Summary
Upon viral infection, host cells mount a concerted innate immune response involving type I interferon and pro-inflammatory cytokines to enable elimination of the pathogen. Recently cGAS and STING have been identified as intracellular sensors that activate the interferon pathway in response to virus infection and thus mediate host defense against a range of DNA and RNA viruses. Here we review how viruses are sensed by the cGAS-STING signaling pathway as well as how viruses modulate this pathway. Mechanisms utilized by viral proteins to inhibit cGAS and/or STING are also discussed. On the flip side, host cells have also evolved strategies to thwart viral immune escape. The balance between host immune control and viral immune evasion is pivotal to viral pathogenesis and we discuss this virus-host stand-off in the context of the cGAS-STING innate immune pathway.
Keywords: Innate immunity, cGAS, STING, virus, IFN
Introduction
Innate immunity is the first and most rapid line of host defense against invading microbial pathogens. Host cells initiate innate immune responses upon recognition of conserved pathogen structures, called pathogen associated molecular patterns (PAMPs), as well as host damage associated molecular patterns (DAMPs) by diverse pattern recognition receptors (PRRs). Upon sensing viral PAMPs and DAMPs released during virus infection, signal cascades are activated to produce type I interferon and/or multiple cytokines and chemokines, which culminate in the synthesis of many antiviral proteins. Antiviral proteins can promote growth arrest or apoptosis, and inhibit cellular protein translation. Cytokines and chemokines help recruit immune cells to the site of viral infection in order to control the spread of the virus and to initiate the adaptive immune response to virus infection. PRRs can also induce antiviral proteins without the requirement of cytokine-dependent autocrine/paracrine stimulation.
Pattern recognition receptors (PRRs) are germ-line encoded receptors present either on the cell surface or within specific cellular compartments of the cytosol, and have been extensively studied over the last two decades. PRRs recognize microbial signatures named pathogen associated molecular patterns (PAMPs). PAMPs are usually conserved molecular components essential for pathogen survival such as nucleic acids, lipopolysaccharide (LPS), lipoproteins, bacterial flagellin, and yeast zymosan. PRRs play a significant role in recognizing invading pathogens and mediating the first steps of host defense. Hence, PRRs are extremely important for a rapid and efficient innate immune response. PRRs include sensors such as Toll-like receptors (TLRs), the nucleotide binding and oligomerization, leucine-rich proteins (NLRs), retinoic acid-like receptors (RLRs), absent in melanoma 2 (AIM2)-like receptors (ALRs) and cytosolic DNA receptors (reviewed in (Brubaker et al., 2015; Medzhitov, 2007; Takeuchi et al., 2010)) (Figure 1). In the context of virus infection, the best characterized PAMP is the viral genome itself and/or viral nucleic acids generated during the replication cycle in the host cell, such as single or double-stranded RNA transcripts or DNA.
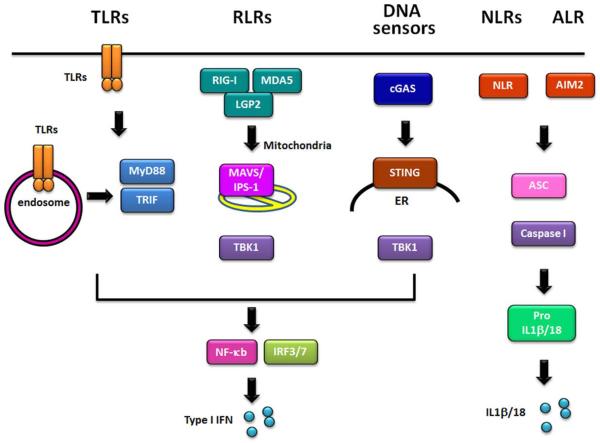
Figure 1. Pattern Recognition Receptors (PRRs) in cells.
Schematic of different PRRs in the cell and the signaling pathways that are activated by each PRR. TLRs are either anchored on the cell surface or are present in endosomal compartments. TLRs utilize myeloid differentiation primary response gene 88 (MYD88) or TIR-domain-containing adapter-inducing interferon (TRIF) as an adaptor protein to recruit downstream molecules, which eventually culminate in the production of pro-inflammatory cytokines and/or type I interferon (IFN). The RLR family comprises of three receptors, RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), each recognizing specific RNA ligands. RLRs signal through the adaptor protein MAVS (also called IPS-1, Cardif, VISA) located on the mitochondria to trigger the production of type I interferons together with NF-κB. NLRs are cytosolic PRRs that contain a nucleotide-binding domain (NBD), an LRR region, and an N-terminal effector which is typically a caspase activation and recruitment domain (CARD) or a pyrin domain (PYD). Several NLR proteins are members of a large complex called the inflammasome, which includes ASC and procaspase I. Inflammasome induction by a number of different PAMPs, results in the activation of IL-1β and IL-18 from pro-IL-1β and pro-IL-18, respectively. AIM2 is a non-NLR family protein which recognizes cytosolic dsDNA. Similar to NLRs, upon stimulation, AIM2 forms an inflammasome with procaspase I and ASC to induce IL-1β and IL-18.
