Abstract
The interferon (IFN) response is a critical arm of the innate immune response and a major host defense mechanism against viral infections. Following microbial encounter, a series of signaling events lead to transcriptional activation of the IFN genes, which in turn leads to significant changes in the cellular transcriptome by altering the expression of hundreds of target genes. Emerging evidence suggest that long non-coding RNAs (lncRNAs) constitute a major subgroup of the IFN target genes, and further, that the IFN response is subject to regulation by a large number of host- and pathogen-derived lncRNAs. While the vast majority of lncRNAs with potential roles in the IFN response remain unstudied, analysis of a very small subset provides a glimpse of the regulatory impact of this class of RNAs on IFN response.
Keywords: IFN, Interferon, lncRNA, long non-coding RNA, NRIR, BISPR
1. Introduction
1.1. The crucial role of the interferon response in innate immunity
The interferon (IFN) response is a pivotal and nearly ubiquitous component of the innate immune response against viruses and other microbial pathogens in mammalian cells. IFNs are grouped into three major groups, with type I (IFN-α, -β, -κ, -ε, and -ω) and type II (IFN-γ) IFNs binding distinct receptors, but nonetheless showing some overlap in their signaling cascades and target genes (Bolen et al., 2014; Hertzog et al., 2011; Pollard et al., 2013; Rusinova et al., 2013). Type III IFNs (IFN-λ1, IFN-λ2, IFN-λ3, also called IL-29, IFN-λ4 IL-28A, and IL-28B, respectively) are predominantly expressed in plasmacytoid dendritic cells and are abundantly expressed in liver and lung tissues. Despite binding a distinct receptor, type III IFNs show significant overlap with type I IFNs in their signaling cascade and target genes (Bolen et al., 2014; Meyer, 2009; Pollard et al., 2013).
The first step in the IFN response involves the induction of expression of IFNs through the activation of a class of cellular sensors that function as initial sensors of pathogen-associated molecular patterns, including the RNA sensors retinoic acid inducible gene I (RIG-I), melanoma differentiation-associated gene-5 (MDA5), membrane-bound Toll-like receptors (TLR 3, 7, or 8) and several DNA sensors (Fig. 1) (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Schneider et al., 2014). These, in turn, activate signal transduction cascades that lead to expression of genes with specific inhibitory functions. In the case of type I IFNs, RIG-I and MDA5 activate the IFN-β promoter stimulator 1 (IPS-1), while TLR3 and TLR7/TLR8 trigger signaling through TRIF and MyD88, respectively (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Schneider et al., 2014). These pathways ultimately lead to phosphorylation and dimerization of transcription factors such as the interferon regulatory factor 3 (IRF-3), and IRF-7, leading to their translocation into the nucleus (Fig. 1) (for a complete discussion of the complex signaling events involved, see Hertzog and Williams, 2013; Hiscott, 2007; Schneider et al., 2014). Once in the nucleus, along with NF-κB and ATF-2/c-jun, they induce expression of antiviral cytokines including type I IFNs (IFN-α and IFN-β) through complex protein-protein interactions with many transcription factors and regulatory proteins including the transcriptional cofactor (CREB)-binding protein (CBP)/p300 (Hiscott, 2007; Jensen and Thomsen, 2012; Kawai and Akira, 2006; Onomoto et al., 2010; Randall and Goodbourn, 2008; Takeuchi and Akira, 2009). While most cells produce IFNβ, hematopoietic cells including monocyte/macrophages and dendritic cells, especially plasmacytoid dendritic cells constitute the predominant producers of IFNα.
Fig. 1. The type I IFN signaling cascade.
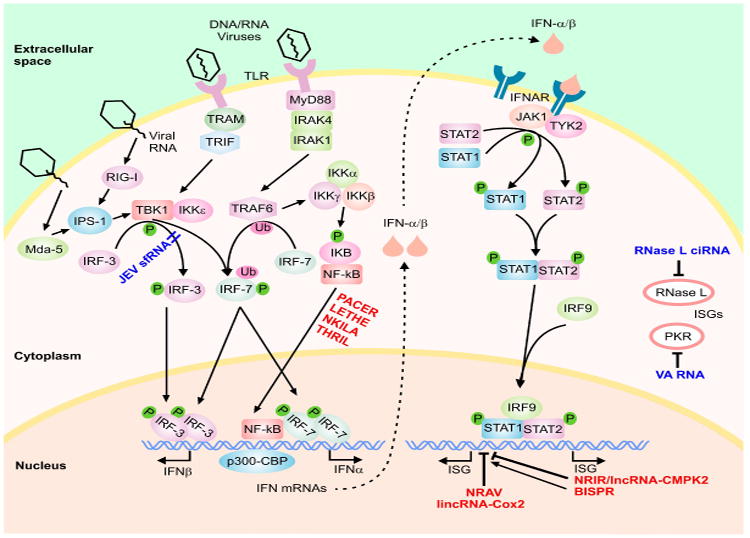
A highly simplified scheme of the signaling cascade is shown. The host-encoded regulatory lncRNAs are shown in red font and the virally-encoded lncRNAs in dark blue font. Phosphorylation and ubiquitination events are marked by small green circles and pink ovals, respectively.
Type I IFNs bind to the IFN-α/β receptor (IFNAR) in an autocrine and paracrine manner, leading to activation of the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling cascade (Fig. 1) (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Schneider et al., 2014). This results in recruitment, phosphorylation, dimerization and nuclear translocation of STAT1 and STAT2. Once in the nucleus, STAT1/STAT2 complex is bound to IRF9/p48, resulting in the formation of the IFN-stimulated gene factor 3 (ISGF3) complex which activates transcription of several hundred IFN-stimulated genes (ISGs) (Fig. 1) (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Schneider et al., 2014). Similarly, IFN-γ, the sole member of the type II IFN family, acts via binding the IFN-γ receptor and signaling through the JAK/STAT pathway leading to STAT1 phosphorylation and nuclear translocation (Hu and Ivashkiv, 2009). Activated STAT1 binds the gamma-activated sequence (GAS) near its target genes as a dimer, resulting in induction of transcription of STAT1-regulated genes. Interestingly, the majority of IFN-γ-regulated genes are also induced/repressed by type I IFN signaling and thus, there is significant overlap in the ISGs induced during the type I and type II IFN response (Hu and Ivashkiv, 2009; Pollard et al., 2013). It has been shown that many ISGs have antimicrobial activity through regulation of cellular gene expression machinery and direct antimicrobial effects, for example several ISGs including RNase L, PKR, IFIT-1, IFIT-2, ISG15, ISG20, IFITM3, BST2/Tetherin and viperin are known to possess antiviral activity (Randall and Goodbourn, 2008). While most cells are competent to mount type I IFN response, cell type and context can strongly affect the magnitude of the response and determine the subset of effector genes activated (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Schneider et al., 2014). In addition to regulation by several cellular signaling pathways, the IFN response is also subject to negative feedback regulation by a number of ISGs (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Porritt and Hertzog, 2015; Schneider et al., 2014; Yoshimura et al., 2007). Ultimately, activation of the IFN response leads to the induction of a cell-intrinsic antimicrobial state in the infected and the neighboring cells, thus limiting the spread of infectious agents. In addition, these potent cytokines modulate the innate immune response by promoting antigen presentation and natural killer cell function, and activate the adaptive immune system by augmenting antibody production in B cells and effector function of T cells (Ivashkiv and Donlin, 2014).
