Abstract
Up to 10% of the mouse genome is comprised of endogenous retrovirus (ERV) sequences, and most represent the remains of ancient germ line infections. Our knowledge of the three distinct classes of ERVs is inversely correlated with their copy number, and their characterization has benefited from the availability of divergent wild mouse species and subspecies, and from ongoing analysis of the Mus genome sequence. In contrast to human ERVs, which are nearly all extinct, active mouse ERVs can still be found in all three ERV classes. The distribution and diversity of ERVs has been shaped by host-virus interactions over the course of evolution, but ERVs have also been pivotal in shaping the mouse genome by altering host genes through insertional mutagenesis, by adding novel regulatory and coding sequences, and by their co-option by host cells as retroviral resistance genes. We review mechanisms by which an adaptive coexistence has evolved. (Part of a Multi-author Review)
Keywords: Virus restriction factors, retroviral receptors, Env-receptor interactions, murine ERV classification
Introduction
Close to 40% of the mouse genome is made up of fossils of transposable elements, of which approximately 10% represent endogenous retrovirus (ERV) sequences [1]. These ERV sequences have resulted from both ancient and modern infections of exogenous retroviruses, which have successfully colonized the germ line of their host [2]. Once a retrovirus becomes endogenous, the provirus survives as part of the host genome rather than as an autonomous infectious agent and thus is subjected to selection pressures acting on the host genome. Although extinction is probably the fate for most ERV lineages, it is clear that certain endogenous viruses are still capable of expression and replication even after millions of years within the host genome. Mouse genomes, in particular, have many active ERVs, which contrasts strikingly with the human genome, where all ERVs are nearly extinct, with the possible exception of HERV-K [3]. Thus, there must be other, poorly defined determinants that influence whether ERVs remain active or become extinct.
The aim of this review is to summarize our current knowledge of the various ERV lineages within the mouse genome and to provide examples that illustrate how elements have evolved together with their host, not only to confer benefits to the host, but also to permit continued coexistence, by modulating viral load and pathogenicity. Such studies in the mouse are particularly valuable because of the key role played by the inbred laboratory mouse strains in studies on mammalian genetics, development, pathology and retrovirology, and also because of the availability of wild mouse species that cover 12 million years of evolutionary history [4]. Emphasis is placed on changes in Env-receptor interactions, which are well studied for the infectious mouse retroviruses, and which have been exploited by host and virus to evolve and evade entry-related restrictions. These receptor interactions may also contribute to horizontal transmission to other species, where ERVs can begin a new cycle of unchecked copy number increase until new mechanisms evolve to limit their spread.
Origin and classification of ERVs
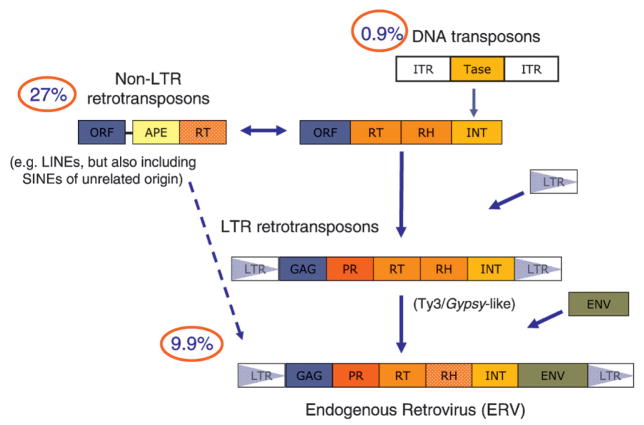
The human and mouse sequencing projects provided definitive evidence that a substantial proportion of both genomes was comprised of transposable elements (46 and 37.5%, respectively). Using the unified classification system proposed by Wicker et al. [5], the transposable elements can be divided into two main classes: retrotransposons and DNA transposons, the former constituting 95% of the transposable element sequences present in the mouse genome (Fig. 1). Whereas DNA transposons amplify without an RNA intermediate, retrotransposons rely on an RNA transcript that is ‘retrotranscribed’ by a reverse transcriptase (RT) before integration into the genome. Retrotransposons can be further divided into five orders, three of which are found in mammals: LINEs (long-interspersed nucleotide elements), SINEs (short-interspersed nucleotide elements), and LTRs (long-terminal direct repeats). LINEs are autonomous and encode at least an RT and a nuclease in their pol ORF [open reading frame] for transposition. Copies of LINE (superfamily L1) form the single largest fraction of interspersed repeat sequence in both human and mouse, with about 4800 full-length copies in mouse, of which 3000 are predicted to be active [6]. The SINE order is also classified within the class I retrotransposons, but is distinct in origin. SINEs originate from accidental retrotranspostion of various polymerase III transcripts and rely on LINEs for trans-acting transposition functions such as RT [7]. Whereas only a single SINE family (Alu) is active in the human lineage, the mouse lineage has been exposed to four distinct SINEs (B1, B2, ID, B4), originally derived from tRNA and 7SL genes [7]. Together they occupy about 27.4% of the mouse genome [1].
Figure 1.
Fossils of transposable elements make up a large proportion of the mouse genome. The percentage of the mouse genome sequences that are derived from one of two types of transposable elements (DNA transposons and retrotransposons) is shown. The retrotransposons are further divided into non-LTR and LTR retrotransposons. LTR retrotransposons in the mouse belong to the ERV superfamily, which is made up of three families. Arrows and fill colors denote potential evolutionary relationships and/or recombination events. Abbreviations used in the figure are defined in the text, with the exception of APE (apurinic endonuclease), found in LINE elements. The figure is adapted from [10] and [5].
The third order is the LTR retrotransposons. LTR retrotransposons are the predominant order of retrotransposons in plants and are generally less abundant in animals; nevertheless, close to 10% of the mouse and human genomes are derived from this order of transposable elements. LTR retrotransposons have a proposed chimeric origin, arising from fusion(s) between a DNA transposon and a non-LTR retrotransposon [8] (Fig. 1); the DNA transposon providing integrase (transposase, Tase) and the requirement for a short inverted terminal repeat at the ends of the element, and the non-LTR retrotransposon contributing the RT and RH (ribonuclease H) enzymatic functions, but also a gag-like ORF domain that provided the basis of the capsid (CA) and nuclear capsid (NC) proteins. Although several other components would be required to complete the formation of a fully functional LTR retrotransposon (e.g. including the evolution of LTRs as the means to overcome the problem of replicating the ends of any DNA molecule), the only additional protein domain required is a aspartic proteinase (PR) domain, which may have originated from the host’s pepsin gene family [9].
In mammals, all LTR retrotransposons are derivatives of the vertebrate-specific ERV superfamily. Retroviruses are generally thought to have evolved from Gypsy-like LTR retrotransposons, which adopted a viral lifestyle through acquisition of an envelope protein (Env) (Fig. 1). Most ERVs show clear homology to one another and to modern exogenous retro-viruses (XRV) (albeit to a lesser extent), especially across the RT gene, which is relatively refractory to nonsynonymous substitution [11, 12]. In addition, shared characteristics such as translational strategy, number of zinc finger proteins in the NC of gag, the presence and location of dUTPase (preventing incoporation of uracil), presence of a GPY/F motif in the C-terminal end of IN, and accessory genes can be used to classify ERVs [11]. There has been a growing tendency to group ERVs into classes according to their similarity to XRVs, which have been classified into seven genera (alpha-, beta-, gamma-, delta, and epsilonretrovirus, lentivirus, and spumavirus), the latter belonging to a distinct subfamily [13]. Using this system of classification, ERVs clustering with gamma- and epsilonretrovirus are termed Class I, those that cluster with lentivirus, alpha-, beta-, and deltaretroviruses are termed Class II, and those that cluster with spumaviruses are termed Class III [11, 14–16]. Notably, intermediates between these different families have been identified, indicating an evolutionary continuum.
Distribution and classes of ERVs in the mouse genome
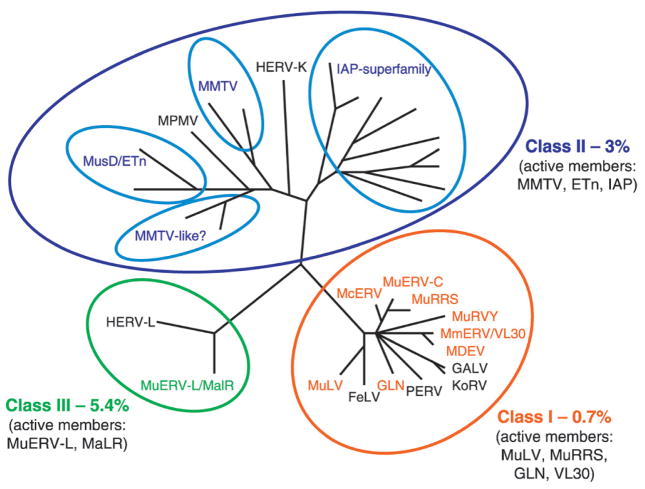
Unveiling of the mouse genome sequence in 2002 allowed the first comprehensive effort to catalogue the diversity of ERVs in the mouse genome [1]. Subsequently, several data-mining programs have been used to both identify novel ERV families, as well as validate earlier genetic analysis [1, 17–20]. As in the human genome, the three different classes of ERVs can be readily distinguished, and together make up close to 10% of their host’s genome (Fig. 2). However, a markedly dissimilar evolutionary history in human and mouse has been noted, both in the distribution and number of ERV families within the different classes, but also in the fact that ERVs are nearly extinct in human, whereas in mouse there are many active members [1, 21]. Although this reduced activity of ERVs in humans reflects in part an unexplained drop in the overall rate of transposition in the human but not mouse genome over the past 40 million years [1, 22], many other factors are clearly involved. Before addressing some of these mechanisms by which ERV activity is maintained or extinguished, an overview of the three major ERV classes found in the mouse genome is warranted.
