Abstract
Background & Aims
Conventional white light colonoscopy aims to reduce the incidence and mortality of colorectal cancer (CRC). CRC has been found to arise from missed polypoid and flat pre-cancerous lesions. We aim to establish proof-of-concept for real time endoscopic imaging of colonic adenomas using a near-infrared peptide that is specific for claudin-1.
Methods
We used gene expression profiles to identify claudin-1 as a promising early CRC target, and performed phage display against the extracellular loop of claudin-1 (amino acids 53–80) to identify the peptide RTSPSSR. With a Cy5.5 label, we characterized binding parameters and demonstrated specific binding to human CRC cells. We collected in vivo near-infrared fluorescence images endoscopically in the CPC;Apc mouse that develops colonic adenomas spontaneously. With immunofluorescence, we validated specific peptide binding to adenomas from proximal human colon.
Results
We found a 2.5-fold increase in gene expression for claudin-1 in human colonic adenomas compared with normal. We demonstrated specific binding of RTSPSSR to claudin-1 in knockdown and competition studies, and measured an affinity of 42 nM and time constant of 1.2 minutes to SW620 cells. In the mouse, we found a significantly higher target-to-background ratio for both polypoid and flat adenomas compared to normal with in vivo images. On immunofluorescence, we found significantly greater intensity (mean±std) for human adenomas (25.5±14.0) versus normal (9.1±6.0) and hyperplastic polyps (3.1±3.7), P=10−5 and 8×10−12, respectively, and for sessile serrated adenomas (20.1±13.3) versus normal and hyperplastic polyps, P=0.02 and 3×10−7, respectively.
Conclusions
Claudin-1 is overexpressed in pre-malignant colonic lesions, and can be detected endoscopically in vivo with a near-infrared labeled peptide.
Keywords: colon cancer, early detection, molecular imaging
Synopsis
Claudin-1 is highly overexpressed in human colonic adenomas. Using a near-infrared labeled fluorescent peptide, we demonstrate real time in vivo images in a mouse model of spontaneous adenomas to show feasibility for future clinical translation to detect pre-cancerous lesions.
Introduction
Colorectal cancer (CRC) is one of the most common causes of cancer-related mortality worldwide.1 Adenomatous polyps are precursor lesions,2 and may express early molecular targets that can be developed for imaging to improve methods of detection and cancer prevention. Conventional white light colonoscopy is the preferred method for screening, and is one of the most frequently performed procedures in the U.S.3 However, the miss rate for grossly visible polyps can be up to 25% or higher.4–6 Also, clinical studies have shown that colonoscopy confers a reduction in mortality from left-sided (distal) lesions but much less so for right-sided (proximal) disease.7–12 Right-sided lesions tend to be smaller in size, have more non-polypoid (flat) features, and are more difficult to visualize.13 Furthermore, flat adenomas may represent >35% of all pre-malignant lesions,14 and may result in preventable cancers.15 Moreover, adenomas, which are pre-malignant, cannot be distinguished from hyperplastic polyps (HP), which have no malignant potential.16 Sessile serrated adenomas (SSA) tend to be flat in appearance, and can result in >17.5% of proximal colon cancers.17
Claudin-1 is an integral membrane protein with four membrane-spanning regions and two extracellular loops that form tight junctions between epithelial cells to maintain cell polarity and regulate paracellular transport.18 This protein is overexpressed in several human cancers, including colorectal,19–22 pancreas,23 cervical,24 squamous cell,25 stomach,26 nasopharyngeal,27 and thyroid.28 From gene expression analysis, claudin-1 is increased by >40-fold in adenocarcinoma compared to normal colon.29 This cell surface target has also been found to be over-expressed in SSA.30 Claudin-1 overexpression in neoplasia is believed to increase cell proliferation, motility, and invasiveness, and may contribute to the loss of cell polarity, abnormal cellular organization, and decreased differentiation.31,32 Claudin-1 has also been found to have increased expression in neoplasia associated with inflammatory bowel disease.33–35
Peptides have shown promise for clinical use to detect overexpressed cell surface targets during endoscopy.36,37 Peptides exhibit high binding affinity with rapid binding onset and are inexpensive to mass manufacture. Also, high peptide concentrations can be used with topical administration to colonic mucosa to maximize the binding interactions and achieve optimal image contrast with minimal risk for toxicity.38 Because of the large diversity of sequences possible, peptide can be very high, and nanomolar binding affinities can be achieved.39 Peptides have flexibility to be optically labeled with a broad range of fluorophores for use in multiplexed imaging strategies to address tumor heterogeneity.40. Peptides with short amino acid sequences have minimal immunogenicity because of their specificity small size (<1 kD), and can be arranged in a multimer configuration to improve detection sensitivity and increase specificity and avidity from a multi-valency effect.41 Here, we aim to demonstrate real time endoscopic imaging of over-expressed claudin-1 in both polypoid and flat adenomas in vivo to establish proof-of-concept for future clinical translation as an early molecular target for detection of CRC.
Methods
Identification of claudin-1 target
We evaluated the GSE41258 gene expression dataset to identify promising early targets for imaging that are overexpressed in colonic adenomas compared to normal.29 We analyzed 22,283 probe-sets using the Affymetrix HG_U133A array platform. Data from n = 52 normal and n = 45 adenomas were selected. Two-sample t-tests and average fold-changes were computed. Data was evaluated based on the following criteria: P-value<10−5, average fold-change >2, and location on plasma membrane using Gene Ontology (GO) terms obtained from Affymetrix (ver na32).
Materials
We used human colorectal adenocarcinoma cell lines, SW620, SW480, and HCT116 (ATCC). SW620 and SW480 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) and HCT116 cells were cultured in McCoy’s 5a Medium using a 37°C humidified incubator with 5% CO2. All cell culture media (Gibco) were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Identification of a peptide specific for claudin-1
We performed phage display with a PhD7 library (New England Biolabs) using the claudin-1 (CLDN1) extracellular loop mimetic peptide CLDN-I53-80 with a biotinylated C-terminus (Biomatik) as the target. Biopanning was performed per manufacturer’s guidelines using 15 mm dishes coated with 0.1 mg/mL streptavidin, washed with TBST (Tris buffered saline (TBS) with 0.1 % Tween-20), and blocked for 1 hour at 4°C with blocking buffer consisting of 0.1M NaHCO3 with 0.5% bovine serum albumin (BSA) and 0.1 µg/mL streptavidin. The phage library (1×1011 pfu containing 1.28×109 unique 7 amino acid sequences with 100 copies) was first cleared of non-specific binders by biopanning against two streptavidin-coated dishes and one uncoated dish for 30 min at room temperature (RT) with agitation. Unbound phages were collected after each clearing step and used in the following rounds. After 3 rounds of clearing, the remaining phages were amplified to 2×1011 for biopanning with the claudin-1 target in a blocked streptavidin-coated dish for 30 min at RT. Biotin at a final concentration of 0.1 mM was added for 5 min to bind any free streptavidin. The dishes were washed 10X with TBST and weak binders were removed by eluting with 0.2 M glycine, pH 2.2, with 1 mg/mL BSA for 2 min. A second elution was performed for 13 min to remove strong binders and incubated with neutralization buffer (1M Tris-HCl, pH 9.1), amplified, and tittered for the next round of biopanning. 3 rounds of biopanning were performed with decreasing concentrations of biotinylated claudin-1 extracellular loop mimetic peptide (75, 50, and 25 nM, respectively) and were incubated with 2×1011 phages for decreasing periods of time (60, 40, and 20 min, respectively) to improve specificity. The concentration of Tween-20 was increased from 0.1% to 0.5% in the washing buffer in rounds 2 and 3. The unamplified eluate from the strong binders in round 3 was tittered overnight and 50 plaques were selected for DNA sequencing.
Peptide synthesis
The RTSPSSR (RTS*) phage was found to be highly enriched (43/50 clones) after 3 rounds. This sequence was scrambled as SPTSSRR (SPT*) for use as control. The peptides were synthesized using standard solid phase Fmoc chemistry, and labeled at the C-terminus with Cy5.5 using a 5 amino acid linker GGGSK. All chemicals and reagents used were analytical grade (Sigma-Aldrich), unless otherwise noted. Reagents for peptide synthesis (Anaspec and AAPPTEC) had >99% purity and were used without further purification. Synthesis of both peptides was performed with a PS3 automatic synthesizer (Protein Technologies, Inc.) using Boc and Fmoc protected L-amino acids before manually labeling with the dye. Upon completion of synthesis, the ivDde side chain was removed with 5% hydrazine in dimethylformamide (DMF) with agitation for 20 min at RT 3X. The resin-linked peptide was then washed 3X with DMF and dichloromethane (DCM). The Cy5.5 fluorophore (Lumiprobe) was added along with di-isopropylethylamine and incubated for 24–48 hours with agitation at RT. The peptide was then cleaved from the resin with chilled trifluoroacetic acid (TFA):triisopropylisilane:water (9.5:0.25:0.25, v/v/v) for 4 hours with agitation at RT. The peptides were separated from the resin and cleavage cocktail was evaporated with N2 gas before precipitating the peptide with diethyl ether in an overnight incubation at −20°C. The precipitate was collected by centrifugation at 1780×g for 5 min and suspended in acetonitrile:water (1:1, v/v). Both peptides were purified to >95% via HPLC (Waters, Milford, MA) using a water (0.1% TFA)-acetonitrile (0.1% TFA) gradient. The mass-to-charge (m/z) ratio of the Cy5.5-labeled peptides was measured using mass spectrometry.
Immunocytochemistry
SW620, SW480, and HCT116 cells were added to 12-well plates at a density of 0.5×106 cells/mL. The following day, they were fixed with ice-cold methanol for 20 min at −20°C, blocked with phosphate buffered saline (PBS) plus 2% BSA for 1 hour at RT, then incubated first with the primary anti-CLDN1 antibody (clone Jay.8, Invitrogen) at 5 µg/mL at 4°C overnight, then with (1:500) goat anti-rabbit secondary antibody labeled with Alexa Fluor 488 (AF488, Life Technologies) for 1 hour at RT. Finally, cells were counterstained with DAPI + Prolong Gold before being mounted on slides and imaged with a Leica SP5x confocal microscope using a 63X (numerical aperture, NA = 1.4) oil-immersion objective.
siRNA knockdown of claudin-1 expression
CLDN1 expression was knocked down using Dharmacon On-Target Plus SMARTpool Human CLDN1 siRNA (Thermo Scientific) per manufacturer’s protocol. SW620 cells were plated overnight on coverslips in a 12-well plate at 0.2×106 cells/mL. The following day, the cells were transfected with 3 µL DharmaFECT1 and either 25 nM CLDN1 or control siRNA. After 72 hours, the transfected cells were analyzed for claudin-1 expression. The cytoplasmic and plasma membrane fractions were extracted using a plasma membrane protein extraction kit (BioVision). The total protein in the cytoplasmic and plasma membrane fractions were quantified using a BCA (bicinchoninic acid) protein assay kit (Pierce), and 3 µg total protein of each fraction were run on a Novex 4–20% 1.5 mm protein gel before transferring to a polyvinylidene (PVDF) membrane. The membrane was blocked with PBS plus 0.1% Tween-20 (PBST) plus 5% non-fat milk for 1 hour at 4°C, stained with either rabbit anti-CLDNl antibody or mouse anti-tubulin antibody (clone 2-28-33, Invitrogen) at 4°C overnight, then with anti-rabbit horseradish peroxidase (HRP) (1:500, for CLDN1) or anti-mouse HRP (1:500, for tubulin) for 1 hour at RT before developing. The transfected cells were lysed with Radio-Immunoprecipitation Assay (RIPA) buffer containing mini-complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor (Roche) for 10 min on ice, collected and spun at 20,000×g for 10 min at 4°C. The total protein concentration was quantified with the BCA assay, and 10 µg protein of each sample was run on a Novex 4–20% 1.5 mm protein gel before transferring to a PVDF membrane. The membrane was blocked with PBST + 5% non-fat milk for 1 hour at 4°C overnight, then with anti-rabbit HRP (1:500, for CLDN1) or anti-mouse HRP (1:500, for tubulin) for 1 hour at RT before developing.
