Abstract
Proteome misfolding and/or aggregation, caused by a thermal perturbation or a related stress, transiently challenges the cellular protein homeostasis (proteostasis) network capacity of cells by consuming chaperone / chaperonin pathway and degradation pathway capacity. Developing protein client-based probes to quantify the cellular proteostasis network capacity in real time is highly desirable. Herein we introduce a small-molecule-regulated fluorescent protein folding sensor based on a thermo-labile mutant of the de novo designed retroaldolase (RA) enzyme. Since RA enzyme activity is not present in any cell, the protein folding sensor is bioorthogonal. The fluorogenic small molecule was designed to become fluorescent when it binds to and covalently reacts with folded and functional RA. Thus, in the first experimental paradigm, cellular proteostasis network capacity and its dynamics is reflected by RA-small molecule conjugate fluorescence, which correlates with the amount of folded and functional RA present, provided that pharmacologic chaperoning is minimized. In the second experimental scenario, the RA-fluorogenic probe conjugate is pre-formed in a cell by simply adding the fluorogenic probe to the cell culture media. Unreacted probe is then washed away before a proteome misfolding stress is applied in a pulse-chase type experiment. Insufficient proteostasis network capacity is reflected by aggregate formation of the fluorescent RA-fluorogenic probe conjugate. Removal of the stress results in apparent RA-fluorogenic probe conjugate refolding, mediated in part by the heat-shock response transcriptional program augmenting cytosolic proteostasis network capacity, and in part, by time dependent RA-fluorogenic probe conjugate degradation by cellular proteolysis.
Keywords: Fluorogenicity, Folding probe, folding sensor
INTRODUCTION
Largely unfolded polypeptides emerging from the ribosome after translation need to properly fold into native three-dimensional structures to perform their physiological functions.1,2 The folding of the proteome within a cell is assisted by the protein homeostasis (proteostasis) network, comprising macro-molecular chaperones and co-chaperones, chaperonins and co-chaperonins, protein degradation machinery and their cellular regulators, as well as other components.3,4 Stresses, such as heat, transiently impair cellular proteostasis capacity through global proteome misfolding and/or aggregation-mediated binding of proteostasis network components, which consumes proteostasis network capacity. A sustained imbalance of proteostasis network capacity can lead to diseases, such as neurodegenerative disorders, cardiomyopathy, and cancers.5–9 Pharmacologic manipulation of proteostasis network capacity is emerging as a therapeutic strategy to ameliorate these diseases.4,10–20
Developing probes to sense the dynamics of cellular proteostasis network capacity in real time within cells is highly desirable, but challenging. Pioneering reports21,22 demonstrate that thermo-labile firefly luciferase mutants can be used as protein client-based sensors to report on the dynamics of cellular proteostasis network capacity. Since proper folding of luciferase mutants (the client) requires sufficient proteostasis network assistance, the solubility of luciferase has been used to quantitatively reflect cellular proteostasis network capacity. Solubility changes upon stress can be directly visualized by imaging the spatio-temporal features of luciferase aggregation in live cells.
This approach requires fusion of a fluorescent protein to the luciferase mutant to visualize its aggregation. Furthermore, global inhibition of protein translation is necessary to avoid contributions from the newly synthesized luciferase-fluorescent protein fusion. A complementary approach would be to use a fluorescence turn-on (fluorogenic)23–26 chemical probe that rapidly binds to and reacts with the folded and functional fraction of a metastable client protein, rendering the conjugate fluorescent.27 Such chemical regulation to turn on the fluorescence signal from the properly folded fraction of the metastable client protein at any desired time point also enables pulse-chase type experiments to study proteostasis network capacity in real time.28–30.
Herein, we introduce a mutant of the de novo designed retroaldolase enzyme27,31 (RA, 29 kDa, Figure 1a) as a thermo-labile client protein. RA is bioorthogonal because it has an enzyme function not shared by any endogenous cellular enzymes, thus its enzymatic activity or lack thereof is not expected to perturb cellular functions.32,33 The RA mutant, RAm1 (E10K:D120V:N124S:L225P),27 is thermo-labile, i.e., it exhibits a loss of function upon thermal stress (heating).
Figure 1.
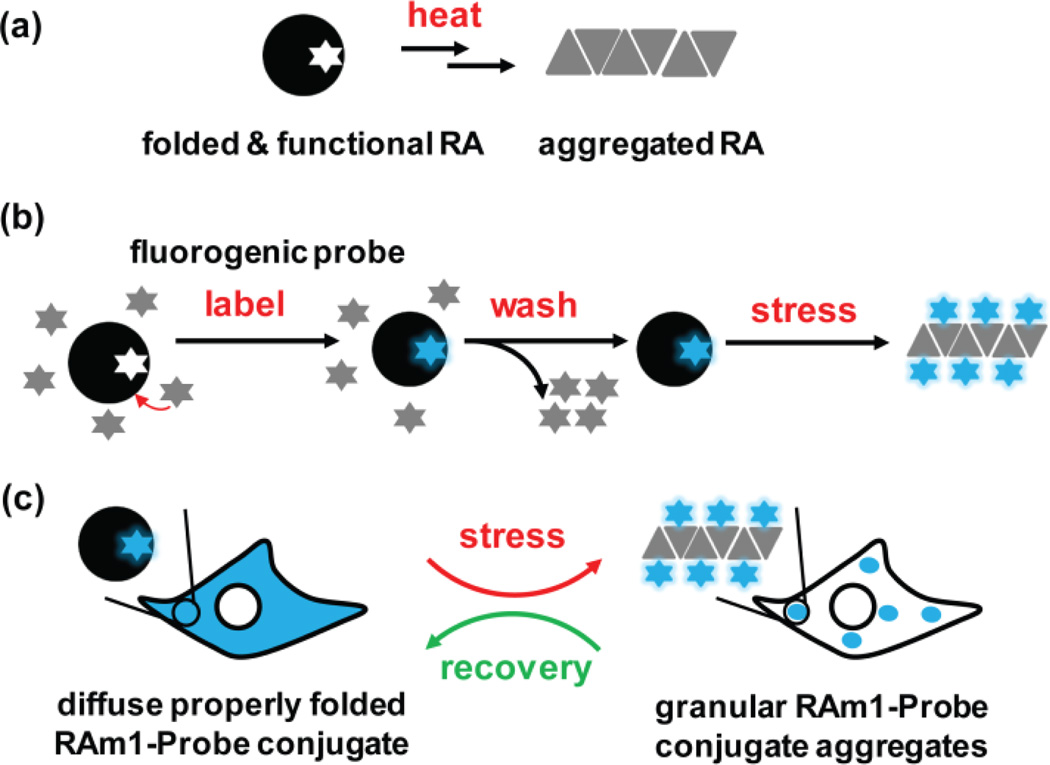
A cellular metastable client protein acts as a proteostasis network capacity sensor. (a) Under physiological conditions, a thermo-labile de novo designed retroaldolase (RA) will be largely folded and functional in the absence of stress, but upon heating, will form aggregates that consume proteostasis network capacity. (b) Folded and functional RA is labeled by a fluorogenic small molecule probe, rendering the covalent conjugate fluorescent. Conjugate fluorescence is retained upon aggregation due to the covalent modification and the chromophore utilized. (c) Preformed RAm1-P1 conjugate can undergo misfolding and aggregation into an aggregated granular state upon application of a heat stress, serving as a sensor of cellular proteostasis network capacity.
We designed a small molecule fluorogenic probe P1 (Figure 1b, 2c) that binds to and reacts with the folded and functional RA/RAm1 fraction selectively, rapidly rendering the conjugate fluorescent and thus reporting quantitatively on the concentration of folded and functional RA/RAm1. Two types of experiments are possible using the thermo-labile RAm1 client protein in combination with the fluorogenic probe P1.
Figure 2.
