Abstract
Purpose
To develop a strategy of achieving targeted collection of endothelial cells (ECs) by endovascular methods and analyzing the gene expression profiles of collected single ECs.
Methods and results
134 ECs and 37 leukocytes were collected from four patients' intra-iliac artery endovascular guide wires by fluorescence activated cell sorting (FACS) and analyzed by single-cell quantitative RT-PCR for expression profile of 48 genes. Compared to CD45+ leukocytes, the ECs expressed higher levels (p < 0.05) of EC surface markers used on FACS and other EC related genes. The gene expression profile showed that these isolated ECs fell into two clusters, A and B, that differentially expressed 19 genes related to angiogenesis, inflammation and extracellular matrix remodeling, with cluster B ECs have demonstrating similarities to senescent or aging ECs.
Conclusion
Combination of endovascular device sampling, FACS and single-cell quantitative RT-PCR is a feasible method for analyzing EC gene expression profile in vascular lesions.
Keywords: Targeted endothelial cell sampling, Single cell quantitative RT-PCR, Gene expression of artery endothelial cells
1. Introduction
Gene expression studies of patient-derived endothelial cells (ECs) provide important information regarding the pathogenesis of many vascular diseases [1–3], in and outside of the central nervous system. Several groups have reported EC enrichment and identification from endovascular guide wires by 2 EC separation methods: micropipette picking-up [4] or CD146 antibody-conjugated magnetic beads [5–7], both followed by either traditional gene expression assays like bulk mRNA reverse transcription (RT) PCR which analyzes RNA extracted from a pool of ECs [4,5], quantitative RT-PCR [6] or quantitative immunofluorescence [7–14]. Because of the limitation set by these conventional methods, only a few (up to 3 or 4) genes can be analyzed. Due to the complexity and heterogeneity of ECs [15], these studies have incurred uncertainty and controversy regarding the purity and functionality of the ECs studied.
Although DNA microarray studies of ECs separated from tissue can provide high throughput EC gene expression information and have indicated that heterogeneity of endothelium exists among different tissues or diseases [15,16], this technique needs bulk mRNA extracted from at least thousands of ECs, numbers difficult to attain using endovascular EC sampling methods. Furthermore, DNA microarray can only analyze gene expression patterns of a group of ECs and not each individual EC. A more complete picture of individual EC functional condition in specific environments needs an assay, which can analyze the expression profile of multiple genes in individual ECs.
Recently we reported that EC candidates attached on guide wires can be collected by fluorescence activated cell sorting (FACS) and laser capture microdissection. The quality of mRNA extracted from the ECs is sufficient for analysis of gene expression using quantitative RT-PCR [17]. Single-cell quantitative RT-PCR combined with high-throughput microfluidic array technology facilitates detection of gene expression profiles of up to 96 genes in 96 individual cells simultaneously [18,19]. Therefore, it is a powerful high throughput tool to characterize gene expression of individual cells.
In this study, we demonstrated that combination of FACS and high throughput microfluidic single-cell quantitative RT-PCR is an efficient and powerful method for analyzing the changes of EC gene expression profiles in vascular lesions.
2. Material and methods
2.1. Case selection and EC harvest
Samples were collected from four patients undergoing routine catheter angiography for assessment of cerebrovascular pathology. The patients provided written consent for the procedure inclusive of the collection and study of tissues for research purposes standard on surgical consent forms. ECs were obtained by inserting a 0.038-inch diameter coaxial curved stainless steel guide wire (Cook Inc., Bloomington, IN) into the right iliac artery as part of routine arterial access. Wires are directed under fluoroscopic visualization so a short (<5 cm) segment of vessel may be specifically contacted. The cells attached on the wires were dislodged by vortexing and centrifuging in a dissociation buffer (Gibco, Grand Island, NY). After lysing RBC by ACK Lysing Buffer (Gibco, Grand Island, NY), and centrifuged at 1500 rpm, the pellets were re-suspended in FACS buffer for incubation of antibodies and sorting. Experiment design is shown in Fig. 1.
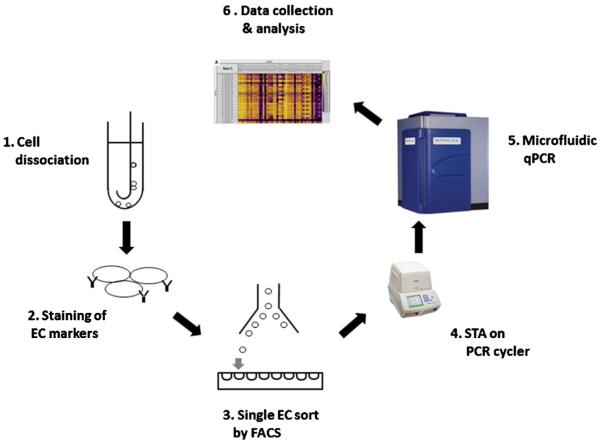
Fig. 1.
Experimental design. Cells were dislodged from the guide wire (1) and were stained by antibodies specific for different cell surface markers (2). Individual ECs were sorted into 96-well plates by FACS (3). Specific gene cDNAs were pre-amplified by thermocycler (4). Quantitative RT-PCR was performed on Biomark HD system (Fluidigm, South San Francisco, CA) (5). Data were collected and analyzed by quantitative RT-PCR analysis software (Fluidigm, South San Francisco, CA) (6).
2.2. EC candidate identification and sorting on FACS
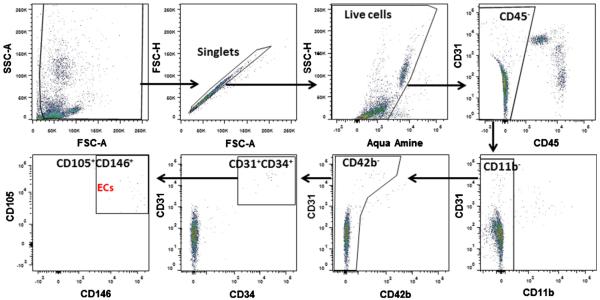
Single EC candidates were identified and sorted by a protocol of seven fluorescently-conjugated monoclonal antibodies on FACS that we described in our previous study [17]. LIVE/DEAD Fixable Dead Cell Stain (Life Technologies, Carlsbad, CA) was used to exclude the dead cells. The antibody information is listed in Table 1. After staining the dislodged cells with these seven antibodies and the Amine Aqua Reactive Dye (AmCyan channel), the debris, doublets and dead cells were excluded before subsequent procedures (Fig. 2). After excluding CD45+ leucocytes, CD11b+ myeloid cells and CD42b+ platelets by three negative gates, the remaining cells were gated by four EC specific surface markers, CD31, CD34, CD105 and CD146. Cells positive for the 4 EC surface markers were collected as EC candidates. CD45+ leucocytes were also collected and used as control. EC candidates and leukocytes were sorted into 96 well plates on a FACS Aria II (BD Biosciences, San Jose, CA) with 100 nm nozzle using single cell sort mode.
Table 1.
