Abstract
Background
While the association of diet and disease is well documented, the biologic mechanisms involved have not been entirely elucidated. In this study we evaluate how dietary intake influences gene expression to better understand the underlying mechanisms through which diet operates.
Methods
We used data from 144 individuals who had comprehensive dietary intake and gene expression data from RNAseq using normal colonic mucosa. Using the DESeq2 statistical package, we identified genes that displayed statistically significant differences in expression between individuals in high intake and low intake categories for several dietary variables of interest adjusting for age and sex. We examined total calories, total fats, vegetable protein, animal protein, carbohydrates, trans-fatty acids, mutagen index, red meat, processed meat, whole grains, vegetables, fruits, fiber, folate, dairy products, calcium, and Prudent and Western Dietary Patterns.
Results
Using a false discovery rate (FDR) of <0.1, meat-related foods were statistically associated with 68 dysregulated genes, calcium with three dysregulated genes, folate with four dysregulated genes, and non-meat related foods with 65 dysregulated genes. With a more stringent FDR of <0.05, there were nine meat-related dysregulated genes and 23 non-meat related genes. Ingenuity pathway analysis (IPA) identified three major networks among genes identified as dysregulated with respect to meat-related dietary variables and three networks among genes identified as dysregulated with respect to non-meat related variables. The top networks (IPA Network Score >30) associated with meat-related genes were 1) cancer, organismal injury and abnormalities, tumor morphology and 2) cellular function and maintenance, cellular movement, cell death and survival. Among genes related to non-meat consumption variables the top networks were 1) hematological system development and function, nervous system development and function, tissue morphology and 2) connective tissue disorders, organismal injury and abnormalities.
Conclusions
Several dietary factors were associated with gene expression in our data. These findings provide insight into possible mechanisms whereby diet may influence disease processes.
Keywords: Diet, meat, vegetables, whole grains, gene expression, folate, calcium, fiber
Introduction
Diet has been associated with several diseases including atherosclerosis [2], inflammatory bowel disease [3], steatosis and non-alcoholic fatty liver disease [4, 5], seizures [6], and various cancers including colon and breast carcinoma [7–9]. Moreover, diet has also been identified as a risk factor for Type 2 diabetes mellitus, obesity, and metabolic syndrome [10]. Beyond its role in disease status, diet has been shown to play an important role in a number of physiologic and metabolic processes including bone remodeling [11] and aspects of the immune response [12]. The biologic mechanisms involved in these processes is less clear.
Studies have evaluated the role of diet in the regulation of specific physiologic and pathologic processes, showing that single dietary metabolites and nutrients play a role in the regulation of certain signaling pathways [11, 13–15] or cytokine profiles [16]. Other studies have focused on the dietary effects on oxidative stress [17] or the role of potentially chemoprotective nutrients [18]. Some of these studies also have looked at localized gene expression within specific hypothesized pathways and processes, suggesting that diet has a role in the regulation of gene expression [7, 8, 16, 19]. Recently the effects of dietary patterns on the entire gene expression profile were investigated, suggesting a differential expression of genes in individuals with a Prudent Dietary Pattern verses a Western Dietary Pattern and identified specific canonical pathways associated with both dietary patterns [20].
In this study we hypothesize that levels of gene expression in normal colonic mucosa will be associated with dietary factors. Dietary factors we consider include sources of calories, including animal protein and vegetable protein, carbohydrates, and total fat. We also consider trans-fatty acids, total red meat, processed meat, mutagen index, dairy products, fruit, vegetables, refined grains, whole grains, sucrose, dietary fiber, calcium, and folate with gene expression profiles. Lastly, we examined the association between Prudent Dietary Pattern and Western Dietary Pattern with gene expression.
Methods
Total RNA was available from normal colonic mucosa for colon cancer cases who were part of the Diet, Activity, and Lifestyle study, an incident, population-based, case-control study of colon cancer from Utah and the Kaiser Permanente Medical Research Program (KPMRP) [21]. Cases were identified using a rapid-reporting-system and had tumor registry verification of a first primary adenocarcinoma of the colon and were diagnosed between October 1991 and September 1994 and tumor tissue blocks (used for RNA) were obtained for 97% of all Utah cases and for 85% of all KPMRP cases [22]. Individuals with known adenomatous polyposis coli (APC), Crohn’s disease, or inflammatory bowel disease were not eligible for the study. The study was approved by the Institutional Review Board of the University of Utah and at KPMRP; all study participants signed informed consent prior to participation.
Dietary Data
Data were collected by trained and certified interviewers using laptop computers shortly after diagnosis. All interviews were audio-taped as previously described and reviewed for quality control purposes [23]. Any interview deemed questionable by the interviewer was reviewed centrally; this allowed us to evaluate the data by distinguishing between an interview that was difficult for the interviewer versus one that was difficult for the participant. Additionally, we reviewed audio tapes for all individuals whose nutrient levels were considered outliers (over the 95%tile or under the 5 percentile of intake); this enabled us to correct any coding errors and determine the quality and face validity of the interview. Dietary information was obtained for the year two years prior to diagnosis using an extensive diet history questionnaire adapted from the validated CARDIA diet history; most individuals were asked to recall usual dietary intake from two to three years ago in order to obtain pre-diagnosis diet [24]. Foods were converted to nutrients using the Nutrition Coding Center Nutrient Data System Version 19 as well as being grouped into categories of similar foods. We assessed both foods and nutrients with gene expression. Foods units were standard servings per day, which was ½ cup of fruit, vegetable, or dairy product; meat servings were 2 to 3 oz of meat; grain products were ½ cup of rice-type grains or one slice of bread. Prudent and Western Dietary Patterns were developed based on principal component program [25]. Our Prudent Dietary Pattern was heavily loaded towards diets high in fruits, vegetables, whole grains, fish, and chicken, while the Western Dietary Pattern was highly loaded towards red meat, processed meats, and refined grains and high-sugar-high-fat foods. Additional questions were asked about meat consumption, doneness, and preparation methods that were combined and used to create a mutagen index score [26].
RNA processing
RNA was extracted from formalin-fixed paraffin-embedded colonic tissues. FFPE tissue has been shown to be a reliable source of RNA for use in conjunction with RNA-seq [27]. The study pathologist reviewed slides to delineate carcinoma and normal colonic mucosa. Total RNA was extracted, isolated, and purified using the RecoverAll Total Nucleic Acid isolation kit (Ambion), RNA yields were determined using a NanoDrop spectrophotometer.
Sequencing Library Preparation
Library construction was performed using the Illumina TruSeq Stranded Total RNA Sample Preparation Kit with Ribo-Zero as previously described [28]. Briefly, ribosomal RNA was removed from 100 ng total RNA using biotinylated Ribo-Zero oligos attached to magnetic beads. Following purification, the rRNA-depleted sample was fragmented and primed with random hexamers. First strand reverse transcription was accomplished using Superscript II Reverse Transcriptase (Invitrogen). Second strand cDNA synthesis was accomplished using DNA polymerase I and Rnase H under conditions in which dUTP is substituted for dTTP, yielding blunt-ended cDNA fragments in which the second strand contains dUTP. An A-base is added to the blunt ends as a means to prepare the cDNA fragments for adapter ligation and block concatamer formation during the ligation step. Adapters containing a T-base overhang were ligated to the A-tailed DNA fragments. Ligated fragments were PCR-amplified (13 cycles) under conditions in which the PCR reaction enables amplification of the first strand cDNA product. The PCR-amplified library was purified using Agencourt AMPure XP beads (Beckman Coulter Genomics).
Sequencing and Data Processing
Sequencing libraries (18 pM) were chemically denatured and applied to an Illumina TruSeq v3 single read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina TruSeq SR Cluster Kit v3-cBot-HS. Following transfer of the flowcell to an Illumina HiSeq instrument, a 50 cycle single-read sequence run was performed using TruSeq SBS v3 sequencing reagents. The single-end 50-base reads from the Illumina HiSeq2500 were aligned to a sequence database containing the human genome (build GRCh37/hg19, February 2009, from genome.ucsc.edu) plus all splice junctions generated using the USeq MakeTranscriptome application (version 8.8.1, available here: http://useq.sourceforge.net/). Alignment was performed using NovoAlign version 2.08.01 available from novocraft.com, which also trimmed any adapter sequence. Genome alignments to splice junctions were translated back to genomic coordinates using the USeq SamTranscriptomeParser application. Alignments were then sorted and indexed using the Picard SortSam application (version 1.100, available here: http://broadinstitute.github.io/picard/). Aligned read counts for each gene were calculated using pysam (https://code.google.com/p/pysam/) and samtools (http://samtools.sourceforge.net/). A python script using the pysam library was given a list of the genome coordinates for each gene, and counts to the exons and UTRs of those genes were calculated. Gene coordinates were downloaded from http://genome.ucsc.edu. Over 17000 genes that were expressed in colon mucosa were analyzed with dietary factors. Rigid quality control procedures were implemented to assure high quality data; participants failing QC were dropped from the analysis.