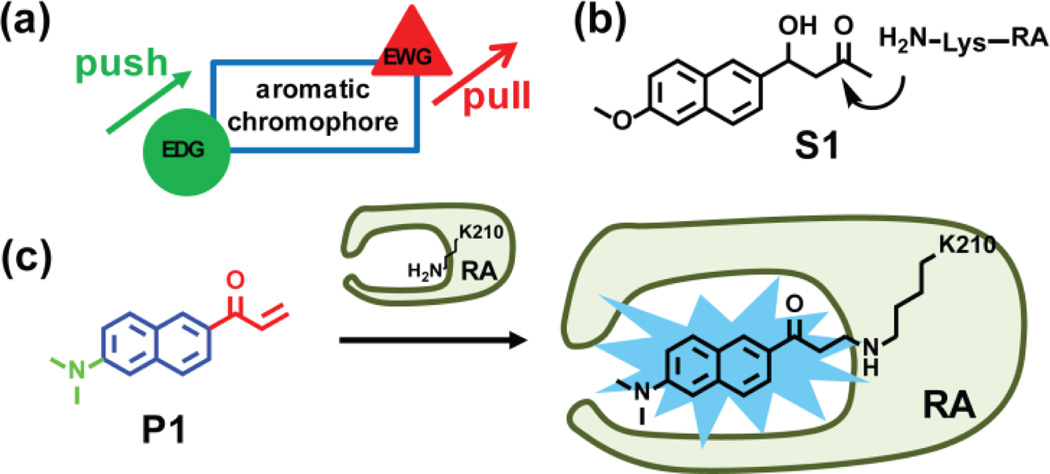
Structure-based design of a fluorogenic probe for folded and functional RA. (a) Schematic of a push-pull environmentally-sensitive fluorophore. EWG = electron-withdrawing group. EDG = electron-donating group. (b) Structure of the retroaldol substrate S1 utilized by the de novo designed RA enzyme.29 RA catalyzes a retroaldol reaction using the pKa-perturbed lysine-210 ε-amine side chain that forms a Schiff base with S1. (c) P1 is a push-pull environmentally-sensitive fluorophore featuring a reactive vinyl ketone (in red) that also serves as an electron-withdrawing group. The dimethyl amino group (in green) serves as an electron-donating group. P1 covalently modifies the pKa-perturbed lysine-210 residue of RA through 1,4-conjugate addition, rendering the RA-P1 covalent conjugate fluorescent.
In the first experimental scenario, a stress is applied before the changes in the folding and function of the thermo-labile RAm1 are probed by P1—the amount of the RA-P1 conjugate formed reports on the alteration of cellular proteostasis network capacity upon thermal stress via the amount of conjugate fluorescence observed when compared to non-stressed control cells. In this scenario, one has to be mindful that probe binding and reaction with RAm1 could change the folded fraction via a pharmacologic chaperoning mechanism.27 In the second experimental paradigm employed predominantly in this paper, P1 is applied to cells for a pulse labeling period, allowing for formation of the RAm1-P1 conjugate. Unreacted P1 is then washed away from the cell media before a stress is applied, and then the fate of the RAm1-P1 conjugate can be monitored in a pulse-chase type experiment. In this scenario, direct visualization of thermal stress in live cells is achieved by imaging the misfolding and aggregation of the preformed RAm1-P1 conjugate vs maintenance of the RAm1-P1 folded and functional state via continuous RAm1-P1 refolding (Figure 1c). In this context, the aggregated RAm1-P1 conjugate, appearing as puncta, remains fluorescent. No fluorescent protein fusion to RA or global inhibition of translation is required for these experiments. Post-stress cellular refolding of the RAm1-P1 conjugate was observed during the recovery period. We also demonstrate the slow, time-dependent cellular degradation of the RAm1-P1 conjugate in the absence of or in the presence of stress in these scenario 2 pulse-chase type experiments. The bipartite thermo-labile RAm1 client protein-P1 fluorogenic probe sensor developed herein appears to be a practical chemical-biological tool to further explore the impact of various cellular stresses on cellular proteostasis network capacity in real time.
RESULTS AND DISCUSSION
Structure-based design of a fluorogenic probe for a metastable retroaldolase
Multiple strategies have been explored to fashion a small molecule that is selective enough to make only one folded protein fluorescent after binding and reacting with it.25,30,34–38 We took advantage of the TIM-barrel structure of RA with its catalytic pKa-perturbed Lys-210 residue buried inside its relatively hydrophobic active site to design a fluorogenic probe35,37–39 that binds and rapidly reacts with only the folded and functional fraction of RA. Environmentally-sensitive push-pull fluorophores, featuring an electron-donating group (EDG) and an electron-withdrawing group (EWG) attached to an aromatic chromophore (Figure 2a), usually are dark in buffer, but fluoresce upon binding to a hydrophobic pocket. In general, a functional group is attached to the environmentally-sensitive fluorophore that keeps the chromophore dark until it reacts with the protein-of-interest.40,41
We proposed a Lys-210 chemoselective fluorogenic probe for RA based on the chemical structure of RA’s retroaldol substrate S1 (Figure 2b). We retained the naphthalene ring (Figure 2c, substructure in blue) responsible for binding selectivity, but converted the β-hydroxy ketone substrate into an electron-withdrawing vinyl ketone (Figure 2c, substructure in red), a Michael acceptor that was envisioned to be reactive towards the pKa-perturbed Lys-210 residue of RA. The alkene substructure typically undergoes photoisomerization after the attached chromophore is excited, keeping the chromophore dark until it reacts with RA, eliminating this non-emissive relaxation pathway.36 Further, we substituted the methoxy group in S1 with a dimethyl amino group (Figure 2c, substructure in green), which is a stronger electron-donating group and one that is compatible with longer wavelength fluorescence.42 Thus we envisioned the probe P1 (Figure 2c), a push-pull type fluorophore that should covalently modify the pKa-perturbed Lys-210 residue of the folded and enzymatically active RA, rendering the RA-P1 conjugate fluorescent. Surprisingly, a literature search revealed that P1, an irreversible inhibitor of RA, was commercially available and is named acrylodan, an environmentally sensitive fluorophore that becomes fluorescent upon slowly reacting with cysteine thiols.43
P1 is a chemoselective and fluorogenic probe for folded and functional RA
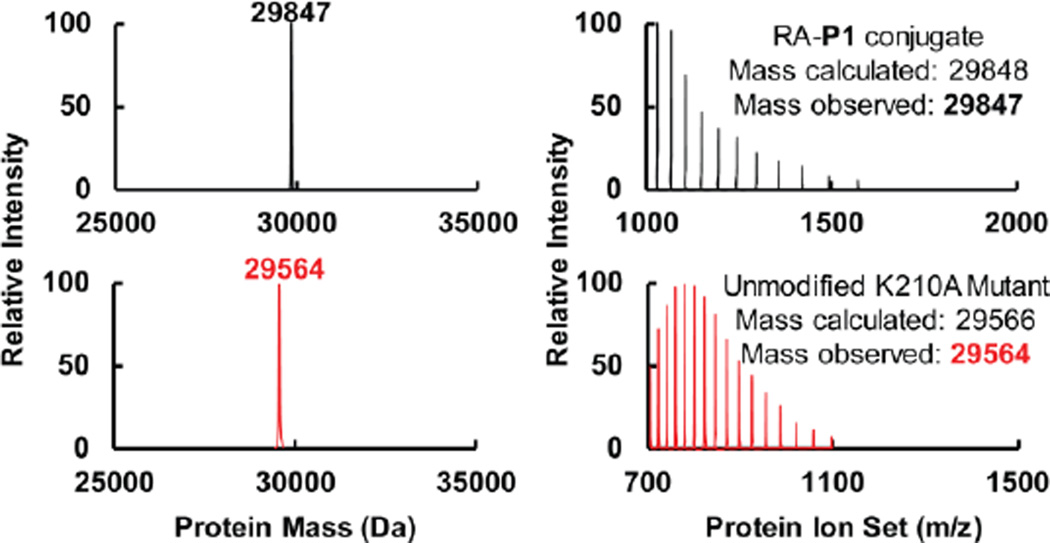
To determine whether P1 is capable of chemoselectively modifying folded and functional RA, we incubated P1 with RA and RA harboring a K210A mutation in buffer, and monitored the covalent modification efficiency by LC-ESI-MS (Figure 3). We found that P1 (50 µM) completely labeled RA (5 µM) at 25 °C within 5 min. The conjugate mass was observed at 29847 Da (apo-RA: 29623 Da, P1: 225 Da). In contrast, folded K210A RA was not modified by P1, providing strong evidence that P1 chemoselectively modifies the pKa-perturbed Lys-210 residue. The excess amount of P1 (50 µM) resulted in no additional RA modification.
Figure 3.