For immunocytochemistry, the transfected cells were further passaged and grown on cover slips. Cells were fixed with ice-cold methanol for 20 min at −20°C, blocked for 1 hour at RT with PBS + 2% BSA, stained with anti-human CLDN1 antibody at 5 µg/mL for 1 hour at RT, then with goat anti-rabbit antibody labeled with AF488 (1:500) for 1 hour at RT before being counterstained with DAPI + Prolong Gold. siRNA-transfected cells were also stained with 5 µM RTS*-Cy5.5 or SPT*-Cy5.5 for 30 min at 4°C, and fixed with 4% PFA for 10 min at RT.
Competition for peptide binding
SW620 cells were plated at ~0.5×106 cells/mL on coverslips in 12-well plates. The following day, the cells were treated first with unlabeled RTS* peptide at 0, 25, 50, 100, 200, or 400 µM for 30 min at 4°C, washed with PBS, and then treated with 5 µM of RTS*-Cy5.5 for 30 min at 4°C. The cells were fixed with 4% PFA for 5 min at RT, washed, and counterstained with DAPI + Prolong Gold. Fluorescence intensities were quantified with custom software developed with Matlab (Mathworks).
Cell Binding Assay
SW620, SW480, and HT29 cells were plated at ~0.5×106 cells/mL on coverslips on 12-well plates. The following day, the cells were treated with either 10 µM of RTS*-Cy5.5 or SPT*-Cy5.5 for 1 hour at 4°C, fixed with 4% PFA for 10 min at RT, then counterstained with DAPI + Prolong Gold. Fluorescence intensities from 10 randomly chosen cells in 3 independent images were quantified using custom software. Statistical analysis was performed using a oneway ANOVA with GraphPad Prism.
Characterization of peptide binding
The apparent dissociation constant of RTS*-Cy5.5 to SW620 cells was measured. SW620 cells were washed 2X with PBS + 0.5% BSA, then ~105 cells were incubated with RTS*Cy5.5 at dilutions ranging from 0–200 nM for 1 hour at 4°C. Cells were then washed of unbound peptide 5X with PBS + 0.5% BSA before analyzing with flow cytometry (FACS Canto, BD). Sample means were used to calculate the equilibrium dissociation constant kd using nonlinear regression analysis with GraphPad Prism software.42
To measure the apparent association time constant of RTS*-Cy5.5, SW620 cells were washed 2X with PBS + 0.5% BSA, then ~105 cells were incubated with 5 µM RTS*-Cy5.5 at 4°C for time intervals ranging from 2–15 min. The cells were immediately washed of unbound peptide 5X with PBS + 0.5% BSA before analyzing with flow cytometry. The mean fluorescence intensity of SW620 cells at the various time points was ratioed with that of untreated cells and used to calculate the rate constant k by fitting the data to a first order kinetics model, y(t)=Imax[1-exp(−kt)], using Origin 6.1 software.41
Transepithelial Electrical Resistance (TEER) measurements
T84 human colon carcinoma cells were grown in a 1:1 mixture of DMEM and Ham’s F-12 culture medium supplemented with 5% FBS. To establish polarized monolayer, the cells were plated on transwell permeable polyester supports (1.12 cm2, pore size 3 µm, Costar) until they reached confluence after ~3–4 days, as determined by an increase in TEER.43 The cells were continually grown on transwell supports until the TEER reached ~2000 Ω-cm2. Then, 5 µM of either RTS-Cy5.5 or control peptides were added. TEER was then measured at 6, 12 and 24 hours. The cells were fixed with 4% PFA for 12 min. After brief washing, 1% SDS in PBS was used to permeabilize the cells. These procedures were followed by 3% goat serum in PBS blocking for 30 min. Mouse anti-zonula occludens-1 (anti-ZO-1) (1:250, Life Technologies) and rabbit anti-claudin-1 (1:200, Life Technologies) antibodies were diluted in block buffer and incubated in humidity box overnight at 4°C and fluorescent secondary antibodies were diluted to 1:1000 and incubated for 1 hour at RT.44 All images were obtained using Nikon Al confocal microscope (MIL, University of Michigan).
In vivo imaging in mouse colon
We used a CPC;Apc mouse model of CRC in which the adenomatous polyposis coli (APC) allele is sporadically deleted by Cre recombinanse in colonic epithelium,45 resulting in spontaneous formation of flat and polypoid colonic adenomas. This model is representative of human disease because APC mutations are found in >80% of sporadic colorectal cancers.46 We used a rigid small animal endoscope (Karl Storz Veterinary Endoscopy-America) to image the distal 2 cm of colon.47 The mouse studies were performed with approval of the University of Michigan Committee on the Use and Care of Animals (UCUCA). The mice were housed in pathogen-free conditions and supplied water ad libitum under controlled conditions of humidity (50±10%), light (12/12 hour light/dark cycle) and temperature (25°C). Anesthesia was induced and maintained via a nose cone with inhaled isoflurane mixed with oxygen at a concentration of 2–4% at a flow rate of −0.5 L/min. Mucus was removed by vigorously rinsing the colon with water. We used white light illumination first to identify adenomas. A 100 µM solution of RTS*-Cy5.5 was administered intra-rectally, and allowed to incubate for 5 min before rinsing away the unbound peptides with water. The colon was then imaged with fluorescence. We recorded the 1) distance between the endoscope tip and the anus and 2) clockwise location of each region of high intensity. Several days later, endoscopy was repeated to confirm that all residual signal from RTS*-Cy5.5 had disappeared, and then the mice were imaged with the SPT*-Cy5.5 control peptide. On the endoscopic images, we determined the average fluorescence intensity from 3 regions of interest (ROI) with dimensions of 20×20 µm2 picked at random from the regions of high fluorescence intensity (target) and adjacent areas of normal colonic mucosa (background) to measure the target-to-background (T/B) ratio.
After completion of imaging, the mice were euthanized, and colon was resected and divided longitudinally to expose the mucosal surface. We first collected white light images using the Xenogen IVIS Spectrum (Caliper Life Sciences, Hopkinton, MA). NIR fluorescence images were then collected using 675 nm excitation and 720 nm emission with 1 sec exposure time. A ruler was placed next to the specimen to determine the distance from the anus for registration with the endoscopy and histology images. The specimen was then processed for histology by cutting sections in the plane parallel to the mucosal surface. Digital images were collected with a Zeiss Axiovision microscope (Thornwood, NY) using 5X magnification, and stitched together using Image Composite Editor (Microsoft, Redmond, WA). A pathologist (SRO) who was blinded to the imaging results reviewed the composite histology, and identified regions of dysplasia and normal colon. Fluorescence intensities from these sites were measured from two concentric ellipses of equal area using Living Image 4.0 software (Caliper Life Sciences; Hopkinton, MA). The inner and outer regions were used to define the target (T) and background (B) values, respectively.
Validation of claudin-1 expression in mouse and human proximal colon specimens
Formalin-fixed specimens from mouse colon were deparaffinized. Antigen retrieval was performed using standard methods. Briefly, the sections were incubated 3X in xylene for 3 min, washed 2X with 100% ethanol for 2 min, and washed 2X with 95% ethanol for 2 min. Rehydration was performed by washing with dH2O for 5 min. Antigen unmasking was performed by boiling the slides in 10 mM sodium citrate buffer with 0.05% Tween at pH 6.0, and then maintaining at a sub-boiling temp for 15 min. The slides were cooled for 30 min. The sections were washed 3X with dH2O for 3 min, and then incubated in 3% H2O2 in methanol for 10 min. The sections were washed 3X in dH2O for 2 min and in PBST for 5 min.
Blocking was performed with protein blocking agent (X0909, Dako) for 15 min at RT. The blocking solution was washed 3X with PBS. For mouse specimens, we used primary rabbit polyclonal anti-claudin-1 antibody (ab 15098, Abcam), and for human specimens, we used primary rabbit polyclonal anti-claudin-1 antibody (clone Jay.8, Invitrogen). The sections were incubated overnight at 4°C in a humidified chamber and then washed 3X in PBST for 5 min. A 1:200 dilution of biotinylated secondary antibody (goat anti-rabbit IgG) was added to each section and incubated for 30 min at RT, and then removed by washing 3X with PBST for 5 min. Pre-mixed Elite Vectastain ABC reagent (Vector Labs) was added to each section and incubated for 30 min at RT. The sections were washed 3X in PBS for 5 min, and developed with 3,3’-diaminobenzidene tetrahydrochloride (DAB) substrate. The reaction was monitored for up to 5 min, and then quenched by immersing the slides in dH2O. Hematoxylin was added as a counterstain for ~20 sec, and the sections were dehydrated in increasing concentrations of ethyl alcohol (2X each at 70%, 80%, 95%, and 100%). Coverslips were mounted using permount mounting medium (#SP15-100, Fisher) in xylene. Serial sections were processed for routine histology (H&E).
Immunofluorescence of proximal human colon with claudin-1 peptide and antibody
Formalin-fixed, paraffin-embedded (FFPE) specimens of adenomas, sessile serrated adenomas, hyperplastic polyps and normal colonic mucosa from human proximal colon were obtained from the archived tissue bank in the Department of Pathology. 5 µm thick sections were cut, and mounted onto glass slides (Superfrost Plus, Fischer Scientific). The tissues were deparaffinized, and antigen retrieval was performed as described above. The sections were blocked with protein serum for 15 min at RT followed by rinsing with PBS. The sections were then stained with RTS*-Cy5.5 at 5 µM concentration for 10 min at RT. The sections were then washed 3X with PBS (3 min each) and incubated overnight with (1:200) anti-claudin-1 antibody (clone Jay.8, Invitrogen). The sections were washed 3X with PBST, and incubated with (1:500) goat anti-rabbit antibody labeled with AF488 (Invitrogen) for 1 hour at RT. The sections were washed again 3X with PBST and mounted with Prolong Gold reagent containing DAPI (Invitrogen) using #1 cover glass (1.5 µm thickness). The images were collected with the same exposure time for all specimens. We placed 3 boxes with dimensions of 20×20 µm2 completely within colonic epithelium in each image, and measured the mean fluorescence intensities for RTS*-Cy5.5 using custom Matlab software. Regions of saturated intensities were avoided. The results were transformed in base-2 log to improve normality and stabilize variance, and then fit with a one-way ANOVA model. Adjacent sections were processed for routine histology (H&E), and reviewed by 2 gastrointestinal pathologists (SRO and HDA).
All authors had access to the study data, and have reviewed and approved the final manuscript.