Interestingly, in addition to responding to the presence of exogenous viral RNA, a class of cellular non-coding transcripts which fall into the broad category of lncRNAs, the retrotransposon-derived RNAs, can also activate the IFN response (Q. Yu et al., 2015). A recent study has shown that activation of the Long Interspersed Element-1 (LINE-1) retroelements leads to an increase in the expression of IFNβ and ISGs (Q. Yu et al., 2015). Pretreatment of cells with IFN resulted in suppression of LINE-1 replication, and conversely, mutations that inactivated different elements in the IFN signaling pathway led to an increase in LINE-1 replication (Q. Yu et al., 2015). While the mechanism through which LINE-1 replication leads to the activation of IFN response is not known, it is plausible that the replication state of LINE elements and other retrotransposons are sensed by the same or similar sensors that detect the replication of exogenous viral RNA in the cell. Available data suggest that the replication of the Small Interspersed Elements (SINEs) similarly leads to activation of the IFN response (Leonova et al., 2013). Interestingly, some studies suggest that LINE-1 elements may contribute to the development of autoimmune diseases such as Systemic Lupus Erythematosus (SLE) (Crow, 2010; Nakkuntod et al., 2011), which are known to be associated with activation of the IFN response (Bronson et al., 2012; Elkon and Wiedeman, 2012). Further investigation of the intriguing connection between the state of activation of the repeat elements and autoimmunity may lead to novel insight into the mechanism of development of autoimmune diseases.
1.2. Long non-coding RNAs as a novel class of cellular regulatory factors
An exciting outcome of the high throughput analyses of higher eukaryotic transcriptomes has been the discovery of thousands of novel RNAs that do not seem to have any protein-coding capacity. These transcripts, the long non-coding RNAs (lncRNAs), are found in both prokaryotes and eukaryotes, however, they are highly abundant in higher eukaryotes including both animals and plants (Morris and Mattick, 2014; Rinn and Chang, 2012). Indeed, while protein-coding sequences occupy <2% of the genome in mammalians, a much larger fraction of the genome is transcribed into lncRNAs (Clark et al., 2013; ENCODE Project Consortium et al., 2012). lncRNAs can be tens of thousands of nucleotides long and although an originally proposed arbitrary lower length limit of 200 nucleotides should not be applied too strictly, it serves to distinguish lncRNAs from the small non-coding RNAs such as snRNAs, snoRNAs, miRNAs etc. (Clark and Mattick, 2011; Mattick and Rinn, 2015; Rinn and Chang, 2012).
Due to the relatively recent availability of high throughput transcriptome analysis technologies, our understanding of the extent of non-coding transcription in the mammalian genomes is highly incomplete. Many lncRNAs are expressed in a strongly cell type- and state-specific manner and their expression is tightly regulated by various cellular signals (Amaral et al., 2013; Rinn and Chang, 2012). Thus, provided there is sufficient sequencing depth, almost every RNA-seq experiment can potentially yield novel lncRNAs that are specific to the cell type and experimental conditions used in the study. In addition, many protein-coding RNAs have alternatively processed isoforms that lack protein-coding capacity and fall into the broad category of lncRNAs (Carninci et al., 2005; Djebali et al., 2012; ENCODE Project Consortium et al., 2012). Even among the lncRNAs that are currently annotated in public databases, the vast majority remain unstudied, thus providing an exciting opportunity for discovery of novel aspects of biological processes.
Emerging evidence from the study of a small subset of lncRNAs point to their involvement in virtually every aspect of cellular function (Amaral et al., 2013; Moran et al., 2012; Rinn and Chang, 2012; Ulitsky and Bartel, 2013; Wapinski and Chang, 2011; Yang et al., 2014), with regulation of nuclear events including the epigenetic state of chromatin and transcription emerging as major themes in lncRNA function (Amaral et al., 2013; Rinn, 2014; Rinn and Chang, 2012). Thus, nuclear localization and association with chromatin modifying complexes and transcription factors provide important clues into the potential function of a lncRNA. In addition, the genomic loci from which lncRNAs originate can yield crucial mechanistic insights into their function (Fig. 2). lncRNA genes can overlap protein-coding genes or other non-coding RNA genes in the sense or antisense orientation by originating from a promoter within an exon or intron of the overlapped gene, or from a promoter located in its 3′ UTR or further downstream. The overlapping genes can affect the biogenesis or function of the other genes in the locus through several mechanisms, including epigenetic regulation of the activity of the locus, transcriptional interference, and masking of functionally critical elements through basepairing to the other transcripts originating from the overlapped locus (Valadkhan and Nilsen, 2010). Another commonly observed conformation of lncRNA loci is transcription from the so-called bidirectional promoters (Fig. 2) (Adachi and Lieber, 2002; Uesaka et al., 2014; Wakano et al., 2012) and in some studied examples, it has been shown that one member of the pair regulates the expression of their promoter-sharing RNA (Uesaka et al., 2014; Wei et al., 2011). In addition, many lncRNAs originate from promoters in enhancer loci and are thought to be needed for the function of the enhancer in which they originate (Fig. 2) (Lam et al., 2014). Finally, many lncRNAs arise from genomic loci that do not overlap with other genes or enhancer elements but are located in vicinity of other genes (Fig. 2). Such vicinal lncRNAs can potentially regulate the expression of their neighboring genes through transcriptional interference or epigenetic regulation (Mattick and Rinn, 2015; Rinn and Chang, 2012; Valadkhan and Nilsen, 2010).
Fig. 2. Diverse genomic origins of lncRNAs.
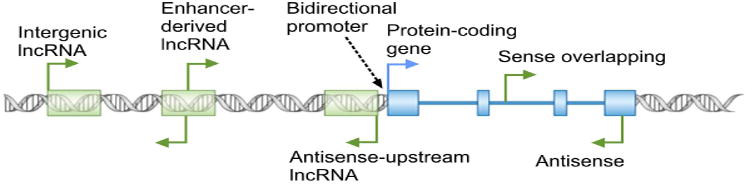
The broken arrows mark the location of transcription start sites and direction of transcription.
In recent years, several pioneering studies have provided evidence for the strong involvement of lncRNAs in regulation of various aspects of the immune response, including several pathways related to innate immunity (Fitzgerald and Caffrey, 2014; Heward and Lindsay, 2014; Imamura and Akimitsu, 2014; Marques-Rocha et al., 2015; Satpathy and Chang, 2015; Sigdel et al., 2015; Stachurska et al., 2014; A. D. Yu et al., 2015, 2015). While our knowledge of the role played by lncRNAs in the immune response is still in its infancy, it is highly likely that similar to miRNAs (Boss and Renne, 2010; Dai and Ahmed, 2011; David, 2010; Forster et al., 2015; Singh et al., 2013; Stachurska et al., 2014), lncRNAs also play a highly prominent role in regulation of immunity-related processes, including the IFN response. Although the main emphasis of this review is on understanding the role of lncRNAs in the IFN response, the mode and mechanism of function of lncRNAs in other immunity-related processes can provide a valuable context for understanding the scope and impact of lncRNA-mediated regulation of the IFN response. Further, many lncRNAs are involved in regulation of immune-related processes that directly or indirectly affect the IFN response. Thus, a brief review of our current state of knowledge of RNA-mediated regulation of the immune system is provided below. Comprehensive reviews of the role of lncRNAs in various aspects of the development and function of the immune response are included elsewhere in this volume (Carpenter, 2015; Aune and Spurlock, 2015).