P1 labels the active site Lys-210 residue of RA chemoselectively. P1 (50 µM) completely labeled RA (5 µM) within 5 min at 25 °C, as shown by LC-ESI-MS. The conjugate mass was observed at 29847 Da (apo-RA: 29623 Da, P1: 225 Da) (top panels). Mutation of the active site Lys-210 residue to alanine eliminated the covalent labeling of RA by P1. The unmodified RA K210A mutant mass was observed at 29564 Da (bottom panels).
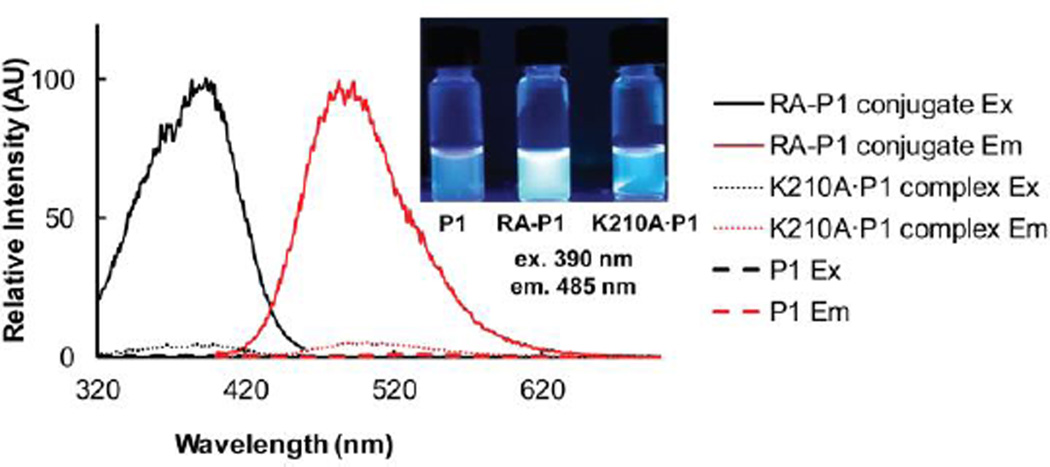
We next explored the fluorogenicity of P1 after reacting with RA. P1 (5 µM) exhibited fluorescence only after binding and reacting with RA (5 µM), whereas P1 alone was dark in buffer (Figure 4). Moreover, P1 was only very weakly fluorescent when it bound to the K210A RA mutant, indicating that the binding of P1 to the RA binding pocket does not contribute significantly to the emergence of the fluorescence. The quantum yields of P1 and the RA-P1 conjugate in buffer were measured respectively as 0 and 0.17, using quinine sulfate as the reference. Importantly, we observed nearly identical conjugate fluorescence resulting from the reaction between P1 and the thermo-labile mutant RAm1 (Figure S1). The conjugate fluorescence seems to arise from the covalent reduction of the double bond of P1 by way of the attack of the pKa-perturbed Lys-210 primary amine of RA and RAm1, eliminating double bond isomerization as a fluorescence quenching mechanism.36
Figure 4.
P1 is fluorogenic upon binding and reacting with RA to form a covalent conjugate. P1 (5 µM) was incubated with RA (5 µM) or the K210A RA mutant (5 µM) for 24 h in buffer at 25 °C resulting in complete covalent modification of RA, except in the case of the K210A mutant which forms a noncovalent K210A RA•P1 complex. P1 (5 µM) is dark in buffer and is only very weakly fluorescent when bound to the K210A RA mutant. In contrast, P1 is strongly fluorescent upon forming a covalent conjugate with RA. Excitation and emission spectra were recorded using an Aviv fluorescence spectrometer. Samples in the inset were photographed under illumination with a hand-held UV lamp.
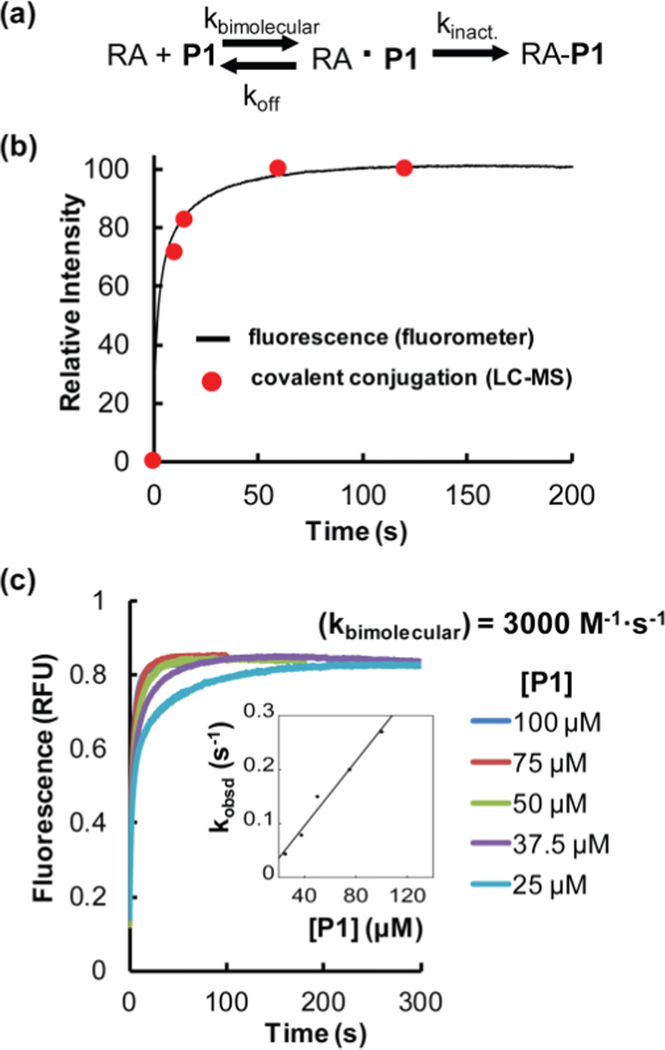
We further scrutinized the origin of P1’s fluorogenicity using a kinetic analysis. P1 binding and reaction with RA can be modeled as a two-step process: pre-equilibrium binding followed by a covalent chemical labeling step (Figure 5a). To examine whether the reversible binding step and/or the irreversible covalent conjugation step is responsible for the observed fluorescence, we recorded and compared the kinetics of covalent conjugation and fluorescence emergence after mixing RA (5 µM) and P1 (50 µM) at 25 °C. The covalent conjugation time course (Figure 5b, red filled circles) was derived from the quantitation of the relative peak intensities of RA-P1 conjugate formation by LC-ESI-MS (Figure S2). The kinetics of fluorescence emergence was generated by recording the fluorescence increase as a function of time using a stopped-flow fluorometer (Figure 5b, black curve). The overlap of these two kinetic curves indicates that the fluorogenicity originates from covalent conjugation, consistent with the lack of fluorescence of the K210A RA-P1 complex (Figure 4). Therefore, the emergence of significant fluorescence requires a chemical reaction between P1 and RA.
Figure 5.
The folded and functional RA-P1 conjugate is fluorescent. (a) RA and P1 conjugation is modeled to proceed by two steps: P1 first binds reversibly to RA. Then the RA1•P1 complex undergoes a reaction to form the fluorescent conjugate. (b) RA (5 µM) was incubated with P1 (50 µM) at 25 °C. The fraction of covalent modification by LC-ESI-MS (red filled circles) correlates with the emergence of fluorescence (black curve) measured by stopped-flow fluorometry using an excitation wavelength of 390 nm and emission wavelength of 485 nm. The extent of covalent modification (second step; red filled circles) was monitored by taking samples from the reaction mixture at the indicated time points, quenching the reaction by acidification with hydrochloric acid, and measuring the relative peak intensity on LC-ESI-MS. (c) Measurement of the bimolecular association rate constant between RA (5 µM) and P1 by stopped-flow fluorometry as a function of the concentration of P1 (indicated).