DNA sensing pathways and sensors
Cytosolic DNA sensors are the least characterized among PRRs, although there is consensus that intracellular detection of pathogen DNA is critical for initiating innate immune responses. While downstream molecules such as TANK binding kinase 1 (TBK1) and the transcriptional regulators interferon, IRF3 and IRF7, have been shown as important for cytosolic sensing pathways, it was not until 2008 that STING (also known as TMEM173, MITA, ERIS, MPYS) was identified as acting upstream of TBK1. Subsequent work characterized this STING-TBK1-IRF3 signaling cascade as critical for DNA sensing pathways that were initiated by multiple DNA sensors. STING was identified as an important molecule for cytoplasmic DNA activated innate immune responses (Ishikawa and Barber, 2008; Jin et al., 2008; Sun et al., 2009; Zhong et al., 2008). Loss of STING completely abolished cytosolic DNA-induced IFNβ production. Correspondingly, STING deficient mice showed great susceptibility to herpes simplex virus 1 (HSV-1) infection, suggesting a STING-dependent cytosolic DNA sensing pathway (Ishikawa et al., 2009).
There are many DNA sensors that have been identified to date. Among the DNA sensors, DNA-dependent activator of IFN-regulatory factors (DAI) was the first cytosolic DNA sensor to be identified. It was shown to be important for HSV-1 triggered IFNβ production (Takaoka et al., 2007). However, DAI-deficient mice displayed a normal response against cytosolic DNA stimulation indicating the possibility of redundancy in cytosolic DNA sensing pathways (Ishii et al., 2008). In 2011, DDX41 was identified as another DNA sensor that plays a role in DNA sensing in dendritic cells. Depletion of DDX41 resulted in a reduction of IFNβ production when cells were stimulated with poly dA:dT or poly dG:dC, and infected with HSV or influenza (Zhang et al., 2011b). However, DDX41 knockout animal studies are absent and these might be needed for deciphering the functional role of DDX41 in innate immune pathways. Besides direct sensing, an indirect method of DNA sensing occurs through RNA polymerase III where B-form DNA can be transcribed by RNA polymerase III and recognized by RIG-I resulting in the induction of interferon responses (Ablasser et al., 2009; Chiu et al., 2009).
IFI16 was identified as a DNA sensor that also signals through STING-TBK1. IFI16 was shown to directly bind vaccinia virus (VACV) DNA and recruit STING to induce IFNβ induction. Knockdown of IFI16 using small interfering RNA showed reduced VACV DNA-stimulated IFNβ signaling (Unterholzner et al., 2010). IFI16 was linked to innate immunity against many viral infections and has been shown to be modulated by viruses to facilitate viral replication. For example, IFI16 has been described as a nuclear viral DNA sensor which is important for HSV-1 triggered IRF3 signaling and induction of inflammasome (Johnson et al., 2013; Morrone et al., 2014; Orzalli et al., 2012). IFI16 was shown to be degraded during HSV-1 infection in a proteasome-dependent manner, and HSV-1 ICP0 protein was identified as an IFI16 binding protein that facilitated IFI16 degradation (Diner et al., 2015b; Orzalli et al., 2013). Upon infection with an ICP0 null virus, IFI16 served as a restriction factor for HSV-1 viral replication and gene expression. Interestingly, IFI16 can be acetylated and can accumulate in the cytoplasm (Ansari et al., 2015; Li et al., 2013). Therefore, it is thought that IFI16 can serve as both a nuclear and cytosolic sensor of DNA. In addition, human cytomegalovirus (HCMV) was also shown to trigger IFI16 induced signaling pathways, and its tegument protein pUL83 inhibits this response by interacting with IFI16 (Li et al., 2013). Additionally, IFI16 was shown to play a role in inducing the inflammasome upon KSHV infection (Kerur et al., 2011). IFIX, a new molecule that restricts herpesvirus replication, was reported to detect viral DNA in both the nucleus and cytoplasm, and bind foreign DNA via its HIN domain in a sequence-non-specific manner. Notably, many human PYHIN proteins, such as AIM2, IFI16, and IFIX have been shown to be important regulators of innate immunity (Diner et al., 2015a).
Moreover, additional proteins such as Cyclic-GMP-AMP (cGAMP) synthase (cGAS) [discussed in detail below], DNA-PK, DHX9, DHX36, DDX60, MRE11 have also been reported to play a role in DNA sensing and trigger innate immune responses (Ferguson et al., 2012; Kim et al., 2010; Kondo et al., 2013; Miyashita et al., 2011; Sun et al., 2013; Zhang et al., 2011a). The question arises as to why so many DNA sensors exist and how these DNA sensors coordinate with each other in the host cell. It is possible that some of these sensors function redundantly or there may be cell type specificity for different sensors. For example, some of the sensors were studied in fibroblast or endothelial cells, e.g. cGAS and IFI16, while others were exclusively studied in dendritic cells, such as DDX41, DHX9 and DHX36. Future discoveries addressing this question might unveil novel signaling mechanisms and elucidate regulatory networks of DNA sensing in innate immunity.