1.3. lncRNAs in development, differentiation and function of the immune response
As mentioned above, recent studies have indicated diverse roles for lncRNAs in the development and function of the immune system. High throughput studies have revealed the presence of widespread changes in the expression of non-coding transcriptome during the differentiation of immune cells, for example during T cell development and differentiation (Spurlock et al., 2015; Hu et al., 2013; Ranzani et al., 2015; Zhang et al., 2014) and the activation of CD8+ T cells (Pagani et al., 2013; Pang et al., 2009). Many of the identified lncRNAs associate with transcription factors known to regulation T cell differentiation such as TBX21, GATA-3, STAT4 and STAT6, and functional analysis of a representative lncRNA, LincR-Ccr2-5′AS, proved the functional importance of the differentially expressed lncRNAs in regulation of T cell differentiation (Hu et al., 2013). Similarly, functional analysis of individual lncRNAs has revealed their role in regulation of granulocyte differentiation (HOTAIRM1 lncRNA) (Zhang et al., 2009); dendritic cell differentiation (lncRNA lnc-DC) (Wang et al., 2014); and regulation of expression of cell surface class I MHC receptors early in the development of NK cells (KIR antisense lncRNA) (Wright et al., 2013). In addition, the involvement of lncRNAs in several immune-related processes has been documented, including regulation of the three-dimensional chromatin structure at the immunoglobulin heavy chain locus that in turn affects V(D)J recombination (Bolland et al., 2004; Verma-Gaur et al., 2012); recruitment of activation-induced cytidine deaminase to its target loci, which is required for somatic hypermutation and antibody diversity (Pefanis et al., 2014); and regulation of T-cell independent B cell responses by transcripts derived from endogenous retroviruses (ERVs) (Zeng et al., 2014). As the studied RNAs represent a very small fraction of the lncRNAs that show differential expression during each of the above processes, it is highly likely that the study of additional lncRNAs will reveal the presence of a complex and widespread lncRNA-mediated regulatory network in immune system differentiation and function.
In addition to regulation of development and differentiation of immune cells, a number of studied lncRNAs play key roles in immunological processes which can affect the IFN response. For example lncRNAs PACER, Lethe, NKILA and THRIL regulate the inflammatory response through binding to the promoter of TNFα gene or subunits of NF-kB (Krawczyk and Emerson, 2014; Liu et al., 2015; Li et al., 2014; Rapicavoli et al., 2013). Another lncRNA that neighbors the TNF gene regulates the repression of the TNF locus by binding to the repressive LRRFIP1 protein (Shi et al., 2014). As mentioned above, NF-kB plays an important role in transcriptional activation of the IFN genes (Hiscott, 2007; Jensen and Thomsen, 2012; Kawai and Akira, 2006; Onomoto et al., 2010; Randall and Goodbourn, 2008; Takeuchi and Akira, 2009). Thus, in addition to their direct effect on the inflammatory response, the above lncRNAs can strongly affect the magnitude of the IFN response through regulation of NF-kB function.
A number of other lncRNAs similarly regulate immune-related processes that indirectly affect the IFN response. For example, lincRNA-Cox2, which is induced in response to stimulation of a subset of Tolllike receptors (TLRs), negatively regulates the expression of a number of immune response genes including ISGs Irf7, Oas1a, Oas1l, Oas2, Ifi204, and Isg15 (Carpenter et al., 2013; Guttman et al., 2009). While the mechanism through which changes in expression of lincRNA-Cox2 affects ISG expression is not known, it is clear that activation of its expression through TLR stimulation can be used to partially dampen the IFN-mediated transcriptional cascade. Similarly, regulation of cytokines and cytokine receptors such as interleukin-7 receptor α-subunit by lnc-IL7R (Cui et al., 2014) and IL1β expression by two neighboring lncRNAs (Ilott et al., 2014) can affect the cellular inflammatory response and thus, indirectly impact the IFN signaling pathway. Finally, a subset of host lncRNAs that are differentially expressed in response to viral infections (Josset et al., 2014; Peng et al., 2010; Yin et al., 2013; Zhang et al., 2013) may facilitate or inhibit viral replication via synergizing with or antagonizing the IFN response.
2. Regulation of the IFN response by host lncRNAs
2.1. Global analysis of lncRNAs induced in response to IFN stimulation
The first global gene expression analysis aiming at detection of the IFN-induced lncRNAs was reported as part of a study of virally-induced host lncRNAs in the mouse (Josset et al., 2014; Peng et al., 2010). The study indicated that a large number of host lncRNAs were induced in response to infection with mouse-adapted H1N1 influenza A virus or the recombinant mouse-adapted severe acute respiratory syndrome coronavirus (SARS-CoV) in mouse lungs in vivo. The differentially expressed lncRNAs accounted for close to 40% of all differentially expressed genes after infection and a higher percentage of them showed downregulation after infection compared to protein-coding genes, which were mostly upregulated. Interestingly, promoter analysis and expression correlation study suggested that many of the induced lncRNAs may be ISGs. Thus, to determine the contribution of the IFN response to the virally-induced lncRNAs, lung tissues were harvested from IFNα-treated or mock treated live mice at 12 hours post-treatment and subjected to RNA-seq. The high throughput analysis revealed the presence of 187 protein-coding genes and 53 lncRNAs that were upregulated after IFN treatment (Josset et al., 2014). Importantly, many of the IFN-induced lncRNAs found in the mouse lung tissue were also induced in cultured mouse embryonic fibroblasts after IFN stimulation. This indicated that at least a fraction of the observed differential expression was not caused by IFN-mediated changes in the cellular composition of the lung tissue, such as infiltration by immune cells, but rather represented bona fide IFN-induced lncRNAs (Josset et al., 2014; Peng et al., 2010).
A second transcriptome-wide analysis of IFNα-induced host lncRNAs was performed using primary human hepatocytes from five donors of different ages and genders before and at three time points of 3, 9 and 24 hours after treatment with IFNα (Kambara et al., 2014). In addition to the expected transcriptional induction of the protein-coding ISGs, close to two hundred lncRNAs showed significant differential expression after IFN stimulation, with roughly half of them being downregulated. The upregulated lncRNAs fell into a number of clusters in terms of the timing of onset and duration of upregulation that resembled the temporal patterns of expression of protein-coding ISGs. Nearly all upregulated lncRNAs were maximally induced 3 or 9 hours after IFN stimulation, with the majority maintaining their elevated cellular level after 24 hours. The remaining lncRNAs showed a sharp reduction in cellular level at 24 hour time point, thus, if they do have a cellular function, it is limited to the early time points after IFN stimulation (Kambara et al., 2014). Analysis of the upregulated lncRNAs indicated that several RNAs originated from bidirectional promoters which they shared with ISGs or protein-coding genes with other immune-related functions, or were otherwise located in very close proximity (<2 kb) of a protein-coding gene with immune-related function (Kambara et al., 2015). In all cases, the lncRNA and the neighboring protein-coding gene showed strongly concordant expression, suggesting the possibility of cis regulatory interactions between these lncRNAs and their protein-coding pair (Kambara et al., 2015).