Figure 2.
Phylogenetic analysis of ERV RT domains [19] demonstrates the three classes of mouse retrotransposons. RT sequences of ERVs from host species other than mouse are included for comparison and are in black letters. The close relationship of Class I ERVs to XRVs and ERVs from other species is clearly shown. Four distinct clades or superfamilies are defined for the Class II ERVs, one of which (MMTV-like) is poorly characterized. Non-autonomous elements, such as the abundant VL30 s (Class I), ETns (Class II), and MaLRs (Class III) are listed with their presumed parental ERVs, as they do not contain RT domains. The phlyogenetic tree was obtained from http://genomebiology.com/2004/5/3/ R14 and modified to include our own analysis of Class I ERVs.
Class III ERVs, which show closest (although distant) homology to the spuma-like genus of retrovirus, make up 5.4% of the mouse genome [1] (Fig. 2). These are probably the most ancient ERVs, accounting for 80% of recognized LTR element copies predating the human-mouse speciation [1, 14]. Interestingly, the human genome contains about the same percentage of class III ERVs as the mouse genome, although in contrast to mouse, no active forms have been detected. This discrepancy most likely reflects a higher nucleotide substitution rate in the mouse, making it more difficult to recognize ancient repeat sequences, and thus giving an underestimate of their actual density [1].
Two types of transposon elements constitute the Class III ERVS: murine ERV-L elements and the non-autonomous MaLRs (mammalian apparent LTR retrotransposons), the latter being the most common retroviral elements in the mouse genome, making up 4.8% of the genome. Based on the 51% similarity between their LTRs, these two types are thought to have a recent common ancestor [19]. The ERV-L family was first identified in humans, but was subsequently found to be in all placental mammals, thus was present in the mammalian common ancestor more than 70 million years ago (Mya) [14]. Accordingly, MuERV-L sequences are found in all characterized species within the Mus subgenus, as well as in mice of the other three subgenera Nannomys, Pyromys, and Coelomys (Fig. 3). Despite its age, this family has maintained some of its elements in an active state in the mouse, as demonstrated by recent amplifications in this species [14, 23]. It has recently also been shown that MuERV-L sequences are responsible for epsilon virus-like particles observed in the early mouse embryo [24], consistent with several reports showing high levels of expression during early embryonic development [25, 26]. Interestingly, the ERV-L family members have gag and pol genes but no detectable env, and an overall length of 6400 bp. Like spumaretroviruses, MuERV-L lacks the Cys-His region in the NC, important for binding to nucleic acids, but does contain the highly conserved major homology region (MHR) in CA, which is lacking in the related XRV family. Notably, both HERV-L and MuERV-L encode dUTPase, in contrast to spumaretrovirus but similar to betaretrovirus and non-primate lentivirus. However, the location of this gene within the viral genome is distinct, suggesting that its acquisition was an independent event [11, 27]. There is little evidence of MuERV-L horizontal transfer between species, an observation that is consistent with its lack of env [14]. The non-autonomous MaLRs are all internally deleted, containing only non-coding repetitive DNA [38]. Nevertheless they have typical LTRs, a primer binding site and a polypurine tract. In the mouse genome there are an estimated 380000 copies of MaLR elements (including solitary LTRs) [1], which belong to one of two types: MT (mouse transposon) and ORR1 (origin-region repeat) MaLRs [38]. They are closely related to the THE-1 (or MstII) family in the human genome. The MT lineage is the most prevalent type of ERV in the mouse genome and has a mean length of 1980 bp. In contrast, members of the ORR1 lineage have a mean length of approximately 2460 bp and are about 10-fold less frequent in the genome. Both member types are still active in the mouse, but died out some 50 Mya in human [38].
Figure 3.
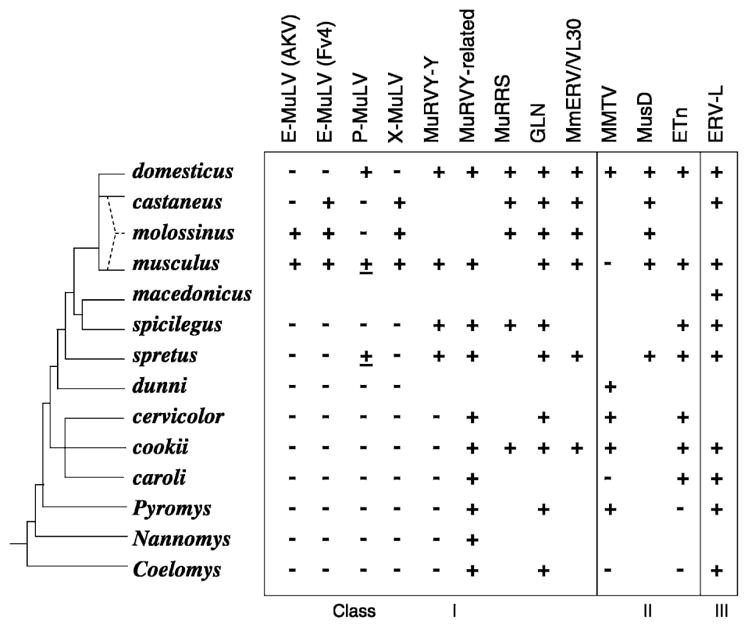
Schematic representation of the evolution of Mus indicating the species distribution of different ERV families. This evolutionary tree is based largely on the synthetic tree of Boursot and Guenet [4, 28]. All species from domesticus through caroli are subgenus Mus. The most recent node of the tree represents the house mouse Mus musculus complex; M. molossinus is a natural hybrid of castaneus and musculus. Species tested for specific ERV sequences by Southern-blot analysis are indicated; cases with a very few detectable copies are underlined [14, 29–37].
Class II ERV elements make up 3.14% of the mouse genome, a 10-fold higher proportion in mouse than in humans [1]. There is no evidence for a Class II ERV that predates the human-mouse speciation. The Class II ERVs include elements that resemble the betaretrovirus XRV genus, typified by the mouse mammary tumor virus (MMTV), which is morphologically classified as a type-B virus, and the Mason-Pfizer Monkey Virus (MPMV), a type-D virus (Fig. 2). Both types are distinguished by their mode of assembly, which initiates in the cytoplasm before transport to the plasma membrane [39]. In addition, betaretroviruses share a translational strategy involving two −1 frame-shifts to translate pro-pol-encoded proteins, the presence of a UTPase coding region upstream of PR, and two zinc-finger motifs in NC [11]. More than 30 endogenous MMTVs are scattered over the laboratory mouse genome, although only two or three full-length, active alleles have been identified [19, 40, 41]. Notably, the human MMTV-like (HML)-2 HERV-K subfamily may be the only ERV family with active elements in the human genome [42, 43].
Related to the MMTV, but more closely to MPMV, is a much larger group of elements known as MusD, with their deleted variants denoted ETn (early transposons) [35, 44]. MusD contains gag-pro-pol genes, but lacks an env gene. Furthermore, its gag-encoded matrix (MA) protein lacks a functional sequence for myristylation and plasma membrane targeting, thereby restricting virions to intracellular compartments, perhaps facilitating its capacity as an intracellular mutagen [45]. Interestingly, due to their high expression levels, ETn genomes are the actual primary mutagen in this family, their retrotransposition being mediated by MusD-encoded viral proteins [46]. ETns, similar to MaLRs, are flanked by LTRs but contain mainly non-retroviral, non-coding sequences of unknown origin. MusD/ETns are widely distributed among the different species of the subgenus Mus but are not found in Rattus, or in the Pyromys or Coelomys subgenera of Mus, which diverged between 5 and 10 Mya [36] (Fig. 3).
The final well-characterized clade within the Class II ERVs is related to IAP [intracisternal A-type particles], which are present in mice at approximately 1000 copies per cell [47]. Phylogenetic analysis of related RT sequences in the mouse genome indicate that this clade of elements can be grouped into seven to eight families (defined by sharing <90% identity at the amino acid level within RT) [19, 47]. They assemble on the endoplasmic reticulum and bud into the cisternae but are not released from the cell. Although originally thought to lack an env gene, two subsets of IAP-related proviruses that encode Env proteins have been identified, with a single provirus being infectious [48, 49].
Class I ERVs comprise the smallest class, making up 0.68% of the mouse genome, but its members are the best-studied ERVs. They are closely related to the gammaretrovirus genus of XRVs, typified by the type-C murine leukemia viruses (MuLV). In contrast to the type-B and -D retroviruses, type-C viruses assemble their genomes at the plasma membrane, with the concurrent formation of the immature viral core and the acquisition of the envelope. Class I ERVs share a similar translation strategy (read-through of the termination codon at the end of gag), the presence of one zinc-finger in NC, and a conserved GPY/F motif in the C-terminus of IN. Although these sequences are fourfold more common in the human than the mouse genome, no active Class I ERVs are known in humans. In contrast, the mouse genome contains many active members of this class, which have been the subject of intense research over the last four decades.
Early work of Todaro et al. based on DNA hybridization and antigen screening defined two major subclasses of type-C retroviruses [50, 51]. Strikingly, these two major subclasses have stood the test of time and technology. The subclass C-I of Todaro includes XRVs and ERVs from several primate sources, including gibbon ape and woolley monkey, as well as isolates from wild Asian mice, such as Mus cervicolor and Mus caroli. In contrast, the subclass C-II is composed of MuLV ERV and XRV isolates from different inbred laboratory strains and species of Mus musculus and includes the related ERV family from cats (FeLV). Sporadic reports over the last two decades have described related but distinct ERV isolates from various mouse strains that belong to one of these two subclasses, but only with the availability of the mouse genome sequence have there been more rigorous attempts to further classify these Class I ERVs. Our attempt is shown in Table 1, and is pictorially demonstrated by a phylogenetic analysis based on RT analysis of different ERV and XRV isolates, which also shows the two major clades (subclasses) of Class I ERVs, first recognized by Todaro (Fig. 2).
