Abstract
Cytoplasmic dyneins 1 and 2 are related members of the AAA+ superfamily (ATPases associated with diverse cellular activities) that function as the predominant minus-end-directed microtubule motors in eukaryotic cells. Dynein 1 controls mitotic spindle assembly, organelle movement, axonal transport, and other cytosolic, microtubule-guided processes, whereas dynein 2 mediates retrograde trafficking within motile and primary cilia. Small-molecule inhibitors are important tools for investigating motor protein-dependent mechanisms, and ciliobrevins were recently discovered as the first dynein-specific chemical antagonists. Here we demonstrate that ciliobrevins directly target the heavy chains of both dynein isoforms and explore the structure-activity landscape of these inhibitors in vitro and in cells. In addition to identifying chemical motifs that are essential for dynein blockade, we have discovered analogs with increased potency and dynein 2 selectivity. These antagonists effectively disrupt Hedgehog signaling, intraflagellar transport, and ciliogenesis, making them useful probes of these and other cytoplasmic dynein 2-dependent cellular processes.
Molecular motors are essential drivers of cellular function, moving cargos along the cytoskeleton and dynamically regulating these filamentous structures. Dyneins are the largest and among the most complex of these mechanoenzymes, having evolved independently from kinesins, myosins, and other nucleotide-binding polypeptides with Ras-like folds.1, 2 Members of the AAA+ superfamily (ATPases associated with diverse cellular activities), these multi-subunit enzymes convert ATP hydrolysis into molecular movement toward the minus ends of microtubules. Axonemal dynein isoforms actuate flagellar and ciliary motility through microtubule crosslinking and sliding,3 and cytoplasmic dyneins 1 and 2 are the primary mediators of minus-end-directed intracellular transport.4–6 For example, dynein 1 regulates spindle assembly and chromatid-microtubule interactions during cell division,7, 8 Golgi formation and positioning,9, 10 vesicular and organelle trafficking,11, 12 retrograde axonal transport,13 and the nuclear translocation of viral capsids.14 Dynein 2 function is more specialized in comparison, driving retrograde intraflagellar transport within motile and primary cilia.5, 6
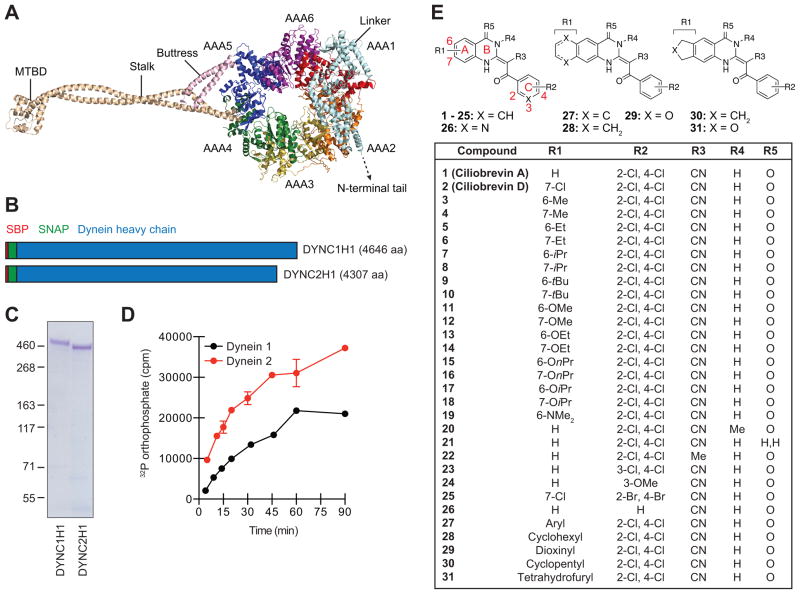
Mutational analyses, electron microscopy, and x-ray crystallography have significantly advanced our mechanistic understanding of dynein function.1 As primarily ascertained through studies of cytoplasmic dynein 1, these microtubule motors are composed of isoform-specific heavy chains (~500 kDa each) that are structurally related to other AAA+ superfamily mechanoenzymes, as well as distinct sets of intermediate (~75 kDa), light intermediate (~50 kDa), and light (~10 kDa) chains. Like other AAA+ proteins, the heavy chains of dyneins 1 and 2 contain six AAA domains (designated as AAA1 to AAA6) to form a ring-shaped structure with ATP hydrolase activity (Figure 1A).15, 16 This C-terminal motor is functionalized with two coiled-coil extensions: a stalk on AAA4 that is terminated with the microtubule-binding domain (MTBD) and a buttress emerging from AAA5 that interacts with the stalk. The motor is also connected to the N-terminal adaptor- and cargo-binding tail through a hinged linker fused to the AAA1 module. Nucleotide-binding sites in AAA+ family members are formed at the interface of adjacent AAA domains, composed of a GXXXGK sequence (Walker A motif; also known as the P-loop), and an arginine that coordinates the phosphate groups (Sensor II), catalytic glutamic acid (Walker B motif), asparagine (Sensor I), and arginine (Arginine Finger) side chains, and non-contiguous residues that interact with the adenosine moiety.17 The highly conserved AAA1 nucleotide-interacting domain (AAA1-AAA2 interface) acts as the primary site of ATP hydrolysis,18 driving conformational changes that alter linker geometry and microtubule binding.15, 19 The more divergent AAA2, AAA3, and AAA4 sites are believed to modulate dynein function in a nucleotide binding- or hydrolysis-dependent manner, varying with the dynein isoform and organism.15, 18, 19
Figure 1. Cytoplasmic dynein heavy chains and ciliobrevin analogs used for structure-activity profiling.