A complementary set of studies in HuH7 human hepatocyte cell line defined gene expression changes occurring at later time points after IFNα2 treatment using microarray analysis (Carnero et al., 2014) and RNA-seq (Barriocanal et al., 2015). In these studies, a high dose of IFNα2 was used to obtain an augmented transcriptional response and global gene expression analysis was performed after 72 hours of stimulation to identify the differentially expressed genes at late time points of the IFN response (Barriocanal et al., 2015; Carnero et al., 2014). As expected, analysis of the differentially expressed protein-coding genes for enriched pathways showed a strong enrichment in IFN-related pathways. In addition to the known protein-coding ISGs, 48 and 890 lncRNAs showed significant differential expression in the microarray and RNA-seq study, respectively (Barriocanal et al., 2015; Carnero et al., 2014). Interestingly, similar to the study by Kambara et al. (2014), the differentially expressed lncRNAs were almost evenly divided to upregulated and downregulated ones in both microarray and RNA-seq datasets, while the vast majority of differentially expressed protein coding genes were upregulated. Among the identified lncRNAs, several were antisense to, overlapping in the sense orientation or seemed to share the same promoter with a protein-coding gene (Barriocanal et al., 2015; Carnero et al., 2014).
Comparison of the above three studies reveals intriguing similarities, including the high proportion of downregulated lncRNAs compared to protein-coding genes which were mostly upregulated. Unfortunately, none of the downregulated lncRNAs have been subjected to functional study to date, however, this difference could point to a fundamental difference in the scope of regulatory function of lncRNAs and protein-coding RNAs. Another important similarity between the two studies in human cells is the presence of a significant number of lncRNAs which are neighboring or overlapping protein-coding immunity-related genes. As mentioned above, the genomic locus of a lncRNA provides important clues into its potential function and thus, these observations suggest that a widespread lncRNA-mediated regulatory network may act in fine-turning the immune response. It will also be of great interest to determine the similarities and differences in the IFN-induced genes between mouse and human cells. Importantly, the above high throughput studies provide a wealth of targets for future functional studies of the role of lncRNAs in the IFN response.
2.2. lncRNA-mediated regulation of the expression of IFN genes
Transcriptional activation of the IFN genes in response to external or internal stimuli presents a unique regulatory opportunity which is regulated through several protein-mediated mechanisms (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Schneider et al., 2014). It is plausible that lncRNA-mediated regulatory mechanisms may also target this important step. Interestingly, evidence for lncRNA-mediated regulation for the IFNG/IFN-γ locus has been reported (Collier, 2014; Collier et al., 2012; Gomez et al., 2013). The involved regulatory lncRNA, IFNG-AS1 (IFNG-antisense-1), also known as Tmevpg1(Collier, 2014; Collier et al., 2012; Vigneau et al., 2003) and NeST (Gomez et al., 2013), is located downstream of the IFNG locus and in human, overlaps this locus. Importantly, its expression shows a strong correlation with that of IFNG (Collier, 2014; Collier et al., 2012; Vigneau et al., 2003). IFNG-AS1 is expressed in CD4+ and CD8+ T cells in addition to natural killer (NK) cells. In CD4+ T cells, the expression of both human IFNG-AS1 lncRNA and its mouse ortholog is dependent on STAT4 and TBX21 (T-BET), transcription factors that drive the differentiation of CD4+ lymphocytes into Th1 polarization (Collier et al., 2014). It has been shown that TBX21 guides the epigenetic remodeling of enhancer elements at the IFNG-AS1 locus, resulting in recruitment of transcriptional activators including NF-kB and transcriptional activation of the IFNG-AS1 gene (Collier et al., 2014). The expression of IFNG-AS1 was not sufficient to induce IFNG expression in the absence of STAT4 and TBX21 in CD4+ cells. However, when it was expressed in the presence of TBX21, the expression of IFNG in CD4+ Th1 polarized T cells could be partially restored in the absence of STAT4 (Collier et al., 2014). Another study in CD8+ T cells using transgenic mice that overexpressed IFNG-AS1 indicated that this lncRNA has a similar positive effect on expression of IFNG after stimulation with PMA (phorbol 12-myristate 13-acetate) and ionomycin (Gomez et al., 2013). Despite overlapping the IFNG locus, IFNG-AS1 seems to act in trans, as in the transgenic animals, IFNG-AS1 transgene can drive the expression of both alleles of IFNG (Gomez et al., 2013). Mechanistic analysis of the function of this lncRNA indicated that it interacts with WDR5, a subunit of the MLL/SET1 histone H3 lysine 4 methyltransferase complex, which is likely guided by the lncRNA to change the methylation status of the IFNG locus, resulting in activation (Gomez et al., 2013). While these results are certainly exciting, it will be important to determine whether any additional cellular factors regulate IFNG-AS1, for example during viral or bacterial infections or other physiological contexts when a strong IFN-γ response is desired.
2.3. IFN-induced lncRNAs as negative regulators of the IFN response
Among the ∼200 IFN-induced lncRNAs identified in primary human hepatocytes by Kambara et al. (2014), one lncRNA showed a dramatic induction after IFN stimulation in diverse cell types from both human and mouse. Similar to protein-coding ISGs, the induction of this RNA, which was located in close proximity of the protein-coding ISG CMPK2, was dependent on the JAK-STAT signaling pathway. Knockdown of this lncRNA, named lncRNA-CMPK2/NRIR (Negative Regulator of the IFN Response) led to a marked reduction in HCV replication in IFN-stimulated hepatocytes, suggesting that it could modulate the antiviral effect of IFN. Indeed, knockdown of lncRNA-CMPK2/NRIR resulted in upregulation of several protein-coding antiviral ISGs. The observed upregulation resulted from an increase in both basal and IFN-stimulated transcription, which could be explained by loss of transcriptional inhibition in knockdown cells. Although the mechanism of function of NRIR was not determined, the nuclear localization of the lncRNA and its impact on transcription of its target genes were consistent with an epigenetic or transcriptional regulatory function. Taken together, these data indicated that the IFN response involves a lncRNA-mediated negative regulatory mechanism (Kambara et al., 2014).