Characterizing the kinetics of P1 binding and reaction with RA and its selectivity in cells
A goal of this study is to utilize the fluorogenic probe P1 inside cells to label folded and functional RA and to visualize what happens to RA-P1 conjugate fluorescence after imposing a thermal stress (a scenario 2 experiment). To realize this goal, the fluorogenic probe has to exhibit fast reaction kinetics and optimal RA binding selectivity. We first measured the bimolecular binding kinetics of P1 to RA in vitro (Figure 5c, ex. λ=390 nm, em. λ=485 nm). We mixed RA (5 µM) with increasing concentrations of P1 (in excess; see concentrations listed in Figure 5c) and recorded individual kinetic curves by monitoring the formation of the fluorescent conjugate (Figure 5c). At the selected concentrations of P1 (25 – 100 µM), the RA-P1 conjugation reaction is rate limited by RA•P1 association and the observed rates of emergence of conjugate fluorescence report on the rate of RA•P1 complex formation (Figure 5c). Therefore, the slope of the initial linear portion of the plot shown as the inset in Figure 5c is equal to the bimolecular association rate constant (kbimolecular). This rate constant (kbimolecular) can be calculated as 3000 M−1•s−1 (for details see supporting information), comparable to the fluorogenic probe used for labeling the SNAP-tag44 (7900 M−1•s−1).
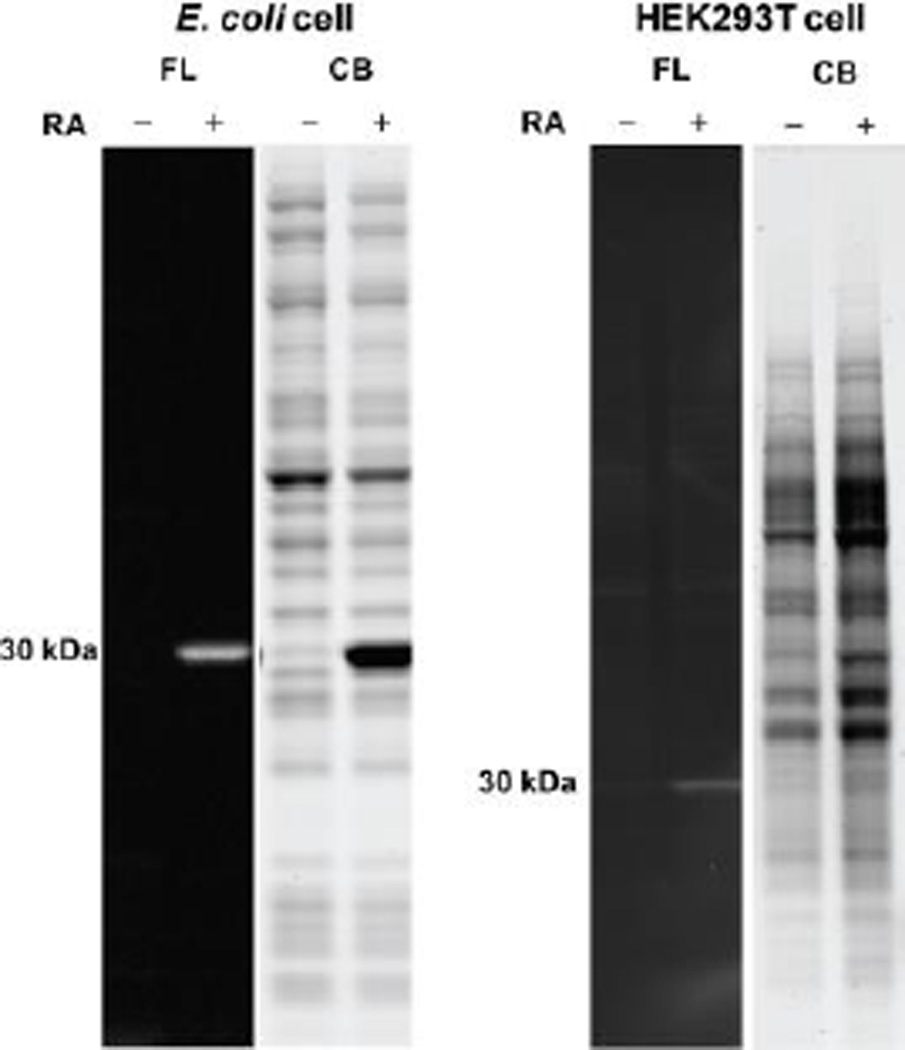
To examine the selectivity of P1 for labeling folded and functional RA inside the cell, we obtained concentrated cell lysates (total protein concentration, 3 mg/mL) by sonication of E. coli or HEK293T cells either lacking or overexpressing RA. We incubated the lysates with P1 (10 µM) for 10 min at 25 °C. P1 was capable of detecting the presence of RA only in the lysates of cells that were transformed/transfected with RA, and importantly no significant off-target bands were observed in the non-transformed/non-transfected controls (Figure 6). Therefore, P1 appears to be a fast, selective, fluorogenic probe for functional RA in cell lysates.
Figure 6.
Selectivity of P1 in E. coli or HEK293T cell lysate lacking or overexpressing RA. Lysates (3 mg/mL) obtained by sonication were incubated with P1 (10 µM) for 10 min at 25 °C. The samples were fractionated on an SDS-PAGE gel and visualized by either a Bio-rad Gel Doc Imager employing UV illumination to see the conjugate fluorescence signal or bright field light to observe the Coomassie staining. No significant off-target bands were observed in lysates of cells lacking or expressing RA. FL = RA-P1 conjugate-associated fluorescence, CB = Coomassie blue.
RAm1 is a quantitative thermal stress sensor, as discerned by monitoring its functionality and solubility changes
The proper folding of proteins exhibiting compromised stability often requires proteostasis network assistance45. Proteostasis network assistance is also required to maintain them in their folded and functional state inside the cell (continuously refold them), especially when the cell is stressed. A rapid elevation of the temperature of cells frequently consumes proteostasis network capacity due to proteome denaturation and aggregation-based binding of chaperones, chaperonins and the like, leading to insufficient proteostasis network capacity to correctly fold or refold metastable proteins. Therefore, a destabilized mutant of RA could be a suitable client-based protein to sense the dynamics of proteostasis network capacity before and after application of a thermal stress. In previous studies, we utilized RAm1 (E10K:D120V:N124S:L225P) as a metastable protein to survey how the proteostasis network components regulate the partitioning of metastable proteins between functional and non-functional states.27 However, whether the cellular functionality and solubility of RAm1 is sensitive to stresses, such as heat, has not been investigated.
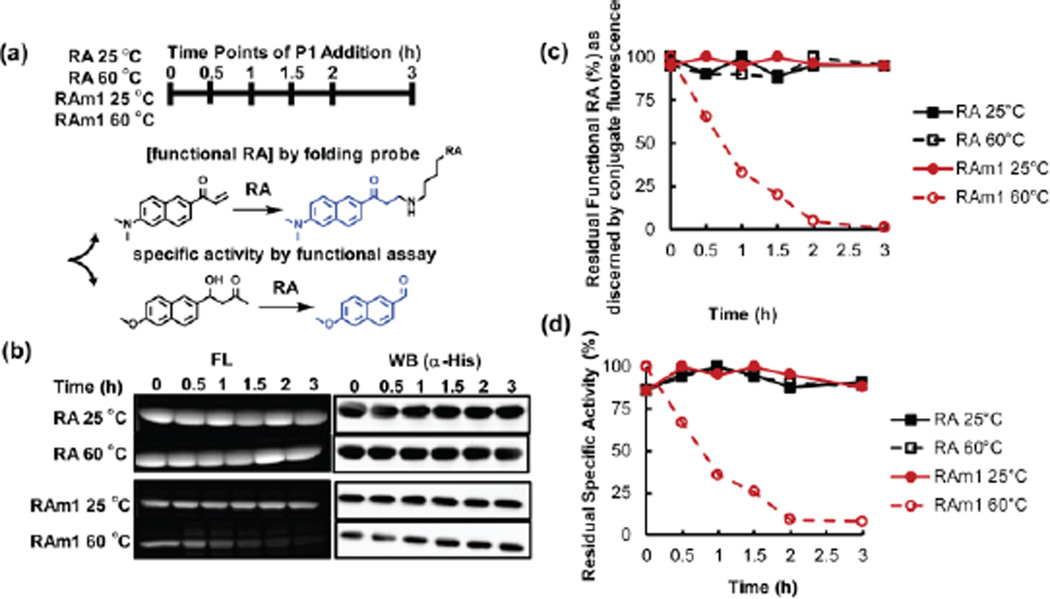
Towards this end, we first examined whether the folding and function of RAm1 is dependent on temperature in E. coli lysate, utilizing a scenario 1 experiment. The soluble lysate was obtained by centrifugation of lysed E. coli K12 cells overexpressing RA or RAm1. Importantly upon cell lysis, ATP was depleted by apyrase treatment, which converts the cellular chaperones and chaperonins to strong holdases.27 Therefore, P1 cannot significantly shift the RA or RAm1 folding equilibria upon binding and conjugate formation because the holdase chaperones/chaperonins retain malfolded RA or RAm1 (Figure S3).27 We examined the functionality of RA or RAm1 in the soluble lysates incubated at 25 °C or 60 °C at the indicated time points (Figure 7a, upper panel). The functionality of RA or RAm1 was examined by directly measuring the folded concentration of RA or RAm1 in the lysates using the fluorogenic folding probe P1 in a scenario 1 experiment (Figure 7b and c). Using the folding probe P1 (100 µM, in excess), we observed that the concentration of folded and functional RA did not change upon heating the lysate from 25 °C to 60 °C, over a time course of 3 h (Figure 7b, upper panel, see Figure 7c for quantification, black curves), whereas there was a time dependent loss of folded and functional metastable RAm1 at 60 °C (Figure 7b, lower panel, see Figure 7c for quantification, open red circles). To validate these results, RA folding and function was scrutinized by quantifying the specific activity of RA or RAm1 using a functional assay. Similarly, we only observed a time dependent loss of function in the destabilized RAm1, but not in RA at 60 °C (Figure 7d, open red circles). It is important to note that the total concentration of RA or RAm1 did not change over the time course of these studies, as shown by immunoblotting (Figure 7b, right panels), indicating that the loss of function was not caused by RAm1 degradation (no ATP present in the lysates, which disables many bacterial proteases).