(A) Cartoon representation of the dynein 2 heavy chain based on crystallographic data for the pre-power stroke conformation (PDB ID: 4RH7). Individual AAA domains within the C-terminal motor are shown, as well as the N-terminal linker, stalk, buttress, and MTBD. (B) Schematic representation of N-terminally SBP- and SNAP-tagged dynein heavy chains. Polypeptide domain lengths are shown to scale. (C) Purified SBP-SNAP-DYNC1H1 and SBP-SNAP-DYNC2H1 proteins resolved by SDS-PAGE and stained with Coomassie Blue. (D) Kinetic analyses of dynein heavy chain activities, as determined by the hydrolysis of γ-32P ATP (17 nM) at 37 °C. Data are the average of two replicates ± s.e.m., and the enzyme reaction curves were used to establish linear assay conditions for the evaluation of ciliobrevin analogs. (E) Structures for the initial set of diverse ciliobrevin analogs profiled in this study.
With speeds of approximately 1 μm/s,20, 21 dynein motors are challenging to study using genetic techniques such as RNA interference and the expression of polypeptide inhibitors, since the perturbation timescales far exceed those of dynein action. Small-molecule modulators with fast kinetics are therefore important tools for interrogating dynein function. However, in contrast to kinesins and myosins, only one class of dynein-specific chemical antagonists has been reported.22 We discovered these benzoyl quinazolinone derivatives in a high-throughput chemical screen for Hedgehog (Hh) pathway antagonists, corroborating the critical role of primary cilia in mammalian Hh signaling.23, 24 Limited structure-activity-relationship (SAR) analyses yielded four analogs that we named ciliobrevins A–D due to their effects on cilium length, and the compounds also induced accumulation of the Hh pathway transcription factor GLI2 in the ciliary distal tip. These functionalized benzoyl quinazolinones abrogate cytoplasmic dynein 1- and 2-dependent cellular processes, allowing real-time assessments of dynein activity in a rapid and reversible manner. Ciliobrevins have been shown to disrupt a variety of dynein-dependent cellular processes, including mitotic spindle pole focusing,22 centrosome repositioning,25, 26 ciliary trafficking,27 organelle transport in axons,28 and viral capsid uncoating.29 While the precise mechanism by which these compounds act remains unknown, biochemical studies with cytoplasmic dynein 1 indicate that they target the heavy chain motor domain and block ATP hydrolysis in a nucleotide-competitive manner.22
The cellular activities of ciliobrevins A–D suggest that they target cytoplasmic dyneins 1 and 2 in a non-preferential manner.22 Given the multiple functions of dynein 1 and the subcellular compartmentalization of dynein 2 activity, isoform-selective antagonists would better discern their individual contributions to specific cellular events. Moreover, they could lead to the development of dynein-targeting therapies, such as anti-cancer drugs that abrogate dynein 2-dependent Hh pathway activation, while avoiding the more pleiotropic effects of dynein 1 inhibition. Toward this goal, we have profiled the activities of structurally diverse ciliobrevin analogs against both dynein isoforms in vitro and in cells. Our studies provide the first biochemical evidence that ciliobrevins directly inhibit cytoplasmic dynein 2 and identify regions in the ciliobrevin pharmacophore that are critical for dynein blockade. Our SAR analyses also uncover a site that influences isoform selectivity, leading to next-generation ciliobrevins that preferentially target cytoplasmic dynein 2 in specific contexts.