Fluorescently conjugated monoclonal antibodies used for EC candidate identification on FACS.
| Target | Format | Dilution | Vendor | Catalog number |
|---|---|---|---|---|
| CD31 | Alexa 647 | 1:500 | BD Biosciences | 561654 |
| CD34 | PE-Cy7 | 1:50 | Biolegend | 343516 |
| CD105 | PE-CF594 | 1:100 | BD Biosciences | 562380 |
| CD146 | PE | 1:50 | BD Biosciences | 561013 |
| CD45 | Alexa 700 | 1:50 | Life technologies | MHCD4529 |
| CD11b | PacBlue | 1:50 | Biolegend | 301324 |
| CD42b | FITC | 1:50 | BD biosciences | 555472 |
Fig. 2.
FACS gating strategy for EC collection. Seven cell surface markers and one viability marker were used to gate the EC candidates. Cells were first gated to exclude debris, doublets and dead cells identified by positive Aqua Amine stain. After gating on the viable single cells, the leukocytes (CD45+), macrophages (CD11b+) and platelets (CD42b+) were eliminated. EC candidates were first selected by CD31 and CD34, and then CD105 and CD146.
2.3. Reverse Transcription and cDNA pre-amplification
Reverse transcription and cDNA pre-amplification were carried out on a PCR thermocycler. Briefly, each EC candidate was sorted directly into one well with 9 μL reverse transcription-specific target amplification (RT-STA) buffer on the 96-well plates (Eppendoff, Hauppauge, NY). The RT-STA buffer contained 5 μL CellsDirect 2× Reaction Mix (Life Technologies, Carlsbad, CA), 0.2 μL SuperScript III RT Platinum Taq Mix (Life Technologies, Carlsbad, CA), 2.8 μL nuclease free water and 1 μL 10× primer mixture (500 nM) that contained a mix of 48 pairs of primers specific to genes listed in Table 2. The primers were custom designed and all expand introns to minimize the genomic DNA fraction (Fluidigm, South San Francisco, CA). The Fluidigm Assay IDs listed in Table 2 can be used to obtain primer sequences.
Table 2.
Genes selected for single gene expression analysis.
| Gene group | Symbol | Gene name |
Description | Function and reference | Fluidigm assay IDb |
|---|---|---|---|---|---|
| Cell marker | PTPRCa | CD45 | Protein tyrosine phosphatase, receptor type C | Leucocyte marker | GEP00055840 |
| PECAM1a | CD31 | platelet endothelial cell adhesion molecule-1 | Adhesion molecular, inflammation [32] | GEP00056436 | |
| CD34a | CD34 | Hematopoietic Progenitor Cell Antigen | EC marker, inflammation [33] | GEA00011907 | |
| ENGa | CD105 | Endoglin | EC marker, angiogenesis [34] | GEP00056632 | |
| MCAMa | CD146 | Melanoma cell adhesion molecule | EC marker, inflammation [35–38] | GEP00056760 | |
| KDR | Flk1 | vascular endothelial growth factor receptor 2 | EC marker, angiogenesis [39,40] | GEA00012361 | |
| FLT1 | VEGFR1 | vascular endothelial growth factor receptor 1 | EC marker, migration [41] | GEP00055864 | |
| TIE1 | Tie1 | tyrosine kinase with Ig-like and EGF-like domains 1 | EC marker, Angiogenesis [39,42] | GEA00012787 | |
| THBD | – | Thrombomodulin | EC marker [43] | GEA00014984 | |
| VWF | vWF | Von Willebrand factor | EC marker, angiogenesis [44] | GEA00013832 | |
| TEK | Tie2 | tyrosine kinase with Ig-like and EGF-like domains 2 | EC marker, angiogenesis [39,40] | GEA00013803 | |
| ACTG2 | α-actin | Actin, gamma-enteric smooth muscle | VSMC marker | GEA00025197 | |
| EPHB2 | EphB2 | Ephrin type-B receptor 2 | Arterial EC marker | GEA00029202 | |
| EPHB4 | EphB4 | Ephrin type-B receptor 4 | Venous EC marker, angiogenesis [45] | GEP00059920 | |
| Angiogenesis | VEGFA | VEGF-A | Vascular endothelial growth factor | Angiogenesis [40,46,47] | GEA00012311 |
| TGFB1 | TGF-b1 | Transforming growth factor beta1 | Modulate angiogenesis [34] | GEA00007272 | |
| PCNA | PCNA | Proliferating Cell Nuclear Antigen | Proliferation marker [48] | GEA00012343 | |
| CAT | – | catalase | Oxidative stress & Proliferation [28,49] | GEA00023106 | |
| SGK1 | SGK | serum-glucocorticoid-induced protein kinase | Proliferation [50] | GEP00060290 | |
| ANGPT1 | – | angiopoietin-1 | angiogenesis [40,51] | GEA00013518 | |
| ANGPT2 | – | angiopoietin-2 | Angiogenesis [52] | GEP00057393 | |
| HIF1A | HIF-1α | Hypoxia-inducible factor 1-alpha | Angiogenesis [53,54] | GEA00012495 | |
| NR4A1 | TR3 | human orphan receptor TR3 | Proliferation [55] | GEA00023496 | |
| ALOX5 | 5-LO | 5-lipoxygenase | Proliferation [56] | GEA00028402 | |
| CD44 | – | Proliferation, angiogenesis [57,58] | GEP00056546 | ||
| ACE | – | Angiotensin-converting enzyme | Angiogenesis [59] | GEP00058643 | |
| Inflammation | IL6 | – | Interleukin 6 | Inflammation [60] | GEA00012521 |
| IL8 | – | Interleukin 8 | Inflammation [61] | GEA00012363 | |
| VCAM1 | VCAM-1 | vascular cell adhesion molecule 1 | Inflammation [32] | GEP00056408 | |
| ICAM1 | ICAM-1 | Intercellular Adhesion Molecule 1 | Inflammation[32] | GEP00056359 | |
| TBXAS1 | THA-2 | thromboxane synthase-A2 | Inflammation [62] | GEP00060291 | |
| NOS3 | eNOS | endothelial nitric oxide synthase | Oxidative stress, Inflammation [63,64] | GEA00032450 | |
| CCL2 | MCP-1 | monocyte chemoattractant protein 1 | Inflammation [61,65,66] | GEP00055652 | |
| SELP | – | P-selectin | Adhesion molecular, Inflammation [32] | GEA00030146 | |
| PTGS1 | COX-1 | Cyclooxygenase-1 | Inflammation [67] | GEA00027133 | |
| PTGS2 | COX-2 | Cyclooxygenase-2 | Inflammation [46] | GEA00007158 | |
| ECM remodeling | MMP2 | MMP-2 | matrix metalloproteinase-2 | ECM metabolism [22,53,68] | GEA00013719 |
| MMP9 | MMP-9 | matrix metalloproteinase-9 | ECM metabolism, inflammation [22] | GEA00013721 | |
| MMP14 | MMP-14 | matrix metalloproteinase-14 | ECM metabolism [68] | GEA00026567 | |
| SERPINE1 | PAI-1 | Plasminogen activator inhibitor-1 | ECM metabolism [69,70] | GEP00056400 | |
| TNF | TNF-α | Tumor necrosis factor-α | ECM metabolism, inflammation [68] | GEP00059924 | |
| ITGA7 | – | Integrin-α | ECM metabolism [71] | GEP00058254 | |
| TIMP1 | TIMP-1 | Tissue inhibitor of metalloproteinase 1 | ECM metabolism, inflammation [72] | GEA00007289 | |
| TIMP2 | TIMP-2 | Tissue inhibitor of metalloproteinase 2 | ECM metabolism, inflammation [73,74] | GEA00020949 | |
| FN1 | – | fibronectin | ECM metabolism [75] | GEA00007778 | |
| TNC | – | Tenasin-C | ECM metabolism [76] | GEA00031358 | |
| SCEL | – | sciellin | ECM metabolism [77] | GEA00031897 | |
| PPL | – | periplakin | ECM metabolism [77] | GEA00032646 |
Genes used in FACS.