Results
Identification of claudin-1 target
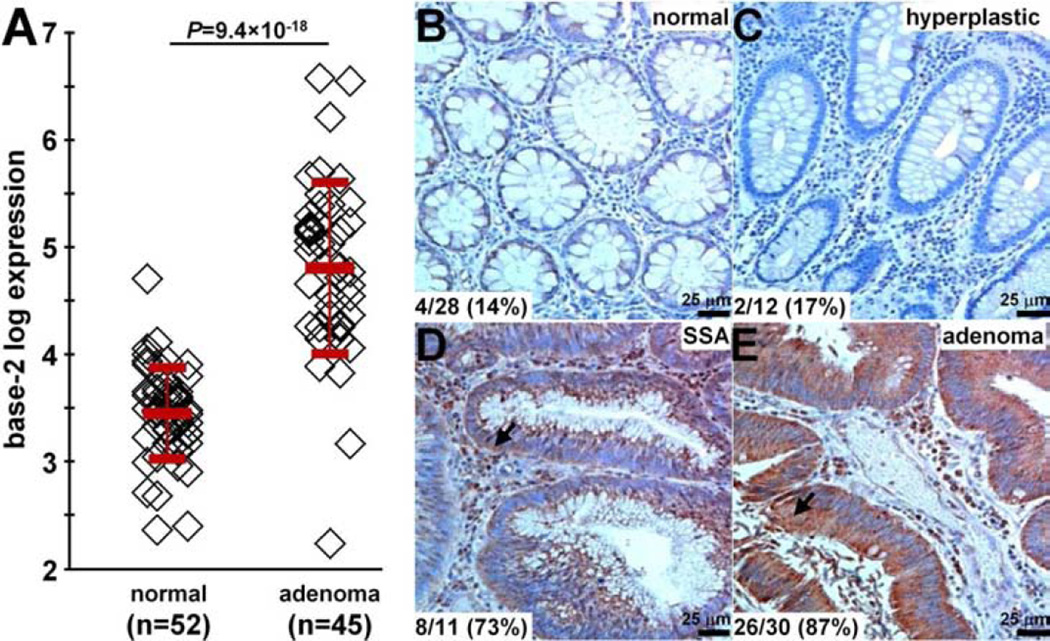
In the GSE41258 dataset,29 we found a 2.5-fold increase in gene expression for claudin-1 in human colonic adenomas (n=45) compared with normal mucosa (n=52), Fig. 1A. On immunohistochemistry (IHC), we evaluated expression of this protein target in archived human specimens from the proximal colon. Figs. 1B and 1C show minimal staining for representative sections of normal and hyperplastic polyps. Figs. 1D and 1E show intense cell surface staining (arrows) for representative sections of sessile serrated adenoma (SSA) and adenomas. Consensus between 2 GI pathologists (SRO, HDA) using a standard IHC scoring system revealed overexpression, defined by either 2+ or 3+ staining, in 14% (4/28) of normal, 17% (2/12) of hyperplastic polyps, 73% (8/11) of SSA, and 87% (26/30) of adenomas. These results support claudin-1 as a promising early target for detection of proximal colon cancers.
Fig. 1. Claudin-1 is an early target for CRC.
A) From the GSE41258 gene expression dataset, the mean (±std) base-2 log level for claudin-1 was 4.8±0.8 and 3.5±0.4 for human adenomas (n=45) and normal (n=52) colonic mucosa, resulting in an average difference of 2.54-fold, P=9.4×10−18 by two-sample t-test. On immunohistochemistry, minimal staining was observed from representative sections of B) normal and C) hyperplastic polyps. Intense cell surface staining (arrows) was seen in representative sections of D) sessile serrated adenoma (SSA) and E) adenomas from human proximal colon specimens. Using a standard IHC scoring system, overexpression (2/3+) of claudin-1 was found in 14% (4/28) of normal, 17% (2/12) of hyperplastic polyps, 73% (8/11) of SSAs, and 87% (26/30) of adenomas.
Identification of a peptide specific for claudin-1
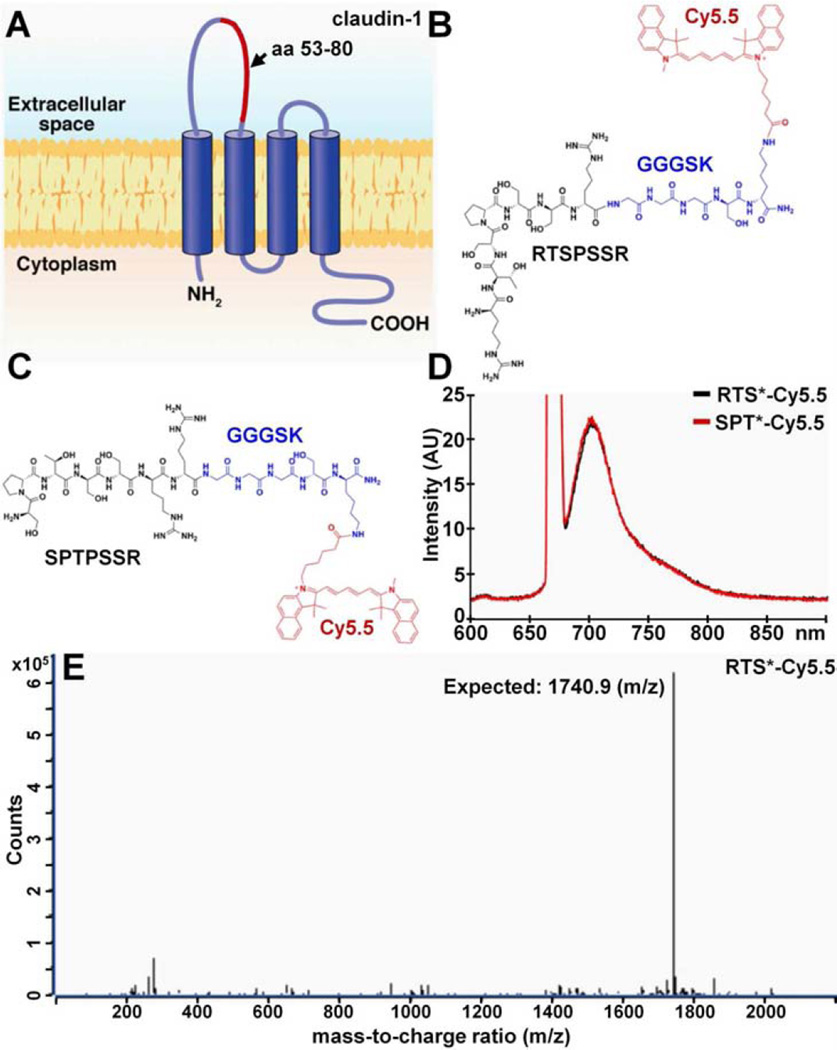
Fig. 2A shows the structure of claudin-1 on the cell surface. We used the 53–80 amino acid (aa) loop (red) of the extra-cellular domain as the biopanning substrate for peptide selection with phage display. After 3 rounds, we found the peptide sequence RTSPSSR to be highly enriched with expression in 43/50 of clones. Fig. 2B shows the synthesized peptide (black) labeled with Cy5.5 (red) via a GGGSK linker (blue) on the C-terminus to prevent steric hindrance. Cy5.5 was chosen for use as the label because of its high quantum yield and photostability.48 Fig. 2C shows the scrambled sequence SPTSSRR used for control. Fig. 2D shows the fluorescence spectra of RTS*-Cy5.5 and SPT*-Cy5.5 with a peak emission in the near-infrared spectrum at λem = 710 nm using excitation at λex = 671 nm. We purified the Cy5.5-labeled peptides to >95% on HPLC, and measured an experimental mass-to-charge (m/z) ratio of 1740.9 for RTS*-Cy5.5 on mass spectrometry which agrees with the expected value, Fig. 2E.
Fig. 2. Peptide specific for claudin-1.
A) The extracellular loop of claudin-1 (CLDN1) consisting of amino acids 53–80 (red) was used to select the B) peptide with sequence RTSPSSR. The chemical structure of the peptide (black) with a GGGSK linker (blue) and Cy5.5 label (red) is shown. C) Scrambled peptide with sequence SPTPSSR labeled with Cy5.5 is used as control. D) Fluorescence spectra with λex = 671 nm excitation shows peak emission in near-infrared at 710 nm for both peptides. E) For RTS*-Cy5.5, the mass-to-charge (m/z) ratio of 1740.9 was measured on mass spectrometry that agrees with the expected value.
Immunocytochemistry
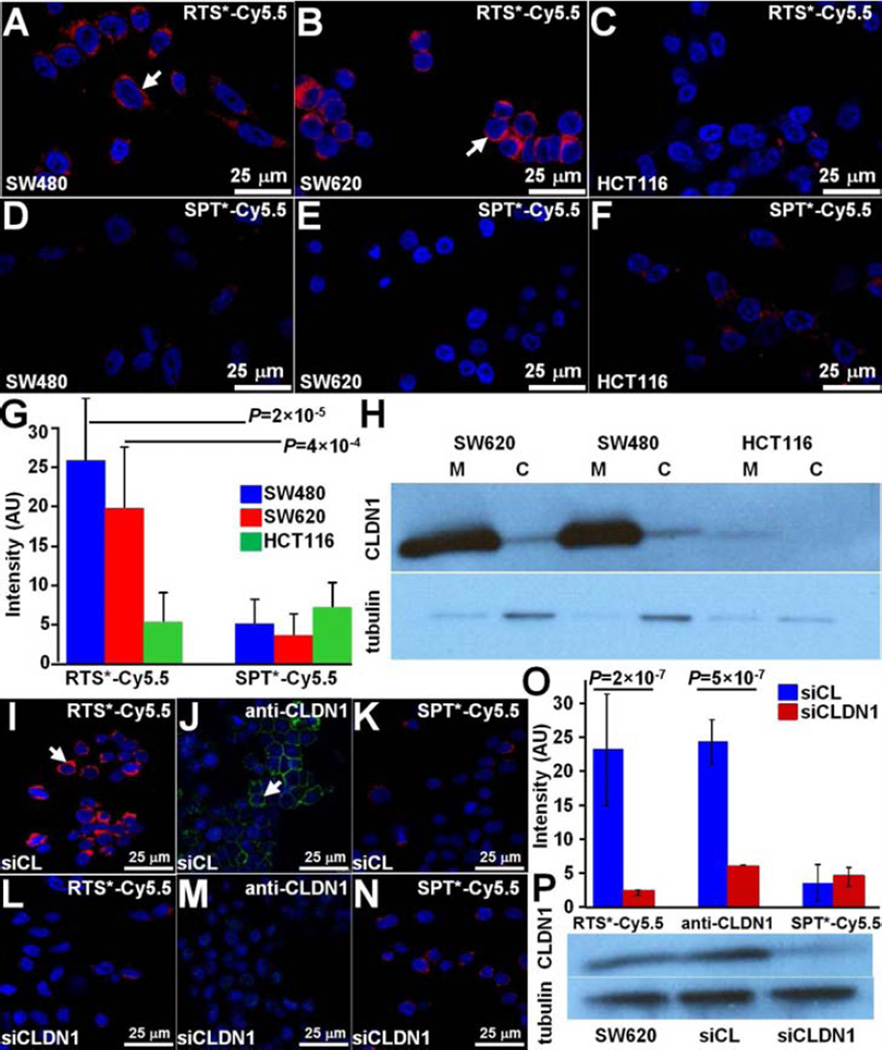
On confocal microscopy, we examined cells with either high (SW620, SW480) or low (HCT116) claudin-1 expression to validate specific binding of the RTS*-Cy5.5 peptide to the plasma membrane. We found greater amounts of RTS*-Cy5.5 bound to the surface (arrows) of SW480 and SW620 cells compared to HCT116 cells, Fig. 3A–C, whereas the SPT*-Cy5.5 control showed minimal binding to all cells, Fig. 3D–F. Fig 3G shows quantified fluorescence intensities for RTS*-Cy5.5 to be significantly greater than that for SPT*-Cy5.5 to SW480 and SW620 but not to HCT116 cells. Also, the RTS*-Cy5.5 versus SPT*-Cy5.5 differences were significantly greater for SW480 and SW620 than for HCT116. A western blot confirmed higher claudin-1 expression for SW480 and SW620 than for HCT116 cells on the plasma membrane (M), Fig. 3H. Claudin-1 expression in the cytoplasmic (C) fraction of each cell line was relatively low compared to the cell surface.