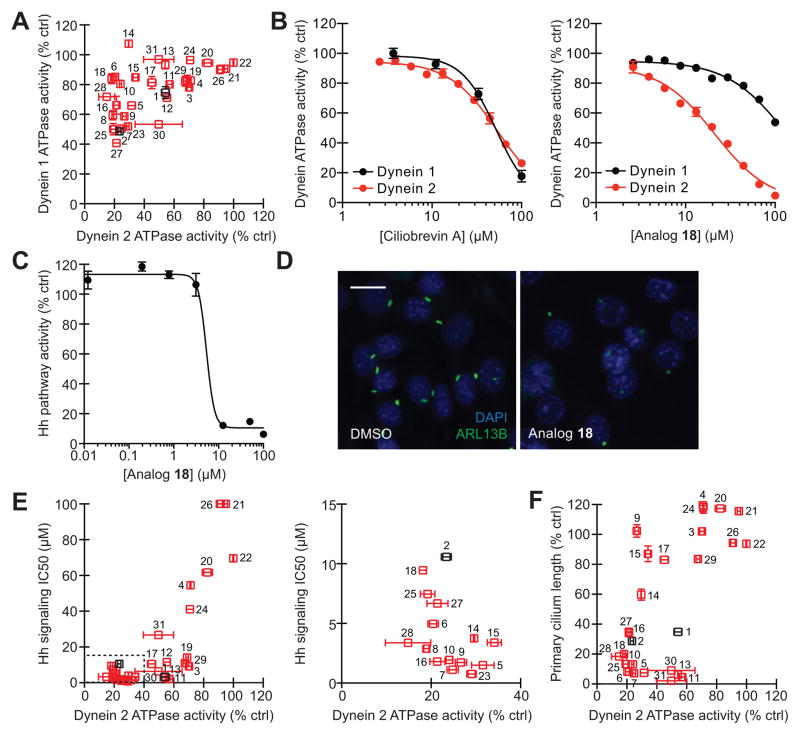
To directly assess the potencies and isoform selectivities of ciliobrevins, we first established assays of human dynein 1 and 2 ATPase activity, utilizing recombinant, full-length heavy chains (DYNC1H1 and DYNC2H1) that are N-terminally fused to a streptavidin-binding peptide (SBP) and SNAP tag.30 The tagged heavy chains were individually expressed in mammalian cells and purified on a streptavidin-Sepharose matrix to yield functional proteins (Figure 1B–C), as gauged by their ability to hydrolyze γ-32P ATP (Figure 1D). Ciliobrevins A and D (Figure 1E, analogs 1 and 2) were then tested for their abilities to inhibit dynein 1- or 2-mediated γ-32P ATP hydrolysis, employing a single, 50-μM compound dose and experimental conditions that maintain linear enzyme kinetics. Both compounds were able to directly inhibit DYNC1H1 and DYNC2H1 function (Figure 2A), corroborating their previously described cellular phenotypes.22, 31 However, neither antagonist demonstrated significant isoform-selectivity, illustrating the need for analogs that can preferentially target individual dynein functions.
Figure 2. Identification of cytoplasmic dynein 2-selective ciliobrevins.
(A) Inhibitory activities of ciliobrevins A and D (1 and 2; black) and structural analogs (3–31; red) in DYNC1H1 and DYNC2H1 ATPase assays. All compounds were tested at a 50-μM dose in the presence of approximately 25 nM γ-32P ATP, and data are the average of duplicate samples ± s.e.m. (B) Dose-dependent inhibition of DYNC1H1 (black) and DYNC2H1 (red) ATPase activities by ciliobrevin A and analog 18. Data are the average of triplicate samples ± s.e.m. Calculated IC50 values: 1, DYNC1H1 = 52 μM and DYNC2H1 = 55 μM; 18, DYNC1H1 = 130 μM and DYNC2H1 = 21 μM. (C) Dose-dependent inhibition of Hh signaling by analog 18. NIH-3T3 fibroblasts stably transfected with a Gli-dependent firefly luciferase reporter were treated ShhN-conditioned medium and the compound for 30 hours, and data are the average of triplicate samples ± s.e.m. (D) Inhibition of ciliogenesis in an NIH-3T3 cell-derived line by analog 18 (30 μM; 24 hours), as visualized by immunostaining of the ciliary GTPase ARL13B. Scale bar: 10 μm. (E) Correlation of analog activities in dynein heavy chain ATPase and Hh signaling assays. IC50 values for Hh pathway inhibition were calculated from dose-response measurements as shown in (C). Tightly clustered analogs (dashed box) are re-plotted in the adjacent graph. (F) Correlation of analog activities in dynein heavy chain ATPase and ciliogenesis assays. Each compound was tested at a 30-μM dose, and primary cilium lengths are the average number of ARL13B-positive pixels/cell in at least 6 separate fields of view ± s.e.m., corresponding to approximately 800 analyzed cells per condition.