Primer sequences for microfluidic qPCR can be traced by these company assay IDs.
The samples were incubated at 50 °C for 15 min for the reverse transcription, 95 °C for 2 min for inactivating reverse transcriptase and activating Taq polymerase, then subjected to 18 PCR cycles (95 °C 15 sec then 60 °C for 4 min for each cycle) for specific targets amplification (STA). To remove the unincorporated primers for best results, each sample was then mixed with 3.6 μL exonuclease treatment buffer composed of 2.52 μL water, 0.36 μL 10× Exonuclease I reaction buffer and 0.72 μL 20 units/μL Exonuclease I (New England BioLabs, Ipswich, MA), incubated at 37 °C for 30 min for digestion and 80 °C for 15 min to inactivate the exonuclease.
2.4. Quantitative RT-PCR
48.48 nanofluidic chips and a BioMark HD system (Fluidigm, South San Francisco, CA) were used. Briefly, each pre-amplified cDNA sample was diluted by 5 fold in TE Buffer (TEKnova, Hollister, CA). Then, 2.25 μL diluted samples were mixed with 2.5 μL 2× SsoFast EvaGreen Supermix with Low ROX (Bio-Rad, Hercules, CA) and 0.25 μL 20× DNA Binding Dye Sample Loading Reagent (Fluidigm, South San Francisco, CA). The pre-mix samples (5 μL each) were loaded into the 48 sample inlets on the 48.48 Dynamic Array (Fluidigm, South San Francisco, CA), which had been primed with control line fluid (Fluidigm, South San Francisco, CA) on IFC Controller MX (Fluidigm, South San Francisco, CA). Assay Mix (5 μl) containing 2.5 μL 2× Assay Loading Reagent (Fluidigm, South San Francisco, CA), 2.25 μL 1× DNA suspension buffer (TEKnova, Hollister, CA) and 0.25 μL primer set (100 μM) were added to the 48 assay inlets on the 48.48 nanofluidic chip (Fluidigm, South San Francisco, CA). After loading both pre-mixed samples and the assay mixtures to the nanochip by IFC Controller MX (Fluidigm, South San Francisco, CA), the chip was loaded into the BioMark HD system (Fluidigm, South San Francisco, CA) for PCR through 35 cycles of 5 sec at 96 °C and 20 sec at 60 °C after a hot start phase of 60 sec at 95 °C. Fluorescence in the EvaGreen channel was detected and collected by a CCD camera placed above the chip and 6-carboxy-X-rhodamine (ROX) intensity was used as normalization.
2.5. Data collection and analysis
Quantitative RT-PCR data of ECs and leukocytes obtained from 4 subjects were analyzed together. Fluidigm quantitative RT-PCR Analysis software (Fluidigm, South San Francisco, CA) was used to process RT-PCR data obtained by Biomark HD system and calculate Ct values. Ct values were further processed in the R statistical language using algorithms provided by SINGuLAR Analysis Toolset 3.5 (Fluidigm, South San Francisco, CA). All Raw Ct values were normalized to the assumed detection Ct level of 24 following the recommendation from this manual. Ct values were converted to relative expression levels using methods described previously [20]. The assumed minimum value of genes without expression was set as 10% lower than the lowest recorded reading. Euclidean distance metric and complete linkage function were used to build the Hierarchical clustering. Mean-centered data were used for principal components analysis (PCA) to avoid bias caused by highly expressed genes.
3. Results
3.1. Selection of genes for profiling EC gene expression
Based on previous EC function studies [21,22], we selected three groups of genes to characterize ECs in this study. They are 19 angiogenesis-related genes, 13 inflammation-related genes and 12 extra-cellular matrix (ECM) remodeling-related genes. To confirm the identity of ECs isolated by FACS, six EC specific marker genes and one vascular smooth muscle cell marker gene (α-actin) were included. We also included the four EC-marker genes and CD45 that were used for FACS selection of EC candidates and leukocytes (Table 2).
3.2. The gene expression profile of EC candidates is distinctively different from that of LCs
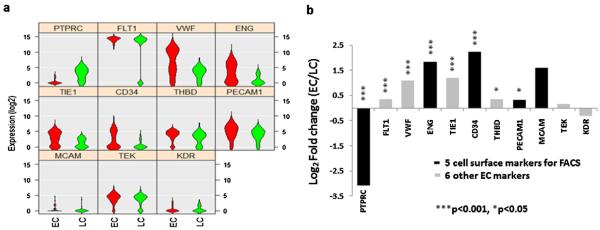
A total of 134 EC candidates and 37 leukocytes (LCs) were collected by FACS through the gating strategy we described previously [17] and shown in Fig. 2. Among these ECs, 64 (48%) expressed three EC markers CD31, CD34 and CD105, and 30 (22%) expressed four EC markers CD31, CD34, CD105 and CD146. Furthermore, we compared gene expression profiles of EC candidates and LCs. Among the 11 marker genes, eight were differentially expressed between the ECs and LCs (Fig. 3). Among the five marker genes used in FACS, the expression of the LC marker CD45 (p = 1.2 × 10−27) was significantly higher in LCs than ECs, and the expression of EC markers, CD31 (p = 0.017), CD34 (p = 3.1 × 10−5) and CD105 (p = 6.3 × 10−7) were significantly higher in the ECs than LCs. The expression of CD146 showed a trend toward higher in ECs than in LCs (p = 0.15). In addition, compared to LCs, ECs expressed higher levels of the other four EC specific genes, VEGFR1 (p = 1.3 × 10−8), vWF (p = 2.3 × 10−7), Tie1 (p = 1.3 × 10−5) and THBD (p = 0.013). These data indicate that the EC candidates isolated by FACS were indeed ECs.
Fig. 3.