The best-characterized ERVs are the members that belong to the subclass C-II MuLV family. This is a homogeneous family that has recently (<1.5 Mya) entered the Mus genome, as indicated by the insertional polymorphism among inbred mouse strains and Mus subspecies and by the presence of infectious members [34, 60, 61] (Fig. 3). MuLVs are generally divided into distinct groups on the basis of the host ranges specified by their env gene (Table 1). The xenotropic (X)-MuLVs are present in about 20 copies per mouse genome, whereas the polytropic (P)-MuLVs, which can be subdivided into two closely related subgroups, are present in about 40 copies per genome [62]. Ecotropic (E)-MuLVs are found in only a few copies in diverse inbred strains [63]. Analysis of wild mouse species indicates that ERVs of all three subgroups entered the Mus germline recently (Fig. 3) [34]. Of the three groups of ERVs, only X-MuLV and E-MuLV have infectious members, although infectious P-MuLVs are generated following recombination with E-MuLVs [64, 65]. Some wild mouse populations carry infectious amphotropic MuLVs (A-MuLVs) or a novel E-MuLV (HoMuLV) in the absence of germline ERVs [66, 67].
Table 1.
Seven families of Class I ERVs and related XRVs with their env variants.
| Families | Prototype viruses in different Mus species1 | Host range2 | Receptor | Related XRVs or ERVs in other species |
|---|---|---|---|---|
| Subclass C-I | ||||
| γ-A | MuRRS | no env | N.A. | GALV, KoRV, PERV |
| γ-B | MURV-Y | not active | unknown | |
| γ-C | MmERV | n.t. | unknown | |
| MDEV (Mus dunni) | multitropic | unknown [52] | ||
| γ-D | McERV(Mus caroli) | monotropic | PLLP [53] | |
| γ-E | MuERV-C | not active | unknown | |
| γ-F | GLN-2 | ecotropic | unknown [18] | |
| Subclass C-II | ||||
| γ-G | P-MuLV | polytropic3 | XPR1[54] | FeLV, BaEV, XMRV |
| X-MuLV | xenotropic | XPR1[54] | ||
| AKR-MuLV | ecotropic | mCAT-1[55] | ||
| CasBrE4 | ||||
| Ho-MuLV (Mus spicilegus)5 | ||||
| 4070A-MuLV5 | amphotropic | PiT2 [56] | ||
| 10A1-MuLV5 | 10A1 | PiT1, PiT2 [57] | ||
| HMEV (Mus spicilegus) | ecotropic | mSMIT [58, 59] | ||
| M813 (Mus cervicolor) | ||||
Prototype ERVs listed have been characterized in inbred mouse strains, unless noted.
Host range refers to the historical nomenclature used to define ‘host’ susceptibility to virus infections, generally tested using cell lines of various origins. Ecotropic refers to the ability of the virus to infect cells of murine origin only; but not all ecotropic viruses use the same receptor. Xenotropic refers to infectivity of cells of non-murine origin only. Poly-, ampho-, 10A1-, and multitropic all refer to the ability of the virus to infect cells from all (or most) species tested, but through different cellular receptors. Monotropic refers to the fact that the virus can infect cells of many species, but only distinct cell types, which reflects the limited expression pattern of the receptor.
Active forms have been found only after recombination events with XRV.
Isolated from Lake Casitas (CA) wild mice of unclear origin. Similar ERVs have been identified in M. musculus and M. castaneus.
These prototypes have only been found as XRVs.
Abbreviations used: N.A., not applicable; n.t., not tested
Somewhat surprisingly, most of the Class I ERVs in the mouse genome actually belong to the lesser-characterized subclass C-I family. At least six distinct families can be recognized (as defined by <90% homology in RT). The first family recognized in this group were the MuRRSs (murine retroviral-related sequences) [37]. This is a family of approximately 30–50 copies with an approximate size of 5.5 kb, with recognizable but highly mutated gag, pol, and env genes. Many of these copies share identical deletions within the pol gene, suggesting that they were mobilized by a nondefective helper virus. In addition to these mutated proviral genomes, there are at least 500 copies of solo LTRs, probably arising through homologous recombination between the two LTRs. Two additional families (MuRV-Y and MuERV-C) are closely related, but are predicted to have been seeded by a separate ‘founder’ retrovirus [68]. Similar to MuRRS, MuERV-C family members are highly mutated but with detectable gag, pol, and env genes, the latter being often extensively deleted. At least 20 copies can be detected in the mouse genome, 10 of which are clustered on the X chromosome. This clustering of integrations suggests that these have been amplified by a non-retroviral mechanism. A similar mechanism is probably responsible for the close to 500 copies of MuRV-Y (murine repeated virus on the Y chromosome), which are almost exclusively found on the Y chromosome, as part of a 25-kb amplicon [69, 70]. In contrast to the MuRRS and MuERV-C members characterized so far, MuRV-Y members have incurred fewer mutations/deletions, but no active forms are known [71]. MuRRS and MuRV-Y are thought to have entered the mouse genome within the past 9 Mya, whereas the multiple Y chromosome MuRV-Y copies are found only in the most recently derived European Mus species, reflecting a more recent amplification [37, 70] (Fig. 3).
In contrast to the families described above, three additional families have been recently recognized for which active ERVs are known. One is the GLN family, named for its glutamine tRNA primer-binding site, in contrast to proline tRNA found in many other Class I ERVs [72]. Of the over 80 copies found in the mouse genome, one copy was recently found both to be intact and capable of releasing virus particles [18]. In view of the fact that related sequences have been found in both Mus subspecies and other rodent (but not nonrodent) genomes, it must have entered a common ancestor prior to the Mus/Rattus split (circa 16–23 Mya) [18, 72] (Fig. 3).
The first member of the next family was first identified in the Mus dunni (also termed M. terricolor) genome as an active ERV (MDEV) [73], but it was not until the sequence of the mouse genome was ‘mined’ for novel ERVs that its counterpart within the C57BL/6 laboratory mouse genome was identified (MmERV) [20]. Among the more than 50+ full-length copies in the mouse genome, at least one provirus has intact reading frames for all three genes, although its activity has not been assessed. Interestingly, both MmERV and MDEV have LTRs that are similar to VL30 elements, which are nonautonomous retrotransposons first identified over 30 years ago [74, 75]. It is thus probable that VL30 s are derived from internally deleted MmERV elements, which utilize various Class I ERVs for transposition. Similar to MmERV and MDEV, VL30 elements generally (but not exclusively) depend on glycine tRNA for priming the reverse transcription. There are approximately 100–200 copies of VL30 elements in the mouse genome. In contrast to other ERV sequences analyzed, where there is generally a 20-fold higher frequency of solitary LTRs than proviral genomes, for VL30 s there are at least 20-fold more full-sized elements than solo LTRs [76]. This probably reflects the high transposition activity of these elements (coupled with increased mutagenesis capacity), which is facilitated by their high expression levels and permissive packaging signals. VL30 (and presumably MmERV) sequences are present in all inbred mice and various Mus species, although the number of copies varies (Fig. 3).
Finally, the last characterized family is actually the first C-I family member to be identified in mice [51]. Recent molecular cloning and analysis of this isolate (dubbed McERV) from Mus caroli fibroblasts has shown that it is closely related but distinct from the MmERV/MDEV family ([77], unpublished observation). Hybridization and sequence analysis has shown that although it is found at a high copy number in Mus caroli cells (ca. 50), highly related proviruses are also found in the inbred C57BL/6 laboratory mouse genome (3 intact but mutated copies). Notably, proviruses with at least two different env genes have been characterized ([77], unpublished observation).
The major lesson learned from mouse ERVs: ‘It takes all the running you can do, to keep in the same place.’ – Lewis Carroll
ERVs are clearly detrimental to their hosts, but are also important functional components of the genome, so opposing mechanisms must be in place to mitigate their effects as well as maintain their presence and function. Two key characteristics of retroviruses are responsible for the production of ERVs: a life cycle that requires integration into the genome of the host cell, and the unique ability to infect germ cells (albeit a rare event). The observation that distinct Mus species contain multiple copies of highly related proviruses prompted the hypothesis that endogenization of a retrovirus occurs in bursts, in which a relatively rapid amplification of ERVs occurs shortly after the initial colonization event, but then declines over time [2]. These initial bursts can be attributed to two factors: 1) The evolution of mechanisms in the host that suppress further infection and thereby prevent the deleterious consequences of ERV activity, as discussed below; and 2) the absence of any strong selection pressure to maintain ERV sequence integrity during host replication, resulting in a decrease in fitness (i.e. replicative ability) of the viral lineage over time. Because the accumulation of mutations from one ERV generation to the next is irreversible, eventually all endogenous viruses become defective, fail to replicate or express any gene products, and the lineage becomes extinct. While this protects the host from the consequences of ERV insertions like disease and mutation, it is becoming clear that these protections are counteracted by mechanisms that work to maintain active ERVs to support their function as an important source of somatic and genomic diversity.
