz washout and CFP sorting. Otherwise, keep the sorted larvae in egg water supplemented with 0.2 mM PTU until ready for harvesting.
-
3.3
Transfer harvested larvae into a 1.5-ml microcentrifuge tube and replace egg water with the fixation solution of 3% formaldehyde in PEM buffer. Incubate the tube on a rotator overnight at 4 °C.
-
3.4
Replace the fixation solution with 1 ml of PBSDT and rotate the tube containing the larvae on the rotator for 5 min.
-
3.5
Transfer the fixed larvae into a 100-mm petri dish containing PBSDT and using forceps, manually remove yolk and pectoral fins under a dissecting microscope. Transfer up tp 20 dissected larvae into a 1.5-ml tube.
-
3.6
Replace PBSDT solution with 1 ml of blocking solution and rock the tube on a rotator at RT for 2 hr.
-
3.7
Replace blocking solution with 100 μl of blocking solution containing primary antibodies. Slowly rock the tube on a rotator overnight at 4°C.
-
3.8
Remove the primary antibody solution and wash with PBSDT 5 times for 10 min each time. Then add 100 ul of blocking solution containing secondary antibodies and rock it on a rotator at RT for 2hr.
-
3.9
Wash 5 times with PBSDT for 10 min each time at RT by rocking on a rotator.
-
3.10
Orient the larvae laterally on a microscope slide and add a drop of mounting media. Carefully cover them with a cover glass and seal the cover glass with a nail polisher. Now, it is ready for confocal microscopy.
Representative Results
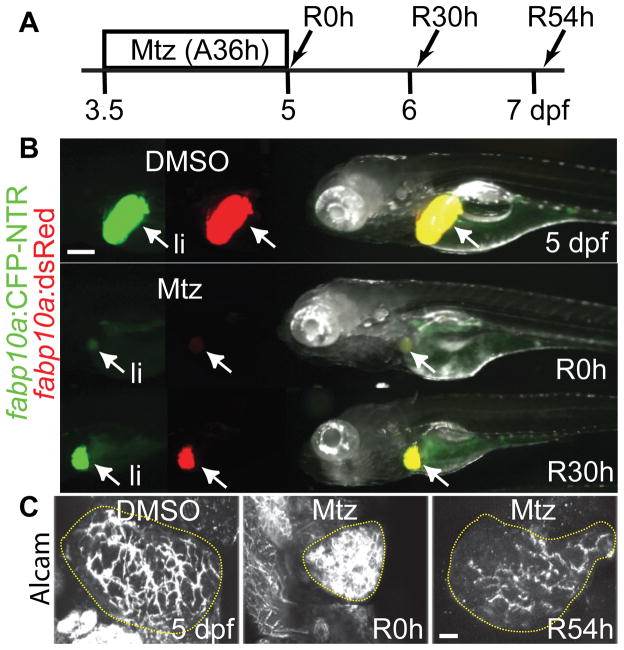
Mtz treatment for 36 hours (A36h, A stands for ablation) from 3.5 to 5 dpf dramatically reduced liver size (Figure 1B). After Mtz washout, considered as the beginning of liver regeneration (R), strong fabp10a:CFP-NTR and fabp10a:dsRed expression reappeared within 30 hours (Figure 1B). The intrahepatic biliary network, as assessed by expression of Alcam that is present in the membrane of BECs22, initially collapsed but was re-established at 54 hours post-washout (R54h) (Figure 1C). These data indicate that liver regeneration and recovery rapidly occur in our liver injury model.
Figure 1. A zebrafish liver injury model.
(A) Scheme illustrating the periods of Mtz treatment and liver regeneration. (B) 36-hour Mtz treatment greatly reduced liver size and fabp10a:CFP and fabp10a:dsRed fluorescence at R0h; however, strong CFP and dsRed fluorescence reappeared at R30h. Arrows point to the liver (li). (C) Confocal projection views of the entire liver immunostained with anti-Alcam antibodies that labelled BECs. The intrahepatic biliary network was collapsed at R0h but rapidly re-established at R54h. Dotted regions outline the liver. Scale bars: 100 (B), 20 (C) μm.