Fig. 3. Validation of specific peptide binding to claudin-1 with human CRC cells.
On confocal microscopy, RTS*-Cy5.5 showed different levels of binding to the cell surface (arrow) of A) SW480, B) SW620, and C) HCT116 cells. D-F) Minimal signal is observed for SPT*-Cy5.5 to each of the cells. G) RTS*-Cy5.5 showed higher fluorescence intensities than SPT*-Cy5.5 on binding to SW480 and SW620 cells with a 7.8 and 4.3 average fold-change, P=2×10−5 and 4×10−4, respectively. A small non-significant increase was observed for HCT116 cells with 0.64 average fold-change, P=0.19. The differences between RTS*-Cy5.5 and SPT*-Cy5.5 were significantly larger for SW480 and SW620 than the same difference for HCT116 with 12.3 and 6.8 average fold-change, P=10−4 and 10−3, respectively. We fit a one-way ANOVA model to log-transformed data with terms for the means of 6 conditions, testing for RTS*-Cy5.5 versus SPT*-Cy5.5 and the difference of those differences between the cell lines. Measurements are an average of 10 randomly chosen cells from each of 3 slides for each condition. H) Western blot shows claudin-1 expression in cytoplasmic fraction (C) and on plasma membrane (M) for each cell. Confocal fluorescence images demonstrate strong binding of I) RTS*-Cy5.5 peptide (red) and J) AF488-labeled anti-CLDN1 antibody (green) to the surface (arrow) of control SW620 cells (transfected with non-targeting siRNA, siCL). K) Binding by the SPT*-Cy5.5 (red) control peptide is minimal. L–N) The fluorescence intensities are significantly reduced in knockdown SW620 cells transfected with CLDN1-targeted siRNA, siCLDN1. O) We fit a two-way ANOVA model with terms for siRNA type (knockdown and control) and targeting moieties (RTS*-Cy5.5, anti-CLDN1, SPT*-Cy5.5) and their interactions to the average intensities on each slide (10 cells per slide with 2 slides per condition). The signal for RTS*-Cy5.5 decreased over 10-fold with siRNA knockdown of CLDN1, P=2×10−7, which was a significantly larger decrease than the same difference for the control peptide, P=10−6. The antibody signal also decreased significantly, P=5×10−7. P) Western blot shown for control (siCL) and knockdown (siCLDN1) SW620 cells.
siRNA knockdown of claudin-1 expression
We performed siRNA knockdown experiments with SW620 cells to support specific binding of the RTS*-Cy5.5 peptide to claudin-1 (CLDN1). We found RTS*-Cy5.5 and AF488-labeled anti-CLDN1 antibody show strong binding to the surface of SW620 cells transfected with a control siRNA (siCL), Figs. 3I and 3J, and reduced binding to cells transfected with siCLDN1 targeted siRNA, Figs. 3L and 3M. SPT*-Cy5.5 produced minimal binding with either siRNA, Figs. 3K and 3N. Fig. 3O shows quantified fluorescence intensities for RTS*-Cy5.5 to decrease >10-fold with CLND1 siRNA knockdown compared with the control knockdown, which was significantly greater than the decrease in signal for SPT*-Cy5.5 (control). Signal from anti-CLDN1 also decreased significantly, demonstrating an effective knockdown. A western blot confirmed the high expression of claudin-1 in SW620 cells as well as for the cells transfected with control siRNA, while expression was reduced by ~44% for cells transfected with siCLDN1, Fig. 3P.
Competition for peptide binding
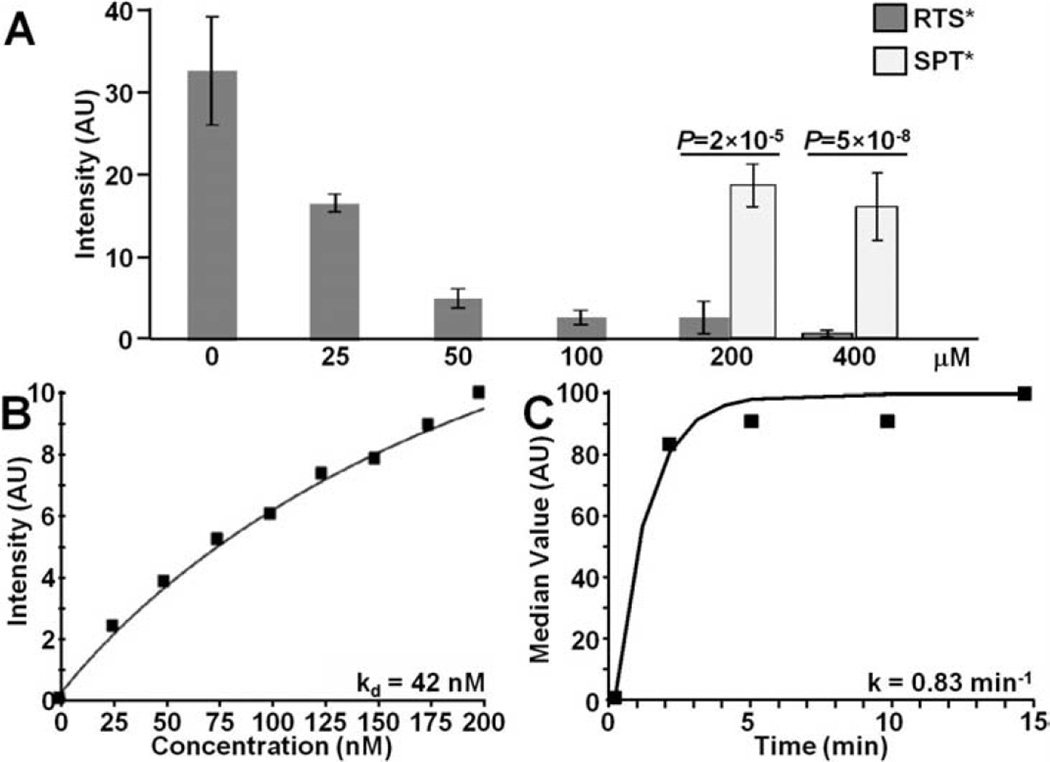
We evaluated binding of RTS*-Cy5.5 to SW620 cells with competition from unlabeled RTS* and SPT* peptides to support specific binding by the RTS* peptide rather than the Cy5.5 label. Fig 4A shows that the addition of 25, 50, 100, 200 and 400 µM of unlabeled RTS* produces a dose-dependent reduction in fluorescence intensity of RTS*-Cy5.5 using confocal microscopy. By comparison, addition of unlabeled SPT* at the higher concentrations of 200 and 400 µM showed a significantly higher RTS*-Cy5.5 fluorescence intensity than with equivalent concentrations of unlabeled RTS*.
Fig. 4. Characterization of claudin-1 peptide binding.
A) On competition, RTS*-Cy5.5 showed less binding to SW620 cells with addition of unlabeled RTS* at concentrations of 25 µM and higher compared to that at 0 µM. With unlabeled RTS* at concentrations of 200 and 400 uM, the signal from RTS*-Cy5.5 was significantly lower than that measured when competing with unlabeled SPT* at the same concentrations, P<2×10−5. Analysis was performed using an ANOVA model with terms for 8 means fit to log-transformed data. Measurements are an average of 10 randomly chosen cells on each of 3 slides at each condition for RTS* and 2 slides for each condition for SPT*. B) We measured an apparent dissociation constant (binding affinity) of kd = 42 nM, R2 = 0.95 for RTS*-Cy5.5 to SW620 cells. C) We measured an apparent association time constant k = 0.83 min−1 which corresponds to <1.2 min. Results for each measurement are representative of 2 independent experiments.
Characterization of peptide binding
We performed flow cytometry experiments with SW620 cells to characterize peptide binding parameters. Fig. 4B shows an apparent dissociation constant for the RTS*-Cy5.5 peptide of kd = 42 nM, R2 = 0.95. This result provides a measure of binding affinity. Fig. 4C shows an apparent association time constant k = 0.83 min−1 for RTS*-Cy5.5. This result provides a time scale of ~1.2 min for binding with topical administration.
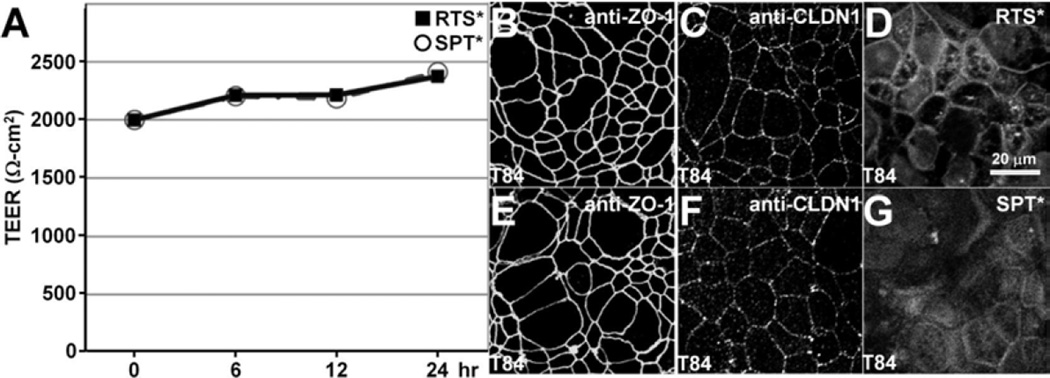
Transepithelial Electrical Resistance (TEER) measurements
We evaluated the effect of peptide binding on tight junction function using a polarized monolayer of T84 cells plated on transwell supports. We found high trans-epithelial electrical resistance (TEER) with either RTS* or SPT* for up to 24 hours, Fig. 5A. On immunofluorescence, we observed antibodies for zonula occludens-1 (ZO-1), Fig. 5B,E, and claudin-1, Fig. 5C,F, to localize to the cell junctions. These results show that neither peptide alters tight junction function or ZO-1 distribution. The RTS* peptide localizes partially to cellular junctions by comparison to SPT*, Fig. 5D,G
Fig. 5. Tight junction function and ZO-1 distribution are not altered by claudin-1 peptide.
A). Confluent T84 monolayers were incubated with either 5 µmol RTS* or SPT* (control) peptides show high trans-epithelial electrical resistance (TEER) for up to 24 hours. Immunofluorescence demonstrates localization of B,E) anti-zonula occludens-1 (anti-ZO-1) and C,F) anti-claudin-1 (anti-CLDN1) antibodies on the apical plasma membrane of tight junctions at 24 hours after peptide incubation. D) RTS* peptide partially localizes to cellular junctions.