Mechanisms supporting the maintenance of active ERVs and de novo endogenization
In the face of host resistance factors and the accumulation of inactivating mutations, ERVs have evolved mechanisms that either postpone inactivation or reestablish replication fitness. Two important mechanisms by which replication fitness is prolonged have been revealed by the analysis of mouse ERVs: 1) the switch from an extra- to intracellular life cycle that does not require receptor interaction and 2) trans complementation by other ERVs or XRVs. Two successful ERVs that have adapted quite efficiently to an intracellular life cycle are IAPs and MusD, members of the Class II ERV family. The transition from an infectious IAPE to intracellular-restricted IAP has been shown to coincide with loss of a functional env gene and the simultaneous gain of a endoplasmic reticulum signal and loss of myristylation signals in the MA protein that determine the site of virus assembly [49, 78, 79]. Similarly, the MA proteins of MusD also have lost plasma-membrane targeting signals [45]; env containing ancestors of MusD have not been identified. Examples of the second common mechanism, trans complementation, are clearly observed in the highly repetitive and non-autonomous MaLR and VL30 elements found in the mouse genome. Such degenerate ERV sequences that do not code for functional pol sequences can continue to replicate provided that their cis-acting elements, permitting packaging and DNA integration, remain intact and the proteins required for replication are supplied by infecting XRVs or by functional, or partially functional, ERVs within the same cell. Importantly, this does not increase the fitness of ERVs – but merely allows defective viruses to continue replicating. It is only through new rounds of endogenization that active ERV proviral genomes can be maintained.
New ERVs may arise within genomes by at least two different mechanisms: retrotransposition from a pre-existing endogenous retrovirus (intraspecific transmission) or infection and integration via an exogenous source virus (horizontal transmission). For defective ERVs, replicative capacity (i.e. full fitness) can be restored via recombination at the RNA level either with XRV or with another expressed ERV. The frequency of these recombinational events is clearly facilitated by the retroviral diploid RNA genome and replication strategy. Analysis of ERV genomes in the mouse genome has revealed multiple examples in which exchanges between different ERV lineages have been observed. Indeed many of the ERV lineages were first discovered due to incorporation of their sequences into other better-characterized ERV lineages, such as the acquisition of LTR sequences from either GLN or MuRRS by VL30 or MuLV elements [72, 80]. Notably, LTR and env sequences have been found to be quite variable in different ERV lineages. Examples of LTR exchanges have been investigated in detail for VL30 elements [81]. Such exchanges would be expected to increase expression rates (and thus increase the chance of de novo endogenization) in particular cell types. Indeed, it is tempting to speculate that the high expression of several mouse ERVs (e.g. IAP and ERV-L) during embryogenesis may increase the likelihood of germ cell infections [26, 82].
The acquisition of novel env sequences is best documented for the Class I ERVs [83]. As mentioned above and depicted in Table 1, members of both MuLV and McERV families have altered host ranges due to genetic mutations or recombination events within their env genes. The acquisition of novel sequences within the variable regions of the SU domain may contribute to overcoming host defence mechanisms that block cell entry (see below) or may result in usage of novel receptors. The latter would be expected to either facilitate infection of different cell types within the host or may increase the host spectrum of the retrovirus, and thereby facilitate infection of other species. It should be noted, however, that there are clearly limits on receptor switches. All six known gammaretrovirus receptors are transporters with multiple transmembrane domains. This common structural feature suggests constraints that may define how these related viruses interact with their receptors to bind, to produce the conformational changes that allow for Env interactions and cell fusion, and to penetrate the cell.
The importance of cross-species transmission in maintaining viable ERV genomes or supporting de novo endogenization has long been recognized based on comparative analysis of host and viral genomes [84–86]. Conceivably, cross-species infections may allow rapid expansion of the XRV in its new host, which may not have developed fine-tuned defence mechanisms against the invading parasite (see [87], this issue). Sequences related to Class I ERVs have been identified in a variety of terrestrial vertebrates suggesting several zoonotic transmissions [86], and several members of this group are also clearly related to infectious retroviruses, such as GALV, KoRV, and FeLV, found in other species (Fig. 2 and Table 1). Although the absence of active ERV elements in the human genome may reflect both changes in host ecology (e.g. decreased exposure to XRV from other animals) plus a strong retroviral defence mechanism, it should be noted that chronic cross-species infection of an X-MuLV in humans (XMRV) has recently been reported [88, 89].
Host mechanisms that suppress retrovirus infection and favor adaptive coexistence
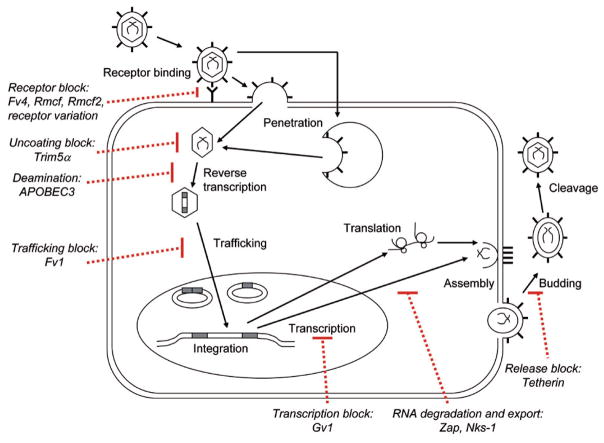
Mice exposed to infectious retroviruses or to unchecked ERV amplification or expression are subject to virus-induced disease or virus-induced genetic mutations. Mice are protected by the innate and acquired immune systems, but have also evolved numerous constitutively expressed antiviral factors that target various stages of the retroviral life cycle, defined largely in studies with the Class I MuLVs. The factors responsible for this intrinsic immunity block virus entry, interfere with specific post-entry stages of virus replication, integration, assembly or release, or mediate endogenization and replicative silencing (Fig. 4). These host resistance factors also produce the selective pressures that favor the outgrowth of virus variants able to circumvent those blocks. This results in a ratchet-like pattern of sequential mutations in both host and virus that generate substantial polymorphism in the critical regions of the responsible genes. This coevolution of virus and host also results in a form of adaptive coexistence in which retroviruses are rarely pathogenic in their natural hosts.
Figure 4.
Retrovirus lifecycle and blocks to replication by host restriction factors.
The best-characterized mechanisms of resistance to mouse retroviruses rely on inhibition of virus entry. Entry can be blocked by mutations in the receptor gene that alter their function or by host resistance genes that interfere with receptor function. To date, seven receptors for mouse retroviruses have been identified (Table 1), of which six represent receptors for Class I gammaretroviruses and four are used by different host range subclasses of MuLVs. Studies on two of these receptors have identified residues critical for virus entry in the E-MuLV CAT1 receptor [90, 91] and in the X/P-MuLV receptor XPR1 [92] (Fig. 5). In both cases, these receptor regions are hypervariable and glycosylated, but it is not clear if the identified critical amino acids actually mediate virus attachment and entry or if they are negative regulators that protect nearby highly conserved receptor determinants [54]. Among mouse species, there are two naturally occurring functional variants of the CAT-1 receptor [55, 93], and four variants of XPR1 [28, 92]. These functional subtypes are distinguished by their ability to mediate entry of different virus isolates (Fig. 5). The evolution of XPR1 can be examined in relation to the appearance and spread of X-MuLVs and P-MuLVs in Mus species [28, 34]. The XPR1 variant with the broadest susceptibility phenotype is widely distributed among the older Asian Mus species. It was not, however, until the appearance of the house mouse (M. musculus) about 1 Mya that mice were exposed to and began to acquire ERVs of X-MuLVs. This acquisition is also coincident with the appearance of restrictive XPR1 variants in M. m. castaneus (cXPR1) and M. m. domesticus (nXPR1). The M. m. domesticus mice, like the laboratory strains they gave rise to, are not infectible by X-MuLVs, but the appearance of this restrictive nXPR1 variant undoubtedly contributed to the evolution of P-MuLVs as a new host range variant able to use this restrictive receptor. This coevolutionary relationship based on the patterns of receptor evolution and the pattern of endogenous X/P-MuLV distribution in wild mouse species is consistent with recent phylogenetic analysis in the sequenced mouse genome. This analysis confirms the close evolutionary relationship of these virus types and suggests X-MuLV is the most recent common ancestor [17].
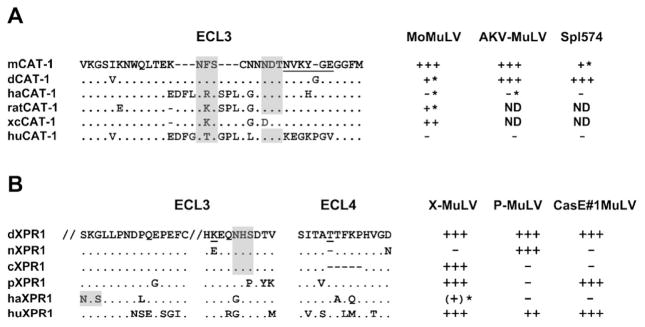
Figure 5.
Amino acid sequences of the extracellular loops (ECLs) of CAT-1 and XPR1 that contain MuLV receptor determinants. (A) Sequences represent NIH 3T3 (mCAT-1), M. dunni (dCAT-1), hamster (haCAT-1), rat (ratCAT-1), rat XC cell (xcCAT-1), and human (huCAT-1) [55, 93, 94]. (B) Sequences represent M. dunni (dXPR1), NIH 3T3 (nXPR1), M. castaneus (cXPR1), M. pahari (pXPR1), hamster (haXPR1), and human (huXPR1) [28, 92]. Shown are residues 416–430 and 499–508 in ECL3. Relative susceptibilities are given on the right for 3 E-MLVs for CAT-1 and for 3 X/P-MLVs for XPR1; asterisks identify infections that can be enhanced by inhibitors of N-linked glycosylation. Critical residues in CAT1 and XPR1 are underlined; N-linked glycosylation sites are shaded. ND, not done.