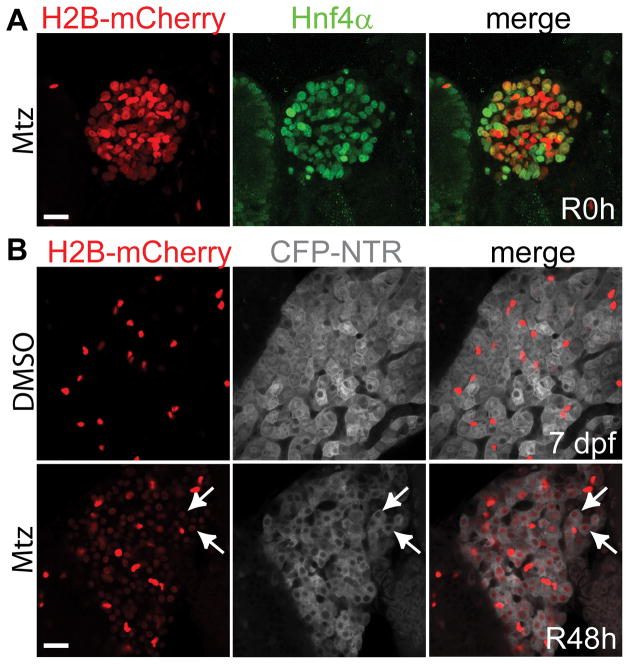
Figure 2 shows that regenerating hepatocytes are derived from BECs. The Tg(Tp1:H2B-mCherry)s939 line expresses a nuclear red fluorescent protein under the control of an element containing 12 RBP-Jκ binding sites23. This Notch responsive element appears to drive gene expression exclusively in BECs in the liver24; however, it is possible that it drives gene expression in LPCs because Notch signaling is closely related to the LPCs25. Therefore, this transgenic line allows for easy detection of BEC, and probably LPC, nuclei. Furthermore, the extended stability of the H2B-mCherry fusion protein permits short-term lineage tracing. Most H2B-mCherry+ cells in the regenerating liver at R0h expressed Hnf4α (Figure 2A), which is expressed in hepatocytes, but not in BECs, in the control liver. Hepatocytes, as assessed by fabp10a:CFP-NTR expression, at R48h still retained Tp1:H2B-mCherry expression (Figure 2B, arrows). These data indicate BEC conversion to hepatocytes upon severe hepatocyte loss, which was further confirmed by permanent lineage tracing19.
Figure 2. BECs give rise to hepatocytes upon severe hepatocyte loss.
(A) Confocal projection views of the liver showing Hnf4α (green) and Tp1:H2B-mCherry (red) expression at R0h. (B) Confocal single-optical section images showing fabp10a:CFP-NTR (grey) and Tp1:H2B-mCherry (red) expression in the liver. mCherry expression was revealed by anti-mCherry immunostaining. Note the weak mCherry expression in the nuclei of CFP+ hepatocytes (arrows). Strong mCherry+ cells are BECs. Scale bars: 20 μm.
Discussion
Severe, but not mild, hepatocyte ablation elicits biliary-driven liver regeneration. Therefore, following Mtz treatment and washout, it is important to examine the liver size of the Mtz-treated larvae, which can be assessed by intrinsic CFP fluorescence from the fabp10a:CFP-NTR transgene. Since a small percentage of the Mtz-treated larvae will display non-ablated (0–5%) or partially ablated (10–20%) livers, it is essential to sort out and only analyze the larvae with a very small liver, which is indicative of severe hepatocyte ablation.
Although in our protocol, we treated zebrafish larvae with Mtz from 3.5 to 5 dpf (a 36-hour treatment period), alternative stages and duration of Mtz treatment are also possible. For example, larvae treated from 5 to 6 dpf (a 24-hour treatment period) also exhibited severe hepatocyte ablation, similar to the aforementioned 36-hour treatment period. Two additional groups independently generated similar fabp10a:NTR transgenic lines and also reported on the process of biliary-driven liver regeneration following hepatocyte-specific ablation. Whereas one group treated larvae with Mtz from 6 to 7 dpf21, another did so from 3.5 to 4.5 dpf, reaching similar conclusions20. Since each fabp10a:NTR transgenic line may exhibit a different level of NTR expression, the duration and dosage of Mtz treatment should be determined separately for each of the transgenic line used. It may also prove useful to generate a transgenic line with an enhanced version of the NTR gene, in which the introduction of three point mutations leads to an increase in its enzymatic activity26. This mutant NTR will result in a faster ablation event than the wild-type NTR26. This is especially advantageous when wild-type NTR kinetics may limit the possibility of complete cell ablation at a desired stage.