Statistical Methods
Of the 197 initial tumor/non-tumor tissue pairs, 22 subjects failed quality control (QC) based on low number of sequence leaving 175 subjects with high quality expression data. Of these, 144 had dietary questionnaire data. For each dietary factor, our analysis centered on contrasting individuals with lower intake to those of individuals with higher intake. Dietary data were evaluated using nutrients per 1000 calories and standard servings of food; sample tertiles were computed for the consumption levels of each dietary factor. For each such dietary variable, we determined which genes displayed statistically significant differential expression between the lowest and highest intake tertile categories after adjusting for age and sex using the Bioconductor package DESeq2 written for the R statistical programming environment; these genes were considered dysregulated. DESeq2 assumes the RNA-Seq counts are distributed according to negative binomial distributions. It utilizes generalized linear modeling and variance reduction techniques for estimated coefficients to test individual null hypotheses of zero log2 fold changes between high and low categories (i.e. no differential expression) for each gene. It employs both an independent-filtering method and the Benjamini and Hochberg (BH) [29] procedure to improve power and control the false discovery rate (FDR). The default DESeq2 options were used, including replacement of outliers, as defined by Cook’s distance, and the Wald test. For further details regarding DESeq2, see Love et al. [30]. In identifying genes with differential expression, an FDR of 0.10 was used. The fold change calculations for differentially expressed genes was determined by DESeq2 and represents the log2 change in expression level (i.e. counts) for the high versus low dietary categories.
Bioinformatics analysis was performed on the list of Ensemble IDs associated with genes identified as differentially expressed between high and low consumption categories for the dietary variables of interest using QIAGEN’s Ingenuity Pathway Analysis (IPA) [31]. We used genes from Ingenuity Knowledge Base and considered both indirect and direct relationships. Causal and interaction networks were generated. Interaction networks were limited to 35 molecules per network and 25 networks per analysis, and excluded endogenous chemicals. We focused on algorithmically derived interaction networks, which are assigned a score based on their relevance to the genes in the input dataset, the number of focus genes (i.e. dysregulated genes in our data that are in that network), and their connectivity [32]. The score is calculated as −log10P, where P is generated using a Fisher’s exact test [33]. Studies have found scores >3 to be significant, with a score of 3 indicating a 1/1000 chance that the focus genes are in a network due to random chance [34–36]. Other studies have opted to utilize more stringent criteria and higher scores to ensure that their discovered networks are highly significant [37, 38]; we utilized highly stringent criteria, only including networks with scores over 20. We applied the BH multiple testing correction to assess pathways in IPA.
Availability of data are restricted to that authorized in patient consent form and in accordance with data transfer agreements and IRB requirements.
Results
The majority of our study population was male and the median age was 65, 13.2% were current smokers, and 37.3% currently took aspirin or non-steroidal anti-inflammatory drugs on a regular basis (Table 1). The median BMI was 29.6. Within this population few people consumed processed meat, with the highest level of intake being less than one serving per day. The highest red meat consumption was less than two servings per day. Over half of study participants had over two servings of vegetables per day and one serving of whole grains per day.
Table 1.
Description of Study population
| N | % | ||
|---|---|---|---|
| Sex | Male | 80 | 55.6% |
| Female | 64 | 44.4% | |
| Smoking Status | Never | 60 | 41.7% |
| Former | 65 | 45.1% | |
| Current | 19 | 13.2% | |
| Recent NSAID Use | No | 89 | 62.7% |
| Yes | 53 | 37.3% | |
| Alcohol Long Term and Current (gm/day) | Low | 53 | 36.8% |
| Mod | 68 | 47.2% | |
| High | 23 | 16.0% | |
| Race/Ethnicity | Non-Hispanic White | 124 | 86.1% |
| Black | 8 | 5.6% | |
| Hispanic | 9 | 6.3% | |
| Other/NA | 3 | 2.1% | |
| Education (years) | <High School | 24 | 16.7% |
| High School | 42 | 29.2% | |
| Some College | 50 | 34.7% | |
| College Grad/Higher | 28 | 19.4% | |
|
| |||
| Median | Interquartile Range (IQR) | ||
|
| |||
| Age (years) | 65.0 | (56, 71) | |
| BMI (kg/m2) | 29.6 | (26.6, 33.2) | |
| Hours Moderate Physical Activity/week | 1.5 | (0.0, 3.9) | |
|
|
|||
| Meat Consumption | |||
| Processed meat (2–3 oz servings/day) | Low1 | 0.05 | (0.01, 0.07) |
| Intermediate | 0.19 | (0.14, 0.23) | |
| High | 0.45 | (0.35, 0.59) | |
| Red Meat (2–3 oz servings/day) | Low | 0.43 | (0.29, 0.61) |
| Intermediate | 0.95 | (0.84, 1.07) | |
| High | 1.58 | (1.41, 1.93) | |
|
|
|||
| Non-Meat Consumption | |||
| Calcium (mg/1000 Kcal) | Low | 300.71 | (261.72, 326.90) |
| Intermediate | 406.47 | (380.31, 445.51) | |
| High | 585.69 | (541.01, 697.95) | |
| Fiber (g/1000 Kcal) | Low | 7.32 | (6.75, 8.09) |
| Intermediate | 10.03 | (9.52, 10.43) | |
| High | 13.06 | (12.15, 15.62) | |
| Folate (mcg/1000 Kcal) | Low | 118.35 | (103.41, 126.46) |
| Intermediate | 151.61 | (142.85, 161.84) | |
| High | 203.63 | (189.98, 247.94) | |
| Vegetables (1/2 cup servings/day) | Low | 1.41 | (1.03, 1.90) |
| Intermediate | 2.85 | (2.55, 3.25) | |
| High | 4.59 | (4.13, 5.74) | |
| Whole Grains (servings/day) | Low | 0.43 | (0.33, 0.63) |
| Intermediate | 1.12 | (1.02, 1.29) | |
| High | 2.14 | (1.82, 2.52) | |
Seven genes were differentially expressed for dietary calcium and folate (Table 2). Sixty-five genes were differentially expressed between high and low intake categories among non-meat variables (i.e. Prudent Dietary Pattern, fruits, vegetables, and whole grains). This can be further broken down to one gene with Prudent Dietary Pattern, one gene with fruit intake, 26 genes with vegetable intake, and 37 genes with whole grain intake, (Table 3, FDR <0.1). Several genes were identified as differentially expressed between consumption categories for multiple dietary variables. TXNDC17 was upregulated for both Prudent Dietary Pattern and vegetable intake; MUC5AC was down regulated for vegetable intake, whole grain intake, and dietary folate. FOXJ2, NECAP1, and C3AR1 were upregulated for both calcium intake and vegetable intake Using a more stringent FDR <0.05, we identify one gene with differential expression between calcium intake categories, one with Prudent Dietary Pattern, 13 with vegetable intake, and eight with whole grain intake (Table 2 and Table 3). Among the genes with an FDR <0.05, FOXJ2 was upregulated with high calcium intake and TXNDC17 was upregualted with high Prudent Dietary Pattern. In contrast, five of the eight genes associated with whole grains were downregulated and four of the 13 genes associated with vegetables were downregulated.
Table 2.
Associations between nutrients and gene expression (FDR<0.1)1
| Gene | Average Adjusted Count | DESeq2 Estimated Log2 Fold Change | P Value | Multiple. Comparison Adjusted P Value | ||
|---|---|---|---|---|---|---|
| Low Tertile | High Tertile | |||||
| Calcium | FOXJ2 | 32.84 | 135.64 | 0.87 | 4.05E-10 | 7.07E-06 |
| NECAP1 | 23.38 | 68.92 | 0.71 | 3.87E-07 | 2.38E-03 | |
| C3AR1 | 3.51 | 21.28 | 0.63 | 4.09E-07 | 2.38E-03 | |
| Folate | A1BG | 3.84 | 8.45 | 0.63 | 2.12E-05 | 9.22E-02 |
| SLC37A2 | 23.04 | 79.47 | 0.61 | 1.83E-05 | 9.22E-02 | |
| CLDN4 | 129.49 | 87.70 | −0.47 | 1.44E-05 | 9.22E-02 | |
| MUC5AC | 58.73 | 5.49 | −0.50 | 1.52E-05 | 9.22E-02 | |
Associations are adjusted for age and sex
Table 3.