In addition to receptor polymorphisms, other mechanisms can inhibit infection at the level of virus entry. First, variable receptor glycosylation can result in resistance. Glycosylation has been found to be responsible for resistance to E-MuLV in rodent cells and X-MuLVs in hamster cells [95–98] (Fig. 5). Second, Env-producing ERVs can interfere with exogenous viruses that use the same receptor. Such interfering Env genes have been found for E-MuLVs (Fv4) [99] and X/P-MuLVs (Rmcf, Rmcf2) [100–102]. All three of these resistance genes are associated with ERVs that have intact env genes. Two of these genes evolved in wild mouse populations exposed to infectious virus; M. castaneus carries Fv4 and Rmcf2 along with infectious E-MuLV and X-MuLV [103]. Other host factors may also inhibit gammaretrovirus infection by targeting Env or affecting other factors needed for binding or entry. For example, mice produce a serum factor, LVIF, which inactivates viruses of the P/X-MuLV host range class but not those of other Env host range groups [104]. Also, although no mouse retro-virus is known to use a coreceptor, the efficiency of entry can be modulated by other factors [105]. For example, neurotropic Friend virus infects brain capillaries more efficiently because of the presence of heparin sulfate [106].
Viruses have evolved mechanisms to evade these entry restrictions. The major determinant of receptor recognition (and thus virus host range) in Class I ERVs is within the variable region of the surface (SU) domain of the Env protein [107]. Mutations within this region can lead to the generation of retroviruses that can use alternative receptors, multiple receptors or even multiple receptor determinants on the same protein; this contributes to virus survival by limiting the impact of host escape mutations. Thus, the closely related phosphate transporters PiT1 or PiT2 both function as gammaretrovirus receptors, and at least one MuLV, mouse 10A1 MuLV, can use either for entry [108]. Also, the XPR1 receptor carries two largely independent determinants in different extracellular loops that can mediate X-MuLV entry and undefined determinants for P-MuLV entry [28, 92, 109]. Several mechanisms also allow viruses to bypass the need for their cognate receptor. In viremic animals, recombination between infectious E-MuLV or A-MuLV and endogenous P-MuLV env genes can generate novel recombinants with P-MuLV host range [110, 111]. These P-MuLVs are also often pseudotyped by E-MuLVs in infected animals. It has also been shown that some viruses can use alternative receptors in the presence of the soluble Env glycoprotein for that receptor. P-MuLVs can be transactivated in this way by E-MuLV Env [112].
In addition to resistance mechanisms that block virus entry, there are three major genes known to inhibit early post entry replication of retroviruses: Fv1, TRIM5α, and APOBEC3 (Fig. 4). Alleles at the Fv1 locus control the relative sensitivities of mouse cells to different subgroups of MuLVs [113]. There are at least five allelic variants of Fv1, four of which produce different patterns of resistance to mouse-tropic viruses, described as N-, B-, NR, or NB-tropic. The Fv1 sequence is related to the gag gene of MuERV-L [114]. The mechanism of restriction is unknown, but Fv1 generally blocks virus replication at or just after reverse transcription to limit proviral integrations [115]. Fv1 targets the viral capsid; the major determinant that distinguishes N- and B-tropic viruses is at CA position 110 [116], but other targets in this same CA region have been identified in studies on NR- and NB-tropism [117]. Fv1 restriction is found only in laboratory mice and laboratory mouse-related Mus species [118, 119], but African pygmy mice carry an unusual post-entry resistance to some ecotropic MuLVs that targets some of the same amino acid residues as Fv1 [96].
While cells of nonrodent species lack Fv1, they carry another restriction gene, TRIM5α, that blocks retrovirus replication before reverse transcription. TRIM5α is a member of the tripartite interaction motif family, and no mouse ortholog has been identified. Primate TRIM5α, however, restricts MuLV infection by targeting amino acids at two of the capsid sites also targeted by Fv1, 110 and 117 [120]. TRIM5α may thus function to limit transpecies transmission by the broadly infectious MuLVs. The evolution of two polymorphic host genes (Fv1 and TRIM5α) targeting the same hypervariable capsid segment also suggests that, like the viral Env and their receptors, virus capsid and host genes that target capsid represent a second major battleground in the coevolution of virus and host.
A third host gene involved in post-entry restriction to retroviral infection is APOBEC3G. This enzyme catalyzes C-to-U deamination during reverse transcription, resulting in G-to-A mutations in the resulting provirus [121]. Although most extensively studied in humans where it inhibits HIV as well as MuLVs, the mouse counterpart, mA3, also blocks HIV-1 and the mouse retroviruses MMTV, IAP, and MusD [122–124]. Although mA3 has now been identified as Friend MuLV resistance gene Rfv3 [125] and a recent study characterized MuLV ERVs in C57BL/6 mice and found that a significant number of the observed mutations in endogenous P-MuLVs, but not X-MuLVs, represented G-to-A mutations and also showed a gradient of G-to-A mutations consistent with mA3 activity [17]. This suggests that mA3 editing may have contributed to endogenization and silencing of these ERVs at the time of integration.
A few mouse genes have been identified that inhibit late stages of retrovirus replication. One such gene, Gv1, is known to broadly restrict transcription of endogenous MuLVs [126]. This restriction was shown to be due to a single host gene that was mapped to chromosome 13 [127], but it has not been characterized further. Transcriptional silencing can also be effected by TRIM28 which targets the MuLV primer binding site [128]. Another late-acting gene, Nks1, encodes an mRNA export factor that suppresses sense-oriented intronic IAP inserts [129]. Also, host genomes can silence ERVs by methylation; failure to methylate new or specific ERVs has been attributed to Dnmt genes [130].
Another factor involved in intrinsic immunity to late stages of retroviral infection is ZAP, a zinc-finger antiviral protein that inhibits translation of MuLVs as well as alphaviruses and filoviruses [131]. ZAP binds viral RNA and leads to its degradation. ZAP is found in multiple species, and sequence comparisons confirm that it has evolved under positive selection. Finally, recent studies have identified a novel factor in human cells that inhibits release of many enveloped viruses, including MuLV [132]. Tetherin binds newly formed virions to cell surfaces, is constitutively expressed by some cells, and can be antagonized by the HIV-1 accessory protein Vpu.
The role of ERVs in shaping the mouse genome
The data from the sequenced mouse genome together with a century of mouse genetic analysis has established that ERVs have played and continue to play an important role in shaping the mouse genome. These ERVs modify the mouse phenome by altering host gene expression or by contributing novel protein-coding sequences. The great majority of ERV-induced changes are insertional mutations that disrupt host protein-coding genes or alter gene expression by affecting splicing or by providing novel signals for initiation, regulation or termination of transcription. ERV insertions in somatic cells have long been studied for their ability to induce neoplastic diseases by activating oncogenes. Insertional mutations also accumulate in the germ line, and a recent review identified 63 examples of ERV-induced mouse mutations and argued that 10–12% of all mutations in the mouse are caused by ERVs [133]. The majority of these germ line mutations are due to IAP and ETn/MusD insertions, which is not surprising as these ERV families are present in high copy numbers and contain many active members. The largest number of these mutational insertions is found in introns, and occasional reversions have been reported which are accompanied by provirus deletion. In addition to insertional mutagenesis, ERV sequences may promote rearrangement of DNA by way of non-allelic homologous recombination between elements [2].
The first example of ERV-induced insertional mutagenesis was identified in the oldest inbred strain of the laboratory mouse, DBA, which was bred to carry three visible mutations all affecting coat color: dilute (d), brown (b), and agouti (a). The single ecotropic MuLV ERV in this strain was linked to the d locus, and it was found that this ERV was lost in rare color revertants [134]. The d locus encodes an unconventional myosin heavy chain gene, Myosin Va, that controls transport of melanosomes into the dendritic processes of melanocytes from where they enter the hair shaft. The ERV integration site is within an intron of this gene and results in abnormally spliced transcripts [135]; this is associated with the failure to produce pigment-filled dendritic processes and an abnormal distribution of pigment in the hair shaft leading to lightened coat color.
The importance of ERV insertions for gene regulation is well recognized. An early and dramatic example was the demonstration that the mouse gene for sex-limited protein requires androgen for its expression because of a hormone-responsive enhancer introduced by ERV insertion [136]. More recent work has provided evidence that MuERV-L and MaLR elements may provide essential functions as ‘early’ promoters in full-grown oocytes and during embryogenesis [26]. The temporal and spatial regulation of ERV elements are controlled not only by the differential expression of transcripton factors that regulate LTR expression, but by epigenetic modifications. ERVs are often preferential sites of methylation and can provide methylation-sensitive promoters for host genes [137]; this can result in variable expression, as is the case for the viable yellow agouti allele that uses an IAP LTR as promoter [138, 139]. Furthermore, it is increasingly appreciated that the stochastic nature of ERV expression modulated by epigenetic modifications during embryogenesis may also impinge on neighboring genes by RNA interference [140, 141].
In addition to altering gene regulation or expression, ERV integrations also insert virus protein-coding genes into host chromosomes, and some of these genes have been co-opted or domesticated by the host for cellular functions. Such genes are rare, and the majority of the recognizable co-opted virus genes serve to protect against or modulate further retroviral infection. The largest set of these genes are the minor lymphocyte-stimulating (Mls) genes. These Mls genes contain integrated MMTVs that express an LTR-encoded sag gene. The sag product is a transmembrane glycoprotein that functions as a superantigen to stimulate T cell proliferation and the deletion of specific T cell subsets. Studies on transgenic mice expressing an exogenous MMTV indicated that Sag-induced T-cell depletion prevents infection by exogenous MMTV of the same sag gene specificity [142]. Subsequent analysis of the 3 endogenous MMTVs of BALB/c mice indicated that Mtv-encoded Sag is needed for exogenous MMTV infection, further suggesting that interfering with sag function alters susceptibility to MMTV infection [143].