Since hundreds of larvae can simultaneously undergo hepatocyte ablation, this zebrafish liver injury model can be applied to chemical screens to identify small molecules that can suppress or augment biliary-driven liver regeneration. For example, using this model, the Shin group at Georgia Tech performed a chemical screen and found several Wnt agonists and Notch antagonists that facilitated liver regeneration20. We also performed a small scale chemical screen and identified several compounds that can suppress liver regeneration (S.K., T.Y.C., J.S., and D.S., in preparation).
Marker analyses in humans suggested that BECs can contribute to regenerated hepatocytes in the diseased liver5,6. Dr. Wanless’ group recently reported that a large percentage of parenchyma in regressed cirrhosis appears to be derived from LPCs/BECs in humans27, implying the importance of BEC-driven liver regeneration in cirrhosis regression. Given the fact that BECs mainly contribute to the regenerating hepatocytes in the zebrafish liver injury model described here, this model will be an invaluable tool for determining the cellular and molecular mechanisms underlying BEC-driven liver regeneration. Understanding such mechanisms will provide significant insights into how to augment innate liver regeneration in patients with severe liver diseases. Furthermore, in combination with chemical screens, this zebrafish model may be useful in identifying a potential drug that can facilitate innate liver regeneration in human patients.
Table 1.
| Name of Reagent/Equipment | Company | Catalog Number | Comments/Description |
|---|---|---|---|
| 3-amino benzoic acid ethyl ester (Tricaine) powder | Sigma-Aldrich | A5040 | Make 0.4% (15 mM) tricaine stock : Bring 0.4 g tricaine to 100 mL with egg water. Adjust to pH 7. |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D8418 | |
| Metronidazole | Sigma-Aldrich | M1547 | |
| Triton X-100 | Sigma-Aldrich | T8532 | |
| Formaldehyde | Sigma-Aldrich | 252549 | |
| Calcium sulfate dihydrate | Sigma-Aldrich | C3771 | |
| N-Phenylthiourea (PTU) | Sigma-Aldrich | P7629 | |
| Mounting media (Vectashield) | Vector Laboratories | H-1000 | |
| 100 × 15 mm petri dish | Denville Scientific Inc. | M5300 | |
| clear nail polish | Fisher Scientific | 50949071 | |
| fine forceps | Fine Science Tools | 11251-20 | Dumont #5 |
| glass slide | Fisher Scientific | 12-544-1 | |
| glass coverslip | Fisher Scientific | 12-540-A and 12-542-A | 18 × 18 mm |
| zn-5 (mouse anti-Alcam) | ZIRC | zn-5 | 1:10 dilution |
| goat anti-Hnf4a | Santa Cruz | sc-6556 (C-19) | 1:50 dilution |
| rat anti-RFP | Allele Biotechnology | ACT-CM-MRRFP10 | 1:300 dilution |
| AlexaFluor 647 AffiniPure Donkey Anti-goat IgG (H+L) | Jackson laboratories | 705-605-147 | 1:500 dilution |
| AlexaFluor 647 AffiniPure Goat Anti-mouse IgG (H+L) | Invitrogen | A21240 | 1:500 dilution |
| Cy3 donkey anti-rat IgG | Jackson laboratories | 712-165-150 | 1:500 dilution |
| dissecting microscope | Leica Microsystems | S6F | |
| epifluorescence microscope | Leica Microsystems | M205FA | |
| confocal microscope | Carl Zeiss | LSM700 | |
| egg water | 0.3 g/L of sea salts ‘Instant Ocean’ and 0.5 mM CaSO4 in distilled water. | ||
| 25.6 g Na2HPO4·7H2O, 80 g NaCl, 2 g KCl, 2 g KH2PO4. Bring to 1 L with distilled water. Adjust pH 7. | |||
| 10X PBS | Autoclave for 20 min at 121°C. | ||
| 1X PBSDT | 0.2% Triton X-100 and 0.2% DMSO in 1 X PBS. | ||
| PEM buffer | 30.2 g PIPES, 761 mg EGTA, 1 mM MgSO4. Bring to 1 L with distilled water. Adjust pH 7. | ||
| Blocking solution | Mix 1 ml of heat-inactivated horse serum with 9 ml of 1X PBSDT. |
Acknowledgments
The authors thank Drs. Hukriede and Tsang for discussions. M.K. was supported by an NIH training grant (T32EB001026). This work was supported in part by grants from the American Liver Foundation, the March of Dimes Foundation (5-FY12-39), and the NIH (DK101426) to D.S.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/52785.
