Abstract
We recently uncovered that glucose is a critical activator of sterol regulatory element-binding proteins (SREBPs). Glucose promotes SREBP-cleavage activating protein (SCAP)/SREBP complex trafficking from the ER to the Golgi and subsequent SREBP activation via N-glycosylation of SCAP. Our study also demonstrated that SCAP plays a critical role in tumor growth.
Keywords: SCAP, SREBPs, lipid metabolism, glucose, glioblastoma, tumor growth, N-glycosylation
Metabolic reprogramming has emerged as a new hallmark of cancer. Growing evidence shows that the alteration of metabolic pathways contributes to the quick generation of bioenergetics and macromolecular building blocks supporting the rapid growth and proliferation of tumor cells.1 It is critical to understand the underlying metabolic regulation of cancer cells as this may lead to the identification of promising strategies to treat malignancies.
In recent years, many talented scientists and physicians have turned their attention to the metabolism field, aiming to identify the Achilles' heel to fight cancer. While glucose and glutamine metabolism in cancer cells has been largely investigated, lipid metabolism is still vastly under-explored in cancer. Although lipid regulation is actively studied in cardiovascular diseases and obesity, there is little crosstalk between the fields of metabolic syndromes and cancer.
Our previous studies revealed that lipid metabolism is altered in glioblastoma (GBM), a most common and deadly brain tumor.2–4 We found that oncogenic epidermal growth factor receptor (EGFR) signaling upregulates sterol regulatory element-binding protein-1 (SREBP-1), a membrane bound transcription factor with a central role in lipid metabolism.5–7 These studies demonstrated that oncogenic signaling hijacks the intrinsic machinery of lipid metabolism, enhancing lipid synthesis and uptake to promote tumor growth. However, the detailed molecular mechanisms by which cancer cells regulate lipid metabolism remain largely elusive.
Intriguingly, increased glucose consumption and enhanced lipogenesis often occur in concert in tumorigenesis.1,2 However, the existence of an intrinsic molecular link between glucose and lipids had not been demonstrated. This important question is what triggered my interest and the ensuing investigations in my laboratory.
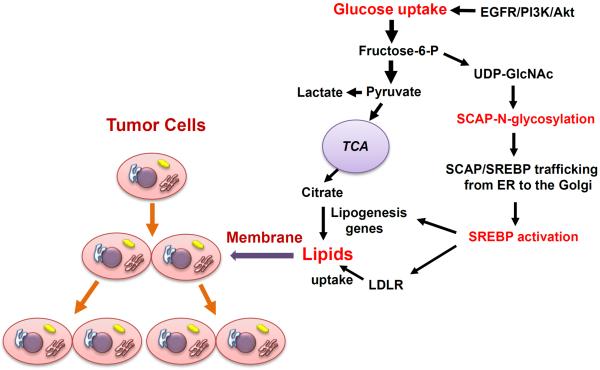
In the last 20 years, Nobel laureates, Brown & Goldstein had developed an elegant model for SREBP regulation where sterols serve as the main regulator of SREBP activation via a negative feedback regulation loop.8 Once sterol levels decrease, SREBP-cleavage activating protein (SCAP) transports SREBPs from the endoplasmic reticulum (ER) to the Golgi for the subsequent proteolytic activation of SREBP.8 Surprisingly, when we started our investigations, our results were completely unexpected and not in line with those published by Brown & Goldstein, as we found that removal of sterols was unable to activate SREBPs when cells were cultured in the absence of glucose.9 Addition of glucose upregulated SCAP, and activated both SREBP-1 and -2.9 These data demonstrated that glucose functions as a critical activator of SREBP function (Figure 1).
Figure 1. Glucose promotes tumor growth as it activates lipid synthesis and uptake upon the N-glycosylation of SCAP.
Glucose-mediated N-glycosylation of SCAP promotes the trafficking of SCAP/SREBP from the ER to the Golgi, leading to subsequent SREBP activation. SREBP acts as a transcription factor to enhance the expression of lipogenesis genes and of the low-density lipoprotein receptor (LDLR), thereby increasing cellular lipids to support tumor growth. Oncogenic EGFR/PI3K/Akt signaling upregulates glucose uptake-SCAP N-glycosylation-SREBP activation pathway to promote tumor growth.