STING: regulation and signaling
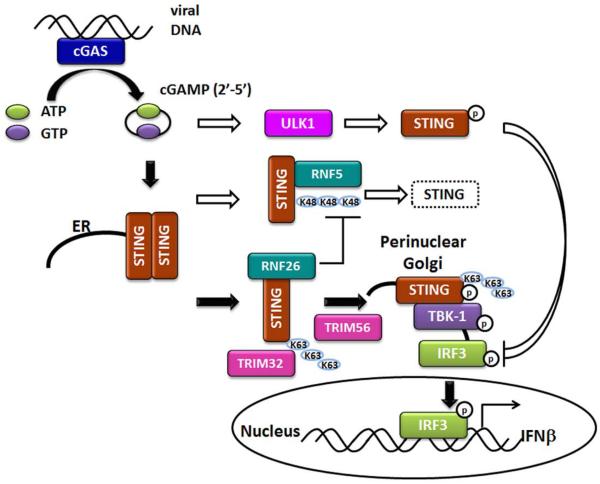
STING is an endoplasmic reticulum (ER) resident membrane protein, and is partially localized to mitochondria and mitochondria-associated membranes (MAMs) through its N-terminal transmembrane domains (Ishikawa et al., 2009). In response to cytosolic DNA, STING dimerizes and translocates to a perinuclear region, which is key for activation of downstream signaling pathways (Ishikawa et al., 2009; Saitoh et al., 2009; Sun et al., 2009). Following STING relocalization, TBK-1 is recruited to STING, leading to the subsequent phosphorylation of STING and IRF-3 by TBK1 (Liu et al., 2015a). This ultimately results in the induction of type I IFN. Interestingly, STING has been shown to directly bind DNA, which suggests that STING can also function as a direct cytosolic DNA sensor. However, physiological conditions of STING sensing DNA and its coordination with other DNA sensors still need to be clarified (Abe et al., 2013).
STING can undergo different modifications for optimal signaling, including ubiquitination and phosphorylation. The ubiquitin ligase TRIM56 was shown to bind STING and mediate K63-linked ubiquitination of STING at lysine 150, which can facilitate STING dimerization as well as binding to TBK1 (Tsuchida et al., 2010). TRIM32 was reported to also function as an E3 ligase of STING capable of adding K63-linked ubiquitin moieties to STING. However, in this study, a K150R mutation in STING only partially reduced TRIM32-mediated STING ubiquitination, and mutation of three additional sites were required to diminish ubiquitination of STING. TRIM32 was shown to interact with STING and is important for STING-TBK1 interaction upon Sendai virus (SeV) or HSV-1 infection (Zhang et al., 2012). STING has also been shown to be negatively regulated by ubiquitin ligase RNF5-mediated K48-linked ubiquitination, which leads to STING degradation. Again, lysine 150 of STING was identified as the target site of RNF5 ubiquitination, which led to STING degradation upon viral infection (Zhong et al., 2009). RNF26 has also been identified as an E3 ligase for K11-linked polyubiquitination of STING at the same K150 residue of STING. RNF26 was shown to attenuate RNF5-directed K48-linked STING ubiquitination at K150, without affecting K63 linked STING ubiquitination, suggesting positive regulation of STING signaling. Curiously, RNF26 also appeared to negatively regulate innate immune signaling in a temporal fashion (Qin et al., 2014). In addition to ubiquitination, STING can also be phosphorylated. ULK1 kinase phosphorylates STING at serine 366 (S366) upon DNA or cGAMP (see below) stimulation, which leads to attenuated IRF3 activation. This suggests a negative-feedback loop is in play to control STING activation (Konno et al., 2013). On the other hand, TBK1 has been shown to phosphorylate STING at this same residue, but positively regulate STING signaling instead (Liu et al., 2015a). This dichotomy suggests that STING is tightly regulated through different mechanisms and additional studies are needed to clarify its regulation.
A role for cGAS-STING in autophagy has also been recently reported. An interaction between cGAS and the autophagy regulator, Beclin-1, represses cGAMP synthesis thereby preventing IFN induction following HSV-1 infection and regulating autophagy. This interaction appears to be an event that limits cGAS activation and prolonged immune stimulation. Furthermore, Akt kinase was shown to phosphorylate cGAS and suppress its activity. During HSV-1 infection, activated Akt kinase leads to a dampening of cGAMP and IFN-β production, resulting in a concomitant increase in HSV-1 replication. Thus, it appears that Akt controls the cGAS-dependent response to HSV-1 (and vaccinia virus) infection (Liang et al., 2014; Seo et al., 2015).