To explore the structure-activity landscape of ciliobrevins, we next synthesized a collection of diverse analogs. The ciliobrevin skeleton is composed of an acrylonitrile backbone and three ring systems: rings A and B comprise the quinazolinone unit, and ring C represents the benzoyl group (Figure 1E). To access modifications to each region, we developed two general synthetic routes. One procedure generated the pharmacophore through sequential condensation of a 2-aminobenzoic acid derivative, 2-cyanothioacetamide, and an acid chloride; the other utilized a 2-benzoyl-3,3-bis(methylthio)acrylonitrile intermediate and bifunctional amines (Supporting Information). Using these methods, we prepared 29 additional derivatives for activity profiling (Figure 1E; analogs 3–31).
Each ciliobrevin derivative was then tested for its ability to block dynein 1- or 2-mediated γ-32P ATP hydrolysis as described above (Figure 2A). Structural elements that were essential for dynein inhibition included the 2,4-substituted C ring, the nitrile group, the quinazolinone carbonyl, and the amide NH. These findings complement our earlier limited SAR analyses of the ciliobrevins that demonstrated the importance of C ring substituents and the acrylonitrile carbonyl.22 Modifications of the A ring significantly enhanced or diminished compound potency, and derivatives functionalized at the C7 position (e.g., analogs 6, 10, 14, 16, 18, and 28) appeared to be selective for dynein 2. We corroborated these observations by obtaining dose-response curves for ciliobrevin A (1) and analog 18 in the in vitro assays of DYNC1H1 and DYNC2H2 function (Figure 2B). While ciliobrevin A inhibited dynein 1 and 2 ATPase activities with comparable potencies, compound 18 exhibited a 6-fold selectivity for dynein 2. We further established that ciliobrevin 18 can disrupt dynein 2 activity in mammalian cells, using Hh signaling and ciliogenesis in murine fibroblasts as functional readouts (Figure 2C–D; human and mouse DYNC2H1 motor domains share 94% identity and 97% similarity). The steep dose-response curve of ciliobrevin 18 in the Hh signaling assay is similar to that previously observed for ciliobrevin A,31 possibly reflecting the collective action of multiple dynein molecules on individual cargos.32, 33
To enhance our understanding of the ciliobrevin activity landscape, we screened the other analogs in these cell-based assays of cytoplasmic dynein 2 function. We observed activity profiles consistent with ciliobrevin disruption of Hh signaling and ciliogenesis through dynein 2 blockade (Figure 2E–F). However, there were a few informative exceptions to this trend. A number of compounds with C6 A-ring substituents abrogated Hh signaling without a commensurate effect on dynein 2 ATPase activity (Figure 2E; e.g., analogs 3, 11, 19, 29, and 30). A distinct but partially overlapping set of C6-substituted derivatives exhibited non-correlated effects on primary cilia length and dynein 2 ATPase activity (Figure 2F; e.g., analogs 9, 11, 13, 15, 30, and 31). Thus, C7 modifications can improve the dynein 2 selectivity of ciliobrevin analogs, but functionalization of the C6 site appears to diminish compound efficacy against dynein while retaining Hh pathway inhibition through mechanisms that remain unclear and may involve additional targets.
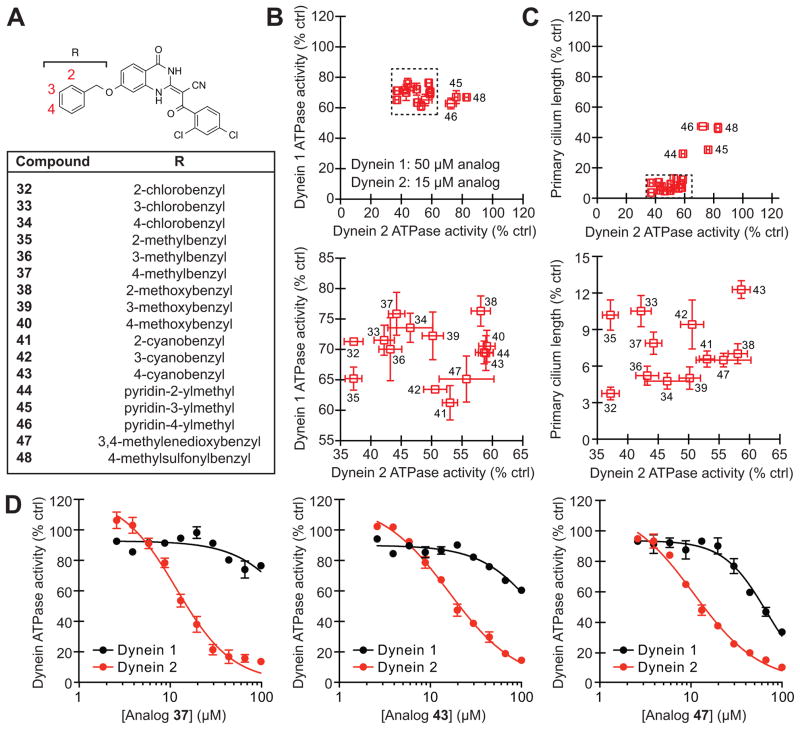
Based on this structure-activity landscape, we turned our attention toward C7 substituents with increased steric bulk and differences in polarity. To facilitate this process, we focused on 17 benzyl ether derivatives that could obtained in a single step by reacting a common C7-hydroxyl intermediate (obtained through demethylation of analog 12) with a variety of commercially available benzyl bromides (Figure 3A and Supporting Information). We tested this focused set of ciliobrevin analogs at 50 μM and 15 μM concentrations in our DYNC1H1 and DYNC2H1 ATPase assays, respectively (a lower dose was utilized for DYNC2H1 due to the increased potencies of C7-benzyl ether analogs against this isoform). The inhibitors clustered into two groups (Figure 3B): a dynein 2-selective subset (analogs 32–44, and 47) and more polar derivatives with diminished isoform specificity (pyridylmethyl and methylsulfonylbenzyl analogs 45, 46, and 48). As with the preceding set of C7-functionalized ciliobrevins, the benzyl ether analogs blocked DYNC2H1-dependent ATP hydrolysis and ciliogenesis in a correlative manner (Figure 3C), and dose-response curves for three representative benzyl derivatives (analogs 37, 43, and 47) confirmed that the inhibitors have improved potencies and selectivities for dynein 2 (Figure 3D).