EGFR, epidermal growth factor (EGF) receptor; PI3K, phosphoinositide 3-kinase; UDP-GlcNAc, Uridine diphosphate (UDP)-N-acetylglucosame (GlcNAc); ER, endoplasmic reticulum; LDLR, low density lipoprotein (LDL) receptor. TCA, tricarboxylic acid cycle.
We next explored how glucose activates SREBPs. After uptake by cells, glucose feeds into distinct metabolic pathways, i.e. glycolysis, oxidative phosphorylation and hexosamine synthesis for glycosylation modification. We first examined the effects of the intermediate metabolites in each pathway on SCAP protein levels and SREBP-1 activation in cells cultured in the absence of glucose. We found that only N-acetlyglucosamine (GlcNAc), the intermediate product of the hexosamine pathway serving as a precursor for N-glycosylation, enhanced SCAP protein levels and activated SREBP-1 similarly as glucose stimulation. We then used inhibitors to block the N-glycosylation or O-glycosylation pathways, and found that inhibition of the N-glycosylation pathway abolished glucose-mediated SCAP upregulation and SREBP-1 activation. Together, these data demonstrated that glucose-mediated N-glycosylation is critical for SCAP stability and promotes SCAP/SREBP trafficking to the Golgi leading to SRBEP activation (Figure 1).
We then investigated how glucose-mediated N-glycosylation regulates SCAP/SREBP activation. In 1998, the Brown & Goldstein laboratory had reported that SCAP is modified by N-glycosylation via three asparagine residues.10 Nohturfft and coworkers demonstrated that mutation of 1 or 2 N-glycosylation sites on SCAP had no noticeable effect on SREBP-2 activation.10 Nevertheless, they had not been able to examine the function of the mutated SCAP protein with all three asparagine (NNN) replaced with glutamine (QQQ) because of its instability.10 As we reasoned that glucose-stimulated SREBP activation might mediate by N-glycosylation of SCAP, we decided to revisit this issue. We were very fortunate to have Drs. Brown, Goldstein and Nohturfft kindly providing us with all the SCAP mutant constructs, including the QQQ-SCAP. We sub-cloned the SCAP mutant fragment into a new plasmid with a GFP-tag driven by the CMV promoter, and thus were able to enrich the mutated QQQ-SCAP protein in sufficient amounts to proceed with our study. The data show that loss of all N-glycosylation sites resulted in the inability of SCAP to move to the Golgi and activate SREBP-1 and -2 under sterol deprivation.
We then demonstrated that N-glycosylation of SCAP reduced its association with insulin-induced gene 1 (Insig-1), an ER-resident protein that blocks SCAP/SREBP exit from the ER, and directed trafficking of the SCAP/SREBP complex to the Golgi. Finally, we demonstrated that EGFR signaling activates SREBP-1 via enhancing glucose uptake and SCAP N-glycosylation (Figure 1).
In summary, our study has expanded our understanding of tumor metabolism and identified the intrinsic link between glucose and lipid metabolism. These data suggest that key players in lipid metabolism may serve as effective therapeutic targets for cancer.
Acknowledgments
Funding This work was supported by NIH/NINDS under Awards RO1 NS079701 and R21 NS072838 (DG), American Cancer Society Research Scholar Grant RSG-14-228-01–CSM (DG).
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Ru P, Williams TM, Chakravarti A, Guo D. Tumor metabolism of malignant gliomas. Cancers (Basel) 2013;5:1469–1484. doi: 10.3390/cancers5041469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncology. 2013;2:289–299. doi: 10.2217/cns.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo D, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo D, Cloughesy TF, Radu CG, Mischel PS. AMPK: A metabolic checkpoint that regulates the growth of EGFR activated glioblastomas. Cell Cycle. 2010;9:211–212. doi: 10.4161/cc.9.2.10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo D, Bell EH, Mischel P, Chakravarti A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr Pharm Des. 2014;20:2619–2626. doi: 10.2174/13816128113199990486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo D, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal. 2009;2:ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo D, et al. An LXR agonist promotes GBM cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 2011;1:442–456. doi: 10.1158/2159-8290.CD-11-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Cheng C, et al. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell. 2015;28:569–581. doi: 10.1016/j.ccell.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nohturfft A, Brown MS, Goldstein JL. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. The Journal of biological chemistry. 1998;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]