The cGAS-STING pathway
Cyclic-GMP-AMP (cGAMP) synthase (cGAS), the most recently identified cytosolic DNA sensor, catalyzes the production of cGAMP from ATP and GTP upon DNA binding. cGAMP then serves as a second messenger, which binds and activates STING for type I IFN production (Ablasser et al., 2013a; Gao et al., 2013; Sun et al., 2013) (Figure 2). With this capacity for DNA sensing, cGAS plays an important role in type I interferon responses against DNA viruses, including HSV-1 and KSHV (Ma et al., 2015; Sun et al., 2013). cGAS deficient mouse cells fail to respond robustly to DNA virus infection and cGAS deficient mice showed higher viral titers and greater susceptibility to HSV-1, vaccinia virus, and MHV68, compared to wild type mice, (Ablasser et al., 2013a; Schoggins et al., 2014). Recently, cGAS was also shown to be partially nuclear in human fibroblasts and keratinocytes, and to contribute to IFI16 stabilization (Orzalli et al., 2015). These results suggest that IFI16 and cGAS cooperate to sense nuclear herpesviral DNA and initiate innate signaling.
Figure 2. The cGAS-STING dependent DNA sensing pathway.
A detailed overview of the cGAS-STING DNA sensing pathway is depicted. Both positive and negative regulators of STING are shown. Black arrows indicate pathways that lead to activation of STING and induction of the type I IFN response. White arrows indicate pathways that negatively regulate STING. Protein ubiquitination is depicted as K48 or K63 linkages and protein phosphorylation is depicted as P.
Interestingly, while cGAS and STING are clearly important in viral DNA sensing, they both also exhibit important functions in host innate immunity against certain positive sense single-stranded RNA viruses with no DNA intermediates in their life cycle. cGAS knockout mice were more susceptible to infection with West Nile virus (WNV), a positive sense single-stranded RNA virus. It is likely that when cGAS is absent, basal transcript levels of certain antiviral genes are decreased, which makes cells more vulnerable to certain RNA viruses like WNV (Schoggins et al., 2014). Furthermore, a recent study demonstrated that RNA:DNA hybrid molecules were able to bind cGAS and lead to cGAMP production in vitro (Mankan et al., 2014). It appears that the DNA component of the RNA:DNA hybrid is essential for cGAMP production. Additional animal studies may be needed to further address cGAS's role in sensing RNA:DNA hybrid molecules.
Of note, STING deficient mice also show greater susceptibility to RNA virus (e.g. VSV) infection and STING deficient cells fail to mount a strong innate immune response against RNA viruses such as VSV and SeV (Ishikawa et al., 2009). Moreover, the 3' UTR of the flavivirus, HCV, can also trigger a STING-dependent response in hepatocytes (Ding et al., 2013), despite the fact that other groups have shown that STING does not directly bind to the dsRNA mimic, poly I:C (Abe et al., 2013). Coimmunoprecipitation data suggest that STING might contribute to the sensing of RNA viruses through the RNA sensor RIG-I, but not the related RLR protein, MDA-5. These data are also correlated with the fact that STING deficiency does not affect the host response to MDA-5 dependent stimuli such as that following infection with the RNA virus, Encephalomyocarditis virus (EMCV)(Ishikawa and Barber, 2008). Notably, STING does not undergo perinuclear translocation after RNA virus infection or cytosolic RNA stimulation, suggesting a distinct functional model for sensing cytosolic DNA (Ishikawa et al., 2009). In conclusion, cGAS and STING can serve as restriction factors of RNA virus infection, via mechanisms that appear distinct from their role in DNA virus sensing, but the precise mechanisms remain to be further delineated.
Interestingly, one study revealed that STING may be activated by the fusion between viral envelopes and target cells, in a DNA- and RNA-independent event. In this study, either cationic liposomes or herpesvirus-derived virus-like particles with no nucleic acids resulted in the translocation of STING to TBK1-containing perinuclear vesicles and type I interferon production through IRF3 (Holm et al., 2012). The innate response to virus-like particles was dependent on STING and not on TLRs or RLRs. This suggests that STING may provide extra protection to cells by sensing an additional danger signal generated during infection of enveloped RNA or DNA viruses.
Modulation of the cGAS-STING pathway by viruses
A number of viruses are equipped to counteract the cGAS-STING pathway. Table 1 summarizes different viral proteins that inhibit cGAS and STING. The different mechanisms by which DNA and RNA viruses circumvent cGAS and STING activation are described and discussed below.
Table 1.