Differential gene expression of ECs and LCs. (a) Violin plots showed the expression of 11 cell-marker genes are different in the ECs (Red) compared with LCs (Green). The gene name is indicated on top of each violin plot and the value on Y-axle represents the gene expression level in the binary logarithm (log2) value. (b) Bar graph shows the values of differential gene expression by fold change of the binary logarithm (log2) in ECs relative to LCs (*p < 0.05; ***p < 0.001). Gene symbols are used in the gures and corresponding gene names can be found in Table 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Two EC clusters were identified based on gene expression profile
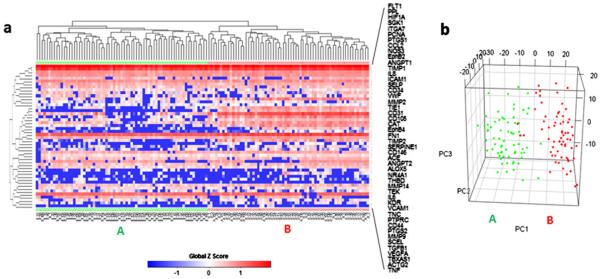
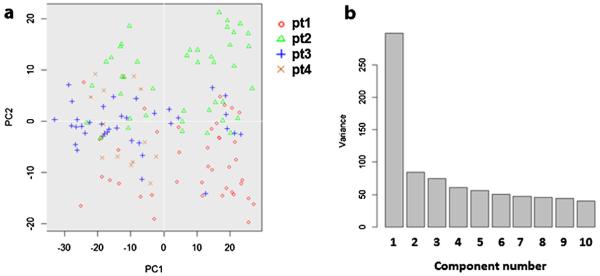
Unsupervised hierarchical clustering separated the 134 ECs into two distinctive clusters according to their expression pattern of the 48 selected functional genes (Fig. 4a). 69 ECs were in cluster A and 65 in cluster B. Principal component analysis (PCA) also showed two distinct populations and was consistent with hierarchical clustering. Only three cluster B cells identified by heat map-based hierarchical clustering were grouped with cluster A cells in PCA, and two cluster A cells were grouped with cluster B cells (Fig. 4b). The correlation of single cell gene expression and different biological donors was also analyzed by PCA. The 2D PCA (Fig. 5a) showed that the ECs from different donors did not overlap and showed no distinguishable cluster. The PCA scree plot (Fig. 5b) showed the contribution of first 10 PCs, which suggested the PC1 which identifies the two clusters gives much more contribution than other PCs.
Fig. 4.
Two EC clusters were identified by gene expression profiles. (a) Heat map and hierarchical clustering separated the 134 ECs into 2 major clusters, A (n = 69, green triangle) and B (n = 65, Red circle), based on their expression pattern of the 48 selected genes. (b) 3D PCA plots confirmed the segregation of these two clusters. Cluster A is annotated by green dots and B by Red dots. Gene symbols are used in the figures and corresponding gene names can be found in Table 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Two clusters identification is stronger identifiers than donor origin. (a) 2D PCA of the 134 ECs from 4 different donors based on their gene expression profile indicated no clear cluster separation among donors. (b) PCA scree plot of the first 10 PCs suggested the PC1 which identifies the two clusters gives much more contribution to the whole variance than other PCs.
3.5. Differential gene expression of the 2 EC subsets
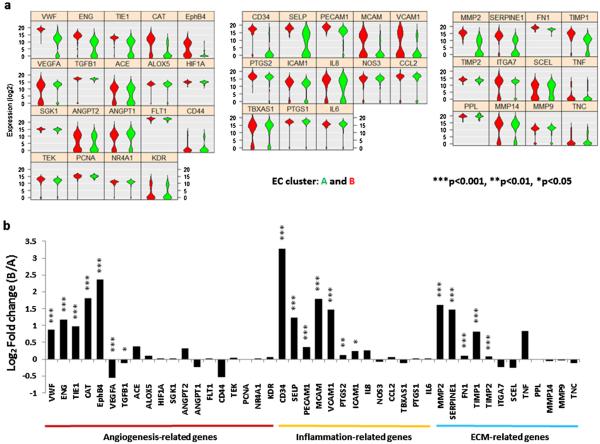
Further analysis showed that seven out of the 19 angiogenesis-related genes were differentially expressed (Fig. 6a Left) by cluster A and B. Among them, five were higher in cluster B [vWF (p < 7.1 × 10−20), CD105 (p < 9.0 × 10−18), TIE1 (p < 1.1 × 10−13), CAT (p < 7.8 × 10−12) and EPHB4 (p < 9.7 × 10−11)], two were higher in cluster A [VEGFA (p < 1.2 × 10−4) and TGFB1 (p < 0.023)]. Compared to cluster A, cluster B express higher levels of seven out of 13 inflammation-related genes (Fig. 6a Middle), [CD34 (p < 9.0 × 10−39), P-selectin (p < 9.0 × 10−22), CD31 (p < 1.2 × 10−9), CD146 (p < 3.1 × 10−7), VCAM-1 (p < 1.4 × 10−4), COX2 (p < 0.008) and ICAM-1 (p < 0.028)], as well as five out of the 12 ECM remodeling-related genes, were also expressed higher by cluster B than cluster A cells (Fig. 6a Right), [MMP2 (p < 5.0 × 10−27), PAI-1 (p < 1.7 × 10−15), FN1 (p < 1.1 × 10−15), TIMP1 (p < 2.9 × 10−11) and TIMP2 (p < 9.5 × 10−6)].
Fig. 6.
Differential gene expression of the two EC clusters. (a) Violin plots. Three functional gene groups are included, 19 angiogensis-related genes (Left), 13 inflammation-related genes (Middle) and 12 ECM remodeling genes (Right) of cluster A (green) and cluster B (Red). The gene name is indicated on top of each violin plot and the value on Y-axle represents the gene expression level in the binary logarithm (log2) value. (b) Bar graph shows the magnitude of differential gene expression by fold change of the binary logarithm (log2) value in cluster B relative to A (*p < 0.05; **p < 0.01; ***p < 0.001). Gene symbols are used in the figures and corresponding gene names can be found in Table 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In this study we demonstrated an innovative strategy for analyzing gene expression profiles of ECs collected from vessels on a single cell level. ECs are collected from endovascular guide wires through FACS. Single cell gene expression is analyzed using high throughput microfluidic quantitative RT-PCR. This method could be used to analyze the changes of EC gene expression at single cell level in vascular lesions. A total of 48 genes in four categories (cell-marker, angiogenesis, inflammation and ECM) were analyzed in this study. Two distinctive ECs clusters were identify from ECs collected from normal iliac arteries, suggesting ECs in normal vessel are heterogeneous.
Researchers who study ECs collected using endovascular techniques encounter a paradox that more EC marker genes need to be detected to identify and characterize the collected ECs, while the EC number harvested from such samples is often too small for such multiple marker detection. The traditional assays such as bulk RT-PCR, real-time RT-PCR or quantitative immunocytochemistry can only detect the mRNA transcription or protein expression of up to 3–4 EC functional genes, which are not enough for this purpose. The combination of single EC sorting and high throughput microfluidic quantitative RT-PCR allowed us to check EC identity through analyzing the expression of both the EC specific markers and the expression of functional genes in individual cells simultaneously. Moreover, because this microfluidic quantitative RT-PCR array technology has 96 gene slots, researchers have much more flexibility to expand the functional genes studied to help characterize ECs in varying disease conditions. This technique also presents a sound basis for comparing EC sampling and characterization data from different research centers.