Figure 3. Benzyl ether-functionalized ciliobrevins with improved cytoplasmic dynein 2 selectivity.
(A) Structures for the focused library of ciliobrevin analogs. (B) Inhibitory activities of the C7-benzyl ether analogs in DYNC1H1 and DYNC2H1 ATPase assays. The compounds were tested in the presence of approximately 25 nM γ-32P ATP, and a lower inhibitor concentration was used in the DYNC2H1 assays since C7-functionalized analogs are generally more potent against this isoform. Data are the average of two experiments ± s.e.m. Tightly clustered analogs (dashed box) are re-plotted in the adjacent graph. (C) Correlation of analog activities in dynein heavy chain ATPase and ciliogenesis assays as described in Figure 2F. Tightly clustered analogs (dashed box) are re-plotted in the adjacent graph. (D) Dose-dependent inhibition of DYNC1H1 (black) and DYNC2H1 (red) ATPase activities by analogs 37, 43, and 47. Data are the average of triplicate samples ± s.e.m. Calculated IC50 values: 37, DYNC1H1 = 280 μM and DYNC2H1 = 11 μM; 43, DYNC1H1 = 158 μM and DYNC2H1 = 16 μM; 47, DYNC1H1 = 130 μM and DYNC2H1 = 11 μM.
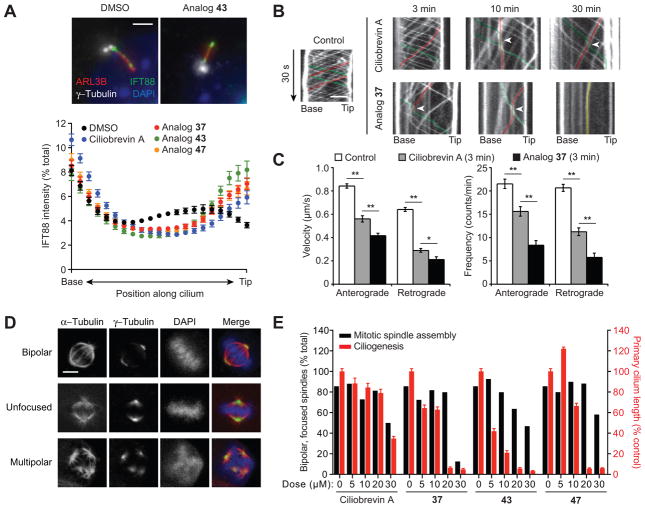
Given the 24-hour regimen of the ciliogenesis assay, we also examined the effects of selected benzyl ether analogs on ciliary trafficking, which can be assessed on much shorter time scales. Cytoplasmic dynein 2 drives the retrograde movement of intraflagellar transport protein 88 (IFT88), which is distributed throughout the ciliary axoneme at steady-state conditions.22, 34 Treatment of cells with ciliobrevins 37, 43 or 47 for only 1 hour induced significant IFT88 accumulation at the tip of primary cilia, and the benzyl ether derivatives exhibited more pronounced effects in comparison to ciliobrevin A (Figure 4A–B). Withdrawal of ciliobrevins from cells restored IFT88 distribution along the primary cilia (Supplementary Figure 1). Using murine renal epithelial cells stably expressing fluorescent protein-tagged IFT88 and real-time imaging, we could further demonstrate the ability of ciliobrevin 37 to disrupt ciliary transport within 3 minutes (Figure 4B–C). These results verify the ability of C7-functionalized ciliobrevins to rapidly and reversibly inhibit cytoplasmic dynein 2 in cells.