Associations between non-meat foods and gene expression (FDR <0.1)1
| Gene | Average Adjusted Count | DESeq2 Estimated Log2 Fold Change | P Value | Multiple. Comparison Adjusted P Value | ||
|---|---|---|---|---|---|---|
| Low Tertile | High Tertile | |||||
| Prudent Diet | TXNDC17 | 6.18 | 16.74 | 0.68 | 4.00E-07 | 6.99E-03 |
| Fruit | HMGN2 | 8.45 | 16.66 | 0.57 | 4.31E-06 | 7.50E-02 |
| Vegetables | ANP32A | 64.67 | 622.05 | 0.86 | 3.22E-10 | 2.90E-06 |
| NECAP1 | 30.12 | 160.72 | 0.76 | 5.98E-08 | 1.94E-04 | |
| HELZ | 639.22 | 10029.11 | 0.66 | 6.49E-08 | 1.94E-04 | |
| SLC2A3 | 50.41 | 263.66 | 0.75 | 9.46E-08 | 1.95E-04 | |
| FOXJ2 | 53.51 | 337.01 | 0.72 | 1.09E-07 | 1.95E-04 | |
| ADAMTS6 | 9.00 | 40.07 | 0.63 | 5.57E-06 | 6.26E-03 | |
| HSPG2 | 285.80 | 190.14 | −0.48 | 4.47E-06 | 6.26E-03 | |
| C3AR1 | 7.36 | 55.37 | 0.57 | 4.98E-06 | 6.26E-03 | |
| TXNDC17 | 6.23 | 14.92 | 0.62 | 9.03E-06 | 9.03E-03 | |
| PXN | 124.57 | 91.47 | −0.39 | 1.54E-05 | 1.26E-02 | |
| MUC5AC | 49.01 | 3.56 | −0.48 | 1.51E-05 | 1.26E-02 | |
| FURIN | 55.18 | 36.93 | −0.45 | 2.09E-05 | 1.56E-02 | |
| DNAH14 | 21.82 | 62.62 | 0.58 | 6.69E-05 | 4.63E-02 | |
| RNF123 | 45.86 | 30.57 | −0.40 | 7.22E-05 | 4.64E-02 | |
| MICALL2 | 81.80 | 57.71 | −0.41 | 9.91E-05 | 5.94E-02 | |
| COL1A1 | 478.40 | 278.35 | −0.53 | 1.09E-04 | 6.15E-02 | |
| COL12A1 | 94.94 | 58.32 | −0.49 | 1.22E-04 | 6.27E-02 | |
| STK36 | 23.04 | 14.94 | −0.43 | 1.25E-04 | 6.27E-02 | |
| REG4 | 75.69 | 30.88 | −0.54 | 1.68E-04 | 7.96E-02 | |
| AMN | 65.79 | 41.60 | −0.47 | 1.92E-04 | 8.62E-02 | |
| PTER | 10.51 | 16.89 | 0.44 | 2.02E-04 | 8.64E-02 | |
| ACAP3 | 56.17 | 37.98 | −0.41 | 2.24E-04 | 9.17E-02 | |
| TAF1C | 45.25 | 31.54 | −0.42 | 2.70E-04 | 9.83E-02 | |
| TTYH3 | 60.34 | 44.08 | −0.39 | 2.58E-04 | 9.83E-02 | |
| EPM2AIP1 | 44.29 | 60.95 | 0.34 | 2.73E-04 | 9.83E-02 | |
| ATP11C | 22.62 | 41.25 | 0.49 | 2.87E-04 | 9.94E-02 | |
| Whole Grains | MUC5AC | 55.79 | 3.17 | −0.62 | 2.52E-06 | 2.47E-02 |
| BCKDHA | 16.28 | 7.89 | −0.69 | 3.59E-06 | 2.47E-02 | |
| BTG1 | 74.46 | 100.65 | 0.42 | 6.99E-06 | 3.20E-02 | |
| ANKRD30BL | 118.66 | 53.70 | −0.70 | 1.05E-05 | 3.61E-02 | |
| ABCC3 | 341.13 | 253.51 | −0.41 | 1.54E-05 | 3.90E-02 | |
| MPEG1 | 33.40 | 49.38 | 0.50 | 2.25E-05 | 3.90E-02 | |
| EPS8L3 | 143.09 | 103.38 | −0.42 | 2.27E-05 | 3.90E-02 | |
| JMJD7 | 11.64 | 6.22 | −0.63 | 1.97E-05 | 3.90E-02 | |
| DVL1 | 39.48 | 25.55 | −0.53 | 5.97E-05 | 5.46E-02 | |
| HELZ2 | 91.24 | 58.30 | −0.52 | 5.32E-05 | 5.46E-02 | |
| GSTM3 | 4.19 | 9.13 | 0.64 | 5.94E-05 | 5.46E-02 | |
| NR1I2 | 56.26 | 39.67 | −0.46 | 4.25E-05 | 5.46E-02 | |
| GMPPB | 54.06 | 40.48 | −0.40 | 4.89E-05 | 5.46E-02 | |
| AMIGO3 | 55.35 | 41.00 | −0.42 | 4.93E-05 | 5.46E-02 | |
| C9orf172 | 10.88 | 6.23 | −0.60 | 5.86E-05 | 5.46E-02 | |
| TPRN | 54.68 | 35.53 | −0.53 | 6.45E-05 | 5.54E-02 | |
| EMB | 14.29 | 24.80 | 0.58 | 6.99E-05 | 5.65E-02 | |
| RASSF7 | 37.58 | 23.88 | −0.51 | 8.00E-05 | 5.98E-02 | |
| KCTD13 | 10.46 | 5.71 | −0.58 | 8.28E-05 | 5.98E-02 | |
| RTEL1-TNFRSF6B | 89.14 | 62.54 | −0.43 | 9.18E-05 | 6.30E-02 | |
| C9orf50 | 11.32 | 2.44 | −0.61 | 9.77E-05 | 6.39E-02 | |
| PFKL | 144.14 | 108.76 | −0.38 | 1.09E-04 | 6.76E-02 | |
| MRPS28 | 3.04 | 7.82 | 0.62 | 1.13E-04 | 6.76E-02 | |
| LTBR | 69.00 | 55.07 | −0.33 | 1.27E-04 | 6.96E-02 | |
| MST1R | 97.85 | 71.06 | −0.41 | 1.26E-04 | 6.96E-02 | |
| TERC | 27.56 | 14.83 | −0.58 | 1.37E-04 | 7.21E-02 | |
| FAM129B | 113.83 | 79.80 | −0.44 | 1.51E-04 | 7.68E-02 | |
| CTC-435M10.3 | 11.71 | 6.39 | −0.57 | 1.57E-04 | 7.70E-02 | |
| FAM83H | 73.50 | 53.33 | −0.41 | 1.68E-04 | 7.86E-02 | |
| CHKB-CPT1B | 70.38 | 48.67 | −0.45 | 1.72E-04 | 7.86E-02 | |
| SEMA3B | 54.89 | 35.79 | −0.50 | 1.95E-04 | 8.37E-02 | |
| DPM2 | 18.77 | 11.53 | −0.51 | 1.97E-04 | 8.37E-02 | |
| CD151 | 79.03 | 57.71 | −0.42 | 2.01E-04 | 8.37E-02 | |
| RNF146 | 13.31 | 19.88 | 0.46 | 2.45E-04 | 9.39E-02 | |
| SEMA4B | 52.73 | 35.60 | −0.45 | 2.39E-04 | 9.39E-02 | |
| NOC2L | 35.57 | 24.81 | −0.44 | 2.46E-04 | 9.39E-02 | |
| EML2 | 21.68 | 13.83 | −0.46 | 2.68E-04 | 9.96E-02 | |
Associations are adjusted for age and sex
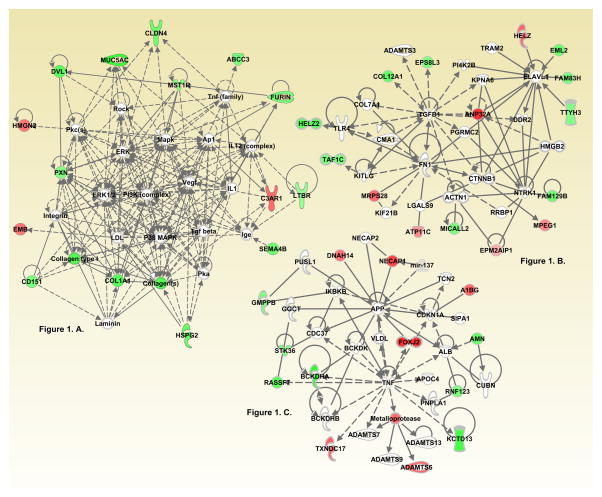
Our IPA analysis found three interaction networks significantly associated with non-meat consumption (IPA Network Score ≥20). These IPA networks were identified as functioning with: cancer, organismal injury and abnormalities, and tumor morphology (Score=31, Focus Molecules=11) (Figure 1A); cellular function, maintenance, cellular movement, cell death and survival (Score=31, Focus Molecules=15) (Figure 1B), and drug metabolism, molecular transport, and small molecule biochemistry (Score=26, Focus Molecules=13) (Figure 1C).
Figure 1. IPA networks associated with dysregulated genes based on level of non-meat-related dietary intake.
Figure 1A. Hematological system development and function, nervous system development and function, tissue morphology (Score 31)
Figure 1B. Connective tissue disorders, organismal injury and abnormalities, cardiovascular system development and function (Score 29)
Figure 1C. Cellular compromise, cellular function and maintenance, protein degradation (Score 21)
Red indicates up-regulation and green indicates down regulation. Node shapes denote enzymes
 ; phosphatases Δ; kinases
; phosphatases Δ; kinases
 ; peptidases
; peptidases
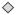 , G-protein coupled receptor
, G-protein coupled receptor
 ; transmembrane receptor
; transmembrane receptor
 ; cytokines
; cytokines
 ; ion channel
; ion channel
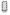 ; transporter
; transporter
 ; translation regulator
; translation regulator
 ; transcription regulator
; transcription regulator
 ;; and complex/group
;; and complex/group

Among genes identified as significantly differentially expressed between high and low meat-related intake categories, 64 genes were differentially expressed with red meat, two genes were differentially expressed with processed meat, and three genes were differentially expressed with a Western Dietary Pattern (Table 4). Of these, eight genes were differentially expressed by level of red meat and LSAMP was differentially expressed by level of Western Dietary Pattern when using a more stringent selection criterion of a maximal FDR of <0.05. The majority of these genes were downregulated by red meat consumption; of all of the genes identified only IL22RA1, SLC17A4, SLC25A25, MUC17, ZFY, FRK, PEX11G, NEDD9, NR3C2, and RPS4Y1 were upregulated by red meat consumption. The two genes identified as being differentially expressed for Western Dietary Pattern also were differentially expressed for red meat; C3 was downregulated by red meat and processed meat.