Based on gene expression profiles, two distinctive clusters were identified in ECs collected from normal iliac arteries. A likely explanation is that ECs in normal conditions undergo turnover. The two EC clusters represent ECs at different functional stages, for example healthy and senescent. ECs are a stable cell type with an average turnover rate of about three years [23]. Senescent endothelium has been reported to have decreased expression of angiogenesis and proliferation genes, attenuated production of dilating factors and increased expression of contracting factors, increased oxidative stress, increased production of leukocyte adhesion-related cytokines or inflammation-related cytokines, and increased apoptosis [23]. Although not typical, cluster B ECs showed an expression pattern reminiscent of senescence-related genes like those of previous studies on EC aging and senescence. These gene expression changes include attenuated gene expression of VEGF and TGFb1 [24], enhanced expression of CD105 (also a EC proliferative marker) [25], COX2 [26,27], catalase [28], VCAM1 and ICAM1 [29], and TIMP2 [30]. Therefore cluster B cells could represent more mature or aged ECs. It is also of interest that cluster B ECs showed enhanced EPHB4 expression compared to cluster A. Although EphB4 is commonly considered a marker for ECs from veins, there is also data indicating EphB4 is expressed on both normal arteries and veins [31]. This gives more supportive evidence that caution should be used when this marker is used to identify venous ECs. We also ran a comparable volume (50ul) blood from each of the same patients on FACS and found no ECs. So, it is unlikely that these ECs came from veins by circulation and attached to the wire.
A noticeable phenomenon in this study is that only a quarter of the FACS sorted ECs expressed the four markers used for sorting and half expressed three. This indicated that FACS sorting cannot give a 100% pure population and a possible solution for this issue is the use of FACs machines with Index sorting capabilities. Several limitations of our study must also be considered. First, the patients selected for cell collection were not matched for their respective diseases necessitating angiography, demographic and co-morbid conditions. Given the small scale and exploratory nature of the study, controlling for such confounders proved difficult. Despite the absence of such analysis, when ECs were analyzed as it related to their patient origin, we noted no significant differences by either PCA or hierarchical clustering. Second, our choice of target genes for microfluidic quantitative RT-PCR was based on literature searches, introducing unavoidable bias. A more objective selection of target genes may be possible by analysis of previous microarray data on ECs. Such analyses are not possible on such small numbers of cells available, though emerging single cell mRNA sequencing may give an unbiased view of the global gene expression and ultimately identify new genes for study. Lastly, EC gene expression profile analysis was based on fewer than 200 ECs from four patients. Further studies of single EC gene expression and transcriptional regulation based on more ECs separated using endovascular cell collection techniques are necessary to investigate differential gene expression in ECs at different vasculature loci and in various vascular lesions.
Acknowledgements
The authors would like to thank Drs. Helen Kim, Tomoki Hashimoto and the other members in the Center for Cerebrovascular Research of UCSF for their technical support and to thank UCSF neurointerventional radiology service particularly its fellows for help in sample collection.
Sources of funding
This study was supported in part by funds to Z. Werb from the National Institutes of Health (R01CA180039), and by grants to H. Su from the National Institutes of Health (R01 NS027713, R01HL122774 and R21 NS083788), the Michael Ryan Zodda Foundation and UCSF Research Evaluation and Allocation Committee (REAC).
Footnotes
Disclosures
None.
References
- [1].Sammons V, Davidson A, Tu J, Stoodley MA. Endothelial cells in the context of brain arteriovenous malformations. J. Clin. Neurosci. 2011;18:165–170. doi: 10.1016/j.jocn.2010.04.045. [DOI] [PubMed] [Google Scholar]
- [2].Chalouhi N, Ali MS, Jabbour PM, Tjoumakaris SI, Gonzalez LF, Rosenwasser RH, et al. Biology of intracranial aneurysms: role of inflammation. J. Cereb. Blood Flow Metab. 2012 Sep;32:1659–1676. doi: 10.1038/jcbfm.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Landmesser U, Drexler H. The clinical significance of endothelial dysfunction. Curr. Opin. Cardiol. 2005 Nov;20:547–551. doi: 10.1097/01.hco.0000179821.11071.79. [DOI] [PubMed] [Google Scholar]
- [4].Feng L, Stern DM, Pile-Spellman J. Human endothelium: endovascular biopsy and molecular analysis. Radiology. 1999 Sep;212:655–664. doi: 10.1148/radiology.212.3.r99au28655. [DOI] [PubMed] [Google Scholar]
- [5].Feng L, Matsumoto C, Schwartz A, Schmidt AM, Stern DM, Pile-Spellman J. Chronic vascular inflammation in patients with type 2 diabetes: endothelial biopsy and RT-PCR analysis. Diabetes Care. 2005 Feb;28:379–384. doi: 10.2337/diacare.28.2.379. [DOI] [PubMed] [Google Scholar]
- [6].Onat D, Jelic S, Schmidt AM, Pile-Spellman J, Homma S, Padeletti M, et al. Vascular endothelial sampling and analysis of gene transcripts: a new quantitative approach to monitor vascular inflammation. J. Appl. Physiol. 2007;103:1873–1878. doi: 10.1152/japplphysiol.00367.2007. [DOI] [PubMed] [Google Scholar]
- [7].Yu SY, Song YM, Li AM, Yu XJ, Zhao G, Song MB, et al. Isolation and characterization of human coronary artery-derived endothelial cells in vivo from patients undergoing percutaneous coronary interventions. J. Vasc. Res. 2009;46:487–494. doi: 10.1159/000200964. [DOI] [PubMed] [Google Scholar]
- [8].Colombo PC, Ashton AW, Celaj S, Talreja A, Banchs JE, Dubois NB, et al. Biopsy coupled to quantitative immunofluorescence: a new method to study the human vascular endothelium. J. Appl. Physiol. 2002 Mar;92:1331–1338. doi: 10.1152/japplphysiol.00680.2001. [DOI] [PubMed] [Google Scholar]