Viral regulators of the cGAS-STING pathway
| Virus Family | Virus | Viral Protein | Target | Proposed Mechanism | Reference |
|---|---|---|---|---|---|
| Herpes viridae | HSV-1 | ICP0, ICP4, US3-PK | STING | Stabilizes STING in HEp-2 cells, which is required for optimal HSV-1 replication | Kalamvoki et al., 2014 |
| KSHV | vIRF1 | STING | STING interaction; Disrupts STING phosphorylation; Disrupt STING binding to TBK1 | Ma et al., 2015 | |
| ORF52 (KicGAS) | cGAS | cGAS interaction; DNA binding; Disrupts cGAS binding to DNA | Wu et al., 2015 | ||
| LANA | cGAS | N-terminally truncated cytoplasmic isoforms of LANA interact with cGAS | Zhang et al., 2016 | ||
| MHV68 | ORF64 | ? | DUB activity loss results in stronger STING dependent signaling | Sun et al., 2015 | |
| ORF52 | cGAS | cGAS interaction; DNA binding; Disrupts cGAS binding to DNA | Wu et al., 2015 | ||
| RRV | ORF52 | cGAS | cGAS interaction; DNA binding; Disrupts cGAS binding to DNA | Wu et al., 2015 | |
| EBV | ORF52 | cGAS | cGAS interaction; DNA binding; Disrupts cGAS binding to DNA | Wu et al., 2015 | |
| Papilloma viridae | HPV | E7 | STING | STING interaction | Lau et al., 2015 |
| Adeno viridae | Adeno virus | E1A | STING | STING interaction | Lau et al., 2015 |
| Hepadna viridae | Hepatitis B virus | Pol | STING | STING interaction; Disrupts K63-linked polyubiquitination of STING | Liu et al., 2015 |
| Flavi viridae | DENV | NS2B/3 | STING | Binds and cleaves hSTING, but not mSTING | Aguirre et al., 2012; Yu et al., 2012 |
| YFV | NS4B | STING | STING interaction | Ishikawa et al., 2009 | |
| HCV | NS4B | STING | Homolog to STING; STING interaction; Disrupts STING signaling complexes | Nitta et al., 2013; Ding et al., 2013 | |
| Corona viridae | HCoV-NL63 | PLP2-TM | STING | STING interaction; Disrupts dimerization and K63-linked polyubiquitination of STING; Disrupts STING signaling complexes | Sun et al., 2012; Clementz et al., 2010 |
| PEDV | PLP2 | STING | STING interaction; Disrupts K63-linked polyubiquitination of STING | Xing et al., 2013 | |
| SARS-CoV | PLpro-TM | STING | STING interaction; Disrupts dimerization and K63-linked polyubiquitination of STING | Sun et al., 2012;Clementz et al., 2010, Chen et al., 2014 |
DNA viruses
Herpesviridae
While the mechanisms by which herpesviruses evade TLR and NLR signaling are well understood (Gregory et al., 2011; Jacobs and Damania, 2012; West et al., 2012), our understanding of herpesvirus evasion of the cGAS-STING pathway is still in its infancy. HSV-1 is the first DNA virus reported to activate STING in vivo and in vitro and is widely used in experimental systems as an activator of the cGAS-STING pathway. STING deficiency in mice led to lethal susceptibility to HSV-1 infection due to the lack of a successful type I interferon response. Kalamvoki and Roizman reported that STING, while critical for cellular innate immunity against HSV-1, is also necessary for HSV-1 replication in certain cell types. While HSV-1 WT virus did not degrade STING in HEp-2 or HeLa cells, an ICP0 or ICP4 deletion mutant of HSV-1 induced STING degradation. Consistent with this, the replication of an ICP0 deleted HSV-1 was attenuated in STING knockdown cells suggesting that STING is important for optimal virus replication under this circumstance (Kalamvoki and Roizman, 2014). However, in human embryonic lung cells, STING was not degraded during infection with either WT or ICP0 deletion viruses and STING depletion in these cells allowed for increased replication of both WT and ICP0 deletion viruses (Kalamvoki and Roizman, 2014). This suggests that the effects of HSV-1 on STING are cell type and context dependent (Kalamvoki and Roizman, 2014).
Kaposi's sarcoma-associated herpesvirus (KSHV) infection has also been reported to activate the cGAS-STING pathway. Knockdown of cGAS or STING in endothelial cells suppressed IFNβ production during KSHV primary infection or during reactivation from latency, which also led to higher KSHV viral gene transcription and viral genome copy number compared to control endothelial cells (Ma et al., 2015). A cGAS-STING screen using ninety KSHV open reading frames (ORFs) was conducted and multiple KSHV viral proteins were found to inhibit cGAS-STING dependent IFNβ promoter activation and IFNβ protein production. The KSHV viral interferon regulatory factor 1 (vIRF1) inhibited cGAS-STING dependent IFNβ induction. vIRF1 was shown to interact with STING and prevent STING from binding to TBK1 (Ma et al., 2015). This led to the inhibition of STING phosphorylation resulting in an overall inhibition of the DNA sensing pathway. Concordantly, knockdown of vIRF1 in the context of KSHV infection led to higher levels of IFNβ induction (Ma et al., 2015). Another KSHV study focused on identifying inhibitors of cGAS (Wu et al., 2015). In this study, ORF52 (also known as KicGAS, KSHV inhibitor of cGAS), an abundant gammaherpesvirus-specific tegument protein, was shown to inhibit cytosolic DNA sensing by directly inhibiting cGAS enzymatic activity through a mechanism involving both cGAS binding and DNA binding. Infection with an ORF52 deletion mutant resulted in higher levels of IRF3 phosphorylation compared to WT KSHV, suggesting that the loss of ORF52 could not protect the virus against host innate immune responses. Importantly, a similar inhibitory effect of ORF52 homologs in the related herpesviruses, MHV68, RRV, and EBV, on the cGAS pathway was observed, implying a conserved mechanism among these viruses (Wu et al., 2015). In addition, N-terminally truncated cytoplasmic isoforms of KSHV latency associated nuclear antigen (LANA) can interact with cGAS and antagonize cGAS-STING dependent signaling, thereby facilitiating the reactivation of KSHV from latency (Zhang et al., 2016). Finally, in murine gammaherpesvirus 68 (MHV68), it was shown that another gene, ORF64, could inhibit type I interferon production in response to STING activation. A MHV68 ORF64 deletion virus displayed a higher STING-dependent innate immune response, identifying a potential role of ORF64 in negatively regulating STING (Sun et al., 2015). Overall, these studies reveal that herpesviruses encode multiple cGAS-STING antagonists utilizing different mechanisms to ensure the successful evasion of host innate immunity. Identification of the strategies these viral proteins use to evade cGAS and STING may be useful for discovering new modulators of this pathway in other viruses as well.