Table 4.
Association between meat-related foods and gene expression (FDR<0.1)1
| Gene | Average Adjusted Count | DESeq2 Estimated Log2 Fold Change | P Value | Multiple. Comparison Adjusted P Value | ||
|---|---|---|---|---|---|---|
| Low Tertile | High Tertile | |||||
| Western Diet | LSAMP | 7.21 | 1.51 | −0.70 | 2.20E-06 | 3.83E-02 |
| TDRD15 | 2.21 | 0.21 | −0.57 | 6.21E-06 | 5.42E-02 | |
| Meat | ONECUT2 | 12.45 | 2.07 | −0.92 | 2.40E-08 | 4.18E-04 |
| TIMP1 | 26.41 | 11.51 | −0.76 | 1.39E-06 | 1.22E-02 | |
| CLDN1 | 12.53 | 2.57 | −0.77 | 3.78E-06 | 1.32E-02 | |
| CMSS1 | 11.46 | 4.35 | −0.77 | 3.34E-06 | 1.32E-02 | |
| LSAMP | 7.47 | 1.56 | −0.79 | 3.58E-06 | 1.32E-02 | |
| SPARC | 118.63 | 74.35 | −0.55 | 8.19E-06 | 2.38E-02 | |
| TDRD15 | 1.98 | 0.14 | −0.64 | 1.54E-05 | 3.84E-02 | |
| COL1A2 | 314.64 | 214.85 | −0.54 | 1.97E-05 | 4.29E-02 | |
| CEMIP | 27.24 | 16.30 | −0.56 | 3.66E-05 | 6.73E-02 | |
| NKD1 | 16.63 | 6.86 | −0.69 | 3.86E-05 | 6.73E-02 | |
| ZFY | 4.11 | 9.63 | 0.65 | 5.84E-05 | 7.01E-02 | |
| FZD3 | 17.37 | 9.06 | −0.63 | 6.32E-05 | 7.01E-02 | |
| COL4A2 | 155.42 | 108.31 | −0.50 | 6.44E-05 | 7.01E-02 | |
| TP53 | 33.67 | 23.40 | −0.44 | 4.87E-05 | 7.01E-02 | |
| SLC17A4 | 46.78 | 78.07 | 0.58 | 6.44E-05 | 7.01E-02 | |
| CCT2 | 27.99 | 18.04 | −0.54 | 5.51E-05 | 7.01E-02 | |
| HSPA5 | 50.38 | 34.01 | −0.50 | 8.23E-05 | 8.24E-02 | |
| TPX2 | 18.50 | 8.63 | −0.61 | 1.47E-04 | 8.24E-02 | |
| FRK | 15.94 | 23.66 | 0.50 | 1.44E-04 | 8.24E-02 | |
| PTK7 | 18.21 | 10.62 | −0.57 | 8.63E-05 | 8.24E-02 | |
| NCL | 109.20 | 80.61 | −0.40 | 1.03E-04 | 8.24E-02 | |
| IL22RA1 | 11.53 | 22.37 | 0.61 | 1.17E-04 | 8.24E-02 | |
| GPR26 | 5.57 | 0.71 | −0.55 | 1.46E-04 | 8.24E-02 | |
| EGFLAM | 4.92 | 1.58 | −0.65 | 1.30E-04 | 8.24E-02 | |
| SLC6A5 | 2.59 | 0.18 | −0.51 | 1.10E-04 | 8.24E-02 | |
| RAB31 | 19.12 | 10.49 | −0.60 | 1.06E-04 | 8.24E-02 | |
| TMEM107 | 37.26 | 23.81 | −0.56 | 1.24E-04 | 8.24E-02 | |
| C1S | 63.12 | 47.50 | −0.43 | 1.37E-04 | 8.24E-02 | |
| C2orf27A | 2.37 | 0.50 | −0.57 | 1.23E-04 | 8.24E-02 | |
| IGHG1 | 171.81 | 74.48 | −0.66 | 1.00E-04 | 8.24E-02 | |
| MUC5AC | 50.46 | 5.63 | −0.54 | 1.30E-04 | 8.24E-02 | |
| HSPA9 | 71.14 | 53.20 | −0.40 | 1.52E-04 | 8.26E-02 | |
| TMEM132B | 6.82 | 2.55 | −0.65 | 1.56E-04 | 8.26E-02 | |
| DCN | 39.88 | 23.71 | −0.55 | 1.98E-04 | 8.40E-02 | |
| SNX10 | 7.08 | 2.27 | −0.64 | 1.99E-04 | 8.40E-02 | |
| AURKA | 10.06 | 3.66 | −0.63 | 2.06E-04 | 8.40E-02 | |
| PEX11G | 1.32 | 3.29 | 0.64 | 1.76E-04 | 8.40E-02 | |
| COL1A1 | 445.08 | 285.52 | −0.55 | 1.87E-04 | 8.40E-02 | |
| NEDD9 | 84.09 | 112.98 | 0.38 | 1.96E-04 | 8.40E-02 | |
| WDR89 | 11.37 | 5.46 | −0.59 | 1.77E-04 | 8.40E-02 | |
| SLC25A25 | 35.34 | 54.48 | 0.46 | 1.73E-04 | 8.40E-02 | |
| NR3C2 | 113.10 | 154.48 | 0.41 | 1.95E-04 | 8.40E-02 | |
| C1R | 44.16 | 25.70 | −0.53 | 2.09E-04 | 8.40E-02 | |
| MUC17 | 107.73 | 269.60 | 0.63 | 2.12E-04 | 8.40E-02 | |
| RPS4Y1 | 8.70 | 19.26 | 0.59 | 2.28E-04 | 8.82E-02 | |
| DOCK3 | 6.29 | 2.66 | −0.62 | 2.86E-04 | 9.55E-02 | |
| CHRAC1 | 14.62 | 8.35 | −0.51 | 2.90E-04 | 9.55E-02 | |
| SPATA17 | 2.21 | 0.55 | −0.58 | 2.82E-04 | 9.55E-02 | |
| NETO1 | 3.40 | 0.46 | −0.52 | 2.86E-04 | 9.55E-02 | |
| TRIL | 7.68 | 4.13 | −0.60 | 2.83E-04 | 9.55E-02 | |
| DDX53 | 1.70 | 0.06 | −0.47 | 2.71E-04 | 9.55E-02 | |
| HIST1H2BN | 5.55 | 1.23 | −0.61 | 2.74E-04 | 9.55E-02 | |
| PEG10 | 6.38 | 2.44 | −0.62 | 2.74E-04 | 9.55E-02 | |
| THY1 | 19.83 | 10.36 | −0.58 | 3.03E-04 | 9.65E-02 | |
| GRM2 | 3.19 | 1.31 | −0.59 | 3.07E-04 | 9.65E-02 | |
| TTC40 | 4.30 | 0.77 | −0.56 | 3.10E-04 | 9.65E-02 | |
| TIMP2 | 94.56 | 68.12 | −0.42 | 3.30E-04 | 9.71E-02 | |
| NOS1 | 6.65 | 0.87 | −0.51 | 3.48E-04 | 9.71E-02 | |
| CLSPN | 12.16 | 4.89 | −0.61 | 3.54E-04 | 9.71E-02 | |
| C3 | 102.46 | 57.73 | −0.56 | 3.62E-04 | 9.71E-02 | |
| CCDC150 | 7.05 | 3.32 | −0.61 | 3.22E-04 | 9.71E-02 | |
| VWDE | 4.91 | 1.77 | −0.61 | 3.50E-04 | 9.71E-02 | |
| CCT3 | 36.04 | 24.11 | −0.44 | 3.37E-04 | 9.71E-02 | |
| HSP90B1 | 142.80 | 105.92 | −0.38 | 3.61E-04 | 9.71E-02 | |
| Processed Meat | C3 | 101.05 | 50.36 | −0.70 | 5.56E-06 | 6.44E-02 |
| KRT1 | 2.97 | 0.08 | −0.60 | 7.40E-06 | 6.44E-02 | |
Associations are adjusted for age and sex
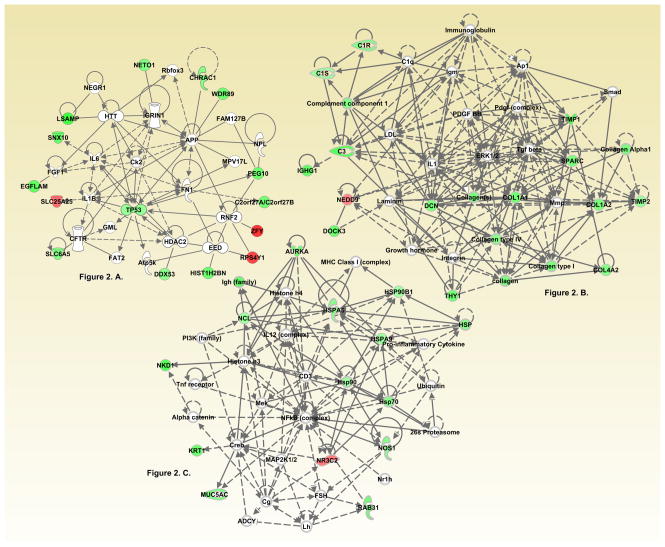
In IPA analysis, three interaction networks were significantly associated (Score ≥20) with meat-related variables (i.e. red meat, processed meat, and Western Dietary Pattern). These IPA networks were identified as Hematological system developmental function, nervous system development and function, Tissue Morphology (IPA Network Score = 31, Focus Molecules 22) (Figure 2A); Connective Tissue Disorders, Organismal Injury and Abnormalities, Cardiovascular System Development and Function (Score=29, Focus Molecules=14) (Figure 2B); and cellular compromise, cellular function and maintenance, protein degradation (Score=21, Focus Molecules 11) (Figure 2C). Online Supplement 1 shows genes up and down regulated in each of these networks. Some genes were differentially expressed between high and low consumption categories for both meat and non-meat dietary variables. COL1A1 was down-regulated for vegetables and meat intake; MUC5AC was down-regulated for folate, vegetables, whole grains, and meat. Foods and nutrients analyzed that were not associated with any differential gene expression (FDR <0.1) were mutagen index, total dairy products, carbohydrates, trans-fatty acids, total dietary fat, dietary fiber, refined grain products, and sucrose. Additionally, we observed no differences in gene expression by level of vigorous physical activity.