Figure 4. Selective inhibition of cellular cytoplasmic dynein 2 function by benzyl ether-functionalized ciliobrevins.
(A) Effects of the designated compounds (50 μM; 1 hour) on ciliary IFT88 transport in an NIH-3T3 cell-derived line, using ARL13B and γ-tubulin staining to define the axoneme and base, respectively. Each axoneme was divided into 21 bins, and the IFT88 signal intensity within each bin was normalized to the total ciliary signal using MatLab R2014A (Mathworks). Data represent the average IFT88 signal intensities for at least 70 cilia ± s.e.m. Representative images of DMSO- or ciliobrevin analog-treated cells are shown. Scale bar: 2 μm. (B) Line scan kymographs demonstrating the movement of mNeonGreen-IFT88 foci in the cilia of IMCD3 cells. Cells were treated with 15 μM ciliobrevin A (1), analog 37, or an equivalent amount of DMSO vehicle for 3, 10, or 30 min. Representative tracks for anterograde (green), retrograde (red), or immobile (yellow) IFT foci are highlighted. Arrowheads indicate velocity changes for anterograde IFT foci, which are rarely observed in control cells but frequently occur when anterograde IFT encounter retrograde or immobile IFT foci in ciliobrevin A- or analog 37-treated cells. Scale bar: 2 μm. (C) Quantification of the average velocity and frequency of mNeonGreen-IFT88 foci movements in control IMCD3 cells and those treated with 15 μM ciliobrevin A or analog 37 for 3 min. Data are the average velocities or frequencies ± s.e.m., and at least 7 cilia from three independent experiments were quantified for each condition. Single and double asterisks indicate P < 0.05 and P < 0.01, respectively. (D) Representative mitotic spindle phenotypes observed in an NIH-3T3 cell-derived line arrested with 15 μM MG132 (90 min) and then treated with either DMSO or ciliobrevin analogs (30 min). Scale bar: 5 μm. (E) Quantification of the mitotic spindle assembly (black bars) and ciliogenesis (red bars) phenotypes. At least 200 mitotic spindles were scored for each condition, and primary cilium lengths were determined as described in Figure 2F.
To conclude our studies, we sought to determine whether an ability to preferentially inhibit DYNC2H1 ATPase activity in vitro would translate into isoform-selective cellular phenotypes. In addition to their effects on dynein 2-dependent ciliogenesis, ciliobrevins A and D have been shown to recapitulate the mitotic phenotypes associated with dynein 1 inhibition, including diffuse spindle structures and disorganized poles.22 We therefore compared the dose-dependent effects of C7-benzyl ether ciliobrevins on mitotic spindle assembly and primary cilium formation. For the spindle assay, murine fibroblasts were pretreated with the proteasome inhibitor MG132 to enrich for M-phase cells,35 incubated with various concentrations of the ciliobrevin analogs 37, 43, and 47, and then scored for spindle morphologies. Ciliogenesis was assessed in a parallel experiment using the same compound doses, with cilium lengths measured as before (Figure 4D–E). Although ciliobrevin A perturbed spindle assembly and ciliogenesis with similar potencies, the benzyl ether analogs preferentially inhibited dynein 2 function in cells.
To further investigate the effects of ciliobrevins on cellular dynein functions, we examined Golgi complex organization, which requires cytoplasmic dynein 1-mediated vesicle transport from the endoplasmic reticulum to the Golgi. Targeted knockout of the dynein 1 heavy chain or siRNA-mediated depletion of dynein 1 subunits causes dispersal of the Golgi apparatus.10, 36 Similar phenotypes can be induced by overexpression of the dynactin subunit dynamitin or microinjection of an anti-dynein 1 intermediate chain antibody.37 We treated murine fibroblasts with ciliobrevin A and analogs 37, 43, and 47 for 4 hours then assessed the resulting Golgi morphologies in fixed cells by immunostaining the Golgi matrix protein GM130. We concurrently stained the cells with antibodies against the ciliary GTPase ARL13B and GLI2 to assess ciliary trafficking under these conditions. All four ciliobrevins induced Golgi organization defects and ciliary GLI2 accumulation in a dose-dependent manner (Supplementary Figure 2). However, the C7-benzyl ether derivatives exhibited activity profiles that were distinct from that of ciliobrevin A, consistent with differing potencies and isoform selectivities. Analogs 37, 43, and 47 inhibited retrograde ciliary GLI2 transport more strongly than ciliobrevin A, including at concentrations that did not cause Golgi dispersal. The C7-benzyl ether analogs did induce Golgi vesiculation at higher doses while unexpectedly decreasing ciliary GLI2 levels.