Another set of co-opted retroviral genes serve an antiviral function through production of MuLV Env glycoproteins. As discussed above, the products of these genes, Fv4, Rmcf, and Rmcf2, interfere with exogenous infection by MuLVs of the same host range subgroup [99, 101, 102]. It is thought that these Env glycoproteins and their receptors associate in the endoplasmic reticulum to prevent processing and transport to the plasma membrane. Of the three identified interfering Env genes, two were found in a single Asian species, M. castaneus, which harbors infectious viruses of the corresponding host range types [103]. Comparable interfering ERV env genes have been found in chickens, sheep, and cats, suggesting that this is a common host resistance mechanism. The oldest known retroviral resistance gene, Fv1, is also a co-opted ERV sequence, [144]. The Fv1 sequence is responsible for resistance to MuLVs and is found in all Mus species [145]. However, the older species carry a non-restrictive null allele, while the restrictive alleles are found only in more recently diverged M. musculus subspecies, animals that are exposed to infectious MuLVs [119].
ERVs have also contributed coding sequences for at least two genes with functions other than virus resistance. The mouse has two syncytin genes, SynA and SynB, that are responsible for the cell fusions that produce the trophoblast during development of the placenta. These genes are ERV env genes [146]. Interestingly, humans also carry two syncytin genes with similar function, but the human and mouse genes are not at orthologous positions in the germ line, suggesting that ERVs were independently co-opted to serve similar functions in these different mammalian lineages.
Conclusions
Mouse XRVs were first identified in 1951 with the discovery that extracts from tumors or embryos could induce leukemia [147]. It was subsequently determined that uninfected mouse embryo cells could produce infectious virus [148]. Liquid DNA hybridization showed that cells carried viral sequences [149], suggesting that these XRVs were the products of ERVs rather than latent exogenous virus infections. Since that time, the number of identified ERV copies has progressively grown along with the number of distinct families, aided by the publication of the mouse genome sequence and increasing use of wild mouse species in addition to inbred laboratory mouse strains. The realization that a staggering proportion of the genome is ERV-related and that some ERVs serve key host functions has revitalized interest in the evolutionary forces that have shaped this host-virus co-dependency. While it is clear that newly emerging XRVs are likely to be mutagenic and pathogenic in their new hosts, long-term associations have clearly produced coevolutionary adaptations that tend to mitigate these deleterious effects. While some domesticated ERVs move toward extinction, others remain active over long evolutionary intervals, suggesting that all this activity is neither necessarily damaging nor entirely superfluous for the host. While ERVs and the transposable elements from which they evolved obviously contribute to genetic diversity on an evolutionary timescale, it can also be argued that these elements may have important roles in development, as shown for example by L1 contributions to neuronal diversity in the developing brain [150] or by the developmentally regulated high level of expression of MuERV-L and MaLR ERVs in early embryogenesis [26, 151]. Because work on murine ERVs and XRVs initially focused on the most pathogenic groups and was limited to studies on the inbred laboratory strains, we are only beginning to identify and characterize the larger and older families present in the mouse germ line, and to describe what may be common and widespread interspecies transmissions of Class I ERVs/XRVs. Further work on these elements should produce novel insights into the evolutionary past and refine our understanding of the present interdependence of these genomic parasites and their hosts.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, NIAID. The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit und soziale Sicherung.
Contributor Information
C. Stocking, Email: stocking@hpi.uni-hamburg.de.
C. A. Kozak, Email: ckozak@niaid.nih.gov.
References
- 1.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Boeke J, Stoye J. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor Laboratory Press; New York: 1997. pp. 343–436. [PubMed] [Google Scholar]
- 3.Stoye J. Endogenous retroviruses: still active after all these years? Curr Biol. 2001;11:R914–R916. doi: 10.1016/s0960-9822(01)00553-x. [DOI] [PubMed] [Google Scholar]
- 4.Guenet JL, Bonhomme F. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 5.Wicker T, Sabot F, Hua-Van A, Bennetzen J, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;9:411–412. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis R, Goodier J, Ostertag E, Kazazian HJ. Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat Genet. 1998;20:288–290. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- 7.Kramerov D, Vassetzky N. Short retroposons in eukaryotic genomes. Int Rev Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- 8.Malik H, Eickbush T. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle R, Feng D, Johnson M, McClure M. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 10.Biémont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 11.Jern P, Sperber G, Blomberg J. Use of endogenous retroviral sequences (ERVs) and structural markers for retroviral phylogenetic inference and taxonomy. Retrovirology. 2005;2:50. doi: 10.1186/1742-4690-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Facquet C, Mayo M, Maniloff J, Desselberger U, Ball L. Virus Taxonomy: The Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press; San Diego, CA: 2004. [Google Scholar]
- 14.Bénit L, Lallemand JB, Casella JF, Philippe H, Heidmann T. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J Virol. 1999;73:3301–3308. doi: 10.1128/jvi.73.4.3301-3308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson D, Mager D, Leong J. Endogenous human retroviruses. In: Levy J, editor. The Retroviridae. Vol. 1. Plenum Press; New York: 1994. [Google Scholar]
- 16.Gifford R, Kabat P, Martin J, Lynch C, Tristem M. Evolution and distribution of class II-related endogenous retroviruses. J Virol. 2005;79:6478–6486. doi: 10.1128/JVI.79.10.6478-6486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jern P, Stoye JP, Coffin JM. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007;3:2014–2022. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribet D, Harper F, Pierron G, Heidmann T. The GLN family of mouse endogenous retroviruses comprises an element competent for infectious viral particle formation. J Virol. 2008;82:4413–4419. doi: 10.1128/JVI.02141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy E, McDonald J. Long-terminal repeat retrotransposons of Mus musculus. Genome Biol. 2004;5:R14. doi: 10.1186/gb-2004-5-3-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromham L, Clark F, McKee J. Discovery of a novel murine type C retrovirus by data mining. J Virol. 2001;75:3053–3057. doi: 10.1128/JVI.75.6.3053-3057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International Human Genome Sequencing Consortium. Initial sequence and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 22.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 23.Costas J. Molecular characterization of the recent intragenomic spread of the murine endogenous retrovirus MuERV-L. J Mol Evol. 2003;56:181–186. doi: 10.1007/s00239-002-2392-3. [DOI] [PubMed] [Google Scholar]
- 24.Ribet D, Louvet-Vallée S, Harper F, de Parseval N, Dewannieux M, Heidmann O, Pierron G, Maro B, Heidmann T. Murine endogenous retrovirus MuERV-L is the progenitor of the ‘orphan’ epsilon viruslike particles of the early mouse embryo. J Virol. 2008;82:1622–1625. doi: 10.1128/JVI.02097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svoboda P, Stein P, Anger M, Bernstein G, Hannon G, Schultz R. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 26.Peaston A, Esikov A, Graber J, de Vries W, Holbrook A, Solter D, Knowles K. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Baldo AM, McClure MA. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J Virol. 1999;73:7710–7721. doi: 10.1128/jvi.73.9.7710-7721.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Y, Knoper RC, Kozak CA. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruses and their receptors elucidate receptor determinants of virus entry. J Virol. 2007;81:10550–10557. doi: 10.1128/JVI.00933-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obata MM, Khan AS. Structure, distribution, and expression of an ancient murine endogenous retrovirus-like DNA family. J Virol. 1988;62:4381–4386. doi: 10.1128/jvi.62.11.4381-4386.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eicher EM, Hutchinson KW, Phillips SJ, Tucker PK, Lee BK. A repeated segment on the mouse Y chromosome is composed of retroviral-related, Y- enriched and Y-specific sequences. Genetics. 1989;122:181–192. doi: 10.1093/genetics/122.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callahan R, Drohan W, Gallahan D, D’Hoostelaere L, Potter M. Novel class of mouse mammary tumor virus-related DNA sequences found in all species of Mus, including mice lacking the virus proviral genome. Proc Natl Acad Sci USA. 1982;79:4113–4117. doi: 10.1073/pnas.79.13.