Figure 2. IPA networks associated with dysregulated genes based on level of meat related dietary intake.
Figure 2A. Cancer, organismal injury and abnormalities, and tumor morphology (Score 31)
Figure 2B. Cellular function and maintenance, cellular movement, cell death and survival (Score 31)
Figure 2C. Drug metabolism, molecular transport, small molecule biochemistry (Score 26)
Red indicates up-regulation and green indicates down regulation. Node shapes denote enzymes
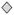 ; phosphatases Δ; kinases
; phosphatases Δ; kinases
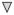 ; peptidases
; peptidases
 , G-protein coupled receptor
, G-protein coupled receptor
 ; transmembrane receptor
; transmembrane receptor
 ; cytokines
; cytokines
 ; ion channel
; ion channel
 ; transporter
; transporter
 ; translation regulator
; translation regulator
 ; transcription regulator
; transcription regulator
 ;; and complex/group
;; and complex/group

Discussion
Our study provides support for associations between level of dietary intake and gene expression in normal colonic mucosa utilizing RNA-Seq which allowed assessment of over 17,000 genes. Some genes were differentially expressed by intake level across multiple dietary variables, generally in the same direction, although there was limited overlap between meat and non-meat variables. We observed three IPA interaction networks highly associated with meat consumption and three IPA interaction networks associated with non-meat consumption. These data suggest that dietary influence on gene expression plays an important role in a number of physiologic and pathologic processes.
The majority of genes that were differentially expressed with high red meat consumption were downregulated. These genes predominately play a role in extracellular matrix (ECM) assembly and structure, calcium binding, and the regulation of signal transduction via the Wnt signaling pathway. The canonical Wnt signaling pathway triggers β-catenin-dependent gene expression, allowing cell cycle progression at the G1/S transition [39]. In particular, we found that high intake of meat correlated with downregulation of FZD3, TMEM132B, CCDC150, EGFLAM, and NKD1. Both TMEM132B and NKD1 are known to downregulate Wnt signaling [40]. Thus meat consumption can contribute to the dysregulation of a proto-oncogenic pathway. At the same time, we found that high red meat consumption was associated with a downregulation of TP53, encoding for the tumor suppressor p53. Both of these findings are consistent with previous findings that red meat intake can induce pro-carcinogenic gene expression changes through the p53- and Wnt signaling pathways [19]. This is possibly due to the fact that red meat is high in dietary heme, a luminal irritant associated with hyperproliferation and hyperplasia through the downregulation of feedback inhibitors, including Wnt inhibitors [17]. These findings add to the literature that suggest red meat increases risk of several chronic diseases including cancer.
High red meat consumption also downregulated the expression of genes associated with ECM. Dysregulation of genes involved in the regulation of the ECM and cell matrix adhesion, such as matrix metalloproteinases (MMPs), collagen-related genes such as COL4A2, COL1A2, and COL1A1, tissue inhibitor of metalloproteinases (TIMPs), and one cut homeobox 2 (ONECUT2) have been implicated in a variety of diseases [41]. Moreover, red meat is associated with both acute oxidative stress and delayed cytotoxic stress [17]. Several genes dysregulated by meat, including THY1, NCL, COL4A2, and KRT1, contribute to metastasis and angiogenesis. We found that diet downregulated a variety of genes associated with ECM assembly and maintenance, including TIMP1, providing an additional mechanistic link between red meat and disease processes.
It is also worth noting that red meat upregulated some genes, namely genes with products involved in phosphate transport (SLC17A4 and SLC25A25) and cytokine signaling (MUC17 and IL22RA1). Diet is known to play an important role in bone health, with meat protein increasing acid load and regulating calcium balance [42]. The upregulation of genes involved in phosphate transport, as well as our observed downregulation of calcium binding genes (SPARC, CEMIP, HSPA5, HSP901B, VWDE, SLC25A25, CIR, CLDN1, and EGFLAM), may help explain the mechanism by which meat consumption modulates bone metabolism, beyond the effects of dietary protein on acid load. Moreover, the upregulation of MUC17 and IL22RA1 may play a role in meat-induced inflammation and inflammation-related diseases. The exact function of MUC17 is unknown; however, other mucins are reportedly involved in the regulation of inflammation [43] and MUC17 expression is related to certain cancers, including pancreatic cancer [44]. IL22 is produced by T lymphocytes and mediates cellular inflammatory responses via the activation of intracellular kinases and has been implicated in a variety of inflammatory diseases, including colon cancer and ulcerative colitis [45], both of which have been associated with red meat consumption [17, 46].
Several genes associated with immune response were down-regulated by high meat consumption, including TRIL and IGFGH1. We found that high red meat consumption decreased expression of C1R and C1S. C1S encodes for a component of C1 in the classic complement pathway; CIR is the R subcomponent of C1. The downregulation of the components of the complement cascade by meat consumptions seems paradoxical; however, evidence suggests that the proximal classical complement components may provide a protective effect against atherosclerosis [47, 48]. While atherosclerosis is an inflammatory disease, Hovland et al. suggest that the classic complement pathway may be important for tissue homeostasis by the clearance of cell debris and immune complexes [48]. Thus it is possible that a downregulation of proximal components of the classic pathway, such as components of C1, could paradoxically contribute to the pro-inflammatory effects of red meat. On the other hand, upregulation of MUC17 by meat could help explain its association with a number of factors, including inflammation [20, 46].
Further evaluation of the genes associated with non-meat related variables showed that genes associated with calcium were upregulated, two of the four genes associated with folate were upregulated, and the TXNDC17 gene associated with Prudent Dietary Pattern was upregulated. The genes associated with calcium consumption were involved in transmembrane transport, regulation of transcription and translation, cell survival and the complement system. Calcium plays a key role in muscle contraction and relaxation, blood coagulation, nerve transmission and keratinocyte differentiation [49]. The genes upregulated by high calcium consumption include FOXJ2, encoding for a transcription activator; NECAP1, whose product is involved in receptor mediated endocytosis; and C3AR1, encoding for a C3A receptor. A Prudent Dietary Pattern was associated with roughly three servings of milk per day in one study [20] and was associated with decreased risk of cancer and the B cell receptor pathways [20]. Our observed calcium-associated upregulation of tumor suppressors and a C3A receptor is consistent with these earlier findings.
Folate-rich foods include green vegetables and citrus fruits, and folate plays a key role as a coenzyme involved in DNA synthesis, amino acid metabolism, and methylations [50]. High versus low folate intake was associated with the upregulation of SLC37A2 and CLDN4 and the downregulation of MUC5AC. The gene products for SLC37A2 and CLDN4 are involved in carbohydrate transport and tight junction organization, respectively. Tight junction proteins, such as claudins, are related to NF-κB, mTOR and Nrf2 [51, 52], while the expression of tight junction proteins is associated with amino acids, such as arginine and tryptophan [51, 52]. As folic acid regulates amino acid metabolism, this could explain its role in modulating the expression of tight junction proteins. Dietary folate also downregulates the expression of MUC5AC, as some mucins have been associated with inflammation [43], this could help explain the reported association between folate and certain cancers and cardiovascular diseases [50]. We also found that a Prudent Dietary Pattern upregulated TXNDC17, which modulates NF-κB. NF-κB signaling plays a role in inflammation, apoptosis and a variety of cellular stress responses [15]. TXNDC17 overexpression has been shown to inhibit TNF-α induced activation of NF-κB [53], thus its upregulation could contribute to the decrease in inflammation and cancer previously noted with a Prudent Dietary Pattern [20].
We found that the majority of genes differentially expressed between intake levels of whole grains and vegetables were down-regulated, especially for vegetable intake. The genes that are differentially expressed with level of whole grain and vegetable consumption are associated with NF-κB signaling, regulation of apoptosis, cytoskeleton dynamics, and carbohydrate metabolism. Carbohydrates are important modulators of insulin action on glucose metabolism [54], which is consistent with our findings regarding PFKL, ABCC3 and GMPPB down-regulated expression with whole grain consumption and HSPG2 and REG4 which were down-regulated by high vegetable consumption. Both fiber and polyphenols attenuate inflammation [46]. Phenolic compounds are major bioactive compounds in whole grains and polyphenols are known to interfere with the NF-κB signaling pathway [55], which is consistent with our data showing a downregulation of genes involved in the NF-κB signaling. Our data suggests that the previously described polyphenolic perturbation of nuclear signaling, and anti-inflammatory effects [56], may be explained by the differential gene expression seen with both whole grain and vegetable levels of intake.