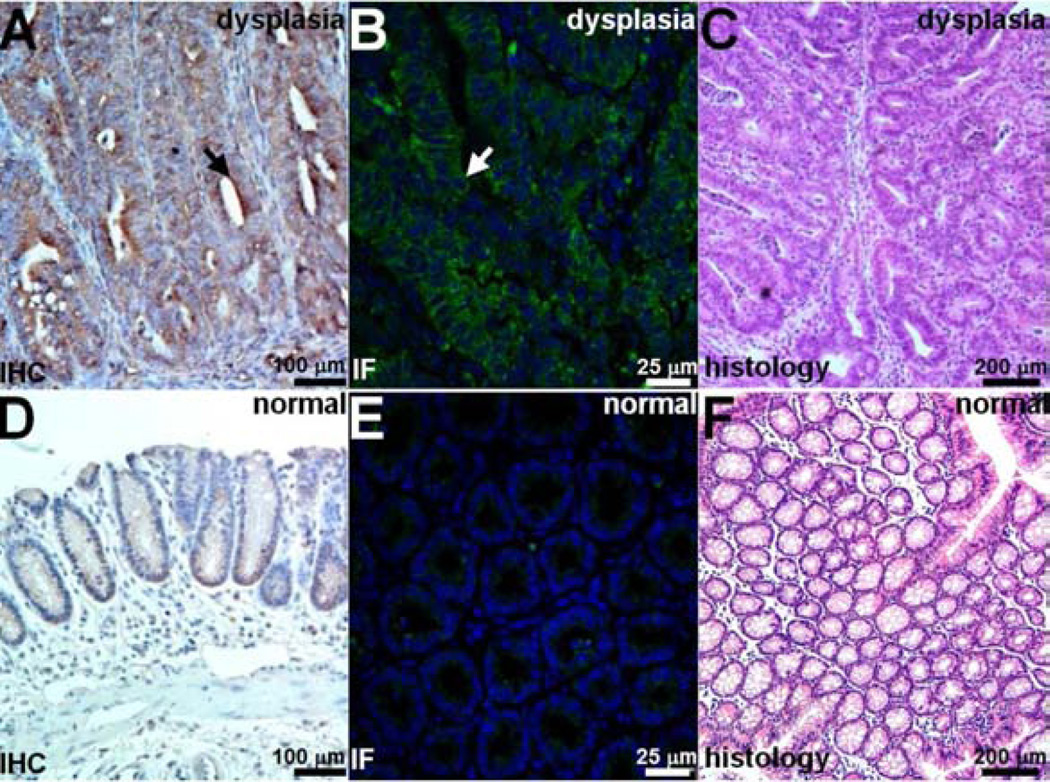
Validation of claudin-1 expression in mouse colon
We performed immunohistochemistry (IHC) and immunofluorescence (IF) to demonstrate overexpression of claudin-1 in dysplasia compared to normal in resected specimens of colon from the CPC;Apc mouse. Figs. 6A and 6B show intense staining using the anti-claudin-1 antibody for dysplasia on IHC and IF, respectively, while Figs. 6D and 6E show minimal signal for normal colonic mucosa on IHC and IF. Figs. 6C and 6F show corresponding histology (H&E) for dysplasia and normal, respectively.
Fig. 6. Increased claudin-1 expression in CPC;Apc mouse colonic adenomas.
Strong staining of claudin-1 in dysplasia is seen with A) immunohistochemistry (IHC) and B) immunofluorescence (IF) using AF488 label. C) Representative histology (H&E). Minimal expression of claudin-1 was observed in normal colonic mucosa on D) immunohistochemistry and E) immunofluorescence. C) Representative histology (H&E).
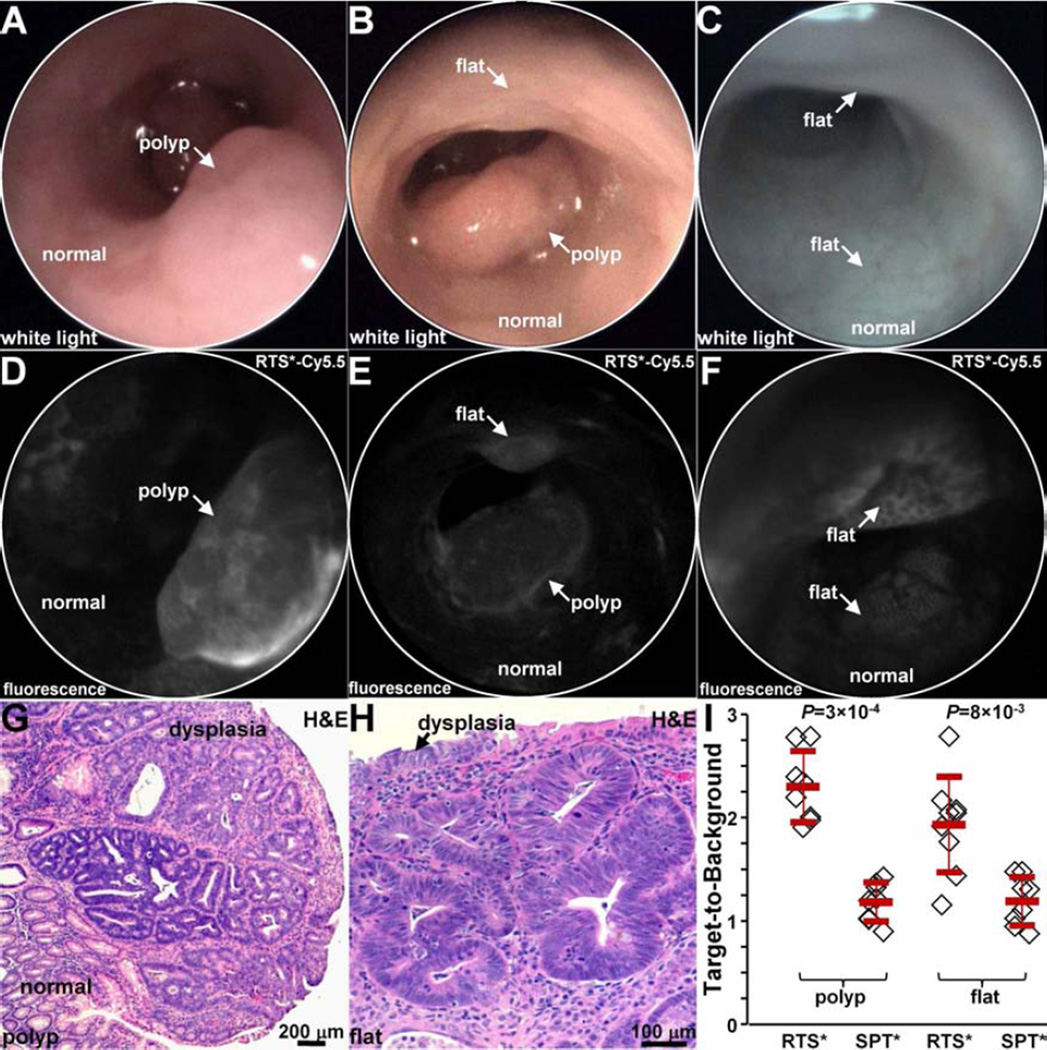
In vivo imaging in mouse colon
We used a small animal endoscope to compare binding between the claudin-1 and control peptides in n = 5 CPC;Apc mice. After collecting white light images, either RTS*-Cy5.5 or SPT*-Cy5.5 was administered intra-rectally, and allowed to incubate for 5 min. The unbound peptides were then rinsed away. On in vivo white light images, Figs. 7A and 7B (video 1) show a sporadic polyp (arrow) and Fig. 7C show apparent normal colonic mucosa with no grossly visible adenomas. On the corresponding fluorescence images after administration of RTS*-Cy5.5, Fig. 6D shows increased intensity from the polyp in a heterogeneous pattern while normal colonic mucosa shows minimal background. Fig. 7E shows the presence of a flat lesion above the polyp that is not apparent on the white light image in Fig. 7B that was found later on pathology to be dysplasia (video 2). Fig. 7F shows two flat regions of increased fluorescence intensity that were confirmed to be dysplasia on pathology. The staining pattern appears to outline dysplastic crypts.
Fig. 7. In vivo imaging in CPC;Apc mouse colon.
White light images show A,B) a spontaneous polyp (arrow) and C) normal appearing mucosa. NIR fluorescence images after topical administration of RTS*-Cy5.5 shows D) increased intensity from polyp (arrow) in A), E) presence of flat lesion above polyp in B), and F) flat lesions not apparent on white light image in C). Representative individual images were extracted from videos recorded at 15 frames per second that showed minimum motion artifact and absence of debris (stool, mucus). Histology (H&E) of G) polyp in A) and H) flat lesion in B) shows features of low-grade dysplasia. I) From n = 5 mice, RTS*-Cy5.5 had a higher mean (±std) T/B ratio than SPT*-Cy5.5 for n=8 polyps (2.3±0.3 and 1.2±0.2, respectively, P=3×10−4 by paired t-test) and n = 9 flat lesions (1.9±0.5 and 1.1±0.2, respectively, P=8×10−3 by paired t-test).
After completion of imaging, the mice were euthanized. The colon was excised and the mucosal surface was exposed to identify polyps. White light and fluorescence images were collected from each specimen using Xenogen IVIS Spectrum. Regions of increased intensity were registered with the endoscopic images using landmarks defined by distance from the anus and clockwise location, and submitted for histology. The pathologist identified n = 8 polyps and n = 9 flat lesions. Fig. 7G shows histology of a polyp with features of low-grade dysplasia, including collections of irregular crypts lined by epithelium with crowded, elongated, and hyperchromatic nuclei. Fig. 7H shows histology of a flat lesion with similar histological features of dysplasia. We found a significantly greater mean T/B ratio for RTS*-Cy5.5 than for SPT*-Cy5.5 for both polyps and flat lesions, Fig. 7I.
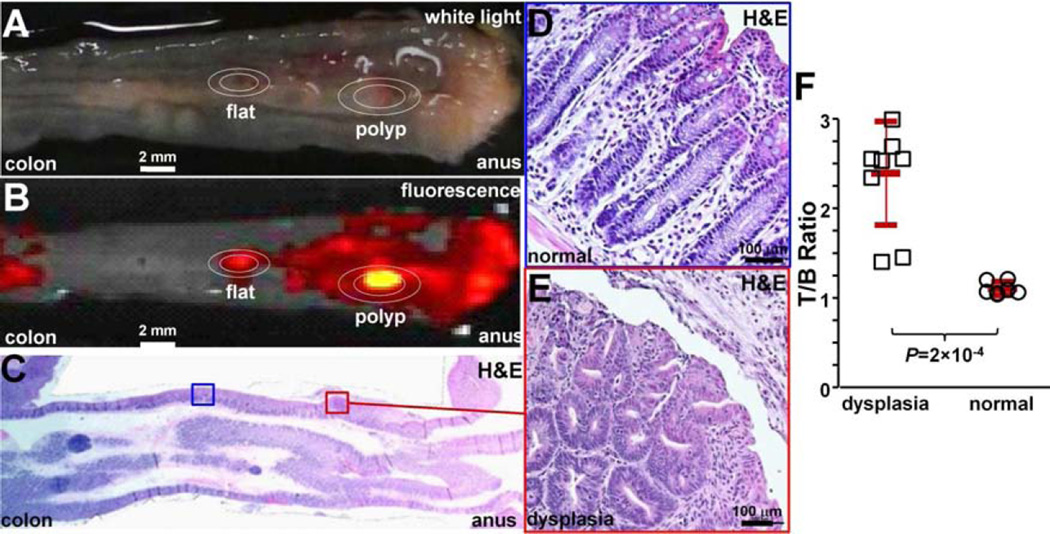
We then compared binding of the claudin-1 peptide between dysplasia and normal colonic mucosa in n = 3 mice at 10 weeks of age when they first begin to form polyps. We administered RTS*-Cy5.5 in vivo. After euthanizing the mice, we collected white light, Fig. 8A, and NIR fluorescence images, Fig. 8B, from the excised colon specimens. The tissues were sectioned along planes parallel to the surface, and the pathologist (SRO) identified regions of dysplasia (red) and normal (blue) on histology, Fig. 8C, while blinded to the imaging results. Magnified regions of normal and dysplasia are shown in Figs. 8D and 8E, respectively. The pathologist identified a total of n = 9 regions of dysplasia and n= 7 sites of normal. The intensities from these regions were measured from the fluorescence images. We found a significantly greater T/B ratio for dysplasia compared to normal, Fig. 8F.