Figure 7.
Loss of function in an ATP-depleted cell lysate upon increasing the temperature to 60 °C occurs in the case of thermo-labile mutant RAm1, but not for RA. (a) Experimental scheme: E. coli lysates expressing RA or RAm1 and depleted of ATP were incubated at 25 °C or 60 °C. (b) & (c) At the indicated time points, the concentration of functional RA was measured in the lysate by adding an excess of folding probe P1 (100 µM, 1 h incubation at 25 °C) and then the samples were subjected to SDS-PAGE (a scenario 1 experiment). Gels were directly visualized using a Bio-rad Gel Doc Imager employing UV illumination to quantify the fluorescence of the conjugate. (d) Using another aliquot, the specific activity of RA and RAm1 were measured by the functional assay at the time points indicated (described in supporting information experimental section (3)). Only the concentration of folded and functional RAm1 decreased upon application of a thermal stress, whereas RA was resistant to heat denaturation. FL = RA / RAm1-P1 conjugate fluorescence, WB = Western immunoblot.
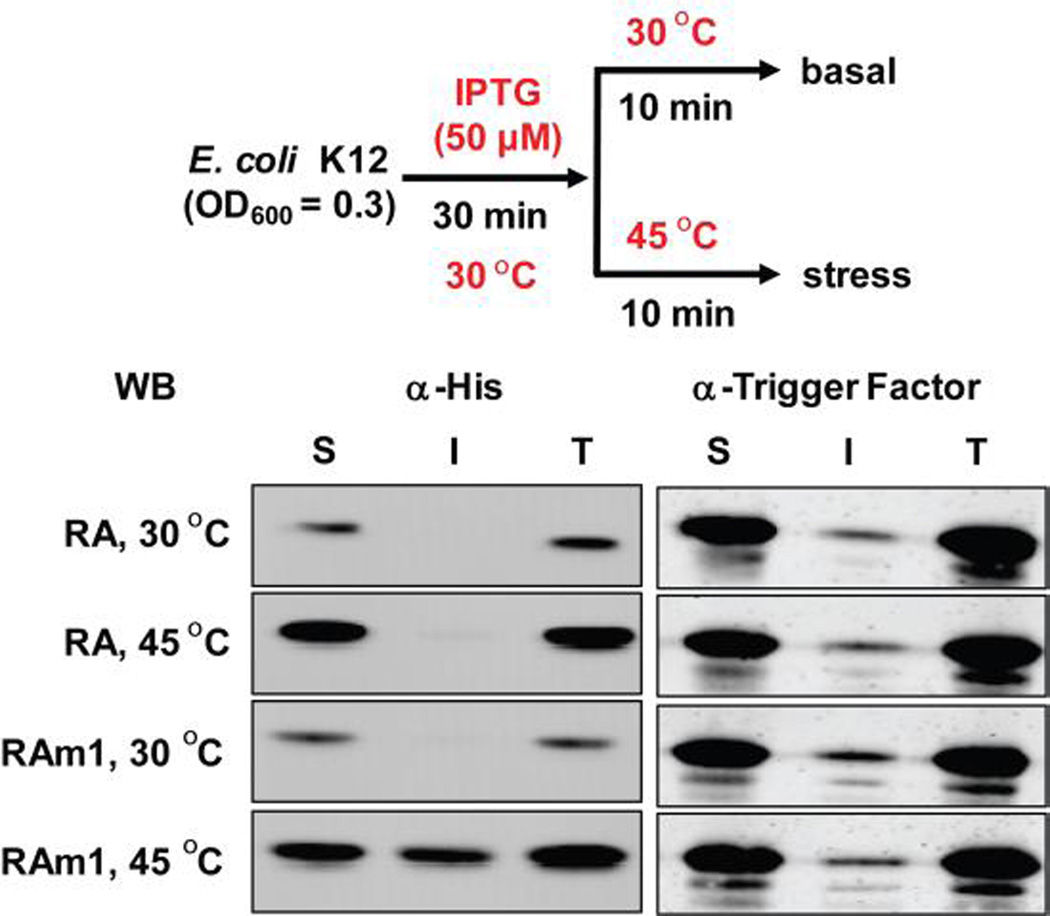
We further tested whether RAm1 is thermo-labile in bacterial cells at 45 °C. For this purpose, we expressed RA or RAm1 in E. coli K12 cells at 30 °C at low levels for 30 min, to mimic endogenous protein expression (see experimental section in supporting information for expression conditions). Half of the cells were then subjected to a 45 °C thermal stress for 10 min, while the remaining half was kept at 30 °C (Figure 8, top panel). After cell lysis and centrifugation, soluble and insoluble fractions were separated. The samples were resolved by SDS-PAGE and visualized by immunoblotting. We observed that after heat shock for 10 min at 45 °C, the destabilized RAm1 partitioned into an insoluble fraction (Figure 8, 4th row, left panel), unlike the stable RA (Figure 8, 2nd row, left panel).
Figure 8.
RAm1 is a sensor of proteostasis network capacity inside living E. coli cells using solubility as an indicator. After the application of a thermal stress that leads to proteome misfolding and aggregation, RAm1 partitioned into an insoluble state in living E. coli cells after 10 min of a heat stress at 45°C, as revealed by an SDS-PAGE gel visualized by immunoblotting. S = soluble fraction, I = insoluble fraction, T = total protein. Trigger factor was used as a loading control.
We next tested whether RAm1 was thermo-labile in mammalian cells at 42 °C employing a scenario 1 experiment. In this experiment, one culture dish of HEK293T cells expressing RAm1 was subjected to a 2 h, 42 °C heat stress prior to labeling with P1 (10 min at 42 °C), whereas the other plate remained at 37 °C before being labeled with P1 for 10 min (Figure S4). Most of the folded and functional RAm1 is converted to a non-native conformation at 42 °C (Figure S4, lower panel, middle lane), unlike the situation at 37 °C (Figure S4, lower panel, left lane). Those cells heated to 42 °C for 2 h can make folded and functional RAm1 from newly synthesized RAm1 by simply reducing the growth temperature back to 37 °C for 4 h (Figure S4, lower panel, right lane).
Direct visualization of the effect of cellular thermal stress on proteostasis network capacity in live cells
The fluorogenicity and the selectivity of P1 reacting with folded and functional RA and RAm1 provide a direct approach to monitor cellular proteostasis network capacity using a scenario 2 experiment, i.e., where the RAm1-P1 conjugate is formed in the cell before a cellular stress is applied. The advantage of using a small-molecule-regulated thermo-labile client protein as a proteostasis network capacity sensor (e.g., RAm1) is the temporal control over the fluorescence signal emerging from the small molecule-protein sensor conjugate. In particular, we can directly visualize the misfolding and aggregation or continuous refolding of RAm1 in live cells by first forming the RAm1-P1 conjugate and then applying the stress to discern its effect on the proteostasis network capacity of the cell at any time point. Continuing RAm1 synthesis post conjugate formation is not problematic, as this does not contribute to RAm1-P1 conjugate fluorescence. In the previous pioneering pulse-chase type experiments, fusion of fluorescent proteins to luciferase is required to monitor the fate of the protein as a function of stress.21,22 In this context, blocking proteome translation is necessary to get time dependent information in the absence of complications from new protein synthesis.