- [9].Colombo PC, Banchs JE, Celaj S, Talreja A, Lachmann J, Malla S, et al. Endothelial cell activation in patients with decompensated heart failure. Circulation. 2005 Jan;111:58–62. doi: 10.1161/01.CIR.0000151611.89232.3B. [DOI] [PubMed] [Google Scholar]
- [10].Eskurza I, Kahn ZD, Seals DR. Xanthine oxidase does not contribute to impaired peripheral conduit artery endothelium-dependent dilatation with ageing. J. Physiol. 2006 Mar;571:661–668. doi: 10.1113/jphysiol.2005.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gates PE, Boucher ML, Silver AE, Monahan KD, Seals DR. Impaired flow-mediated dilation with age is not explained by L-arginine bioavailability or endothelial asymmetric dimethylarginine protein expression. J. Appl. Physiol. 2007 Jan;102:63–71. doi: 10.1152/japplphysiol.00660.2006. [DOI] [PubMed] [Google Scholar]
- [12].Silver AE, Beske SD, Christou DD, Donato AJ, Moreau KL, Eskurza I, et al. Overweight and obese humans demonstrate increased vascular endothelial NAD(p)H oxidase-p47(phox) expression and evidence of endothelial oxidative stress. Circulation. 2007 Feb;115:627–637. doi: 10.1161/CIRCULATIONAHA.106.657486. [DOI] [PubMed] [Google Scholar]
- [13].Donato AJ, Eskurza I, Silver AE, Levy AS, Pierce GL, Gates PE, et al. Direct evidence of endothelial oxidative stress with aging in humans: relation to impaired endothelium-dependent dilation and upregulation of nuclear factor-kappaB. Circ. Res. 2007 Jun;100:1659–1666. doi: 10.1161/01.RES.0000269183.13937.e8. [DOI] [PubMed] [Google Scholar]
- [14].Silver AE, Christou DD, Donato AJ, Beske SD, Moreau KL, Magerko KA, et al. Protein expression in vascular endothelial cells obtained from human peripheral arteries and veins. J. Vasc. Res. 2010;47:1–8. doi: 10.1159/000231715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell. 2013 Jul;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Harris LW, Wayland M, Lan M, Ryan M, Giger T, Lockstone H, et al. The cerebral microvasculature in schizophrenia: a laser capture microdissection study. PLoS One. 2008;3:e3964. doi: 10.1371/journal.pone.0003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sun Z, Su H, Long B, Sinclair E, Hetts SW, Higashida RT, et al. Endothelial cell high-enrichment from endovascular biopsy sample by laser capture microdissection and fluorescence activated cell sorting. J. Biotechnol. 2014 Oct;192PA:34–39. doi: 10.1016/j.jbiotec.2014.07.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].White AK, VanInsberghe M, Petriv OI, Hamidi M, Sikorski D, Marra MA, et al. High-throughput microfluidic single-cell RT-qPCR. Proc. Natl. Acad. Sci. U.S.A. 2011 Aug;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR, Wu JC. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat. Protoc. 2012 May;7:829–838. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].MacArthur BD, Sevilla A, Lenz M, Muller FJ, Schuldt BM, Schuppert AA, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat. Cell Biol. 2012 Nov;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lehoux S, Tedgui A. Cellular mechanics and gene expression in blood vessels. J. Biomech. 2003 May;36:631–643. doi: 10.1016/s0021-9290(02)00441-4. [DOI] [PubMed] [Google Scholar]
- [22].Anwar MA, Shalhoub J, Lim CS, Gohel MS, Davies AH. The effect of pressure-induced mechanical stretch on vascular wall differential gene expression. J. Vasc. Res. 2012;49:463–478. doi: 10.1159/000339151. [DOI] [PubMed] [Google Scholar]
- [23].Brandes RP, Fleming I, Busse R. Endothelial aging. Cardiovasc. Res. 2005 May;66:286–294. doi: 10.1016/j.cardiores.2004.12.027. [DOI] [PubMed] [Google Scholar]
- [24].Sadoun E, Reed MJ. Impaired angiogenesis in aging is associated with alterations in vessel density, matrix composition, inflammatory response, and growth factor expression. J. Histochem. Cytochem. 2003 Sep;51:1119–1130. doi: 10.1177/002215540305100902. [DOI] [PubMed] [Google Scholar]
- [25].Blanco FJ, Grande MT, Langa C, Oujo B, Velasco S, Rodriguez-Barbero A, et al. S-endoglin expression is induced in senescent endothelial cells and contributes to vascular pathology. Circ. Res. 2008 Dec;103:1383–1392. doi: 10.1161/CIRCRESAHA.108.176552. [DOI] [PubMed] [Google Scholar]
- [26].Heymes C, Habib A, Yang D, Mathieu E, Marotte F, Samuel J, et al. Cyclo-oxygenase-1 and -2 contribution to endothelial dysfunction in ageing. Br. J. Pharmacol. 2000 Oct;131:804–810. doi: 10.1038/sj.bjp.0703632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mukai Y, Shimokawa H, Higashi M, Morikawa K, Matoba T, Hiroki J, et al. Inhibition of renin-angiotensin system ameliorates endothelial dysfunction associated with aging in rats. Arterioscler. Thromb. Vasc. Biol. 2002 Sep;22:1445–1450. doi: 10.1161/01.atv.0000029121.63691.ce. [DOI] [PubMed] [Google Scholar]
- [28].Han J, Shuvaev VV, Muzykantov VR. Catalase and superoxide dismutase conjugated with platelet-endothelial cell adhesion molecule antibody distinctly alleviate abnormal endothelial permeability caused by exogenous reactive oxygen species and vascular endothelial growth factor. J. Pharmacol. Exp. Ther. 2011 Jul;338:82–91. doi: 10.1124/jpet.111.180620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Morisaki N, Saito I, Tamura K, Tashiro J, Masuda M, Kanzaki T, et al. New indices of ischemic heart disease and aging: studies on the serum levels of soluble intercellular adhesion molecule-1 (ICAM-1) and soluble vascular cell adhesion molecule-1 (VCAM-1) in patients with hypercholesterolemia and ischemic heart disease. Atherosclerosis. 1997 May;131:43–48. doi: 10.1016/s0021-9150(97)06083-8. [DOI] [PubMed] [Google Scholar]
- [30].Koike T, Vernon RB, Gooden MD, Sadoun E, Reed MJ. Inhibited angiogenesis in aging: a role for TIMP-2. J. Gerontol. A Biol. Sci. Med. Sci. 2003 Sep;58:B798–B805. doi: 10.1093/gerona/58.9.b798. [DOI] [PubMed] [Google Scholar]
- [31].Diehl S, Bruno R, Wilkinson GA, Loose DA, Wilting J, Schweigerer L, et al. Altered expression patterns of EphrinB2 and EphB2 in human umbilical vessels and congenital venous malformations. Pediatr. Res. 2005 Apr;57:537–544. doi: 10.1203/01.PDR.0000155761.70710.C4. [DOI] [PubMed] [Google Scholar]
- [32].Krieglstein CF, Granger DN. Adhesion molecules and their role in vascular disease. Am. J. Hypertens. 2001 Jun;14:44S–54S. doi: 10.1016/s0895-7061(01)02069-6. [DOI] [PubMed] [Google Scholar]