One possible explanation for these results is that Golgi organization and GLI2 trafficking are not completely dynein isoform-specific processes. Whether cytoplasmic dynein 2 contributes to the regulation of Golgi dynamics remains controversial. Although siRNA-induced knockdowns of DYNC2H1 and the associated light chain DYNC2LI1 were not found to disrupt Golgi structure,36 both dynein subunits have been localized to the Golgi apparatus by immunocytochemistry,38, 39 and antibodies to DYNC2H1 can cause Golgi dispersal.39 Conversely, we speculate that a role for dynein 1 in transporting GLI2 from the cytoplasm to the primary cilium base could account for loss of ciliary GLI2 at the highest ciliobrevin doses. Comparable ciliobrevin concentrations still induce distal accumulation of IFT88 (see Figure 4A), suggesting that this transport protein localizes to primary cilia through a different mechanism.
Taken together, our findings provide new insights into the structural elements required for cytoplasmic dynein inhibition and reveal modifications that influence target specificity. In particular, the ability of analogs with bulky C7 A-ring substituents to preferentially inhibit cytoplasmic dynein 2 in vitro and in cells enhances the utility of these dynein antagonists. Given the impact of A-ring modifications on target interactions, we anticipate that additional modifications of the C7 site and possibly the C5 and C8 positions could further enhance ciliobrevin potency and/or isoform specificity. In contrast, C6 functionalization appears to decrease ciliobrevin efficacy through mechanisms yet to be determined. We note that while the effects of C7-substituted ciliobrevins on mitotic spindle assembly, ciliary function, and Golgi organization are separable, they are diminished in comparison to the isoform selectivities observed in vitro. The reasons for these disproportionate effects are unclear, but they could reflect differences in ciliobrevin subcellular localization and effective ATP concentrations. Consistent with this idea, inhibition of basal dynein ATPase activity by ciliobrevins is sensitive to ATP concentration (Supplementary Figure 3).22 Dynein heavy chain-interacting proteins could also influence ciliobrevin activity, and specific cellular processes could require different levels of dynein 1 or 2 function.
Our results also provide clues about the mechanism by which ciliobrevins abrogate dynein function. The nucleotide-sensitive activities of these inhibitors and their heterocyclic structures suggest that they engage one of the four adenine-binding regions in the cytoplasmic dynein heavy chain, inducing conformation changes that suppress motor activity. Although the AAA1 nucleotide-interacting site is the primary driver of ATP hydrolysis and motor function, its adenine-binding surface is completely conserved between cytoplasmic dyneins 1 and 2 (Supplementary Figure 4). Key residues that recognize the triphosphate group and catalyze its hydrolysis are also maintained in both isoforms. It is therefore difficult to conceive how C7-substituted ciliobrevin analogs might discriminate between the two enzyme isoforms. Of the other three nucleotide-interacting regions, the AAA2 site has the most sequence-divergent adenine-binding surface between the two cytoplasmic dynein isoforms and is structurally open, making it a plausible target of isoform-selective ciliobrevins with bulky substituents. Moreover, the AAA2 site appears to bind ATP with high affinity.16, 19 Alternatively, ciliobrevins might engage the AAA4 site since it also has isoform-specific, adenine-interacting residues within a solvent-accessible pocket, whereas the AAA3 site is perhaps less likely due to its comparatively closed, well-conserved structure. Determining the specific ciliobrevin-binding site will help reveal the molecular basis for these new dynein 2-selective inhibitors and advance future efforts to optimize the ciliobrevin pharmacophore.
METHODS AND MATERIALS
Detailed procedures for synthesizing ciliobrevin analogs and assessing their biological activities are available in Supporting Information.
Supplementary Material
Acknowledgments
We thank Y. Toyoshima for cDNAs encoding N-terminally tagged DYNC1H1 and DYNC2H1, C. Chao and Shenzhen Shengjie Biotech Co. Ltd. for assistance with compound syntheses, S. van Dorp for imaging analysis scripts, and V. Gelfand for helpful discussions. This work was supported by the NIH (R01 CA136574 to J.K.C.; R01 GM098579 to T.M.K.; R01 GM089933 to M.V.N.); the Medical Research Council, UK (MC_UP_A025_1011 to A.P.C.); a Stanford SPARK/Spectrum Pilot Grant to J.K.C.; an NWO Rubicon Postdoctoral Fellowship to S.H.; Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional Medical Scientist Training Program Grant T32 GM007739 support to J.B.S.; and a Damon Runyon Cancer Foundation Postdoctoral Fellowship (DRG-2222-15) to R.M.M.