Some genes were up-regulated by high vegetable intake, including ANP32A, NECAP1, and HELZ. ANP32A, encodes for a tumor suppressor [57]; HELZ, encodes an RNA helicase, which has been shown in conjunction with other genes to regulate downstream genes as a transcription factor containing histone methyltransferase activity [58]. In association with SMYD3, HELZ plays a role in the proliferation of cancer cells [58]. It has also been suggested that HELZ plays a wider role in global translation activation and the phosphorylation (activation) of Ribosomal protein S6, which in turn activates eIF4b [59]. This indicates that the upregulation of HELZ, could help explain the association of vegetables with several factors, including inflammation [20, 46] through the translational activation of numerous genes within the inflammatory process.
IPA found three interaction networks with meat consumption and three interaction networks associated with non-meat consumption. This correlates with earlier IPA studies showing that Western Dietary Pattern is associated with five canonical pathways related to cancer, six pathways related to immune and/or inflammatory responses and three pathways related to cardiovascular signaling and that Prudent Dietary Pattern is associated with nine IPA canonical pathways associated with immune/inflammatory response, and six pathways associated with cancer [20]. It has previously been noted that Western Dietary Patterns are associated with a pro-inflammatory gene expression profile and that Prudent Dietary patterns are associated with an anti-inflammatory gene expression profile [60], which would be consistent with our described networks. Moreover our networks suggest that while both meat and non-meat related dietary variables are associated with the cellular functions of intracellular transport and ECM regulation, the signal transduction pathways differ between meat and non-meat networks. We found meat consumption to be associated with TP53, NFκB1, and MAPK signaling through the ERK1/2 signaling pathway. The Ras-MAPK system is involved in cell proliferation [61]; moreover, our findings in this network are consistent with earlier findings that high fat diets, the cooked meat carcinogen 2-Amino-1-Methyl-6-phenylimidazo[4,5-b]-pyridine (PhiP), and sodium nitrate can activate the MAPK pathways [9, 16, 62]. Non-meat consumption appears to play a greater role in the regulation of the PI3K complex, TNF-signaling, and TGFB1-signaling pathways. All of these pathways have been identified as major signaling pathways for colon cancer [63–66].
To the best of our knowledge this is one of the few studies to examine the role of diet on gene expression using data from a population-based study and comprehensive gene expression data. Our study has a number of strengths including using RNA-Seq which produces global gene expression data for each RNA sample and is an ideal method to undertake a discovery study such as ours [67, 68]. However, it is important to keep in mind that the gene expression profile is most relevant to current diet exposure. The time between tissue ascertainment and referent period for diet in our study could be from several months to three or possibly four years. While lack of findings could indicate a disparity in time between exposure and tissue sample acquirement, finding associations would imply that the exposure is recent enough, in that dietary patterns are consistent over time, to alter the expression. Additionally, we have utilized normal colonic mucosa, so genes would have to be expressed in colon tissue for detection. It should be recognized that different platforms carry different technical strengths and weaknesses that can influence results. Thus, it is essential to validate these findings in other populations using the methods we used here as well as other platforms to better understand associations between diet and gene expression. We utilized DESeq2 to assess gene expression data adjusting for age and sex. We previously have shown associations between cigarette smoking and alcohol and gene expression in these samples [69]. Likewise, we report statistically significant differential expression as an indicator of dysregulated genes, however, it is not clear the level of dysregulation that is necessary to result in functional significance.
Although this study was conducted in a rigorous manner, there are limitations. Our dietary data were collected shortly after diagnosis. Although our dietary questionnaire allowed for reporting of over 800 food items and we implemented extremely rigid quality control procedures, it is possible that recall could have been influenced by the disease status. While this is a possibility, it should be noted that our risk estimates for diet and colon cancer are almost identical to those reported by large cohort studies [70]. Additionally, these data reflect foods consumed in the early 1990s. While we believe that people still eat the foods that we report here, different dietary patterns may be more important today than the Western and Prudent diet that we report.
Our data support the hypothesis that diet is associated with gene expression. Our data showed that diet was involved in de-regulation of genes involved in numerous pathways and functions such as Wnt-signaling, MAPK, NF-κB, TP53, ECM maintenance, cytoskeletal structure, and cell cycle regulation. These data suggest that differential gene expression associated with dietary intake is a possible mechanism by which diet can influence a variety of biological processes and functions. Focused functionality studies evaluating dietary influence on gene expression are needed.
Supplementary Material
Supplemental Table 1. Top genes associated with meat and non-meat networks from IPA
Acknowledgments
This study was funded by NCI grants CA48998. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. We would like to acknowledge the contributions of Dr. Bette Caan, Judy Morse and Donna Schaffer and the Kaiser Permanente Medical Research Program, and Sandra Edwards for data collection and organization, Jennifer Herrick for data management, Erica Wolff and Michael Hoffman for RNA extraction, Wade Samowitz for slide review, and Brett Milash at the Bioinformatics Core Facility at the University of Utah.
Abbreviations
- KPMRP
Kaiser Permanente Medical Research Program
- APC
adenomatous polyposis coli
- BH
Benjamini and Hochberg procedure
- FDR
false discovery rate
- IPA
Ingenuity Pathway Analysis
- Wnt
Wingless Type
- ECM
Extracellular Matrix
- MMP
matrix metalloproteinase
- TIMPs
tissue inhibitor of metalloproteinases
- FGF1
acidic fibroblast growth factor
- IL
interleukin
- NF-κB
nuclear factor kappa b
- mTOR
mechanistic target of rapamycin
- Nrf2
NF-E2-related factor 2
- PAK
p21-activated kinase
- MAPK
mitogen activated protein kinase
- PhiP
2-Amino-1-Methyl-6-phenylimidazo[4,5-b]-pyridine
Footnotes
Competing Interests and
All authors declare that they have no competing financial interests
Author’s Contributions
AP compiled and interpreted the data and wrote the manuscript; DP conducted statistical analysis, edited manuscript and approved final version of manuscript; LM did bioinformatics analysis and approved final manuscript; RW oversaw RNAseq data collection and approved final manuscript. MS obtained funding, collected data for study, assisted in data interpretation and analysis, edited and approved final manuscript.
Contributor Information
Andrew J. Pellatt, Email: apellatt@tulane.edu.
Martha L. Slattery, Email: Marty.Slattery@hsc.utah.edu.
Lila E. Mullany, Email: Lila.Mullany@hsc.utah.edu.
Roger K. Wolff, Email: Roger.Wolff@hsc.utah.edu.
Daniel F. Pellatt, Email: Daniel.Pellatt@hsc.utah.edu.