- [33].Nielsen JS, McNagny KM. Novel functions of the CD34 family. J. Cell Sci. 2008 Nov;121:3683–3692. doi: 10.1242/jcs.037507. [DOI] [PubMed] [Google Scholar]
- [34].Park S, Dimaio TA, Liu W, Wang S, Sorenson CM, Sheibani N. Endoglin regulates the activation and quiescence of endothelium by participating in canonical and non-canonical TGF-beta signaling pathways. J. Cell Sci. 2013 Mar;126:1392–1405. doi: 10.1242/jcs.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Solovey AN, Gui L, Chang L, Enenstein J, Browne PV, Hebbel RP. Identification and functional assessment of endothelial P1H12. J. Lab. Clin. Med. 2001 Nov;138:322–331. doi: 10.1067/mlc.2001.118519. [DOI] [PubMed] [Google Scholar]
- [36].Kratzer A, Chu HW, Salys J, Moumen Z, Leberl M, Bowler R, et al. Endothelial cell adhesion molecule CD146: implications for its role in the pathogenesis of COPD. J. Pathol. 2013 Aug;230:388–398. doi: 10.1002/path.4197. [DOI] [PubMed] [Google Scholar]
- [37].Bardin N, George F, Mutin M, Brisson C, Horschowski N, Frances V, et al. S-Endo 1, a pan-endothelial monoclonal antibody recognizing a novel human endothelial antigen. Tissue Antigens. 1996 Nov;48:531–539. doi: 10.1111/j.1399-0039.1996.tb02666.x. [DOI] [PubMed] [Google Scholar]
- [38].Bardin N, Reumaux D, Geboes K, Colombel JF, Blot-Chabaud M, Sampol J, et al. Increased expression of CD146, a new marker of the endothelial junction in active inflammatory bowel disease. Inflamm. Bowel Dis. 2006 Jan;12:16–21. doi: 10.1097/01.mib.0000194181.46930.88. [DOI] [PubMed] [Google Scholar]
- [39].Zheng W, Christensen LP, Tomanek RJ. Stretch induces upregulation of key tyrosine kinase receptors in microvascular endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 2004 Dec;287:H2739–H2745. doi: 10.1152/ajpheart.00410.2004. [DOI] [PubMed] [Google Scholar]
- [40].Satchell SC, Anderson KL, Mathieson PW. Angiopoietin 1 and vascular endothelial growth factor modulate human glomerular endothelial cell barrier properties. J. Am. Soc. Nephrol. 2004 Mar;15:566–574. doi: 10.1097/01.asn.0000115397.22519.03. [DOI] [PubMed] [Google Scholar]
- [41].Orecchia A, Lacal PM, Schietroma C, Morea V, Zambruno G, Failla CM. Vascular endothelial growth factor receptor-1 is deposited in the extracellular matrix by endothelial cells and is a ligand for the alpha 5 beta 1 integrin. J. Cell Sci. 2003 Sep;116:3479–3489. doi: 10.1242/jcs.00673. [DOI] [PubMed] [Google Scholar]
- [42].Porat RM, Grunewald M, Globerman A, Itin A, Barshtein G, Alhonen L, et al. Specific induction of tie1 promoter by disturbed flow in atherosclerosis-prone vascular niches and flow-obstructing pathologies. Circ. Res. 2004 Feb;94:394–401. doi: 10.1161/01.RES.0000111803.92923.D6. [DOI] [PubMed] [Google Scholar]
- [43].Martin FA, Murphy RP, Cummins PM. Thrombomodulin and the vascular endothelium: insights into functional, regulatory, and therapeutic aspects. Am. J. Physiol. Heart Circ. Physiol. 2013 Jun;304:H1585–H1597. doi: 10.1152/ajpheart.00096.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Randi AM, Laffan MA, Starke RD. Von Willebrand factor, angiodysplasia and angiogenesis, Mediterr. J. Hematol. Infect. Dis. 2013;5:e2013060. doi: 10.4084/MJHID.2013.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yang C, Guo Y, Jadlowiec CC, Li X, Lv W, Model LS, et al. Vascular endothelial growth factor-A inhibits EphB4 and stimulates delta-like ligand 4 expression in adult endothelial cells. J. Surg. Res. 2013 Jul;183:478–486. doi: 10.1016/j.jss.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Akarasereenont PC, Techatraisak K, Thaworn A, Chotewuttakorn S. The expression of COX-2 in VEGF-treated endothelial cells is mediated through protein tyrosine kinase. Mediators Inflamm. 2002 Feb;11:17–22. doi: 10.1080/09629350210311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Sho E, Komatsu M, Sho M, Nanjo H, Singh TM, Xu C, et al. High flow drives vascular endothelial cell proliferation during flow-induced arterial remodeling associated with the expression of vascular endothelial growth factor. Exp. Mol. Pathol. 2003 Aug;75:1–11. doi: 10.1016/s0014-4800(03)00032-7. [DOI] [PubMed] [Google Scholar]
- [48].Hurley NE, Schildmeyer LA, Bosworth KA, Sakurai Y, Eskin SG, Hurley LH, et al. Modulating the functional contributions of c-Myc to the human endothelial cell cyclic strain response. J. Vasc. Res. 2010;47:80–90. doi: 10.1159/000235928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Frye SR, Yee A, Eskin SG, Guerra R, Cong X, McIntire LV. cDNA microarray analysis of endothelial cells subjected to cyclic mechanical strain: importance of motion control. Physiol. Genomics. 2005 Mar;21:124–130. doi: 10.1152/physiolgenomics.00029.2003. [DOI] [PubMed] [Google Scholar]
- [50].Ferrelli F, Pastore D, Capuani B, Lombardo MF, Blot-Chabaud M, Coppola A, et al. Serum glucocorticoid inducible kinase (SGK)-1 protects endothelial cells against oxidative stress and apoptosis induced by hyperglycaemia. Acta Diabetol. 2014 Jun;:55–64. doi: 10.1007/s00592-014-0600-4. [DOI] [PubMed] [Google Scholar]
- [51].Kim I, Kim HG, So JN, Kim JH, Kwak HJ, Koh GY. Angiopoietin-1 regulates endothelial cell survival through the phosphatidylinositol 30 -Kinase/Akt signal transduction pathway. Circ. Res. 2000 Jan;86:24–29. doi: 10.1161/01.res.86.1.24. [DOI] [PubMed] [Google Scholar]
- [52].Daly C, Pasnikowski E, Burova E, Wong V, Aldrich TH, Griffiths J, et al. Angiopoietin-2 functions as an autocrine protective factor in stressed endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 2006 Oct;103:15491–15496. doi: 10.1073/pnas.0607538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Milkiewicz M, Haas TL. Effect of mechanical stretch on HIF-1li and MMP-2 expression in capillaries isolated from overloaded skeletal muscles: laser capture microdissection study. Am. J. Physiol. Heart Circ. Physiol. 2005 Sep;289:H1315–20. doi: 10.1152/ajpheart.00284.2005. [DOI] [PubMed] [Google Scholar]
- [54].Milkiewicz M, Doyle JL, Fudalewski T, Ispanovic E, Aghasi M, Haas TL. HIF-1alpha and HIF-2alpha play a central role in stretch-induced but not shear-stress-induced angiogenesis in rat skeletal muscle. J. Physiol. 2007 Sep;583:753–766. doi: 10.1113/jphysiol.2007.136325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Arkenbout EK, van Bragt M, Eldering E, van Bree C, Grimbergen JM, Quax PH, et al. TR3 orphan receptor is expressed in vascular endothelial cells and mediates cell cycle arrest. Arterioscler. Thromb. Vasc. Biol. 2003 Sep;23:1535–1540. doi: 10.1161/01.ATV.0000084639.16462.7A. [DOI] [PubMed] [Google Scholar]