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtney MG, Elder PK, Steffen DL, Getz MJ. Evidence for an early evolutionary origin and locus polymorphism of mouse VL30 DNA sequences. J Virol. 1982;43:511–518. doi: 10.1128/jvi.43.2.511-518.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inaguma Y, Miyashita N, Moriwaki K, Huai WC, Mei-Lei J, Xinqiao H, Ikeda H. Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations. J Virol. 1991;65:1796–1802. doi: 10.1128/jvi.65.4.1796-1802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozak C, O’Neil R. Diverse wild mouse origins of xenotropic, mink-cell focus-forming, and two types of ecotropic proviral genes. J Virol. 1987;61:3082–3388. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mager D, Freeman J. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J Virol. 2000;74:7221–7229. doi: 10.1128/jvi.74.16.7221-7229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonigo P, Wain-Hobson S, Bougueleret L, Tiollais P, Jacob F, Brûlet P. Nucleotide sequence and evolution of ETn elements. Proc Natl Acad Sci USA. 1987;84:3768–3771. doi: 10.1073/pnas.84.11.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt M, Wirth T, Krçger B, Horak I. Structures and genomic organization of a new family of murine retrovirus-related DNA sequences (MuRRS) Nucleic Acids Res. 1985;13:3461–3470. doi: 10.1093/nar/13.10.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit AFA. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogt V. Retroviral virions and genomes. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor Laboratory Press; New York: 1997. pp. 343–436. [PubMed] [Google Scholar]
- 40.Kozak CA, Ruscetti S. Retroviruses in rodents. In: Levy J, editor. The Retroviridae. Vol. 3. Plenum Press; New York: 1994. [Google Scholar]
- 41.Michalides R, van Nie R, Nusse R, Hynes N, Groner B. Mammary tumor induction loci in GR and DBAf mice contain one provirus of the mouse mammary tumor virus. Cell. 1981;23:165–173. doi: 10.1016/0092-8674(81)90281-6. [DOI] [PubMed] [Google Scholar]
- 42.Medstrand P, Mager D. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belshaw R, Dawson A, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brûlet P, Kaghad M, Xu Y, Croissant O, Jacob F. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci USA. 1983;80:5641–5645. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ribet D, Harper F, Dewannieux M, Pierron G, Heidmann T. Murine MusD retrotransposon: structure and molecular evolution of an ‘intracellularized’ retrovirus. J Virol. 2007;81:1888–1898. doi: 10.1128/JVI.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baust C, Gagnier L, Baillie G, Harris M, Juriloff D, Mager D. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J Virol. 2003;77:11448–11458. doi: 10.1128/JVI.77.21.11448-11458.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuff E, Lueders K. The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res. 1988;51:183. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- 48.Reuss F. Expression of intracisternal A-particle-related retroviral element-encoded envelope proteins detected in cell lines. J Virol. 1992;66:1915–1923. doi: 10.1128/jvi.66.4.1915-1923.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T. An infectious progenitor for the murine IAP retrotransposon: emergence of an intra-cellular genetic parasite from an ancient retrovirus. Genome Res. 2008;18:597–609. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benveniste R, Callahan R, Sherr C, Chapman V, Todaro G. Two distinct endogenous type C viruses isolated from the asian rodent Mus cervicolor: conservation of virogene sequences in related rodent species. J Virol. 1977;21:849–862. doi: 10.1128/jvi.21.3.849-862.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lieber M, Sherr C, Todaro G, Benveniste R, Callahan R, Coon H. Isolation from the asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc Natl Acad Sci USA. 1975;72:2315–2319. doi: 10.1073/pnas.72.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volgamot G, Bonham L, Miller A. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL/30 gibbon ape leukemia virus-like structure and a distinct envelope. J Virol. 1998;72:7459–7466. doi: 10.1128/jvi.72.9.7459-7466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller A, Bergholz U, Ziegler M, Stocking C. Identification of the myelin protein plasmolipin as the cell-entry receptor for Mus caroli endogenous retrovirus. J Virol. 2008;82:6862–6868. doi: 10.1128/JVI.00397-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tailor CS, Nouri A, Lee CG, Kozak CA, Kabat D. A cell surface receptor for xenotropic and polytropic murine leukemia viruses and characterized. Proc Natl Acad Sci USA. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albritton L, Tseng L, Scadden D, Cunningham J. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 56.Miller D, Edwards R, Miller A. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller D, Miller A. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hein S, Prassolov V, Zhang Y, Ivanov D, Lçhler J, Ross S, Stocking C. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cervicolor M813 murine leukemia virus. J Virol. 2003;77:5926–5932. doi: 10.1128/JVI.77.10.5926-5932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tipper C, Bencsics C, Coffin J. Characterization of Hortulanus endogenous murine leukemia virus, and endogenous provirus that encodes an infectious murine leukemia virus of a novel subgroup. J Virol. 2005;79:8316–8329. doi: 10.1128/JVI.79.13.8316-8329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kozak C, Hartley J, Morse H., 3rd Laboratory and wild-derived mice with multiple loci for production of xenotropic murine leukemia virus. J Virol. 1984;51:77–80. doi: 10.1128/jvi.51.1.77-80.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levy J, Pincus T. Demonstration of biological activity of a murine leukemia virus of New Zealand black mice. Science. 1970;170:326–327. doi: 10.1126/science.170.3955.326. [DOI] [PubMed] [Google Scholar]
- 62.Frankel WN, Stoye JP, Taylor BA, Coffin JM. A linkage map of endogenous murine leukemia proviruses. Genetics. 1990;124:221–236. doi: 10.1093/genetics/124.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenkins NA, Copeland NG, Taylor BA, Lee BK. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alamgir A, Owens N, Lavignon M, Malik F, Evans L. Precise identification of endogenous proviruses of NFS/N mice participating in recombination with Moloney ecotropic murine leukemia virus (MuLV) to generate polytropic MuLVs. J Virol. 2005;79:4664–4671. doi: 10.1128/JVI.79.8.4664-4671.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khan A. Nucleotide sequence analysis establishes the role of endogenous murine leukemia virus DNA segments in formation of recombinant mink cell focus-forming murine leukemia viruses. J Virol. 1984;50:864–871. doi: 10.1128/jvi.50.3.864-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voytek P, Kozak C. HoMuLV: a novel pathogenic ecotropic virus isolated from the European mouse, Mus hortulanus. Virology. 1988;165:469–475. doi: 10.1016/0042-6822(88)90590-9. [DOI] [PubMed] [Google Scholar]
- 67.O’Neill R, Hartley J, Repaske R, Kozak C. Amphotropic proviral envelope sequences are absent from the Mus germ line. J Virol. 1987;61:2225–2231. doi: 10.1128/jvi.61.7.2225-2231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao Y, Jacobs C, Wang L, Hardies S. MuERVC: a new family of murine retrovirus-related repetitive sequences and its relationship to previously known families. Mamm Genome. 1999;10:477–448. doi: 10.1007/s003359901026. [DOI] [PubMed] [Google Scholar]
- 69.Phillips S, Birkenmeier E, Callahan R, Eicher E. Male and female mouse DNAs can be discriminated using retroviral probes. Nature. 1982;297:241–243. doi: 10.1038/297241a0. [DOI] [PubMed] [Google Scholar]
- 70.Hutchison K, Eicher EM. An amplified endogenous retroviral sequence on the murine Y chromosome related to the murine leukemia viruses and viruslike 30S sequences. J Virol. 1989;63:4043–4046. doi: 10.1128/jvi.63.9.4043-4046.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fennelly J, Harper K, Laval S, Wright E, Plumb M. Co-amplification of tail-to-tail copies of MuRVY and IAPE retroviral genomes on the Mus musculus Y chromosome. Mamm Genome. 1996;7:31–36. doi: 10.1007/s003359900008. [DOI] [PubMed] [Google Scholar]
- 72.Itin A, Keshet E. A novel retroviruslike family in the mouse DNA. J Virol. 1986;59:301–307. doi: 10.1128/jvi.59.2.301-307.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolgamot G, Bonhan L, Miller A. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J Virol. 1998;72:7459–7466. doi: 10.1128/jvi.72.9.7459-7466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Howk R, Troxler D, Lowy D, Duesberg P, Scolnick E. Identification of a 30S RNA with properties of a defective type C virus in murine cells. J Virol. 1978;25:115–123. doi: 10.1128/jvi.25.1.115-123.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.French NS, Norton JD. Structure and functional properties of mouse VL30 retrotransposons. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 76.Rotman G, Itin A, Keshet E. ‘Solo’ large terminal repeats (LTR) of an endogenous retrovirus-like gene family (VL30) in the mouse genome. Nucleic Acids Res. 1984;12:2273–2282. doi: 10.1093/nar/12.5.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reference removed in proof.