Fig. 8. Ex vivo validation of peptide binding to colonic dysplasia.
A) Representative white light image of excised distal 2 cm of mouse colonic mucosa accessed by endoscopy after RTS*-Cy5.5 was topically administered in vivo. B) NIR image shows regions of increased fluorescence intensity. C) Histology (H&E) sectioned parallel to mucosal surface was evaluated for presence of dysplasia by expert GI pathologist. Expanded views of D) normal and E) dysplasia, 20X magnification. F) From n = 3 mice, the mean (±std) T/B ratio was significantly higher for n=9 regions of dysplasia compared to n=7 normal (2.4±0.6 versus 1.1±0.1, respectively, P=2×10−4 by unpaired t-test).
Binding of claudin-1 peptide to human proximal colonic lesions
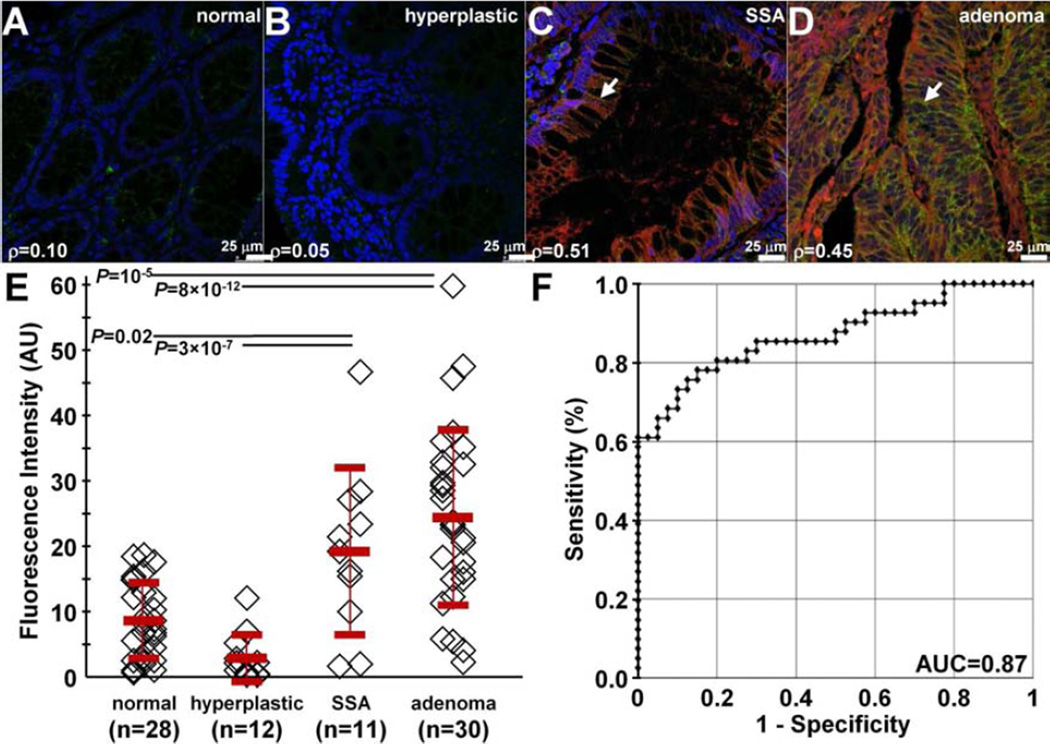
We demonstrate potential for clinical translation of this peptide by examining specific binding on formalin-fixed, paraffin-embedded (FFPE) specimens of human proximal colon. On confocal microscopy, we observed minimal fluorescence intensity for both RTS*-Cy5.5 (red) and AF488-labeled anti-CLDN1 antibody (green) on representative specimens of human normal and hyperplastic polyps, respectively, Figs. 9A and 9B. By comparison, we observed bright fluorescence from representative specimens of sessile serrated adenomas (SSA) and adenomas, Figs. 9C and 9D, respectively. These results demonstrate strong binding of the RTS*-Cy5.5 peptide to claudin-1 at the cell surface (arrows) of SSAs and adenomas. We measured the fluorescence intensities from binding of the RTS*-Cy5.5 peptide in a set of 3 boxes with dimensions of 20×20 µm2 located at random on cells in the epithelium. We found greater mean intensities for adenomas versus normal and hyperplastic polyps and for SSA versus normal and hyperplastic polyps. Fig. 8E shows a mean fold-difference of 2.8 and 2.2 for adenoma and SSA versus normal, respectively.
Fig. 9. Binding of claudin-1 peptide to human proximal colonic neoplasia.
On confocal microscopy, there was minimal binding of RTS*-Cy5.5 peptide (red) and AF488-labeled anti-claudin-1 antibody (green) to human A) normal colonic mucosa and B) hyperplastic polyps. DAPI (blue) identifies nuclei. Strong staining with both peptide and antibody was observed for representative specimens of C) sessile serrated adenomas (SSA) and D) adenomas from the proximal colon. The extent of co-localization of peptide and antibody binding is characterized by the Pearson’s correlation coefficient ρ. Representative images were selected from n = 28 normal, n = 12 hyperplastic polyps, n = 11 SSA, and n = 30 adenomas. E) We found a significantly greater mean (±std) intensity for adenomas (25.5±14.0) versus normal (9.1±6.0) and hyperplastic polyps (3.1±3.7), P=10−5 and 8×10−12, respectively, as well as for SSA (20.1±13.3) versus normal and hyperplastic polyps, P=0.02 and 3×10−7, respectively. Analysis used an ANOVA models with means for 4 groups, fit to log-transformed data. The fluorescence intensities from 3 boxes (20×20 µm2) located randomly on cells within each specimen were measured and averaged. F) ROC curve shows an area under the curve of AUC = 0.87.
Discussion
On gene expression profiles, we found claudin-1 to be overexpressed in human colonic adenomas by 2.54-fold compared with normal mucosa. This result was confirmed by expression of the protein target in 73% (8/11) of SSAs and 87% (26/30) of adenomas from proximal human colon on immunohistochemistry. Significantly reduced staining of this cell surface target was observed in hyperplastic polyps and normal mucosa. Because claudin-1 is expressed early in the development of CRC, it may be useful for detecting either polypoid or flat pre-cancerous lesions that are difficult to visualize.13 We identified the peptide RTSPSSR, and showed that it binds specifically to claudin-1 in knockdown and competition experiments. We found this peptide to have adequate binding affinity of kd = 42 nM and rapid binding within <1.2 min (k = 0.83 min−1) with topical administration in vivo. We demonstrated specific peptide binding to spontaneous colonic adenomas in mice that were either polypoid or flat in morphology. Finally, we found significantly greater fluorescence intensity from peptide binding to SSA and adenomas from human proximal colon compared to normal and hyperplastic polyps.
Although widespread use of colonoscopy has resulted in reduced incidence and mortality from CRC, this procedure has been shown to be much less effective than expected.4–12 Cases of CRC diagnosed after colonoscopy are common, and as many as 1 in 10 CRCs are found in patients who have completed this procedure.49–51 Although interval cancers may occur for a variety of reasons, most tumors are thought to arise from prevalent lesions that were missed by colonoscopy. Efforts to improve quality have focused on increasing instrument withdrawal time, adenoma detection rate, and bowel prep quality.52 Yet, reports of interval cancers in subjects undergoing careful endoscopic examination in clinical studies show that conventional white light colonoscopy can be ineffective even under optimal conditions.53,54 Advanced endoscopic techniques such as narrowband imaging (NBI)55–57 and chromoendoscopy58–60 are being investigated to improve polyp visualization, but these technologies are limited by non-specific detection mechanisms and have not shown improvement in the adenoma detection rate or in patient outcomes.61,62 Thus, a targeted approach, such as with use of peptides, may be more effective.
Use of fluorescently-labeled imaging agents that are specific for early targets may improve cancer surveillance in high-risk populations, such as those with multiple polyps, family history of CRC, Lynch syndrome, or inflammatory bowel disease.63,64 In particular, pre-malignant lesions found sporadically in the proximal colon may be more difficult to detect because of a flat appearance. Multimodal endoscopes that are sensitive to either white light or fluorescence have been developed for clinical use.65 A peptide specific for c-Met and labeled with Cy5 has recently been used in humans with intravenous administration.66 An increase in fluorescence was found for adenomas with either polypoid or flat morphology on back-to-back examinations using white light alone followed by white light with peptide. With topical administration, peptides can be delivered in high concentrations directly to mucosa at risk of harboring disease to maximize binding interactions and achieve high image contrast with little risk for toxicity. This approach results in rapid and predictable binding with minimal background, and avoids undesired biodistribution of the exogenous agent to other tissues, such as what occurs with intravenous administration. Because of their small size, peptides have reduced immunogenicity and lower costs than antibodies for mass manufacture. This method of contrast application is similar to that used in chromoendoscopy, which is now recommended by an international panel of experts for cancer surveillance in patients with ulcerative colitis,67 but with the added benefit of molecular specificity.
Future development of this peptide will require in vivo clinical validation in human studies. While we found promising results with this peptide alone, disease heterogeneity in a broad patient population may require use of additional targets using multiplexed imaging methods.40 We have previously demonstrated a peptide VRPMPLQ that was identified from human colonic polyps obtained via biopsy using phage display.36 This peptide was labeled with FITC, and specific binding was validated in vivo on dysplastic polyps with confocal endomicroscopy.36 Because this peptide was selected empirically, the target is unknown and its clinical use may not be widely generalizable. Our claudin-1 peptide, on the other hand, was selected based on a known target identified from a human gene expression dataset. Furthermore, this peptide can be used in a multimer configuration to detect multiple targets concurrently and potentially detect disease at lower levels of molecular expression.41 Claudin-1 is over-expressed in human colonic adenomas, and represents a promising early target for detection of CRC using a near-infrared labeled fluorescence peptide.
Supplementary Material
A sporadic polyp is seen surrounded by normal appearing colonic mucosa.
After topical administration of the claudin-1 peptide RTS*-Cy5.5, increased intensity is seen from the polyp with minimal background from normal mucosa. A flat lesion above the polyp can also be seen that was not apparent on the white light video, and was found later on pathology to be dysplasia.
Acknowledgments
Grant support – Funding was provided in part by National Institutes of Health R01 CA142750 (T.D.W.), R01 CA200007 (T.D.W.), R01 EB020644 (T.D.W.), and Mary L. Petrovich.