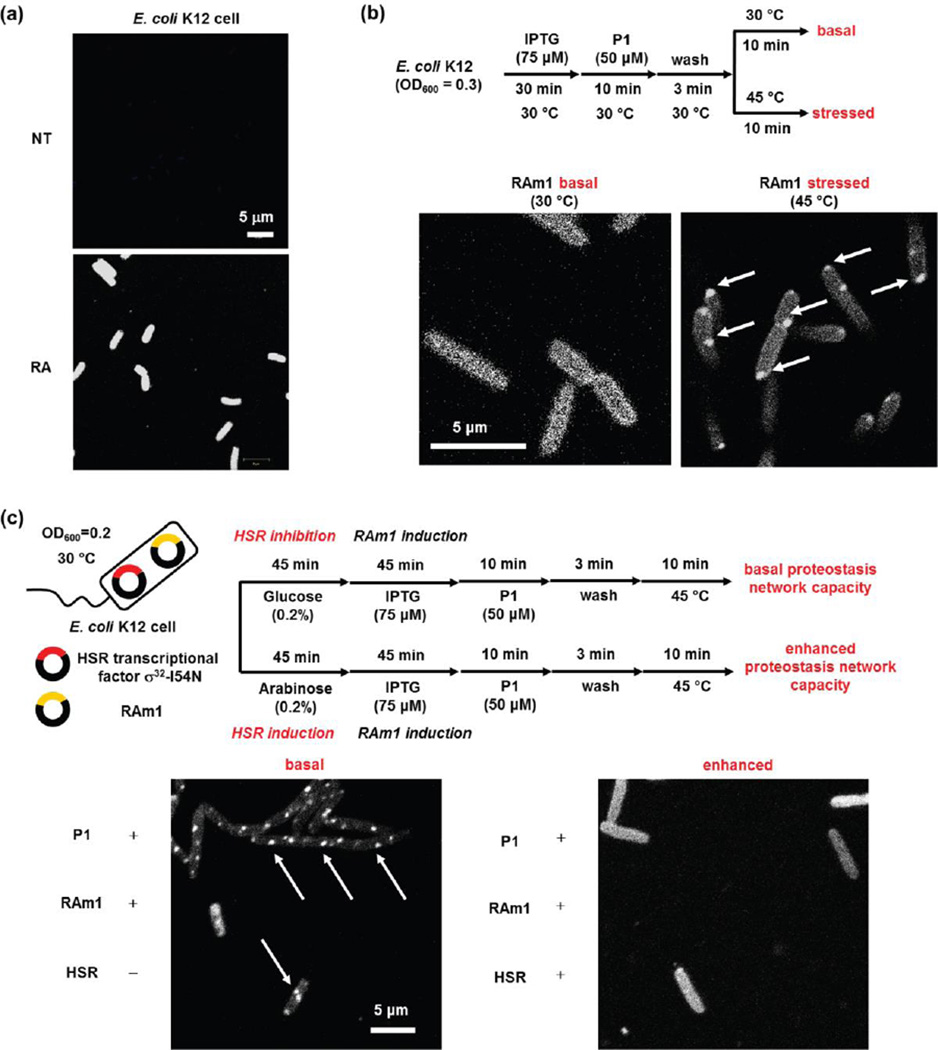
To demonstrate the feasibility of direct visualization of RA / RAm1-P1 conjugate misfolding and aggregation as a consequence of thermal stress in live cells (a scenario 2 experiment), we first tested whether P1 can selectively label RA in live E. coli K12 cells (for further details on cell culture and imaging conditions see supplemental experimental section). We treated both non-transformed and transformed cells with P1 for 10 min, washed both cell cultures to rid the media of P1, and observed uniform RA fluorescence only in the transformed cells (Figure 9a). We then examined how the fluorescent RAm1-P1 conjugate behaved after a 10-min thermal stress at 45 °C, relative to keeping the cells at 30 °C. We observed granular structures across all cells heated to 45 °C for 10 min (Figure 9b, right panel, white arrows) along with diffuse conjugate fluorescence, indicating incomplete aggregation. Strictly analogous results were observed in the E. coli DE3 Star strain (Figure S5). These experiments also demonstrated that the RAm1-P1 conjugate remained fluorescent after aggregating (Figures 9, S5 and S6).
Figure 9.
The RAm1-P1 fluorescent conjugate is a cellular client-based thermo-labile sensor of proteostasis network capacity in E. coli. (a) P1 selectively binds to and reacts with RA in E. coli affording the RAm1-P1 conjugate. Thus, only cells transformed with RA exhibit conjugate fluorescence in the confocal image. (b) The pre-formed RAm1-P1 conjugate formed granular aggregate structures as observed by confocal fluorescence imaging (white arrows) after heating at 45 °C for 10 min, serving as a sensor of proteostasis network capacity insufficiency. (c) Transcriptional reprogramming of E. coli by over-expressing the σ32-I54N heat shock response transcription factor enhances the proteostasis network capacity of the cytosol protecting the preformed RAm1-P1 conjugate from aggregating upon application of thermal stress, as reflected by the lack of granular structures in rightmost confocal image in comparison to the image on the left where aggregation is observed because cytosolic proteostasis network capacity was not preemptively enhanced. Sample preparation and imaging details are described in experimental section of the supporting information. NT: Non-transformed. Images were taken using a Zeiss LSM710 confocal microscope.
We further examined whether our preformed RAm1-P1 client-based fluorescent proteome stress sensor responds to a proteostasis network capacity change afforded by transcriptionally reprograming the bacteria. We transcriptionally reprogramed E. coli K12 cells by over-expressing the heat shock factor σ32-I54N to enhance the cytosolic proteostasis network capacity (Figure 9c).46 The I54N variant of σ32 was chosen because previous data shows that it is resistant to negative feedback regulation.46,47 With enhanced cytosolic proteostasis network capacity mediated by σ32-I54N expression, the bacterial cells exhibited minimal granular aggregated RAm1-P1 structures upon heating at 45 °C for 10 min (Figure 9c, right panel), indicating that transcriptional reprogramming creates sufficient proteostasis network capacity to continuously refold RAm1-P1 and protect it from aggregation, unlike the situation in the control cells at 45 °C wherein RAm1-P1 predominantly aggregated (Figure 9c, left panel). This experiment is distinct from the preceding experiments, because it shows that in spite of a thermal stress (known to misfold and aggregate the RAm1-P1 conjugate), the increased cytosolic proteostasis network capacity keeps the RAm1 soluble and presumably properly folded.