Hepadnaviridae
The Hepatitis B virus (HBV) polymerase was found to be the only HBV-encoded protein that inhibited STING-triggered IFNβ promoter activation (Liu et al., 2015b). Further dissection revealed that either the reverse transcriptase (RT) or the RNase H (RH) domains of the HBV polymerase protein was sufficient to suppress STING signaling. HBV polymerase bound to STING and attenuated K63-linked polyubiquitination and function, without affecting STING protein levels (Liu et al., 2015b).
Papillomaviridae and Adenoviridae
Human papillomavirus (HPV) is a ubiquitous DNA tumor virus. A recent study provided the link between the HPV oncoprotein E7 and the cGAS-STING pathway. HPV E7 was identified as a potent inhibitor of the DNA, but not RNA, activated antiviral response. E7 bound to STING, and the E7 LXCXE motif that is involved in Rb-binding was also necessary for antagonizing cGAS-STING activation. Furthermore, the absence of E7 resulted in a significant induction of type I interferon (Lau et al., 2015). In the same study, adenovirus E1A was also shown to function as a STING antagonist. E1A also bound to STING to block its activity. Like E7, E1A also contains an LXCXE motif, which was necessary for antagonizing DNA sensing (Lau et al., 2015). This is in agreement with another study demonstrating that the cGAS/STING pathway is critical for type I interferon induction in response to all serotypes of adenovirus tested (Lam and Falck-Pedersen, 2014). However, Lam and Falck-Pedersen did not find evidence for suppression of cGAS/STING-dependent TBK1/IRF3 activation at least up to 6 hours post-adenovirus infection. Moreover, viral replication did not seem to be affected by the lack of cGAS or STING in Hela or THP1 cells at either 6 hours or 24 hours (Lam and Falck-Pedersen, 2014). Interestingly, E1A has been shown to block STAT1 activation after IFNβ is produced, which might explain why adenovirus can replicate without being affected by upstream cGAS/STING dependent signaling, as long as E1A is expressed (Look et al., 1998). Nevertheless, the finding that E1A acts as a STING antagonist suggests another step of the pathway where adenovirus can interfere with the innate immune response, which may further ensure adenoviral evasion of the host antiviral response.
RNA viruses
Despite the fact that the role of cGAS-STING in sensing of DNA viruses is more clearly elucidated than for RNA viruses, many RNA viruses have been shown to be restricted in either a cGAS and/or STING dependent manner and RNA viral proteins can function as antagonists of the cGAS-STING pathway. Interestingly, in some cases, cGAS-STING seems to restrict virus infection independent of type I interferon. For example, type I IFN is only partially blocked upon dengue virus (DENV) infection (Aguirre et al., 2012).
Flaviviridae
The non-structural protein NS4B of yellow fever virus (YFV) was shown to colocalize and interact with STING and block STING and RIG-I dependent signaling. Bioinformatic analyses indicate that the NS4B proteins from YFV, DENV, and hepatitis C virus (HCV) display certain regions with limited homology to STING (Ishikawa et al., 2009). As a result of this initial study, two other groups found that HCV NS4B disrupted the STING signaling complex and NS4B could bind to both overexpressed and endogenous STING. This interaction was further supported by the observation that STING colocalized with NS4B although NS4B did not affect endogenous STING protein levels or STING oligomerization (Ding et al., 2013; Nitta et al., 2013). Each group proposed a different mechanism for STING inhibition. One paper reported that the HCV NS4B colocalization with STING in the ER attenuated the STING-TBK1 interaction (Ding et al., 2013), and the other paper suggested that NS4B and STING colocalization in the mitochondrial associated membranes (MAM) where MAVS and STING are colocalized, impaired the STING-MAVS interaction (Nitta et al., 2013). Irrespective of mechanism, the two studies identified a viral protein mimic of STING that disrupts optimal signaling of this pathway by interfering with protein-protein interactions.