- 78.Rhee S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 79.Fehrmann F, Jung M, Zimmermann R, Kräusslich H. Transport of the intracisternal A-type particle Gag polyprotein to the endoplasmic reticulum is mediated by the signal recognition particle. J Virol. 2003;77:6293–6304. doi: 10.1128/JVI.77.11.6293-6304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khan A, Martin M. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci USA. 1983;80:2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Itin A, Keshet E. Diverse long terminal repeats are associated with murine retroviruslike (VL30) elements. Mol Cell Biol. 1986;6:1276–1282. doi: 10.1128/mcb.6.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poznanski A, Calarco P. The expression of intracisternal A particle genes in the preimplantation mouse embryo. Dev Biol. 1991;143:271–281. doi: 10.1016/0012-1606(91)90077-g. [DOI] [PubMed] [Google Scholar]
- 83.Tailor C, Lavillette D, Marin M, Kabat D. Cell surface receptors for gammaretroviruses. Curr Top Microbio Immunol. 2003;281:29–106. doi: 10.1007/978-3-642-19012-4_2. [DOI] [PubMed] [Google Scholar]
- 84.Benveniste R, Todaro G. Evolution of C-type viral genes: inheritance of exogenously acquired viral genes. Nature. 1974;252:456–459. doi: 10.1038/252456a0. [DOI] [PubMed] [Google Scholar]
- 85.Benveniste R, Todaro G. Homology between type-C viruses of various species as determined by molecular hybridization. Proc Natl Acad Sci USA. 1973;70:3316. doi: 10.1073/pnas.70.12.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin J, Herniou E, Cook J, O’Neill RW, Tristem M. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J Virol. 1999;73:2442–2449. doi: 10.1128/jvi.73.3.2442-2449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Young P. Biology and evolution of the endogenous Koala retrovirus. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8499-y. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Urisman A, Molinaro R, Fischer N, Plummer S, Casey G, Klein E, Malathi K, Magi-Galluzzi C, Tubbs R, Ganem D, Silverman R, et al. Identification of a novel gammaretrovirus in prostate tumors pf patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein E, Ganem D, Derisi J, Chow S, Silverman R. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci USA. 2007;104:1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Albritton LM, Kim JW, Tseng L, Cunningham JM. Envelope-binding domain in the cationic amino acid transporter determined the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshimoto T, Yoshimoto E, Meruelo D. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J Virol. 1993;67:1310–1314. doi: 10.1128/jvi.67.3.1310-1314.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marin M, Tailor CS, Nouri A, Kozak SL, Kabat D. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J Virol. 1999;73:9362–9368. doi: 10.1128/jvi.73.11.9362-9368.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eiden M, Farell K, Warsowe J, Mahan L, Wilson C. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J Virol. 1993;67:4056–4061. doi: 10.1128/jvi.67.7.4056-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kubo Y, Ono T, Ogura M, Ishimoto A, Amanuma H. A glycosylation-defective variant of the ecotropic murine retrovirus receptor is expressed in rat XC cells. Virology. 2002;303:338–344. doi: 10.1006/viro.2002.1641. [DOI] [PubMed] [Google Scholar]
- 95.Eiden MV, Farrell K, Wilson CA. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J Virol. 1994;68:626–631. doi: 10.1128/jvi.68.2.626-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan Y, Jung YT, Wu T, Kozak CA. Role of receptor polymorphism and glycosylation in syncytium induction and host range variation of ecotropic mouse gammaretroviruses. Retrovirology. 2008;5:2. doi: 10.1186/1742-4690-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miller DG, Miller AD. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tavoloni N, Rudenholz A. Variable efficiency of murine leukemia retroviral vector on mammalian cells: role of cellular glycosylation. Virology. 1997;229:49–56. doi: 10.1006/viro.1996.8412. [DOI] [PubMed] [Google Scholar]
- 99.Ikeda H, Laigret F, Martin MA, Repaske R. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol. 1985;55:768–777. doi: 10.1128/jvi.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bassin RH, Ruscetti S, Ali I, Haapala DK, Rein A. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology. 1982;123:139–151. doi: 10.1016/0042-6822(82)90301-4. [DOI] [PubMed] [Google Scholar]
- 101.Jung YT, Lyu MS, Buckler-White A, Kozak CA. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J Virol. 2002;76:8218–8224. doi: 10.1128/JVI.76.16.8218-8224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu T, Yan Y, Kozak CA. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J Virol. 2005;79:9677–9684. doi: 10.1128/JVI.79.15.9677-9684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chattopadhyay SK, Oliff AI, Linemeyer DL, Lander MR, Lowy DR. Genomes of murine leukemia viruses isolated from wild mice. J Virol. 1981;39:777–791. doi: 10.1128/jvi.39.3.777-791.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu T, Lee CG, Buckler-White A, Kozak CA. Genetic control of a mouse serum lipoprotein factor that inactivates murine leukemia viruses: evaluation of apolipoprotein F as a candidiate. J Virol. 2002;76:279–2286. doi: 10.1128/jvi.76.5.2279-2286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tailor C, Nouri A, Kabat D. Cellular and species resistance to murine amphotropic, gibbon ape, and feline subgroup C leukemia viruses is strongly influenced by receptor expression levels and by receptor masking mechanisms. J Virol. 2000;74:9797–9801. doi: 10.1128/jvi.74.20.9797-9801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jinno-Oue A, Oue M, Ruscetti S. A unique heparin-binding domain in the envelope protein of the neuropathogenic PVC-211 murine leukemia virus may contribute to its brain capillary endothelial cell tropism. J Virol. 2001;75:12439–12445. doi: 10.1128/JVI.75.24.12439-12445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Battini JL, Heard J, Danos O. Receptor choice determinants in the envelope glycoprotein of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilson CA, Farrell KB, Eiden MV. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Hoeven NS, Miller AD. Use of different but overlapping determinants in a retrovirus receptor accounts for non-reciprocal interference between xenotropic and polytropic murine leukemia viruses. Retrovirology. 2005;2:76. doi: 10.1186/1742-4690-2-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fischinger PJ, Nomura S, Bolognesi DP. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc Natl Acad Sci USA. 1975;72:5150–5155. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hartley JW, Wolford NK, Old LJ, Rowe WP. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci USA. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wensel DL, Li W, Cunningham JM. A virus-virus interaction circumvents the virus receptor requirement for infection by pathogenic retroviruses. J Virol. 2003;77:3460–3469. doi: 10.1128/JVI.77.6.3460-3469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pincus T, Hartley JW, Rowe WP. A major genetic locus affecting resistance to infection with murine leukemia viruses. I Tissue culture studies of naturally occurring viruses. J Exp Med. 1971;133:1219–1233. doi: 10.1084/jem.133.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Best S, LeTossier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 115.Sveda MM, Soeiro R. Host restriction of Friend leukemia virus: synthesis and integration of the provirus. Proc Natl Acad Sci USA. 1976;73:2356–2360. doi: 10.1073/pnas.73.7.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kozak CA, Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- 117.Stevens A, Bock M, Ellis S, LeTissier P, Bishop KN, Yap MW, Taylor W, Stoye JP. Retroviral capsid determinants of Fv1 NB and NR tropism. J Virol. 2004;78:9592–9598. doi: 10.1128/JVI.78.18.9592-9598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yan Y, Kozak CA. Novel post-entry resistance to AKV ecotropic mouse gammaretroviruses in the African pygmy mouse, Mus minutoides. J Virol. 2008 doi: 10.1128/JVI.00202-08. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kozak CA. Analysis of wild-derived mice for the Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J Virol. 1985;55:281–285. doi: 10.1128/jvi.55.2.281-285.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5a protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goila-Gaur R, Strebel K. HIV-1 vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 123.Okeoma CM, Lovsin N, Peterlin MB, Ross SR. APOBEC3 inhibits mouse mammary tumor virus replication in vivo. Nature. 2007;445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 124.Esnault C, Neidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 125.Santiago ML, Montano M, Benitez RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, Greene WC. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Levy DE, Lerner RA, Wilson MC. The Gv-1 locus coordinately regulates the expression of multiple endogenous murine retroviruses. Cell. 1985;44:289–299. doi: 10.1016/0092-8674(85)90082-0. [DOI] [PubMed] [Google Scholar]
- 127.Oliver PL, Stoye JP. Genetic analysis of Gv1, a gene controlling transcription of endogenous murine polytropic proviruses. J Virol. 1999;73:8227–8234. doi: 10.1128/jvi.73.10.8227-8234.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wolf D, Hug K, Goff SP. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc Natl Acad Sci USA. 2008;105:12521–12526. doi: 10.1073/pnas.0805540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Floyd JA, Gold DA, Concepcion D, Poon TH, Wang X, Keithley E, Chen D, Ward EJ, Chinn SB, Friedman RA, et al. A natural allele of Nxf1 suppresses retroviurs insertional mutations. Nat Genet. 2003;35:221–228. doi: 10.1038/ng1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 131.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCG-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 132.Neil SJD, Zang T, Bieniasz P. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 133.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jenkins NA, Copeland NG, Taylor BA, Lee BK. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature. 1981;293:370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- 135.Separack PK, Mercer JA, Stobel MC, Copeland NG, Jenkins NA. Retroviral sequences located within an intron of the dilute gene alter dilute expression in a tissue-specific manner. EMBO J. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stavenhagen JB, Robins DM. An ancient provirus has imposed androgen regulation on the adjacent mouse sex-limited protein gene. Cell. 1988;55:247–254. doi: 10.1016/0092-8674(88)90047-5. [DOI] [PubMed] [Google Scholar]
- 137.Maksakova I, Mager D. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J Virol. 2005;79:13865–13874. doi: 10.1128/JVI.79.22.13865-13874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morgan H, Sutherland H, Martin D, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 139.Gaudet F, Rideout W, 3rd, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol. 2004;24:1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jähner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 141.Whitelaw E, Martin D. Retrotransposons as epigenetic mediators of phenotpyic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 142.Golovkina TV, Chervonsky A, Dudley JP, Ross SR. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 143.Bhadra S, Lozano MM, Payne SM, Dudley JP. Endogenous MMTV proviruses induce susceptibility to both viral and bacterial pathogens. PLoS Pathogens. 2006;2:1134–1143. doi: 10.1371/journal.ppat.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bénit L, De Parseval N, Casella J, Callebaut I, Cordonnier A, Heidmann T. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol. 1997;71:5652–5657. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qi CF, Bonhomme F, Buckler-White A, Buckler C, Orth A, Lander MR, Chattopadhyay SK, Morse HC., III Molecular phylogeny of Fv1. Mamm Genome. 1998;9:1049–1055. doi: 10.1007/s003359900923. [DOI] [PubMed] [Google Scholar]
- 146.Dupressoir A, Marceau G, Vernochet C, Bénit L, Kanellopoulos C, Sapin V. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gross L. ‘Spontaneus’ leukemia developing in C3H mice following inoculation in infancy, with Ak-leukemic extracts, or AK-embryos. Proc Soc Exp Biol Med. 1951;76:27–32. [PubMed] [Google Scholar]
- 148.Aaronson SA, Hartley JW, Todaro GJ. Mouse leukemia virus: ‘spontaneous’ release by mouse embryo cells after long-term in vitro cultivation. Proc Natl Acad Sci USA. 1969;64:87–94. doi: 10.1073/pnas.64.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Harel L, Harel J, Huppert J. Partial homology between RNA from Rauscher mouse leukemia virus and cellular DNA. Biochem Biophys Res Commun. 1967;28:44–49. doi: 10.1016/0006-291x(67)90403-2. [DOI] [PubMed] [Google Scholar]
- 150.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 151.Peaston A, Knowles B, Hutchison K. Genome plasticity in the mouse oocyte and early embryo. Biochem Soc Trans. 2007;35:618–622. doi: 10.1042/BST0350618. [DOI] [PubMed] [Google Scholar]