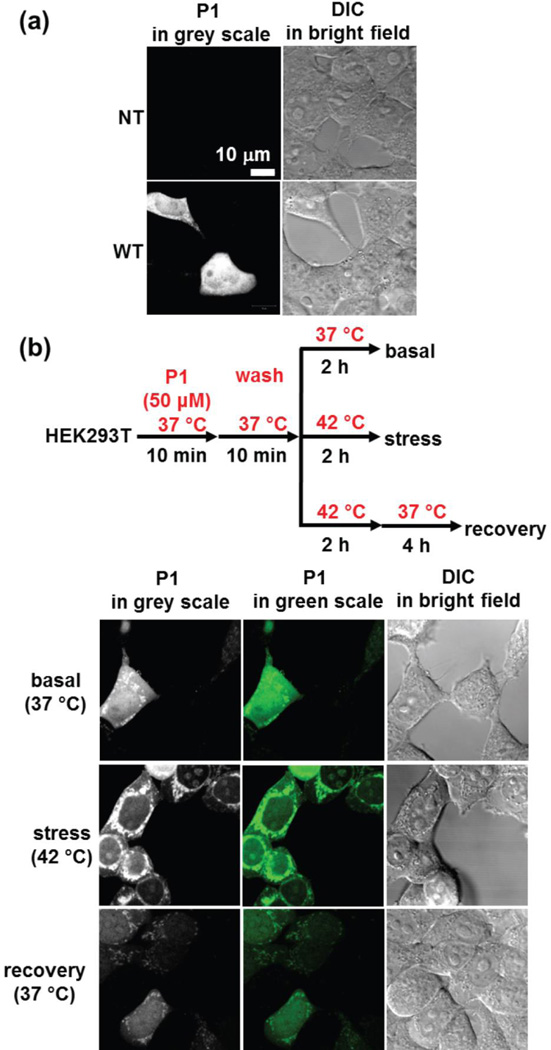
We next explored whether we could extend this scenario 2 experimental approach to mammalian cells. Since protein expression tends to be significantly lower in mammalian cells relative to E. coli, we first had to demonstrate that P1 is sensitive enough to selectively label RA / RAm1 in mammalian cells. Thus, we expressed RA in both HEK293T cells (Figure 10a) and in HeLa cells (Figure S7) and then treated the cells with P1 (10 µM) for 10 min, followed by cell washing to remove excess P1 (for more details see supporting information). Importantly, no off-target fluorescence was observed in the non-transfected (NT) HEK293T cells (Figure 10a, upper panel) or HeLa cells (Figure S7, upper panel). Selective labeling of RA in isolated transfected cells was clear from the observed fluorescence (Figures 10a and S7, bottom panels). Importantly, we also observed unlabeled cells in the transfected samples (Figure 10a, lower panel, Figure S7, lower panel), due to incomplete transfection, which is typical. To further validate that the emergence of fluorescence originates from the covalent labeling of RA by P1, we transfected HEK293T cells with an RA-RFP fusion protein to monitor the co-localization of RA-P1 conjugate fluorescence and RFP fluorescence at distinct wavelengths (Figure S8). In the field shown, we only observed one transfected cell with both blue fluorescence emerging from P1 labeling of RA and red fluorescence coming from the RA-RFP fusion protein (Figure S8a). The co-localization coefficient was 0.91 (Figure S8b), indicating that the fluorescence labeling by P1 originates from selective modification of functional RA in the cells. We also examined the RAm1 labeling kinetics by P1 (10 µM) under the mammalian cell conditions used in this study and observed saturated labeling after 10 min, indicating complete labeling of RAm1 on this time scale (Figure S9).
Figure 10.
The RAm1-P1 fluorescent conjugate is a cellular client-based thermo-labile cellular proteostasis network capacity sensor in HEK293T cells. (a) P1 selectively binds to and reacts with RA and exhibits fluorescence only in HEK293T cells transfected with RA, as discerned from the confocal fluorescence images. (b) The confocal fluorescence images show that the pre-formed RAm1-P1 conjugate retained predominant solubility at 37 °C (first row), however aggregates predominated upon heating at 42 °C for 2 h (second row). Notably, reduction of the temperature from 42 °C to 37 °C for an additional 4 h after thermal stress eliminates the RAm1-P1 granular aggregate structures in the cell (third row), presumably as a con-sequence of the heat shock response transcriptional program-enabled refolding of RAm1-P1 and partial degradation of the RAm1-P1 conjugate (see main text). Images were taken using a Zeiss LSM710 confocal microscope.
We next examined whether RAm1, when used with fluorogenic probe P1, is a sensor of thermally induced loss of cellular proteostasis network capacity with regard to the RAm1 client protein. We tested this hypothesis by first imaging the fate of the pre-formed RAm1-P1 conjugate in HEK293T cells (a scenario 2 experiment). We treated two independent samples of cells with P1 (50 µM) at 37 °C for 10 min, followed by washing the cells to rid the media of excess P1 (Figure 10b flow chart). One sample of the cells was shifted to 42 °C for 2 h, while the other population remained at 37 °C for 2 h (Figure 10b, flow chart). We observed soluble and granular aggregate RAm1-P1 conjugate structures at both growth temperatures (Figure 10b, 1st row and 2nd row). However, the granular structures were much more prominent at 42 °C (Figure 10b, 2nd row), presumably as a consequence of thermal proteome denaturation-associated consumption of proteostasis network capacity and a higher propensity for RAm1 to misfold and aggregate at the elevated temperature. This suggests that the proteostasis network capacity is not sufficient to maintain the correct folding / refolding of the RAm1-P1 conjugate, especially at 42 °C. If the 42 °C cells were allowed to recover at 37 °C for an additional 4 h, the majority of the granular RAm1-P1 conjugate aggregate structures were refolded and resolubilized or degraded (Figure 10b, 3rd row). To confirm the apparent clearance of RAm1-P1 aggregates in HEK293T cells, as suggested by the imaging results in Figure 10b, bottom row, we monitored the fate of the RAm1-P1 fluorescent conjugate as a function of time by SDS-PAGE employing fluorescence detection (Figure S10). We observed a decreasing amount of the RAm1-P1 conjugate as a function of time that we attribute to continuous RAm1-P1 conjugate degradation. The presence of RAm1-P1 conjugate degradation was observed even in the absence of thermal stress (Figure S11); in fact, it appeared to be faster. Thus, it appears that both transcriptional heat shock response reprogramming induced by a thermal stress and partial degradation of the RAm1-P1 conjugate re-establish sufficient cytosolic proteostasis network capacity to refold the RAm1-P1 conjugate, rendering it soluble and diffuse. In bacteria, we showed above that preemptive enhancement of the cytosolic proteostasis network capacity through transcriptional reprogramming (simulating the heat shock response without heating) protects the RAm1-P1 conjugate from misfolding and aggregation even at 45 °C (Figure 9c, right panel), where the RAm1-P1 conjugate is inherently thermo-labile (Figure 9c, left panel). Considering all the data, it is clear that the thermo-lability of the fluorescent RAm1-P1 conjugate provides a direct approach to visualize the cellular proteostasis network capacity through the distribution of fluorescence signals in a scenario 2 experiment, using confocal fluorescence imaging.
The RAm1-P1 conjugate folding sensor reports on alterations in cellular proteostasis capacity due to other stresses
We next asked whether other stresses, like the production of reaction oxygen species (ROS), could reduce the cellular proteostasis network capacity and thus lead to more extensive misfolding and aggregation of the preformed RAm1-P1 conjugate in HEK293T cells. In this scenario 2 experiment, we triggered oxidative stress by treatment of HEK293T cells with tert-butyl hydrogen peroxide (TBHP). Confocal fluorescent images show that the preformed RAm1-P1 conjugate folding sensor was completely transformed to aggregates after ROS induction (Figure S12a, right panel), relative to a mixture of aggregated and properly folded RAm1-P1 conjugate in the absence of stress (Figure S12a, left panel). We hypothesize that ROS production (confirmed by the CellROX® Green Reagent–which becomes fluorescent upon ROS-mediated oxidation; Figure S12b) compromises protein homeostasis in the cell by causing proteome misfolding, consuming the proteostasis network capacity that is required for the continuous refolding of RAm1. Thus, the RAm1-P1 conjugate folding sensor should be useful for reporting on other stresses that compromise cellular proteostasis network capacity.
CONCLUSIONS
In summary, we have demonstrated how we utilized the thermo-labile de novo designed RAm1 enzyme in combination with its complementary fluorogenic folding probe P1 to form the RAm1-P1 conjugate fluorescent folding sensor to monitor the cellular proteostasis network capacity after thermal stress in a scenario 2 pulse-chase-like experiment. The time-resolved nature of these experiments (Figures 9 and 10) enable the re-establishment of proteostasis to be studied without the need for translational inhibition, which can be problematic for long duration experiments as it constitutes an additional stress that can lead to cell death. The client-based cellular proteostasis network capacity sensor developed herein (RAm1-P1 fluorescent conjugate) can be further explored to monitor the dynamics of proteostasis network capacity in response to other distinct stresses in real time in relevant cells (such as oxidative stress; Figure S12).
Supplementary Material
Acknowledgments
This work was supported by the Skaggs Institute for Chemical Biology, the Lita Annenberg Hazen Foundation, and by National Institutes of Health grants AG046495 (to JWK). Y.L.T. was supported by a predoctoral fellowship from the Agency of Science, Technology and Research (A*STAR). X.Z. was a Howard Hughes Medical Institute Fellow of the Helen Hay Whitney Foundation, and is currently supported by the Burroughs Wellcome Fund Career Award at the Scientific Interface.