References
- 1.McDaniel J, Askew W, Bennett D, Mihalopoulos J, Anantharaman S, Fjeldstad AS, Rule DC, Nanjee NM, Harris RA, Richardson RS. Bison meat has a lower atherogenic risk than beef in healthy men. Nutrition research. 2013;33(4):293–302. doi: 10.1016/j.nutres.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres N, Guevara-Cruz M, Velazquez-Villegas LA, Tovar AR. Nutrition and Atherosclerosis. Archives of medical research. 2015 doi: 10.1016/j.arcmed.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Sarbagili-Shabat C, Sigall-Boneh R, Levine A. Nutritional therapy in inflammatory bowel disease. Current opinion in gastroenterology. 2015;31(4):303–308. doi: 10.1097/MOG.0000000000000178. [DOI] [PubMed] [Google Scholar]
- 4.Yang CQ, Shu L, Wang S, Wang JJ, Zhou Y, Xuan YJ, Wang SF. Dietary Patterns Modulate the Risk of Non-Alcoholic Fatty Liver Disease in Chinese Adults. Nutrients. 2015;7(6):4778–4791. doi: 10.3390/nu7064778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia Caraballo SC, Comhair TM, Houten SM, Dejong CH, Lamers WH, Koehler SE. High-protein diets prevent steatosis and induce hepatic accumulation of monomethyl branched-chain fatty acids. The Journal of nutritional biochemistry. 2014;25(12):1263–1274. doi: 10.1016/j.jnutbio.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Jeong EA, Jeon BT, Shin HJ, Kim N, Lee DH, Kim HJ, Kang SS, Cho GJ, Choi WS, Roh GS. Ketogenic diet-induced peroxisome proliferator-activated receptor-gamma activation decreases neuroinflammation in the mouse hippocampus after kainic acid-induced seizures. Experimental neurology. 2011;232(2):195–202. doi: 10.1016/j.expneurol.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Zhu QC, Gao RY, Wu W, Guo BM, Peng JY, Qin HL. Effect of a high-fat diet in development of colonic adenoma in an animal model. World journal of gastroenterology: WJG. 2014;20(25):8119–8129. doi: 10.3748/wjg.v20.i25.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauber SN, Ali S, Gooderham NJ. The cooked food derived carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine is a potent oestrogen: a mechanistic basis for its tissue-specific carcinogenicity. Carcinogenesis. 2004;25(12):2509–2517. doi: 10.1093/carcin/bgh268. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Pei H, Kaeck M, Lu J. Mammary cancer promotion and MAPK activation associated with consumption of a corn oil-based high-fat diet. Nutrition and cancer. 1999;34(2):140–146. doi: 10.1207/S15327914NC3402_3. [DOI] [PubMed] [Google Scholar]
- 10.Marangoni F, Corsello G, Cricelli C, Ferrara N, Ghiselli A, Lucchin L, Poli A. Role of poultry meat in a balanced diet aimed at maintaining health and wellbeing: an Italian consensus document. Food & nutrition research. 2015;59:27606. doi: 10.3402/fnr.v59.27606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HJ, Yoon HJ, Kim SY, Yoon YR. A medium-chain fatty acid, capric acid, inhibits RANKL-induced osteoclast differentiation via the suppression of NF-kappaB signaling and blocks cytoskeletal organization and survival in mature osteoclasts. Molecules and cells. 2014;37(8):598–604. doi: 10.14348/molcells.2014.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rafi MM, Shafaie Y. Dietary lutein modulates inducible nitric oxide synthase (iNOS) gene and protein expression in mouse macrophage cells (RAW 264. 7) Molecular nutrition & food research. 2007;51(3):333–340. doi: 10.1002/mnfr.200600170. [DOI] [PubMed] [Google Scholar]
- 13.Cao HH, Cheng CY, Su T, Fu XQ, Guo H, Li T, Tse AK, Kwan HY, Yu H, Yu ZL. Quercetin inhibits HGF/c-Met signaling and HGF-stimulated melanoma cell migration and invasion. Molecular cancer. 2015;14:103. doi: 10.1186/s12943-015-0367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee WJ, Chen WK, Wang CJ, Lin WL, Tseng TH. Apigenin inhibits HGF-promoted invasive growth and metastasis involving blocking PI3K/Akt pathway and beta 4 integrin function in MDA-MB-231 breast cancer cells. Toxicol Appl Pharmacol. 2008;226(2):178–191. doi: 10.1016/j.taap.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Haar L, Ren X, Liu Y, Koch SE, Goines J, Tranter M, Engevik MA, Nieman M, Rubinstein J, Jones WK. Acute consumption of a high-fat diet prior to ischemia-reperfusion results in cardioprotection through NF-kappaB-dependent regulation of autophagic pathways. American journal of physiology Heart and circulatory physiology. 2014;307(12):H1705–1713. doi: 10.1152/ajpheart.00271.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Gayyar MM, Al Youssef A, Sherif IO, Shams ME, Abbas A. Protective effects of arjunolic acid against cardiac toxicity induced by oral sodium nitrite: effects on cytokine balance and apoptosis. Life sciences. 2014;111(1–2):18–26. doi: 10.1016/j.lfs.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Ijssennagger N, Rijnierse A, de Wit NJ, Boekschoten MV, Dekker J, Schonewille A, Muller M, van der Meer R. Dietary heme induces acute oxidative stress, but delayed cytotoxicity and compensatory hyperproliferation in mouse colon. Carcinogenesis. 2013;34(7):1628–1635. doi: 10.1093/carcin/bgt084. [DOI] [PubMed] [Google Scholar]
- 18.Guo X, Yin S, Dong Y, Fan L, Ye M, Lu J, Hu H. Enhanced apoptotic effects by the combination of curcumin and methylseleninic acid: potential role of Mcl-1 and FAK. Mol Carcinog. 2013;52(11):879–889. doi: 10.1002/mc.21933. [DOI] [PubMed] [Google Scholar]
- 19.Hebels DG, Sveje KM, de Kok MC, van Herwijnen MH, Kuhnle GG, Engels LG, Vleugels-Simon CB, Mares WG, Pierik M, Masclee AA, et al. Red meat intake-induced increases in fecal water genotoxicity correlate with pro-carcinogenic gene expression changes in the human colon. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association. 2012;50(2):95–103. doi: 10.1016/j.fct.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 20.Bouchard-Mercier A, Paradis AM, Rudkowska I, Lemieux S, Couture P, Vohl MC. Associations between dietary patterns and gene expression profiles of healthy men and women: a cross-sectional study. Nutr J. 2013;12:24. doi: 10.1186/1475-2891-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slattery ML, Potter J, Caan B, Edwards S, Coates A, Ma KN, Berry TD. Energy balance and colon cancer--beyond physical activity. Cancer Res. 1997;57(1):75–80. [PubMed] [Google Scholar]
- 22.Slattery ML, Edwards SL, Palmer L, Curtin K, Morse J, Anderson K, Samowitz W. Use of archival tissue in epidemiologic studies: collection procedures and assessment of potential sources of bias. Mutat Res. 2000;432(1–2):7–14. doi: 10.1016/s1383-5726(99)00010-2. [DOI] [PubMed] [Google Scholar]
- 23.Edwards S, Slattery ML, Mori M, Berry TD, Caan BJ, Palmer P, Potter JD. Objective system for interviewer performance evaluation for use in epidemiologic studies. Am J Epidemiol. 1994;140(11):1020–1028. doi: 10.1093/oxfordjournals.aje.a117192. [DOI] [PubMed] [Google Scholar]
- 24.Liu K, Slattery M, Jacobs D, Jr, Cutter G, McDonald A, Van Horn L, Hilner JE, Caan B, Bragg C, Dyer A, et al. A study of the reliability and comparative validity of the cardia dietary history. Ethn Dis. 1994;4(1):15–27. [PubMed] [Google Scholar]
- 25.Slattery ML, Boucher KM, Caan BJ, Potter JD, Ma KN. Eating patterns and risk of colon cancer. Am J Epidemiol. 1998;148(1):4–16. doi: 10.1093/aje/148.1.4-a. [DOI] [PubMed] [Google Scholar]
- 26.Kampman E, Slattery ML, Bigler J, Leppert M, Samowitz W, Caan BJ, Potter JD. Meat consumption, genetic susceptibility, and colon cancer risk: a United States multicenter case-control study. Cancer Epidemiol Biomarkers Prev. 1999;8(1):15–24. [PubMed] [Google Scholar]
- 27.Li P, Conley A, Zhang H, Kim HL. Whole-Transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC Genomics. 2014;15:1087. doi: 10.1186/1471-2164-15-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slattery ML, Herrick JS, Mullany LE, Gertz J, Wolff RK. Improved survival among colon cancer patients with increased differentially expressed pathways. BMC Med. 2015;13:75. doi: 10.1186/s12916-015-0292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.QIAGEN’s Ingenuity Pathway Analysis. 2014 [Google Scholar]
- 32.Savli H, Szendroi A, Romics I, Nagy B. Gene network and canonical pathway analysis in prostate cancer: a microarray study. Experimental & molecular medicine. 2008;40(2):176–185. doi: 10.3858/emm.2008.40.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Carrillo JA, Ding Y, He Y, Zhao C, Zan L, Song J. Ruminal Transcriptomic Analysis of Grass-Fed and Grain-Fed Angus Beef Cattle. PLoS One. 2015;10(6):e0116437. doi: 10.1371/journal.pone.0116437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, Uitdehaag BM, Kappos L, Gene MSAC, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Human molecular genetics. 2009;18(11):2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan-Fang T, Dong W, Li P, Wen-Li Z, Jun L, Na W, Jian W, Xing F, Yan-Hong L, Jian N, et al. Analyzing the gene expression profile of pediatric acute myeloid leukemia with real-time PCR arrays. Cancer cell international. 2012;12(1):40. doi: 10.1186/1475-2867-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naito Y, Kuroda M, Mizushima K, Takagi T, Handa O, Kokura S, Yoshida N, Ichikawa H, Yoshikawa T. Transcriptome Analysis for Cytoprotective Actions of Rebamipide against Indomethacin-Induced Gastric Mucosal Injury in Rats. Journal of clinical biochemistry and nutrition. 2007;41(3):202–210. doi: 10.3164/jcbn.2007029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyes-Gibby CC, Yuan C, Wang J, Yeung SC, Shete S. Gene network analysis shows immune-signaling and ERK1/2 as novel genetic markers for multiple addiction phenotypes: alcohol, smoking and opioid addiction. BMC systems biology. 2015;9:25. doi: 10.1186/s12918-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia P, Kao CF, Kuo PH, Zhao Z. A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC systems biology. 2011;5(Suppl 3):S12. doi: 10.1186/1752-0509-5-S3-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stolz A, Bastians H. Fresh WNT into the regulation of mitosis. Cell cycle. 2015:0. doi: 10.1080/15384101.2015.1064569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lv ZD, Zhang L, Liu XP, Jin LY, Dong Q, Li FN, Wang HB, Kong B. NKD1 down-regulation is associated with poor prognosis in breast invasive ductal carcinoma. International journal of clinical and experimental pathology. 2015;8(4):4015–4021. [PMC free article] [PubMed] [Google Scholar]