Another lncRNA, named NRAV (Negative Regulator of Antiviral Response), was discovered in a microarray screen for genes induced in response to influenza virus H1N1 infection in A549 human alveolar epithelium cell line (Ouyang et al., 2014). NRAV overlapped the first intron of the dynein light chain gene DYNLL1 in antisense orientation and was both spliced and polyadenylated. The cellular level of NRAV was strongly reduced after infection with a number of viruses in addition to the influenza virus and the reduced level was present in a number of cell lines. Interestingly, forced overexpression and knock down studies indicated that the cellular level of NRAV showed a direct correlation with viral reproduction both in cell lines and in transgenic mice. Microarray analysis of NRAV-overexpressing cell lines pointed to reduction in the level of a large number of ISGs, and additional studies indicated that NRAV, similar to NRIR (Kambara et al., 2014), can partially block IFN-induced expression of its target ISGs, likely through transcriptional regulation or epigenetic mechanisms (Ouyang et al., 2014). Considering the very small number of lncRNAs studied in the context of the immune response, the discovery of NRIR and NRAV suggests that lncRNAs play a critical role in negative regulation of the IFN response and likely other signaling cascades involved in innate and adaptive immunity. Defining the interplay of the negative regulatory lncRNAs with the known protein factors involved in negative regulation of IFN response (Hertzog and Williams, 2013; Ivashkiv and Donlin, 2014; Porritt and Hertzog, 2015; Schneider et al., 2014; Yoshimura et al., 2007) will provide a unified picture of the built-in mechanisms that mediate the termination of IFN signaling cascade.
2.4. Regulation of biogenesis or function of immune-related genes by IFN-induced lncRNAs
Of the several lncRNAs showing differential expression in response to IFN-α2 stimulation in HuH7 human hepatocyte cell line (see above) (Barriocanal et al., 2015; Carnero et al., 2014), five lncRNAs were chosen for further study. They originated from loci neighboring BST2 and ISG15 (both ISGs), GBP1, IRF1 and IL6 genes, all of which play key roles in the immune response, including regulatory impacts on the IFN response. These lncRNAs were predominantly nuclear in localization and showed concordant expression with their protein-coding neighbors. The above data point to the possibility that the lncRNAs may play a role in the regulation, or have a function related to that of their neighboring genes. Further, the lncRNAs are transcriptionally induced in response to a number of viral infections including Influenza virus and upregulated in a subset of liver samples from HCV-infected patients, which further suggests that they may have a function during the antiviral response in vivo (Barriocanal et al., 2015; Carnero et al., 2014). The two lncRNAs that neighbored ISG15 and BST2 originated from bidirectional promoters, and the induction of BST2 and its neighboring lncRNA gene after IFN stimulation was dependent on the JAK/STAT pathway, suggesting that the lncRNA, similar to BST2, was a true ISG. siRNA-mediated knockdown of the lncRNA neighbor of BST2 led to reduced cellular level of BST2 gene itself, pointing to the presence of a regulatory mechanism that coordinated the expression of BST2 with that of its neighboring lncRNA (Barriocanal et al., 2015).
The BST2 promoter-sharing lncRNA was also one of the most differentially expressed lncRNAs in primary human hepatocytes after IFN-α stimulation (Kambara et al., 2014). An independent study of this RNA by Kambara et al. (2015) confirmed that both BST2 and its promoter-sharing lncRNA, which was named BISPR (BST2 IFN-Stimulated Positive Regulator), were induced in response to IFNα in diverse cell types, including cells of lymphocytic and monocyte origins through the activation of the JAK–STAT pathway (Kambara et al., 2015). Similar to the results of Barriocanal and colleagues (2015) in hepatocytes, RNAi-mediated knock-down of BISPR resulted in transcriptional down-regulation of BST2 in THP1 monocytes (Kambara et al., 2015). On the other hand, forced overexpression of BISPR from a transgene resulted in up-regulation of BST2, indicating that BISPR regulates BST2 expression through interactions involving the BISPR RNA itself, rather than through the impact of its transcription on the local chromatin environment (Kambara et al., 2015). Interestingly, after stimulation by IFN-α, transcriptional activation of BISPR preceded the induction of BST2, suggesting that expression of BISPR induced or facilitated the initiation of transcription in its paired protein-coding gene. Since several protein-coding ISGs and immunity-related genes originate from bidirectional promoters that also give rise to lncRNAs, it is likely that at least a subset of them are regulated through a mechanism similar to the one described above (Barriocanal et al., 2015; Kambara et al., 2015; Carnero and Fortes, 2015; Landeras-Bueno and Ortin, 2015).
3. Regulation of the IFN response by virally-encoded lncRNAs
Many viruses have evolved strategies to either evade or suppress the host immune response, including the IFN response (Table 1) (Cox et al., 2015; Diamond, 2009; Gale and Sen, 2009). Through diverse mechanisms that range from inducing the degradation of various proteins involved in IFN signaling, inhibition of phosphorylation of key transcription factors, blocking their dimerization and interaction with their co-activator and preventing recruitment of RNA polymerase II, viruses have successfully blocked the launch of an effective IFN response by their host (Cox et al., 2015; Diamond, 2009; Gale and Sen, 2009). This is often accomplished by virally encoded protein products, although recent studies suggest that many virally-encoded miRNAs also play a key role in this process (Cox et al., 2015). Emerging data suggest that in addition to miRNAs and protein-coding RNAs, virally-encoded lncRNAs are also major participants in blocking of the IFN response during viral infections.
Table 1.
lncRNAs with a role in regulation of the IFN response.
| LncRNA name | Studied system |
|---|---|
| IFNG-AS1 | Multiple human and mouse cell types |
| NRIR | Multiple human and mouse cell types |
| NRAV | Multiple human and mouse cell types |
| BISPR | Multiple human cell types |
| PAN RNA | Kaposi's Sarcoma-associated Herpes Virus |
| West Nile Virus sfRNA | West Nile Virus |
| Japanese encephalitis virus sfRNA | Japanese encephalitis virus |
| Dengue virus sfRNAs | Dengue virus |
| RNase L ciRNA | Group C enteroviruses |
| VA RNAs | Adenoviruses |
A number of studied virally-encoded lncRNAs seem to target the transcription factors that mediate the induction of expression of IFN genes. An example of such a mechanism comes from studies in Kaposi's Sarcoma-associated Herpes Virus (KSHV), which have led to the identification of a nuclear-localized virally-encoded lncRNA named PAN RNA (Polyadenylated Nuclear RNA) (Rossetto et al., 2013; Rossetto and Pari, 2011; Sun et al., 1996). In vitro studies have suggested that PAN RNA interacts with a number of virus- and host-encoded factors, including interferon regulatory factor 4 (IRF4). Expression of PAN RNA, which occurs during the lytic phase of KSHV infection, leads to a decrease in expression of IFNγ, IFNα, and the key ISG RNase L, likely through modulation of the IRF4-mediated transcriptional activation (Rossetto et al., 2013; Rossetto and Pari, 2011).
Another viral immune-inhibitory lncRNA is encoded within the 3′ UTR of the arthropod-borne flaviviruses, a subgroup of flaviviridae which includes several human pathogens. The 3′ UTR of these viruses contains stable secondary structure elements which stall its degradation by the host exonuclease XRN1 (Lin et al., 2004; Pijlman et al., 2008; Schuessler et al., 2012; Silva et al., 2010; Urosevic et al., 1997). The partially degraded product, a virally-encoded lncRNAs that is 0.3-0.5 kb in length, is named the subgenomic flavivirus RNAs (sfRNAs) and has been shown to play an important role in viral pathogenicity (Pijlman et al., 2008). To define the role of sfRNAs in viral virulence, Schuessler and colleagues (Schuessler et al., 2012) showed that sfRNA-deficient West Nile Virus (WNV) loses its virulence and has severely reduced viral replication after IFNα administration. However, in cell lines or mice that lacked IRF-3, IRF-7 or the type I alpha/beta IFN receptor and thus cannot launch an IFN response, the sfRNA-deficient virus showed normal replication (Schuessler et al., 2012). Interestingly, transfection of an RNA species corresponding to the 3′ UTR of WNV improved the replication level of the IFN-sensitive Semliki Forest virus in the presence of exogenously added IFN (Schuessler et al., 2012). While the cellular impact of the sfRNA on the transcription of IFN genes and ISGs was not determined, these results suggest that WNV sfRNAs likely inhibit the signaling cascade downstream of the IFN receptor, or specifically inhibits certain key ISGs.