Abbreviations
- AU
arbitrary unit
- NIR
near-infrared
- ROI
regions of interest
- RT
room temperature
- SSA
sessile serrated adenoma
- T/B
target-to-background
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures – EFR, BPJ, and TDW are co-inventors on a provisional patent submitted to the University of Michigan on the peptide presented in this manuscript
Transcript Profiling – N/A
Writing Assistance – none
Author Contributions – EFR, BPJ, TDW designed research; EFR, BPJ, JZ, AP, TDW performed research; EFR, BPJ, XD, TDW contributed new reagents or analytic tools; EFR, BPJ, AS, RK, SRO, HDA, TDW analyzed data; EFR, BPJ, TDW wrote manuscript.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. PMID: 21351269. [DOI] [PubMed] [Google Scholar]
- 2.Levine JS, Ahnen DJ. Clinical practice. Adenomatous polyps of the colon. N Engl J Med. 2006;355:2551–2557. doi: 10.1056/NEJMcp063038. PMID: 17167138. [DOI] [PubMed] [Google Scholar]
- 3.Seeff LC, Richards TB, Shapiro JA, Nadel MR, Manninen DL, Given LS, Dong FB, Winges LD, McKenna MT. How many endoscopies are performed for colorectal cancer screening? Results from CDC’s survey of endoscopic capacity. Gastroenterology. 2004;127:1670–1677. doi: 10.1053/j.gastro.2004.09.051. PMID: 15578503. [DOI] [PubMed] [Google Scholar]
- 4.Heresbach D, Barrioz T, Lapalus MG, Coumaros D, Bauret P, Potier P, Sautereau D, Boustière C, Grimaud JC, Barthélémy C, Sée J, Serraj I, D’Halluin PN, Branger B, Ponchon T. Miss rate for colorectal neoplastic polyps: a prospective multicenter study of back-to-back video colonoscopies. Endoscopy. 2008;40:284–290. doi: 10.1055/s-2007-995618. PMID: 18389446. [DOI] [PubMed] [Google Scholar]
- 5.Rex DK, Cutler CS, Lemmel GT, Rahmani EY, Clark DW, Helper DJ, Lehman GA, Mark DG. Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology. 1997;112:24–28. doi: 10.1016/s0016-5085(97)70214-2. PMID: 8978338. [DOI] [PubMed] [Google Scholar]
- 6.O’brien MJ, Winawer SJ, Zauber AG, Bushey MT, Sternberg SS, Gottlieb LS, Bond JH, Waye JD, Schapiro M National Polyp Study Workgroup. Flat adenomas in the National Polyp Study: is there increased risk for high-grade dysplasia initially or during surveillance? Clin Gastroenterol Hepatol. 2004;2:905–911. doi: 10.1016/s1542-3565(04)00392-1. PMID: 15476154. [DOI] [PubMed] [Google Scholar]
- 7.Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–1105. doi: 10.1056/NEJMoa1301969. PMCID: PMC3840160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh H, Nugent Z, Demers AA, Kliewer EV, Mahmud SM, Bernstein CN. The reduction in colorectal cancer mortality after colonoscopy varies by site of the cancer. Gastroenterology. 2010;139:1128–1137. doi: 10.1053/j.gastro.2010.06.052. PMID: 20600026. [DOI] [PubMed] [Google Scholar]
- 9.Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of colonoscopy and death from colorectal cancer. Ann Intern Med. 2009;150:1–8. doi: 10.7326/0003-4819-150-1-200901060-00306. PMID: 19075198. [DOI] [PubMed] [Google Scholar]
- 10.Lakoff J, Paszat LF, Saskin R, Rabeneck L. Risk of developing proximal versus distal colorectal cancer after a negative colonoscopy: a population-based study. Clin Gastroenterol Hepatol. 2008;6:1117–1121. doi: 10.1016/j.cgh.2008.05.016. PMID: 18691942. [DOI] [PubMed] [Google Scholar]
- 11.Bressler B, Paszat LF, Chen Z, Rothwell DM, Vinden C, Rabeneck L. Rates of new or missed colorectal cancers after colonoscopy and their risk factors: a population-based analysis. Gastroenterology. 2007;132:96–102. doi: 10.1053/j.gastro.2006.10.027. PMID: 17241863. [DOI] [PubMed] [Google Scholar]
- 12.Singh H, Turner D, Xue L, Targownik LE, Bernstein CN. Risk of developing colorectal cancer following a negative colonoscopy examination: evidence for a 10-year interval between colonoscopies. JAMA. 2006;295:2366–2373. doi: 10.1001/jama.295.20.2366. PMID: 16720822. [DOI] [PubMed] [Google Scholar]
- 13.Rondagh EJ, Bouwens MW, Riedl RG, Winkens B, de Ridder R, Kaltenbach T, Soetikno RM, Masclee AA, Sanduleanu S. Endoscopic appearance of proximal colorectal neoplasms and potential implications for colonoscopy in cancer prevention. Gastrointest Endosc. 2012;75:1218–1225. doi: 10.1016/j.gie.2012.02.010. PMID: 22482917. [DOI] [PubMed] [Google Scholar]
- 14.Rembacken BJ, Fujii T, Cairns A, Dixon MF, Yoshida S, Chalmers DM, Axon AT. Flat and depressed colonic neoplasms: a prospective study of 1000 colonoscopies in the UK. Lancet. 2000;8(355):1211–1214. doi: 10.1016/s0140-6736(00)02086-9. PMID: 10770302. [DOI] [PubMed] [Google Scholar]
- 15.le Clercq CM, Bouwens MW, Rondagh EJ, Bakker CM, Keulen ET, de Ridder RJ, Winkens B, Masclee AA, Sanduleanu S. Postcolonoscopy colorectal cancers are preventable: a population-based study. Gut. 2013 doi: 10.1136/gutjnl-2013-304880. PMID: 23744612. [DOI] [PubMed] [Google Scholar]
- 16.Bond JH. Polyp guideline: diagnosis, treatment, and surveillance for patients with colorectal polyps. Practice Parameters Committee of the American College of Gastroenterology. Am J Gastroenterol. 2000;95:3053–3063. doi: 10.1111/j.1572-0241.2000.03434.x. PMID: 11095318. [DOI] [PubMed] [Google Scholar]
- 17.Patil DT, Shadrach BL, Rybicki LA, Leach BH, Pai RK. Proximal colon cancers and the serrated pathway: a systematic analysis of precursor histology and BRAF mutation status. Mod Pathol. 2012;25:1423–1431. doi: 10.1038/modpathol.2012.98. PMID: 22684223. [DOI] [PubMed] [Google Scholar]
- 18.Mrsny RJ, Brown GT, Gerner-Smidt K, Buret AG, Meddings JB, Quan C, Koval M, Nusrat A. A key claudin extracellular loop domain is critical for epithelial barrier integrity. Am J Pathol. 2008;172:905–915. doi: 10.2353/ajpath.2008.070698. PMID: 18349130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, Neff J, Washington MK, Beauchamp RD. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–1776. doi: 10.1172/JCI24543. PMID: 15965503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12:469–476. doi: 10.3727/096504001108747477. PMID: 11939410. [DOI] [PubMed] [Google Scholar]
- 21.de Oliveira SS, de Oliveira IM, De Souza W, Morgado-Díaz JA. Claudins upregulation in human colorectal cancer. FEBS Lett. 2005;579:6179–6185. doi: 10.1016/j.febslet.2005.09.091. PMID: 16253248. [DOI] [PubMed] [Google Scholar]
- 22.Ersoz S, Mungan S, Cobanoglu U, Turgutalp H, Ozoran Y. Prognostic importance of Claudin-1 and Claudin-4 expression in colon carcinomas. Pathol Res Pract. 2011;207:285–289. doi: 10.1016/j.prp.2011.01.011. PMID: 21493012. [DOI] [PubMed] [Google Scholar]
- 23.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, van Heek T, Ashfaq R, Meyer R, Walter K, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. PMID: 11943709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JW, Lee SJ, Seo J, Song SY, Ahn G, Park CS, Lee JH, Kim BG, Bae DS. Increased expressions of claudin-1 and claudin7 during the progression of cervical neoplasia. Gynecol Oncol. 2005;97:53–59. doi: 10.1016/j.ygyno.2004.11.058. PMID: 15790437. [DOI] [PubMed] [Google Scholar]
- 25.Morita K, Tsukita S, Miyachi Y. Tight junction-associated proteins (occludin, ZO-1, claudin-1, claudin4) in squamous cell carcinoma and Bowen’s disease. Br J Dermatol. 2004;151:328–334. doi: 10.1111/j.1365-2133.2004.06029.x. PMID: 15327539. [DOI] [PubMed] [Google Scholar]
- 26.Resnick MB, Gavilanez M, Newton E, Konkin T, Bhattacharya B, Britt DE, Sabo E, Moss SF. Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol. 2005;36:886–892. doi: 10.1016/j.humpath.2005.05.019. PMID: 16112005. [DOI] [PubMed] [Google Scholar]
- 27.Hsueh C, Chang YS, Tseng NM, Liao CT, Hsueh S, Chang JH, Wu IC, Chang KP. Expression pattern and prognostic significance of claudins 1, 4, and 7 in nasopharyngeal carcinoma. Hum Pathol. 2010;41:944–950. doi: 10.1016/j.humpath.2010.01.005. PMID: 20334898. [DOI] [PubMed] [Google Scholar]
- 28.Fluge Ø, Bruland O, Akslen LA, Lillehaug JR, Varhaug JE. Gene expression in poorly differentiated papillary thyroid carcinomas. Thyroid. 2006;16:161–175. doi: 10.1089/thy.2006.16.161. PMID: 16676402. [DOI] [PubMed] [Google Scholar]
- 29.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA, Domany E. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. PMID: 19359472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caruso M, Fung KY, Moore J, Brierley GV, Cosgrove LJ, Thomas M, Cheetham G, Brook E, Fraser LM, Tin T, Tran H, Ruszkiewicz A. Claudin-1 Expression Is Elevated in Colorectal Cancer Precursor Lesions Harboring the BRAF V600E Mutation. Transl Oncol. 2014;7:456–463. doi: 10.1016/j.tranon.2014.05.009. PMID: 24954356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinugasa T, Huo Q, Higashi D, Shibaguchi H, Kuroki M, Tanaka T, Futami K, Yamashita Y, Hachimine K, Maekawa S, Nabeshima K, Iwasaki H, Kuroki M. Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res. 2007;27:3729–3734. PMID: 17970035. [PubMed] [Google Scholar]
- 32.Gröne J, Weber B, Staub E, Heinze M, Klaman I, Pilarsky C, Hermann K, Castanos-Velez E, Röpcke S, Mann B, Rosenthal A, Buhr HJ. Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of claudin-1, 8 and 12. Int J Colorectal Dis. 2007;22:651–659. doi: 10.1007/s00384-006-0197-3. PMID: 17047970. [DOI] [PubMed] [Google Scholar]
- 33.Mees ST, Mennigen R, Spieker T, Rijcken E, Senninger N, Haier J, Bruewer M. Expression of tight and adherens junction proteins in ulcerative colitis associated colorectal carcinoma: upregulation of claudin-1, claudin-3, claudin-4, and beta-catenin. Int J Colorectal Dis. 2009;24:361–368. doi: 10.1007/s00384-009-0653-y. PMID: 19184060. [DOI] [PubMed] [Google Scholar]
- 34.Weber CR, Nalle SC, Tretiakova M, Rubin DT, Turner JR. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest. 2008;88:1110–1120. doi: 10.1038/labinvest.2008.78. PMID: 18711353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinugasa T, Akagi Y, Yoshida T, Ryu Y, Shiratuchi I, Ishibashi N, Shirouzu K. Increased claudin-1 protein expression contributes to tumorigenesis in ulcerative colitis-associated colorectal cancer. Anticancer Res. 2010;30:3181–3186. PMID: 20871038. [PubMed] [Google Scholar]