Disclosures
The authors declare that they have no competing financial interests.
Contributor Information
Tae-Young Choi, Email: choity@pitt.edu, Department of Developmental Biology, McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, USA.
Mehwish Khaliq, Email: mek118@pitt.edu, Department of Developmental Biology, McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, USA.
Sungjin Ko, Email: sungjin@pitt.edu, Department of Developmental Biology, McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, USA.
Juhoon So, Email: juhoon@pitt.edu, Department of Developmental Biology, McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, USA.
Donghun Shin, Email: donghuns@pitt.edu, Department of Developmental Biology, McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, USA, 1-412-624-2144.
References
- 1.Riehle KJ, Dan YY, Campbell JS, Fausto N. New concepts in liver regeneration. J Gastroen Hepatol. 2011;26:203–212. doi: 10.1111/j.1440-1746.2010.06539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mechanisms of Development. 2003;120:117–130. doi: 10.1016/S0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 3.Duncan AW, Dorrell C, Grompe M. Stem Cells and Liver Regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michalopoulos GK. Liver regeneration: Alternative epithelial pathways. Int J Biochem Cell B. 2011;43:173–179. doi: 10.1016/j.biocel.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falkowski O, et al. Regeneration of hepatocyte ‘buds’ in cirrhosis from intrabiliary stem cells. Journal of hepatology. 2003;39:357–364. doi: 10.1016/S0168-8278(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 6.Yoon SM, et al. Epithelial Cell Adhesion Molecule (EpCAM) Marks Hepatocytes Newly Derived from Stem/Progenitor Cells in Humans. Hepatology. 2011;53:964–973. doi: 10.1002/Hep.24122. [DOI] [PubMed] [Google Scholar]
- 7.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalopoulos GK. Liver Regeneration after Partial Hepatectomy Critical Analysis of Mechanistic Dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanger K, et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell stem cell. 2014;15:340–349. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a Stem Cell Origin of New Hepatocytes in a Common Mouse Model of Chronic Liver Injury. Cell reports. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dovey M, et al. Topoisomerase II alpha is required for embryonic development and liver regeneration in zebrafish. Molecular and cellular biology. 2009;29:3746–3753. doi: 10.1128/MCB.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goessling W, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Developmental Biology. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- 13.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox AG, et al. S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell reports. 2014;6:56–69. doi: 10.1016/j.celrep.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.North TE, et al. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc Natl Acad Sci USA. 2010;107:17315–17320. doi: 10.1073/pnas.1008209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curado S, et al. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Developmental Dynamics. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 17.Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curado S, Stainier DYR, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–954. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang M, et al. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology. 2014 doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789–800 e788. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 22.Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninov N, Borius M, Stainier DYR. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/Dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh T, Miyajima A. Liver Regeneration by Stem/Progenitor Cells. Hepatology. 2014;59:1617–1626. doi: 10.1002/Hep.26753. [DOI] [PubMed] [Google Scholar]
- 26.Mathias JR, Zhang Z, Saxena MT, Mumm JS. Enhanced cell-specific ablation in zebrafish using a triple mutant of Escherichia coli nitroreductase. Zebrafish. 2014;11:85–97. doi: 10.1089/zeb.2013.0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stueck AE, Wanless IR. Regression of cirrhosis: The maturation sequence of buds arising from hepatocyte progenitor cells. Hepatology. 2013;58:235A. doi:210.1002/hep.26800. [Google Scholar]