Another study on the impact of sfRNAs in the Japanese encephalitis virus (JEV) by Chang and colleagues (Chang et al., 2013) showed that forced overexpression of the sfRNA from a transgene in JEV-infected cells reduced IFNβ promoter activity and mRNA levels. They could show that this at least partially results from a reduced phosphorylation of IRF-3, which led to a partial block in its nuclear localization. This inhibition is analogous to what has been previously observed with the cysteine protease domain of the non-structural protein 2 (NS2) of porcine reproductive and respiratory syndrome virus (Li et al., 2010). Interestingly, the JEV sfRNA binds to the non-structural protein 5 (NS5) of JEV, potentially acting as an RNP (Chang et al., 2013). While this study suggests a different mechanism of action for sfRNAs compared to the study conducted in WNV (Schuessler et al., 2012), it is possible that both mechanisms are active in the two studied systems, with the dominant mechanism being different in each system. Alternatively, sfRNAs from different viruses may have evolved to perform distinct functions. For example, sfRNAs from dengue virus repress the expression of IFN-β and this effect is more pronounced with sfRNAs from epidemiologically dominant clades (Manokaran et al., 2015). While the mechanism behind this repressive function is not completely understood, it may involve interacting with the RNA-binding protein TRIM25 and preventing its activation, which is required for sustained RIG-I signaling (Manokaran et al., 2015). Dengue virus sfRNAs also repress translation of a number of ISGs, likely through binding to a number of proteins involved in post-transcriptional regulation of ISG expression (Bidet et al., 2014). Additional examples of proposed functions for sfRNAs include suppression of siRNA- and miRNA-mediated silencing pathways in both mammalian and insect cells by WNV and dengue virus sfRNAs (Schnettler et al., 2012) and altering mRNA stability by inhibiting XRN1 by sfRNAs from Dengue or Kunjin viruses (Moon et al., 2012).
In addition to inhibiting the induction of IFN expression and the IFN response cascade, virally-encoded RNAs can directly target key ISGs. A conserved kissing loop RNA structure within group C enteroviruses, named the RNase L competitive inhibitor RNA (RNase L ciRNA), is resistant to and inhibits the endoribonuclease activity of the ISG RNase L by binding the endoribonuclease domain as a competitive inhibitor (Han et al., 2007; Townsend et al., 2008a, 2008b). Finally, a number of virally-encoded RNAs such as the adenoviral VA (virus-associated) RNAs target the ISG PKR (protein kinase R), which regulates several signal transduction cascades as part of the IFN response (Langland et al., 2006; Sullivan, 2008).
It should be mentioned that in addition to inhibiting the IFN response, virally-encoded lncRNAs affect additional aspects of host antiviral response, such as regulation of protein synthesis through interacting with dsRNA-activated inhibitor of protein synthesis (DAI) by Epstein-Barr virus-encoded RNA EBER (Clarke et al., 1991), blocking apoptotic stimuli via interacting with the mitochondrial enzyme complex I by cytomegalovirus encoded β2.7 RNA (Reeves et al., 2007) or through interaction with TIAR (TIA∼1∼related) by Sendai virus trailer RNA (Iseni et al., 2002), inhibition of RNA interference by adenoviral VA1 RNA (Lu and Cullen, 2004) and regulation of viral latency by an HIV-1-encoded antisense RNA (Lazar et al., 2015; Saayman et al., 2015, 2014). Together, these lncRNA-mediated inhibitory mechanisms play a crucial role in viral pathogenicity by debilitating the host antiviral response, and present promising targets for future antiviral therapeutic development.
4. Concluding remarks
The rapid progress in our understanding of function of lncRNAs has revealed the presence of a large and nearly ubiquitous lncRNA-mediated regulatory network in higher eukaryotes that has revolutionized our understanding of many aspects of cellular function. While the study of this class of RNAs in the immune response in general and in the IFN response in particular is in its infancy, the significant number of lncRNAs that show differential expression in response to studied immunological stimuli make it highly likely that this class of RNAs are involved in regulation of most, if not all, steps in signaling and transcriptional events in innate and adaptive immunity. Functional study of a handful of IFN-induced lncRNAs suggest that negative regulation may be a major function of these RNAs, providing a novel class of therapeutic targets for boosting the endogenous IFN response. Similarly, the virally-encoded immune-inhibitory lncRNAs provide highly suitable targets for future antiviral therapies. Finally, the presence of SNPs or dysregulation in several lncRNAs and Endogenous Retroviral- (ERV) derived transcripts has been observed in a number of autoimmune diseases (Shirasawa et al., 2004; Sigdel et al., 2015; Tugnet et al., 2013), raising the possibility of a causative effect for immune modulatory lncRNAs in at least a subset of diseases of the immune system and further adding to the impact of studies on the function of this class of RNAs.
Highlights.
A large number of lncRNAs are differentially expressed in response to IFN stimulation.
Two IFN-induced lncRNAs act as negative regulators of the IFN response.
Another IFN-induced lncRNA positively regulates the expression of its neighboring gene, BST2/Tetherin.
Several virally-encoded lncRNAs increase viral pathogenicity by suppressing the IFN response.
Acknowledgments
Funding for this study was provided by CFAR grant number P30-AI036219.
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Saba Valadkhan, Email: saba.valadkhan@case.edu.
Lalith S. Gunawardane, Email: lsg48@case.edu.