Footnotes
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
Supplementary figures, synthetic procedures and compound characterization data for ciliobrevins 1–48, and methods for biochemical and cell-based assays of cytoplasmic dynein function. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. doi: 10.1038/nrm3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibbons BH, Gibbons IR. The effect of partial extraction of dynein arms on the movement of reactivated sea-urchin sperm. J Cell Sci. 1973;13:337–357. doi: 10.1242/jcs.13.2.337. [DOI] [PubMed] [Google Scholar]
- 4.Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330:181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- 5.Pazour GJ, Dickert BL, Witman GB. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter ME, Bower R, Knott JA, Byrd P, Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corthesy-Theulaz I, Pauloin A, Pfeffer SR. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroer TA, Steuer ER, Sheetz MP. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 13.Schnapp BJ, Reese TS. Dynein is the motor for retrograde axonal transport of organelles. Proc Natl Acad Sci U S A. 1989;86:1548–1552. doi: 10.1073/pnas.86.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell. 2002;13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8 A crystal structure of the dynein motor domain. Nature. 2012;484:345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-A crystal structure. Nat Struct Mol Biol. 2012;19:492–497. S491. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter AP. Crystal clear insights into how the dynein motor moves. J Cell Sci. 2013;126:705–713. doi: 10.1242/jcs.120725. [DOI] [PubMed] [Google Scholar]
- 18.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2015;518:435–438. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paschal BM, Shpetner HS, Vallee RB. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol. 1987;105:1273–1282. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 22.Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O’Donnell M, Gelfand VI, Kapoor TM, Chen JK. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 24.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi J, Wu X, Chung AH, Chen JK, Kapoor TM, Hammer JA. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J Cell Biol. 2013;202:779–792. doi: 10.1083/jcb.201301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Kapoor TM, Chen JK, Huse M. Diacylglycerol promotes centrosome polarization in T cells via reciprocal localization of dynein and myosin II. Proc Natl Acad Sci U S A. 2013;110:11976–11981. doi: 10.1073/pnas.1306180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye F, Breslow DK, Koslover EF, Spakowitz AJ, Nelson WJ, Nachury MV. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. Elife. 2013;2:e00654. doi: 10.7554/eLife.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sainath R, Gallo G. The dynein inhibitor Ciliobrevin D inhibits the bidirectional transport of organelles along sensory axons and impairs NGF-mediated regulation of growth cones and axon branches. Dev Neurobiol. 2015;75:757–777. doi: 10.1002/dneu.22246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee I, Miyake Y, Nobs SP, Schneider C, Horvath P, Kopf M, Matthias P, Helenius A, Yamauchi Y. Influenza A virus uses the aggresome processing machinery for host cell entry. Science. 2014;346:473–477. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 30.Ichikawa M, Watanabe Y, Murayama T, Toyoshima YY. Recombinant human cytoplasmic dynein heavy chain 1 and 2: observation of dynein-2 motor activity in vitro. FEBS Lett. 2011;585:2419–2423. doi: 10.1016/j.febslet.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 31.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, Sun M, Rack PG, Sinha S, Wu JJ, Solow-Cordero DE, Jiang J, Rowitch DH, Chen JK. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci U S A. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendricks AG, Holzbaur EL, Goldman YE. Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc Natl Acad Sci U S A. 2012;109:18447–18452. doi: 10.1073/pnas.1215462109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torisawa T, Ichikawa M, Furuta A, Saito K, Oiwa K, Kojima H, Toyoshima YY, Furuta K. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol. 2014;16:1118–1124. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- 34.Robert A, Margall-Ducos G, Guidotti JE, Bregerie O, Celati C, Brechot C, Desdouets C. The intraflagellar transport component IFT88/polaris is a centrosomal protein regulating G1-S transition in non-ciliated cells. J Cell Sci. 2007;120:628–637. doi: 10.1242/jcs.03366. [DOI] [PubMed] [Google Scholar]
- 35.Wojcik C, Schroeter D, Stoehr M, Wilk S, Paweletz N. An inhibitor of the chymotrypsin-like activity of the multicatalytic proteinase complex (20S proteasome) induces arrest in G2-phase and metaphase in HeLa cells. Eur J Cell Biol. 1996;70:172–178. [PubMed] [Google Scholar]
- 36.Palmer KJ, Hughes H, Stephens DJ. Specificity of cytoplasmic dynein subunits in discrete membrane-trafficking steps. Mol Biol Cell. 2009;20:2885–2899. doi: 10.1091/mbc.E08-12-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grissom PM, Vaisberg EA, McIntosh JR. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol Biol Cell. 2002;13:817–829. doi: 10.1091/mbc.01-08-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaisberg EA, Grissom PM, McIntosh JR. Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J Cell Biol. 1996;133:831–842. doi: 10.1083/jcb.133.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.