Flaviviruses have evolved distinct mechanisms to block STING activation. The discovery of DENV NS2B/3, which consists of the non-structural proteins NS3 and its cofactor NS2B, as an antagonist of type I IFN production, was reported before STING was fully characterized (Rodriguez-Madoz et al., 2010). However, it was not until 2012 that the mechanism of inhibition was linked to its ability to bind and cleave STING independently (Aguirre et al., 2012; Yu et al., 2012). STING cleavage and degradation were specifically observed in cells with ectopically expressed NS2B/3, or endogenously expressed NS2B/3 upon DENV infection. However, catalytically inactive NS2B/3 was not able to cleave STING, which correlated with the previous finding that protease activity is required for type I IFN inhibition. NS2B/3 was found to target the residues 93–96 (LRRG) of human STING, a sequence that is not found in murine STING (Aguirre et al., 2012; Yu et al., 2012). Consequently, NS2B/3 was neither able to cleave murine STING, nor block IFNβ production, which might explain why murine cells are not very susceptible to DENV (Aguirre et al., 2012; Yu et al., 2012).
Coronaviridae
Human coronavirus (HCoV) NL63 has been recently described as an inhibitor of type I IFN production through STING. Specifically, the membrane anchored PLP domain (PLP2-TM) protein colocalized and interacted with STING at the endoplasmic reticulum (ER) (Sun et al., 2012). PLP2-TM disrupted STING dimerization and STING-TBK1 interaction. In addition, PLP2-TM attenuated K63-linked polyubiquitination of STING, which does not necessarily require PLP2-TM's deubiquitinase (DUB) activity. Therefore, the insufficient STING polyubiquitination might be due to PLP2-TM's ability to disrupt STING binding to TBK1 rather than its DUB activity (Clementz et al., 2010; Sun et al., 2012). Interestingly, the PLP2 protein of porcine epidemic diarrhea virus (PEDV) has also been shown to interact with STING and repress K63-linked polyubiquitination of STING (Xing et al., 2013). However, the DUB activity of PEDV PLP2 seems to be required for inhibition. The difference between human and porcine viruses needs further clarification.
SARS-CoV has been also reported to suppress type I IFNβ production. One of the SARS-CoV proteins, papain-like protease (PLpro), was shown to inhibit STING activation (Clementz et al., 2010; Sun et al., 2012). The transmembrane domain of PLpro was co-immunoprecipitated with STING, as well as with the binding partners of STING such as RIG-I, TBK1, and IRF3. Similar to HCoV NL63, PLpro also negatively affected STING dimerization and K63-linked polyubiquitination, suggesting a conserved function of papain-like protease domain containing proteins from the Coronaviridae family (Clementz et al., 2010; Sun et al., 2012).
Host defense strategies: Utilization of innate sensors to counter viral infection
Host cells have also developed strategies to counter viral immune evasion mechanisms. For example, STING was identified in both exosomes and virions from HSV-1 infected cells, resulting in STING being transferred to adjacent cells (Kalamvoki et al., 2014). Thus, STING packaged into virions may allow the host cell to limit cell to cell spread of herpesviruses.
Similarly, cGAMP, the second messenger which binds to STING, was shown in two independent studies to be incorporated into viral particles from lentiviruses and herpesviruses. These virions transferred cGAMP to newly infected cells and induced STING-dependent antiviral responses (Bridgeman et al., 2015; Gentili et al., 2015). Distinct from previously reported STING transfer, these effects were independent of exosomes and viral nucleic acids, suggesting a novel mode by which the host protects itself from the virus. Most importantly, this also implies that cGAMP can initiate innate immune signaling without the need for cGAS activation, which would allow for a faster and more efficient host response against viral infection (Bridgeman et al., 2015; Gentili et al., 2015). Although it has been previously been reported that cGAMP can be transferred to bystander cells independent of virus infection (Ablasser et al., 2013b), these findings indicate a more specific host strategy that is utilized against virus infection by only targeting the de novo infected cell. Induction of a second messenger like cGAMP is also important under conditions where viruses inhibit the translation of host proteins e.g. IFNβ, since cGAMP can be transmitted to neighboring cells in lieu of IFNβ and elicit innate immune responses from neighboring cells.
Conclusions
In summary, while host cells have developed various strategies to detect pathogens and initiate robust immune responses, viruses are also evolving over time to modulate or escape the host innate immune system. While there is still a lot to be uncovered with respect to cGAS and STING function within an infected cell, the many different immune evasion strategies employed by both RNA and DNA viruses are indicative of the importance of this pathway for sensing viral pathogens. Some of the RNA and DNA viruses use similar mechanisms to inhibit STING-TBK1 interactions and STING function, while others use more distinct mechanisms involving STING cleavage and degradation. Currently, only STING antagonists from positive-sense RNA viruses have been discovered but not negative-sense RNA viruses. Future identification of viral inhibitors from the latter group will be highly informative. Notably, mechanisms of cGAS inhibition remain to be further elucidated. Both RNA and DNA viruses that exhibit either acute lytic infection or establish lifelong persistent, latent infections in the host were found to target the cGAS-STING pathway, indicating that this immune response is likely important for both lytic and latent phases of the viral lifecycle. In conclusion, a better understanding of the battle between the virus and the human host will most certainly benefit the prevention and treatment of all viral diseases.