- 36.Hsiung P, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nature Medicine. 2008;14:454–458. doi: 10.1038/nm1692. PMID: 18345013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturm MB, Joshi BP, Lu S, Piraka C, Khondee S, Elmunzer BJ, Kwon RS, Beer DG, Appelman HD, Turgeon DK, Wang TD. Targeted endoscopic imaging of Barrett’s neoplasia with specific fluorescent-labeled peptide: first in-human results. Science Translational Medicine. 2013;5 doi: 10.1126/scitranslmed.3004733. 184ra61. PMID: 23658246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturm MB, Piraka C, Elmunzer BJ, Kwon RS, Joshi BP, Appelman HD, Turgeon DK, Wang TD. In vivo molecular imaging of Barrett’s esophagus with confocal laser endomicroscopy. Gastroenterology. 2013;145:56–58. doi: 10.1053/j.gastro.2013.05.008. PMID: 23684943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M, Anastassiades CP, Joshi B, Komarck CM, Piraka C, Elmunzer BJ, Turgeon DK, Johnson TD, Appelman H, Beer DG, Wang TD. Affinity peptide for targeted detection of dysplasia in Barrett’s esophagus. Gastroenterology. 2010;139:1472–1480. doi: 10.1053/j.gastro.2010.07.007. PMID: 20637198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joshi BP, Miller SJ, Lee CM, Seibel EJ, Wang TD. Multispectral endoscopic imaging of colorectal dysplasia in vivo. Gastroenterology. 2012;143:1435–1437. doi: 10.1053/j.gastro.2012.08.053. PMID: 23041325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi BP, Liu Z, Elahi SF, Appelman HD, Wang TD. Near-infrared-labeled peptide multimer functions as phage-mimic for high affinity, specific targeting of colonic adenomas in vivo. Gastrointestinal Endoscopy. 2012;76:1197–1206. doi: 10.1016/j.gie.2012.07.017. PMID: 23022051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas R, Chen J, Roudier MM, Vessella RL, Lantry LE, Nunn AD. In vitro binding evaluation of 177Lu-AMBA, a novel 177Lu-labeled GRP-R agonist for systemic radiotherapy in human tissues. Clin Exp Metastasis. 2009;26:105–119. doi: 10.1007/s10585-008-9220-0. PMID: 18975117. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Nusrat A, Schnell FJ, Reaves TA, Walsh S, Pochet M, Parkos CA. Human junction adhesion molecule regulates tight junction resealing in epithelia. J Cell Sci. 2000;113(Pt 13):2363–2374. doi: 10.1242/jcs.113.13.2363. [DOI] [PubMed] [Google Scholar]
- 44.Fan S, Fogg V, Wang Q, Chen XW, Liu CJ, Margolis B. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. J Cell Biol. 2007;178:387–398. doi: 10.1083/jcb.200609096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, Fearon ER. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67:9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. PMID: 17942902. [DOI] [PubMed] [Google Scholar]
- 46.Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer WF, Tomlinson IP. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci U S A. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. PMCID: PMC16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Joshi BP, Duan X, Pant A, Kuick R, Owens SR, Wang TD. EGFR overexpressed in colonic neoplasia can be detected on wide-field endoscopic imaging. Clinical Translational Gastroenterology. 2015;6:e101. doi: 10.1038/ctg.2015.28. PMID: 26181290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo S, Zhang E, Su Y, Cheng T, Shi C. A review of NIR dyes in cancer targeting and imaging. Biomaterials. 2011;32:7127–7138. doi: 10.1016/j.biomaterials.2011.06.024. PMID: 21724249. [DOI] [PubMed] [Google Scholar]
- 49.Sanduleanu S, Masclee AM, Meijer GA. Interval cancers after colonoscopy-insights and recommendations. Nat Rev Gastroenterol Hepatol. 2012;9:550–554. doi: 10.1038/nrgastro.2012.136. [DOI] [PubMed] [Google Scholar]
- 50.Robertson DJ, Lieberman DA, Winawer SJ, Ahnen DJ, Baron JA, Schatzkin A, Cross AJ, Zauber AG, Church TR, Lance P, Greenberg ER, Martínez ME. Colorectal cancers soon after colonoscopy: a pooled multicohort analysis. Gut. 2014;63:949–956. doi: 10.1136/gutjnl-2012-303796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farrar WD, Sawhney MS, Nelson DB, Lederle FA, Bond JH. Colorectal cancers found after a complete colonoscopy. Clin Gastroenterol Hepatol. 2006;4:1259–1264. doi: 10.1016/j.cgh.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Rex DK, Schoenfeld PS, Cohen J, Pike IM, Adler DG, Fennerty MB, Lieb JG2nd, Park WG, Rizk MK, Sawhney MS, Shaheen NJ, Wani S, Weinberg DS. Quality indicators for colonoscopy. Am J Gastroenterol. 2015;110:72–90. doi: 10.1038/ajg.2014.385. PMID: 25448873. [DOI] [PubMed] [Google Scholar]
- 53.Shergill AK, Conners EE, McQuaid KR, Epstein S, Ryan JC, Shah JN, Inadomi J, Somsouk M. Protective association of colonoscopy against proximal and distal colon cancer and patterns in interval cancer. Gastrointest Endosc. 2015;82:529–537. doi: 10.1016/j.gie.2015.01.053. e1. PMCID: 4540647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.le Clercq CM, Winkens B, Bakker CM, Keulen ET, Beets GL, Masclee AA, Sanduleanu S. Metachronous colorectal cancers result from missed lesions and non-compliance with surveillance. Gastrointest Endosc. 2015;82:325–333. doi: 10.1016/j.gie.2014.12.052. e2. [DOI] [PubMed] [Google Scholar]
- 55.Adler A, Aschenbeck J, Yenerim T, Mayr M, Aminalai A, Drossel R, Schroder A, Scheel M, Wiedenmann B, Rosch T. Narrow-band versus white-light high definition television endoscopic imaging for screening colonoscopy: a prospective randomized trial. Gastroenterology. 2009;136:410–416. doi: 10.1053/j.gastro.2008.10.022. e1; quiz 715. [DOI] [PubMed] [Google Scholar]
- 56.East JE, Ignjatovic A, Suzuki N, Guenther T, Bassett P, Tekkis PP, Saunders BP. A randomized, controlled trial of narrow-band imaging vs high-definition white light for adenoma detection in patients at high risk of adenomas. Colorectal Dis. 2012;14:e771–e778. doi: 10.1111/codi.12014. [DOI] [PubMed] [Google Scholar]
- 57.Pasha SF, Leighton JA, Das A, Harrison ME, Gurudu SR, Ramirez FC, Fleischer DE, Sharma VK. Comparison of the yield and miss rate of narrow band imaging and white light endoscopy in patients undergoing screening or surveillance colonoscopy: a meta-analysis. Am J Gastroenterol. 2012;107:363–370. doi: 10.1038/ajg.2011.436. [DOI] [PubMed] [Google Scholar]
- 58.Kahi CJ, Anderson JC, Waxman I, Kessler WR, Imperiale TF, Li X, Rex DK. High-definition chromocolonoscopy vs. high-definition white light colonoscopy for average-risk colorectal cancer screening. Am J Gastroenterol. 2010;105:1301–1307. doi: 10.1038/ajg.2010.51. [DOI] [PubMed] [Google Scholar]
- 59.Lapalus MG, Helbert T, Napoleon B, Rey JF, Houcke P, Ponchon T. Does chromoendoscopy with structure enhancement improve the colonoscopic adenoma detection rate? Endoscopy. 2006;38:444–448. doi: 10.1055/s-2006-925265. [DOI] [PubMed] [Google Scholar]
- 60.Le Rhun M, Coron E, Parlier D, Nguyen JM, Canard JM, Alamdari A, Sautereau D, Chaussade S, Galmiche JP. High resolution colonoscopy with chromoscopy versus standard colonoscopy for the detection of colonic neoplasia: a randomized study. Clin Gastroenterol Hepatol. 2006;4:349–354. doi: 10.1016/j.cgh.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 61.Dinesen L, Chua TJ, Kaffes AJ. Meta-analysis of narrow-band imaging versus conventional colonoscopy for adenoma detection. Gastrointest Endosc. 2012;75:604–611. doi: 10.1016/j.gie.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 62.Rex DK, Clodfelter R, Rahmani F, Fatima H, James-Stevenson TN, Tang JC, Kim HN, McHenry L, Kahi CJ, Rogers NA, Helper DJ, Sagi SV, Kessler WR, Wo JM, Fischer M, Kwo PY. Narrow-band imaging versus white light for the detection of proximal colon serrated lesions: a randomized, controlled trial. Gastrointest Endosc. 2015 doi: 10.1016/j.gie.2015.03.1915. [DOI] [PubMed] [Google Scholar]
- 63.Kaminski MF, Hassan C, Bisschops R, Pohl J, Pellise M, Dekker E, Ignjatovic-Wilson A, Hoffman A, Longcroft-Wheaton G, Heresbach D, Dumonceau JM, East JE. Advanced imaging for detection and differentiation of colorectal neoplasia: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2014;46:435–449. doi: 10.1055/s-0034-1365348. [DOI] [PubMed] [Google Scholar]
- 64.Committee ASoP, Shergill AK, Lightdale JR, Bruining DH, Acosta RD, Chandrasekhara V, Chathadi KV, Decker GA, Early DS, Evans JA, Fanelli RD, Fisher DA, Fonkalsrud L, Foley K, Hwang JH, Jue TL, Khashab MA, Muthusamy VR, Pasha SF, Saltzman JR, Sharaf R, Cash BD, DeWitt JM. The role of endoscopy in inflammatory bowel disease. Gastrointest Endosc. 2015;81:1101–1121. doi: 10.1016/j.gie.2014.10.030. e1–13. [DOI] [PubMed] [Google Scholar]
- 65.Joshi BP, Duan X, Kwon RS, Piraka C, Elmunzer BJ, Lu S, Rabinsky E, Beer DG, Appelman HD, Owens SR, Kuick R, Dogushi N, Turgeon DK, Wang TD. Multimodal endoscope can quantify wide-field fluorescence detection of Barrett’s neoplasia. Endoscopy. 2016;48:1–15. doi: 10.1055/s-0034-1392803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burggraaf J, Kamerling IM, Gordon PB, Schrier L, de Kam ML, Kales AJ, Bendiksen R, Indrevoll B, Bjerke RM, Moestue SA, Yazdanfar S, Langers AM, Swaerd-Nordmo M, Torheim G, Warren MV, Morreau H, Voorneveld PW, Buckle T, van Leeuwen FW, Ødegårdstuen LI, Dalsgaard GT, Healey A, Hardwick JC. Detection of colorectal polyps in humans using an intravenously administered fluorescent peptide targeted against c-Met. Nat. Med. 2015;21:955–961. doi: 10.1038/nm.3641. [DOI] [PubMed] [Google Scholar]
- 67.Laine L, Kaltenbach T, Barkun A, McQuaid KR, Subramanian V, Soetikno R SCENIC Guideline Development Panel. SCENIC international consensus statement on surveillance and management of dysplasia in inflammatory bowel disease. Gastroenterology. 2015;148:639–651. doi: 10.1053/j.gastro.2015.01.031. e28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A sporadic polyp is seen surrounded by normal appearing colonic mucosa.
After topical administration of the claudin-1 peptide RTS*-Cy5.5, increased intensity is seen from the polyp with minimal background from normal mucosa. A flat lesion above the polyp can also be seen that was not apparent on the white light video, and was found later on pathology to be dysplasia.