References
- Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics. 2013;12:254–278. doi: 10.1093/bfgp/elt016. [DOI] [PubMed] [Google Scholar]
- Aune TM, Spurlock CF. Long non-coding RNAs in innate and adaptive immunity. Virus Res. 2015 Jul 9; doi: 10.1016/j.virusres.2015.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriocanal M, Carnero E, Segura V, Fortes P. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Front Immunol. 2015;5:655. doi: 10.3389/fimmu.2014.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 2014;10:e1004242. doi: 10.1371/journal.ppat.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen CR, Ding S, Robek MD, Kleinstein SH. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland DJ, Wood AL, Johnston CM, Bunting SF, Morgan G, Chakalova L, Fraser PJ, Corcoran AE. Antisense intergenic transcription in V(D)J recombination. Nat Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- Boss IW, Renne R. Viral miRNAs: tools for immune evasion. Current Opinion in Microbiology, Host–Microbe Interactions: Fungi/Parasites/Viruses. 2010;13:540–545. doi: 10.1016/j.mib.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson PG, Chaivorapol C, Ortmann W, Behrens TW, Graham RR. The genetics of type I interferon in systemic lupus erythematosus. Curr Opin Immunol. 2012;24:530–537. doi: 10.1016/j.coi.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Carnero E, Barriocanal M, Segura V, Guruceaga E, Prior C, Börner K, Grimm D, Fortes P. Type I Interferon Regulates the Expression of Long Non-Coding RNAs. Front Immunol. 2014;5:548. doi: 10.3389/fimmu.2014.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero E, Fortes P. HCV infection, IFN response and the coding and non-coding host cell genome. doi: 10.1016/j.virusres.2015.10.001. In press. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest ARR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Gatta GDella, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SPT, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schönbach C, Sekiguchi K, Semple CAM, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y FANTOM Consortium, RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carpenter S. Long noncoding RNA: novel links between gene expression and innate immunity. Virus Res. 2015 Sep 8; doi: 10.1016/j.virusres.2015.08. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O'Neill LAJ, Moore MJ, Caffrey DR, Fitzgerald KA. A Long Noncoding RNA Mediates Both Activation and Repression of Immune Response Genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RY, Hsu TW, Chen YL, Liu SF, Tsai YJ, Lin YT, Chen YS, Fan YH. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet Microbiol. 2013;166:11–21. doi: 10.1016/j.vetmic.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Clarke PA, Schwemmle M, Schickinger J, Hilse K, Clemens MJ. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucl Acids Res. 1991;19:243–248. doi: 10.1093/nar/19.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Choudhary A, Smith MA, Taft RJ, Mattick JS. The dark matter rises: the expanding world of regulatory RNAs. Essays Biochem. 2013;54:1–16. doi: 10.1042/bse0540001. [DOI] [PubMed] [Google Scholar]
- Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Collier SP. TMEVPG1, a long noncoding RNA within the immune system [WWW Document] [accessed 6.29.15];2014 URL http://etd.library.vanderbilt.edu/available/etd-03182014-144429/
- Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SP, Henderson MA, Tossberg JT, Aune TM. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol. 2014;193:3959–3965. doi: 10.4049/jimmunol.1401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JE, McClure LV, Goga A, Sullivan CS. Pan-viral-microRNA screening identifies interferon inhibition as a common function of diverse viruses. Proc Natl Acad Sci USA. 2015;112:1856–1861. doi: 10.1073/pnas.1417891112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Crow MK. Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity. 2010;43:7–16. doi: 10.3109/08916930903374865. [DOI] [PubMed] [Google Scholar]
- Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, Liu G. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol. 2014;44:2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M. Interferons and microRNAs. J Interferon Cytokine Res. 2010;30:825–828. doi: 10.1089/jir.2010.0080. [DOI] [PubMed] [Google Scholar]
- Diamond MS. Mechanisms of evasion of the type I interferon antiviral response by flaviviruses. J Interferon Cytokine Res. 2009;29:521–530. doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon KB, Wiedeman A. Type I IFN system in the development and manifestations of SLE. Curr Opin Rheumatol. 2012;24:499–505. doi: 10.1097/BOR.0b013e3283562c3e. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol. 2014;26C:140–146. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster SC, Tate MD, Hertzog PJ. MicroRNA as type I interferon-regulated transcripts and modulators of the innate immune response. Front Immunol. 2015;334 doi: 10.3389/fimmu.2015.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M, Sen GC. Viral evasion of the interferon system. J Interferon Cytokine Res. 2009;29:475–476. doi: 10.1089/jir.2009.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ. A Phylogenetically Conserved RNA Structure in the Poliovirus Open Reading Frame Inhibits the Antiviral Endoribonuclease RNase L. J Virol. 2007;81:5561–5572. doi: 10.1128/JVI.01857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog P, Forster S, Samarajiwa S. Systems biology of interferon responses. J Interferon Cytokine Res. 2011;31:5–11. doi: 10.1089/jir.2010.0126. [DOI] [PubMed] [Google Scholar]
- Hertzog PJ, Williams BRG. Fine tuning type I interferon responses. Cytokine Growth Factor Rev. 2013;24:217–225. doi: 10.1016/j.cytogfr.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends in Immunology. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J. Triggering the Innate Antiviral Response through IRF-3 Activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, Donnelly LE, Sims D, Lindsay MA. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun. 2014;5 doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Akimitsu N. Long non-coding RNAs involved in immune responses. Front Immunol. 2014;5:573. doi: 10.3389/fimmu.2014.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseni F, Garcin D, Nishio M, Kedersha N, Anderson P, Kolakofsky D. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 2002;21:5141–5150. doi: 10.1093/emboj/cdf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josset L, Tchitchek N, Gralinski LE, Ferris MT, Eisfeld AJ, Green RR, Thomas MJ, Tisoncik-Go J, Schroth GP, Kawaoka Y, Manuel de Villena FP, Baric RS, Heise MT, Peng X, Katze MG. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014;11:875–890. doi: 10.4161/rna.29442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Gunawardane L, Zebrowski E, Kostadinova L, Jobava R, Krokowski D, Hatzoglou M, Anthony DD, Valadkhan S. Regulation of interferon-stimulated gene BST2 by a lncRNA transcribed from a shared bidirectional promoter. Front Immunol. 2015;5:676. doi: 10.3389/fimmu.2014.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, Carnero E, Barriocanal M, Fortes P, Anthony DD, Valadkhan S. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Krawczyk M, Emerson BM. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MTY, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeras-Bueno S, Ortín J. Regulation of influenza virus infection by long non-coding RNAs. Virus Res. 2015 Aug 29; doi: 10.1016/j.virusres.2015.08. [DOI] [PubMed] [Google Scholar]
- Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Research, Translational Control During Virus Infections. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Lazar DC, Morris KV, Saayman SM. The emerging role of long non-coding RNAs in HIV infection. Virus Res. 2015 doi: 10.1016/j.virusres.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, Sen GC, Komarova EA, Gudkov AV. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. PNAS. 2013;110:E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zheng Z, Zhou P, Zhang B, Shi Z, Hu Q, Wang H. The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. J Gen Virol. 2010;91:2947–2958. doi: 10.1099/vir.0.025205-0. [DOI] [PubMed] [Google Scholar]