Acknowledgements
Due to space limitations, we sincerely apologize for not being able to cite all papers related to this topic. We thank Damania lab members for helpful discussions. BD is supported by NIH grants DE018281, AI107810, AI109965, CA019014 and CA096500. BD is a Leukemia & Lymphoma Society Scholar and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease.
References
- Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. STING recognition of cytoplasmic DNA instigates cellular defense. Molecular cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature immunology. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013a;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013b;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS pathogens. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari MA, Dutta S, Veettil MV, Dutta D, Iqbal J, Kumar B, Roy A, Chikoti L, Singh VV, Chandran B. Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-beta Responses. PLoS pathogens. 2015;11:e1005019. doi: 10.1371/journal.ppat.1005019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgeman A, Maelfait J, Davenne T, Partridge T, Peng Y, Mayer A, Dong T, Kaever V, Borrow P, Rehwinkel J. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 2015;349:1228–1232. doi: 10.1126/science.aab3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annual review of immunology. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, et al. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. Journal of virology. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, Cristea IM. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Molecular systems biology. 2015a;11:787. doi: 10.15252/msb.20145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Lum KK, Javitt A, Cristea IM. Interactions of the Antiviral Factor Interferon Gamma-Inducible Protein 16 (IFI16) Mediate Immune Signaling and Herpes Simplex Virus-1 Immunosuppression. Molecular & cellular proteomics : MCP. 2015b;14:2341–2356. doi: 10.1074/mcp.M114.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Cao X, Lu J, Huang B, Liu YJ, Kato N, Shu HB, Zhong J. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. Journal of hepatology. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M, Kowal J, Tkach M, Satoh T, Lahaye X, Conrad C, Boyron M, Lombard B, Durand S, Kroemer G, et al. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 2015;349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- Gregory SM, Davis BK, West JA, Taxman DJ, Matsuzawa S, Reed JC, Ting JP, Damania B. Discovery of a viral NLR homolog that inhibits the inflammasome. Science. 2011;331:330–334. doi: 10.1126/science.1199478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nature immunology. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Damania B. NLRs, inflammasomes, and viral infection. Journal of leukocyte biology. 2012;92:469–477. doi: 10.1189/jlb.0312132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Molecular and cellular biology. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. Journal of virology. 2013;87:5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Du T, Roizman B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4991–4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Roizman B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E611–617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell host & microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Falck-Pedersen E. Unabated adenovirus replication following activation of the cGAS/STING-dependent antiviral response in human cells. Journal of virology. 2014;88:14426–14439. doi: 10.1128/JVI.02608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell host & microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell host & microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015a;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu Y, Li J, Chen J, Li Y, Wang W, Du X, Song W, Zhang W, Lin L, Yuan Z. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. Journal of virology. 2015b;89:2287–2300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look DC, Roswit WT, Frick AG, Gris-Alevy Y, Dickhaus DM, Walter MJ, Holtzman MJ. Direct suppression of Stat1 function during adenoviral infection. Immunity. 1998;9:871–880. doi: 10.1016/s1074-7613(00)80652-4. [DOI] [PubMed] [Google Scholar]
- Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E4306–4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. The EMBO journal. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. TLR-mediated innate immune recognition. Seminars in immunology. 2007;19:1–2. doi: 10.1016/j.smim.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Molecular and cellular biology. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E62–71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, Kiyohashi K, Murakawa M, Nishimura-Sakurai Y, Azuma S, et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology. 2013;57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1773–1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4492–4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3008–3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, Zhong B, Shu HB. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS pathogens. 2014;10:e1004358. doi: 10.1371/journal.ppat.1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, Fernandez-Sesma A. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. Journal of virology. 2010;84:9760–9774. doi: 10.1128/JVI.01051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Yang A, Tan B, Kim S, Liang Q, Choi Y, Yuan W, Feng P, Park HS, Jung JU. Akt Kinase-Mediated Checkpoint of cGAS DNA Sensing Pathway. Cell reports. 2015;13:440–449. doi: 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, et al. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol. 2015;194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, Clementz MA, Banach BS, Li K, Baker SC, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PloS one. 2012;7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Fujiwara N, Ido A, Oono M, Takeuchi Y, Tateno M, Suzuki K, Takahashi R, Tooyama I, Taniguchi N, et al. Induction of protective immunity by vaccination with wild-type apo superoxide dismutase 1 in mutant SOD1 transgenic mice. Journal of neuropathology and experimental neurology. 2010;69:1044–1056. doi: 10.1097/NEN.0b013e3181f4a90a. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Gregory SM, Damania B. Toll-like receptor sensing of human herpesvirus infection. Frontiers in cellular and infection microbiology. 2012;2:122. doi: 10.3389/fcimb.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, et al. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell host & microbe. 2015;18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, Baker SC, Feng L, Chen Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. The Journal of general virology. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS pathogens. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. The Journal of biological chemistry. 2012;287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011a;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011b;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]