- 41.Piccardi B, Palumbo V, Nesi M, Nencini P, Gori AM, Giusti B, Pracucci G, Tonelli P, Innocenti E, Sereni A, et al. Unbalanced Metalloproteinase-9 and Tissue Inhibitors of Metalloproteinases Ratios Predict Hemorrhagic Transformation of Lesion in Ischemic Stroke Patients Treated with Thrombolysis: Results from the MAGIC Study. Frontiers in neurology. 2015;6:121. doi: 10.3389/fneur.2015.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannan MT, Tucker KL, Dawson-Hughes B, Cupples LA, Felson DT, Kiel DP. Effect of dietary protein on bone loss in elderly men and women: the Framingham Osteoporosis Study. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2000;15(12):2504–2512. doi: 10.1359/jbmr.2000.15.12.2504. [DOI] [PubMed] [Google Scholar]
- 43.Ng GZ, Menheniott TR, Every AL, Stent A, Judd LM, Chionh YT, Dhar P, Komen JC, Giraud AS, Wang TC, et al. The MUC1 mucin protects against Helicobacter pylori pathogenesis in mice by regulation of the NLRP3 inflammasome. Gut. 2015 doi: 10.1136/gutjnl-2014-307175. [DOI] [PubMed] [Google Scholar]
- 44.Shibahara H, Higashi M, Yokoyama S, Rousseau K, Kitazono I, Osako M, Shirahama H, Tashiro Y, Kurumiya Y, Narita M, et al. A comprehensive expression analysis of mucins in appendiceal carcinoma in a multicenter study: MUC3 is a novel prognostic factor. PloS one. 2014;9(12):e115613. doi: 10.1371/journal.pone.0115613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nardinocchi L, Sonego G, Passarelli F, Avitabile S, Scarponi C, Failla CM, Simoni S, Albanesi C, Cavani A. Interleukin-17 and interleukin-22 promote tumor progression in human nonmelanoma skin cancer. European journal of immunology. 2015;45(3):922–931. doi: 10.1002/eji.201445052. [DOI] [PubMed] [Google Scholar]
- 46.Andersen V, Olsen A, Carbonnel F, Tjonneland A, Vogel U. Diet and risk of inflammatory bowel disease. Digestive and liver disease: official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2012;44(3):185–194. doi: 10.1016/j.dld.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Vlaicu SI, Tatomir A, Rus V, Mekala AP, Mircea PA, Niculescu F, Rus H. The role of complement activation in atherogenesis: the first 40 years. Immunologic research. 2015 doi: 10.1007/s12026-015-8669-6. [DOI] [PubMed] [Google Scholar]
- 48.Hovland A, Jonasson L, Garred P, Yndestad A, Aukrust P, Lappegard KT, Espevik T, Mollnes TE. The complement system and toll-like receptors as integrated players in the pathophysiology of atherosclerosis. Atherosclerosis. 2015;241(2):480–494. doi: 10.1016/j.atherosclerosis.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 49.Jiang Y, Liao L, Shrestha C, Li D, Li M, Mu Y, Crumrine D, Wang L, Xie Z. Inhibition of 4-nitroquinoline-1-oxide-induced oral carcinogenesis by dietary calcium. International journal of clinical and experimental pathology. 2015;8(4):3529–3542. [PMC free article] [PubMed] [Google Scholar]
- 50.Monch S, Netzel M, Netzel G, Ott U, Frank T, Rychlik M. Folate bioavailability from foods rich in folates assessed in a short term human study using stable isotope dilution assays. Food & function. 2015;6(1):242–248. doi: 10.1039/c4fo00658e. [DOI] [PubMed] [Google Scholar]
- 51.Wang B, Feng L, Jiang WD, Wu P, Kuang SY, Jiang J, Tang L, Tang WN, Zhang YA, Liu Y, et al. Copper-induced tight junction mRNA expression changes, apoptosis and antioxidant responses via NF-kappaB, TOR and Nrf2 signaling molecules in the gills of fish: preventive role of arginine. Aquatic toxicology. 2015;158:125–137. doi: 10.1016/j.aquatox.2014.10.025. [DOI] [PubMed] [Google Scholar]
- 52.Jiang WD, Wen HL, Liu Y, Jiang J, Kuang SY, Wu P, Zhao J, Tang L, Tang WN, Zhang YA, et al. The tight junction protein transcript abundance changes and oxidative damage by tryptophan deficiency or excess are related to the modulation of the signalling molecules, NF-kappaB p65, TOR, caspase-(3,8,9) and Nrf2 mRNA levels, in the gill of young grass carp (Ctenopharyngodon idellus) Fish & shellfish immunology. 2015 doi: 10.1016/j.fsi.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Jeong W, Chang TS, Boja ES, Fales HM, Rhee SG. Roles of TRP14, a thioredoxin-related protein in tumor necrosis factor-alpha signaling pathways. The Journal of biological chemistry. 2004;279(5):3151–3159. doi: 10.1074/jbc.M307959200. [DOI] [PubMed] [Google Scholar]
- 54.Tremblay F, Lavigne C, Jacques H, Marette A. Dietary cod protein restores insulin-induced activation of phosphatidylinositol 3-kinase/Akt and GLUT4 translocation to the T-tubules in skeletal muscle of high-fat-fed obese rats. Diabetes. 2003;52(1):29–37. doi: 10.2337/diabetes.52.1.29. [DOI] [PubMed] [Google Scholar]
- 55.Kim YH, Kim JL, Lee EJ, Park SH, Han SY, Kang SA, Kang YH. Fisetin antagonizes cell fusion, cytoskeletal organization and bone resorption in RANKL-differentiated murine macrophages. The Journal of nutritional biochemistry. 2014;25(3):295–303. doi: 10.1016/j.jnutbio.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Farzaei MH, Abdollahi M, Rahimi R. Role of dietary polyphenols in the management of peptic ulcer. World journal of gastroenterology: WJG. 2015;21(21):6499–6517. doi: 10.3748/wjg.v21.i21.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buddaseth S, Gottmann W, Blasczyk R, Huyton T. Dysregulation of cell cycle control caused by overexpression of the oncogene pp32r1 (ANP32C) and the Tyr>His mutant pp32r1Y140H. Biochimica et biophysica acta. 2013;1833(5):1212–1221. doi: 10.1016/j.bbamcr.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nature cell biology. 2004;6(8):731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 59.Hasgall PA, Hoogewijs D, Faza MB, Panse VG, Wenger RH, Camenisch G. The putative RNA helicase HELZ promotes cell proliferation, translation initiation and ribosomal protein S6 phosphorylation. PloS one. 2011;6(7):e22107. doi: 10.1371/journal.pone.0022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Dijk SJ, Feskens EJ, Bos MB, Hoelen DW, Heijligenberg R, Bromhaar MG, de Groot LC, de Vries JH, Muller M, Afman LA. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am J Clin Nutr. 2009;90(6):1656–1664. doi: 10.3945/ajcn.2009.27792. [DOI] [PubMed] [Google Scholar]
- 61.Walczak K, Turski WA, Rajtar G. Kynurenic acid inhibits colon cancer proliferation in vitro: effects on signaling pathways. Amino acids. 2014;46(10):2393–2401. doi: 10.1007/s00726-014-1790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Creton SK, Zhu H, Gooderham NJ. The cooked meat carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine activates the extracellular signal regulated kinase mitogen-activated protein kinase pathway. Cancer research. 2007;67(23):11455–11462. doi: 10.1158/0008-5472.CAN-07-2821. [DOI] [PubMed] [Google Scholar]
- 63.Slattery ML, Lundgreen A, Herrick JS, Caan BJ, Potter JD, Wolff RK. Associations between genetic variation in RUNX1, RUNX2, RUNX3, MAPK1 and eIF4E and riskof colon and rectal cancer: additional support for a TGF-beta-signaling pathway. Carcinogenesis. 2011;32(3):318–326. doi: 10.1093/carcin/bgq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slattery ML, Lundgreen A, Herrick JS, Wolff RK. Genetic variation in RPS6KA1, RPS6KA2, RPS6KB1, RPS6KB2, and PDK1 and risk of colon or rectal cancer. Mutat Res. doi: 10.1016/j.mrfmmm.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slattery MLHJ, Lundgreen A, Wolff RK. Genetic variation in the TGF-Beta-signaling pathway and colon and rectal cancer risk. Cancer Epidemiology Biomarkers and Prevention. 2010 doi: 10.1158/1055-9965.EPI-10-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Slattery ML, Lundgreen A, Bondurant KL, Wolff RK. Tumor necrosis factor-related genes and colon and rectal cancer. Int J Mol Epidemiol Genet. 2011;2(4):328–338. [PMC free article] [PubMed] [Google Scholar]
- 67.Dorr C, Wu B, Guan W, Muthusamy A, Sanghavi K, Schladt DP, Maltzman JS, Scherer SE, Brott MJ, Matas AJ, et al. Differentially expressed gene transcripts using RNA sequencing from the blood of immunosuppressed kidney allograft recipients. PLoS One. 2015;10(5):e0125045. doi: 10.1371/journal.pone.0125045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo Y, Sheng Q, Li J, Ye F, Samuels DC, Shyr Y. Large scale comparison of gene expression levels by microarrays and RNAseq using TCGA data. PLoS One. 2013;8(8):e71462. doi: 10.1371/journal.pone.0071462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Slattery ML, Pellatt DF, Mullany LE, Wolff RK. Differential Gene Expression in Colon Tissue Associated With Diet, Lifestyle, and Related Oxidative Stress. PLoS One. 2015;10(7):e0134406. doi: 10.1371/journal.pone.0134406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Figueiredo JC, Hsu L, Hutter CM, Lin Y, Campbell PT, Baron JA, Berndt SI, Jiao S, Casey G, Fortini B, et al. Genome-wide diet-gene interaction analyses for risk of colorectal cancer. PLoS Genet. 2014;10(4):e1004228. doi: 10.1371/journal.pgen.1004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Top genes associated with meat and non-meat networks from IPA