- Lin KC, Chang HL, Chang RY. Accumulation of a 3′-Terminal Genome Fragment in Japanese Encephalitis Virus-Infected Mammalian and Mosquito Cells. J Virol. 2004;78:5133–5138. doi: 10.1128/JVI.78.10.5133-5138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci USA. 2014;111:1002–1007. doi: 10.1073/pnasnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, Harris E, Garcia-Blanco MA, Ooi EE. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015 doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015 doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- Meyer O. Interferons and autoimmune disorders. Joint Bone Spine. 2009;76:464–473. doi: 10.1016/j.jbspin.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucl Acids Res. 2012;40:6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakkuntod J, Avihingsanon Y, Mutirangura A, Hirankarn N. Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin Chim Acta. 2011;412:1457–1461. doi: 10.1016/j.cca.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Onomoto K, Onoguchi K, Takahasi K, Fujita T. Type I interferon production induced by RIG-I-like receptors. J Interferon Cytokine Res. 2010;30:875–881. doi: 10.1089/jir.2010.0117. [DOI] [PubMed] [Google Scholar]
- Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF, Wang G, Chen JL. NRAV, a Long Noncoding RNA, Modulates Antiviral Responses through Suppression of Interferon-Stimulated Gene Transcription. Cell Host & Microbe. 2014;16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJP, Rossi RL, Geginat J, Abrignani S. Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation. Immunol Rev. 2013;253:82–96. doi: 10.1111/imr.12055. [DOI] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-Wide Identification of Long Noncoding RNAs in CD8+ T Cells. J Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, Basu U. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, Bouzek HK, Haynor DR, Frieman MB, Heise M, Raymond CK, Baric RS, Katze MG. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. A Highly Structured, Nuclease-Resistant, Noncoding RNA Produced by Flaviviruses Is Required for Pathogenicity. Cell Host & Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Pollard KM, Cauvi DM, Toomey CB, Morris KV, Kono DH. Interferon-gamma and Systemic Autoimmunity. Discovery Medicine. 2013;16:123–131. [PMC free article] [PubMed] [Google Scholar]
- Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends in Immunology. 2015;36:150–160. doi: 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJP, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, De Francesco R, Geginat J, Bodega B, Abrignani S, Pagani M. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol. 2015;16:318–325. doi: 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I Binding by a Virally Encoded RNA Regulates Mitochondria-Induced Cell Death. Science. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- Rinn JL. lncRNAs: Linking RNA to Chromatin. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto CC, Pari GS. Kaposi's sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J Virol. 2011;85:13290–13297. doi: 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P, Pari GS. Regulation of viral and cellular gene expression by Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA. J Virol. 2013;87:5540–5553. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S, Ackley A, Turner AMW, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther. 2014;22:1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S, Roberts TC, Morris KV, Weinberg MS. HIV Latency and the noncoding RNA therapeutic landscape. Adv Exp Med Biol. 2015;848:169–189. doi: 10.1007/978-1-4939-2432-5_9. [DOI] [PubMed] [Google Scholar]
- Satpathy AT, Chang HY. Long Noncoding RNA in Hematopoiesis and Immunity. Immunity. 2015;42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu Rev Immunol. 2014 doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. West Nile Virus Noncoding Subgenomic RNA Contributes to Viral Evasion of the Type I Interferon-Mediated Antiviral Response. J Virol. 2012;86:5708–5718. doi: 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Song L, Fitzgerald M, Maurer K, Bagashev A, Sullivan KE. Noncoding RNAs and LRRFIP1 regulate TNF expression. J Immunol. 2014;192:3057–3067. doi: 10.4049/jimmunol.1302063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa S, Harada H, Furugaki K, Akamizu T, Ishikawa N, Ito K, Ito K, Tamai H, Kuma K, Kubota S, Hiratani H, Tsuchiya T, Baba I, Ishikawa M, Tanaka M, Sakai K, Aoki M, Yamamoto K, Sasazuki T. SNPs in the promoter of a B cell-specific antisense transcript, SAS-ZFAT, determine susceptibility to autoimmune thyroid disease. Hum Mol Genet. 2004;13:2221–2231. doi: 10.1093/hmg/ddh245. [DOI] [PubMed] [Google Scholar]
- Sigdel KR, Cheng A, Wang Y, Duan L, Zhang Y. The Emerging Functions of Long Noncoding RNA in Immune Cells: Autoimmune Diseases. J Immunol Res. 2015;2015:848790. doi: 10.1155/2015/848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva PAGC, Pereira CF, Dalebout TJ, Spaan WJM, Bredenbeek PJ. An RNA Pseudoknot Is Required for Production of Yellow Fever Virus Subgenomic RNA by the Host Nuclease XRN1. J Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, Mc Curdy DK, Sharma S, Wong D, Hahn BH, Rehimi H. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12:1160–1165. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Spurlock CF, Tossberg JT, Guo Y, Collier SP, Crooke PS, Aune TM. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachurska A, Zorro MM, van der Sijde MR, Withoff S. Small and long regulatory RNAs in the immune system and immune diseases. Front Immunol. 2014;5:513. doi: 10.3389/fimmu.2014.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet. 2008;9:503–507. doi: 10.1038/nrg2349. [DOI] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA. 2008a;14:1026–1036. doi: 10.1261/rna.958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Silverman RH, Barton DJ. A putative loop E motif and an H-H kissing loop interaction are conserved and functional features in a group C enterovirus RNA that inhibits ribonuclease L. RNA Biol. 2008b;5:263–272. doi: 10.4161/rna.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugnet N, Rylance P, Roden D, Trela M, Nelson P. Human Endogenous Retroviruses (HERVs) and Autoimmune Rheumatic Disease: Is There a Link? Open Rheumatol J. 2013;7:13–21. doi: 10.2174/1874312901307010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesaka M, Nishimura O, Go Y, Nakashima K, Agata K, Imamura T. Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genomics. 2014;15:35. doi: 10.1186/1471-2164-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urosevic N, van Maanen M, Mansfield JP, Mackenzie JS, Shellam GR. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and - resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol. 1997;78(Pt 1):23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Nilsen TW. Reprogramming of the non-coding transcriptome during brain development. J Biol. 2010;9:5. doi: 10.1186/jbiol197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci USA. 2012;109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a Candidate Gene for the Control of Theiler's Virus Persistence, Could Be Implicated in the Regulation of Gamma Interferon. J Virol. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakano C, Byun JS, Di LJ, Gardner K. The dual lives of bidirectional promoters. Biochim Biophys Acta. 2012;1819:688–693. doi: 10.1016/j.bbagrm.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Järvelin AI, Steinmetz LM. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PW, Huehn A, Cichocki F, Li H, Sharma N, Dang H, Lenvik TR, Woll P, Kaufman D, Miller JS, Anderson SK. Identification of a KIR antisense lncRNA expressed by progenitor cells. Genes Immun. 2013;14:427–433. doi: 10.1038/gene.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Guan D, Fan Q, Su J, Zheng W, Ma W, Ke C. lncRNA expression signatures in response to enterovirus 71 infection. Biochem Biophys Res Commun. 2013;430:629–633. doi: 10.1016/j.bbrc.2012.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- Yu AD, Wang Z, Morris KV. Long noncoding RNAs: a potent source of regulation in immunity and disease. Immunol Cell Biol. 2015;93:277–283. doi: 10.1038/icb.2015.2. [DOI] [PubMed] [Google Scholar]
- Yu Q, Carbone CJ, Katlinskaya YV, Zheng H, Zheng K, Luo M, Wang PJ, Greenberg RA, Fuchs SY. Type I interferon controls propagation of long interspersed element-1. J Biol Chem. 2015;290:10191–10199. doi: 10.1074/jbc.M114.612374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Hu Z, Shi X, Li X, Zhan X, Li XD, Wang J, Choi JH, Wang K, Purrington T, Tang M, Fina M, DeBerardinis RJ, Moresco EMY, Pedersen G, McInerney GM, Karlsson Hedestam GB, Chen ZJ, Beutler B. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 2014;346:1486–1492. doi: 10.1126/science.346.6216.1486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang Q, Chen CY, Yedavalli VSRK, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4:e00596–00512. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, Gu W, Sun B. TH1/TH2 cell differentiation and molecular signals. Adv Exp Med Biol. 2014;841:15–44. doi: 10.1007/978-94-017-9487-9_2. [DOI] [PubMed] [Google Scholar]