ABBREVIATIONS
- LC-ESI-MS
liquid chromatography electrospray ionization mass spectrometry
- NT
non-transfected
- RA
retroaldolase
- RFP
red fluorescent protein
- ROS
reactive oxygen species
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TBHP
tert-butyl hydrogen peroxide
Footnotes
ASSOCIATED CONTENT
Supporting information. Supporting figures S1– S12 and Experimental Procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Anfinsen CB. Science. 1973;181:223. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Hartl FU, Hayer-Hartl M. Nat Struct Mol Biol. 2009;16:574. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 3.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Annu Rev Biochem. 2013;82:323. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 4.Balch WE, Morimoto RI, Dillin A, Kelly JW. Science. 2008;319:916. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 5.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Science. 2006;311:1471. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Zvi A, Miller EA, Morimoto RI. Proc Natl Acad Sci U S A. 2009;106:14914. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hipp MS, Park SH, Hartl FU. Trends Cell Biol. 2014;24:506. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Hutt D, Balch WE. Science. 2010;329:766. doi: 10.1126/science.1194160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, Michalopoulos G, Perlmutter DH. Science. 2010;329:229. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 10.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Annu Rev Biochem. 2009;78:959. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 11.Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Cell. 2008;134:769. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang AM, Miyata Y, Klinedinst S, Peng HM, Chua JP, Komiyama T, Li X, Morishima Y, Merry DE, Pratt WB, Osawa Y, Collins CA, Gestwicki JE, Lieberman AP. Nat Chem Biol. 2013;9:112. doi: 10.1038/nchembio.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westerheide SD, Bosman JD, Mbadugha BNA, Kawahara TLA, Matsumoto G, Kim SJ, Gu WX, Devlin JP, Silverman RB, Morimoto RI. J. Biol. Chem. 2004;279:56053. doi: 10.1074/jbc.M409267200. [DOI] [PubMed] [Google Scholar]
- 14.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. Elife. 2015;4:e05033. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinwal UK, Miyata Y, Koren J, 3rd, Jones JR, Trotter JH, Chang L, O'Leary J, Morgan D, Lee DC, Shults CL, Rousaki A, Weeber EJ, Zuiderweg ER, Gestwicki JE, Dickey CA. J Neurosci. 2009;29:12079. doi: 10.1523/JNEUROSCI.3345-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das I, Krzyzosiak A, Schneider K, Wrabetz L, D'Antonio M, Barry N, Sigurdardottir A, Bertolotti A. Science. 2015;348:239. doi: 10.1126/science.aaa4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers JE, Dalton LE, Clarke HJ, Malzer E, Dominicus CS, Patel V, Moorhead G, Ron D, Marciniak SJ. Elife. 2015;4:e04872. doi: 10.7554/eLife.04872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, Saldanha SA, Hodder P, Tait BD, Garza D, Balch WE, Morimoto RI. Nat Chem Biol. 2012;8:185. doi: 10.1038/nchembio.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooley CB, Ryno LM, Plate L, Morgan GJ, Hulleman JD, Kelly JW, Wiseman RL. Proc Natl Acad Sci U S A. 2014;111:13046. doi: 10.1073/pnas.1406050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Nature. 2010;467:179. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler J, Seybert A, Konig L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B. EMBO J. 2010;29:910. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta R, Kasturi P, Bracher A, Loew C, Zheng M, Villella A, Garza D, Hartl FU, Raychaudhuri S. Nat. Methods. 2011;8:879. doi: 10.1038/nmeth.1697. [DOI] [PubMed] [Google Scholar]
- 23.Lavis LD, Raines RT. ACS Chem. Biol. 2014;9:855. doi: 10.1021/cb500078u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm JB, Sung AJ, Legant WR, Hulamm P, Matlosz SM, Betzig E, Lavis LD. ACS Chem. Biol. 2013;8:1303. doi: 10.1021/cb4000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing CR, Cornish VW. ACS Chem. Biol. 2013;8:1704. doi: 10.1021/cb300657r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Tan YL, Zhang X, Bhabha G, Ekiert DC, Genereux JC, Cho Y, Kipnis Y, Bjelic S, Baker D, Kelly JW. Proc Natl Acad Sci U S A. 2014;111:4449. doi: 10.1073/pnas.1323268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizukami S, Watanabe S, Akimoto Y, Kikuchi K. J. Am. Chem. Soc. 2012;134:1623. doi: 10.1021/ja208290f. [DOI] [PubMed] [Google Scholar]
- 29.Bojkowska K, de Sio FS, Barde I, Offner S, Verp S, Heinis C, Johnsson K, Trono D. Chem Biol. 2011;18:805. doi: 10.1016/j.chembiol.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Fan J, Wang J, Zhang S, Dou B, Peng X. J. Am. Chem. Soc. 2013;135:11663. doi: 10.1021/ja4056905. [DOI] [PubMed] [Google Scholar]
- 31.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giger L, Caner S, Obexer R, Kast P, Baker D, Ban N, Hilvert D. Nat Chem Biol. 2013;9:494. doi: 10.1038/nchembio.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Zhang X, Tan YL, Bhabha G, Ekiert DC, Kipnis Y, Bjelic S, Baker D, Kelly JW. J. Am. Chem. Soc. 2014;136:13102. doi: 10.1021/ja5056356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukinavicius G, Reymond L, D'Este E, Masharina A, Gutfert F, Ta H, Guether A, Fournier M, Rizzo S, Waldmann H, Blaukopf C, Sommer C, Gerlich DW, Arndt HD, Hell SW, Johnsson K. Nat. Methods. 2014;11:731. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 35.Baranczak A, Connelly S, Liu Y, Choi S, Grimster NP, Powers ET, Wilson IA, Kelly JW. Biopolymers. 2014;101:484. doi: 10.1002/bip.22407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S, Ong DST, Kelly JW. J. Am. Chem. Soc. 2010;132:16043. doi: 10.1021/ja104999v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimster NP, Connelly S, Baranczak A, Dong JJ, Krasnova LB, Sharpless KB, Powers ET, Wilson IA, Kelly JW. J. Am. Chem. Soc. 2013;135:5656. doi: 10.1021/ja311729d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh EH, Liu Y, Connelly S, Genereux JC, Wilson IA, Kelly JW. J. Am. Chem. Soc. 2013;135:17869. doi: 10.1021/ja408230k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi S, Kelly JW. Bioorgan Med Chem. 2011;19:1505. doi: 10.1016/j.bmc.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longstreet AR, Jo M, Chandler RR, Hanson K, Zhan N, Hrudka JJ, Mattoussi H, Shatruk M, McQuade DT. J. Am. Chem. Soc. 2014;136:15493. doi: 10.1021/ja509058u. [DOI] [PubMed] [Google Scholar]
- 41.Baranczak A, Liu Y, Connelly S, Du WG, Greiner ER, Genereux JC, Wiseman RL, Eisele YS, Bradbury NC, Dong J, Noodleman L, Sharpless KB, Wilson IA, Encalada SE, Kelly JW. J. Am. Chem. Soc. 2015;137:7404. doi: 10.1021/jacs.5b03042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grimm JB, English BP, Chen JJ, Slaughter JP, Zhang ZJ, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, Lavis LD. Nat. Methods. 2015;12:244. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibbs RE, Talley TT, Taylor P. J. Biol. Chem. 2004;279:28483. doi: 10.1074/jbc.M403713200. [DOI] [PubMed] [Google Scholar]
- 44.Komatsu T, Johnsson K, Okuno H, Bito H, Inoue T, Nagano T, Urano Y. J. Am. Chem. Soc. 2011;133:6745. doi: 10.1021/ja200225m. [DOI] [PubMed] [Google Scholar]
- 45.Cho Y, Zhang X, Pobre KF, Liu Y, Powers DL, Kelly JW, Gierasch LM, Powers ET. Cell Rep. 2015;11:321. doi: 10.1016/j.celrep.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Liu Y, Genereux JC, Nolan C, Singh M, Kelly JW. ACS Chem. Biol. 2014;9:1945. doi: 10.1021/cb5004477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yura T, Guisbert E, Poritz M, Lu CZ, Campbell E, Gross CA. Proc Natl Acad Sci U S A. 2007;104:17638. doi: 10.1073/pnas.0708819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.