- [56].Walker JL, Loscalzo J, Zhang YY. 5-Lipoxygenase and human pulmonary artery endothelial cell proliferation. Am. J. Physiol. Heart Circ. Physiol. 2002 Feb;282:H585–H593. doi: 10.1152/ajpheart.00003.2001. [DOI] [PubMed] [Google Scholar]
- [57].Trochon V, Mabilat C, Bertrand P, Legrand Y, Smadja-Joffe F, Soria C, et al. Evidence of involvement of CD44 in endothelial cell proliferation, migration and angiogenesis in vitro. Int. J. Cancer. 1996 May;66:664–668. doi: 10.1002/(SICI)1097-0215(19960529)66:5<664::AID-IJC14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- [58].Tsuneki M, Madri JA. CD44 regulation of endothelial cell proliferation and apoptosis via modulation of CD31 and VE-cadherin expression. J. Biol. Chem. 2014 Feb;289:5357–5370. doi: 10.1074/jbc.M113.529313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Silva EA, Eseonu C, Mooney DJ. Endothelial cells expressing low levels of CD143 (ACE) exhibit enhanced sprouting and potency in relieving tissue ischemia. Angiogenesis. 2014 Jul;17:617–630. doi: 10.1007/s10456-014-9414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kobayashi S, Nagino M, Komatsu S, Naruse K, Nimura Y, Nakanishi M, et al. Stretch-induced IL-6 secretion from endothelial cells requires NF-kappaB activation. Biochem. Biophys. Res. Commun. 2003 Aug;308:306–312. doi: 10.1016/s0006-291x(03)01362-7. [DOI] [PubMed] [Google Scholar]
- [61].Okada M, Matsumori A, Ono K, Furukawa Y, Shioi T, Iwasaki A, et al. Cyclic stretch upregulates production of interleukin-8 and monocyte chemotactic and activating factor/monocyte chemoattractant protein-1 in human endothelial cells. Arterioscler. Thromb. Vasc. Biol. 1998 Jun;18:894–901. doi: 10.1161/01.atv.18.6.894. [DOI] [PubMed] [Google Scholar]
- [62].Zhao H, Hiroi T, Hansen BS, Rade JJ. Cyclic stretch induces cyclooxygenase-2 gene expression in vascular endothelial cells via activation of nuclear factor kappa-beta. Biochem. Biophys. Res. Commun. 2009 Nov;389:599–601. doi: 10.1016/j.bbrc.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Matsushita H, Chang E, Glassford AJ, Cooke JP, Chiu CP, Tsao PS. eNOS activity is reduced in senescent human endothelial cells: preservation by hTERT immortalization. Circ. Res. 2001 Oct;89:793–798. doi: 10.1161/hh2101.098443. [DOI] [PubMed] [Google Scholar]
- [64].Hoffmann J, Haendeler J, Aicher A, Rossig L, Vasa M, Zeiher AM, et al. Aging enhances the sensitivity of endothelial cells toward apoptotic stimuli: important role of nitric oxide. Circ. Res. 2001 Oct;89:709–715. doi: 10.1161/hh2001.097796. [DOI] [PubMed] [Google Scholar]
- [65].Wung BS, Cheng JJ, Hsieh HJ, Shyy YJ, Wang DL. Cyclic strain-induced monocyte chemotactic protein-1 gene expression in endothelial cells involves reactive oxygen species activation of activator protein 1. Circ. Res. 1997 Jul;81:1–7. doi: 10.1161/01.res.81.1.1. [DOI] [PubMed] [Google Scholar]
- [66].Wung BS, Cheng JJ, Shyue SK, Wang DL. NO modulates monocyte chemotactic protein-1 expression in endothelial cells under cyclic strain. Arterioscler. Thromb. Vasc. Biol. 2001 Dec;21:1941–1947. doi: 10.1161/hq1201.099428. [DOI] [PubMed] [Google Scholar]
- [67].Tailor A, Wood KC, Wallace JL, Specian RD, Granger DN. Roles of platelet and endothelial cell COX-1 in hypercholesterolemia-induced microvascular dysfunction. Am. J. Physiol. Heart Circ. Physiol. 2007 Dec;293:H3636–42. doi: 10.1152/ajpheart.01105.2006. [DOI] [PubMed] [Google Scholar]
- [68].Wang BW, Chang H, Lin S, Kuan P, Shyu KG. Induction of matrix metalloproteinases-14 and -2 by cyclical mechanical stretch is mediated by tumor necrosis factor-alpha in cultured human umbilical vein endothelial cells. Cardiovasc. Res. 2003 Aug;59:460–469. doi: 10.1016/s0008-6363(03)00428-0. [DOI] [PubMed] [Google Scholar]
- [69].Cavallaro U, Castelli V, Del Monte U, Soria MR. Phenotypic alterations in senescent large-vessel and microvascular endothelial cells. Mol. Cell Biol. Res. Commun. 2000 Aug;4:117–121. doi: 10.1006/mcbr.2000.0263. [DOI] [PubMed] [Google Scholar]
- [70].Foreman KE, Tang J. Molecular mechanisms of replicative senescence in endothelial cells. Exp. Gerontol. 2003 Nov-Dec;38:1251–1257. doi: 10.1016/j.exger.2003.09.005. [DOI] [PubMed] [Google Scholar]
- [71].Gao B, Saba TM, Tsan MF. Role of alpha(v) beta(3)-integrin in TNF-alpha-induced endothelial cell migration. Am. J. Physiol. Cell Physiol. 2002 Oct;283:C1196–205. doi: 10.1152/ajpcell.00064.2002. [DOI] [PubMed] [Google Scholar]
- [72].Forough R, Koyama N, Hasenstab D, Lea H, Clowes M, Nikkari ST, et al. Overexpression of tissue inhibitor of matrix metalloproteinase-1 inhibits vascular smooth muscle cell functions in vitro and in vivo. Circ. Res. 1996 Oct;79:812–820. doi: 10.1161/01.res.79.4.812. [DOI] [PubMed] [Google Scholar]
- [73].Saunders WB, Bohnsack BL, Faske JB, Anthis NJ, Bayless KJ, Hirschi KK, et al. Coregulation of vascular tube stabilization by endothelial cell TIMP-2 and pericyte TIMP-3. J. Cell. Biol. 2006 Oct;175:179–191. doi: 10.1083/jcb.200603176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Stetler-Stevenson WG, Seo DW. TIMP-2: an endogenous inhibitor of angiogenesis. Trends Mol. Med. 2005 Mar;11:97–103. doi: 10.1016/j.molmed.2005.01.007. [DOI] [PubMed] [Google Scholar]
- [75].Kumazaki T. Modulation of gene expression during aging of human vascular endothelial cells. Hiroshima J. Med. Sci. 1993 Jun;42:97–100. [PubMed] [Google Scholar]
- [76].Alves TR, da Fonseca AC, Nunes SS, da Silva AO, Dubois LG, Faria J, et al. Tenascin-C in the extracellular matrix promotes the selection of highly proliferative and tubulogenesis-defective endothelial cells. Exp. Cell Res. 2011 Sep;317:2073–2085. doi: 10.1016/j.yexcr.2011.06.006. [DOI] [PubMed] [Google Scholar]
- [77].Pyle AL, Li B, Maupin AB, Guzman RJ, Crimmins DL, Olson S, et al. Biomechanical stress induces novel arterial intima-enriched genes: implications for vascular adaptation to stress. Cardiovasc. Pathol. 2010 Mar-Apr;19:e13–e20. doi: 10.1016/j.carpath.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]