Abstract
Classically, model-based segmentation procedures match magnetic resonance imaging (MRI) volumes to an expertly labeled atlas using nonlinear registration. The accuracy of these techniques are limited due to atlas biases, misregistration, and resampling error. Multi-atlas-based approaches are used as a remedy and involve matching each subject to a number of manually labeled templates. This approach yields numerous independent segmentations that are fused using a voxel-by-voxel label-voting procedure. In this article, we demonstrate how the multi-atlas approach can be extended to work with input atlases that are unique and extremely time consuming to construct by generating a library of multiple automatically generated templates of different brains (MAGeT Brain). We demonstrate the efficacy of our method for the mouse and human using two different nonlinear registration algorithms (ANIMAL and ANTs). The input atlases consist a high-resolution mouse brain atlas and an atlas of the human basal ganglia and thalamus derived from serial histological data. MAGeT Brain segmentation improves the identification of the mouse anterior commissure (mean Dice Kappa values (κ = 0.801), but may be encountering a ceiling effect for hippocampal segmentations. Applying MAGeT Brain to human subcortical structures improves segmentation accuracy for all structures compared to regular model-based techniques (κ = 0.845, 0.752, and 0.861 for the striatum, globus pallidus, and thalamus, respectively). Experiments performed with three manually derived input templates suggest that MAGeT Brain can approach or exceed the accuracy of multi-atlas label-fusion segmentation (κ = 0.894, 0.815, and 0.895 for the striatum, globus pallidus, and thalamus, respectively).
Keywords: segmentation, label-fusion, multi-atlas, subcortical anatomy, striatum, globus pallidus, thalamus, small animal imaging, mouse imaging, atlases, nonlinear registration
INTRODUCTION
Manually derived delineations of human neuroanatomical structures are often considered the “gold-standard” for the segmentation of magnetic resonance images [Burk et al., 2004; Schulz et al., 1999; Seeck et al., 2005]. Similarly, many semi-automated techniques exist for neuroanatomical phenotyping of wild-type and transgenic murine brain anatomy using magnetic resonance images (MRI) [Bock et al., 2006; Ma et al., 2005]. However, several factors create a need for accurate and robust segmentation methods that are fully automated. These include the advent of high-throughput phenotyping of mouse models of human disease and behavior accompanied by advances in imaging methodology [Lerch et al., 2011a; Nieman et al., 2007]. Likewise, in human imaging there is a vast and ever-increasing amount of structural MRI data [Mazziotta et al., 1995, 2001a,b; Pausova et al., 2007], which make manual segmentation increasingly problematic. On a practical level, manual or semiautomated methods are too time consuming to properly deal with such extensive quantities of data and are fraught with issues of inter- and intra-rater variability and observer subjectivity [Chakravarty et al., 2008, 2009a,b; Shattuck et al., 2008].
One of the earliest methods devised for overcoming the limitations of manual or semiautomated procedures was model-based segmentation. Classically, these algorithms have relied on using a well-defined template derived from the manual segmentations of a single rater or multiple raters. The automated segmentation can then be accomplished through the estimation of a subject-to-template nonlinear transformation, and then applying the inverse transformation to the labels defined on the template [Bajcsy et al., 1983; Collins et al., 1995; Miller et al., 1997]. Many of these methods have since been adapted to the animal imaging literature [Ali et al., 2005; Bae et al., 2009; Dorr et al., 2008; Frey et al., 2011]. While this methodology provides a reasonable strategy for customizing these predefined labels onto the anatomy of individual subjects, it is constrained by a number of limitations:
Errors in the estimation of the nonlinear subject-to-template transformation will cause errors in the final labeling of the subject.
The neuroanatomy of the template may not be representative of the underlying neuroanatomy of the subjects being analyzed.
Resampling errors are often present after applying the transformation to the labels defined on the template.
There are several studies which have evaluated the choice of registration procedure [Chakravarty et al., 2008, 2009a,b; Hellier et al., 2003; Klein et al., 2009] and the parameters of a registration algorithm [Avants et al., 2011; Robbins et al., 2004] in the context of segmentation accuracy. Most of these evaluations estimate how well nonlinear registration algorithms are able to recover anatomical differences between subjects and between a subject and a neuroanatomical template [Grabner et al., 2006; Holmes et al., 1998; Mazziotta et al., 2001a], but do little in the way of accounting for other sources of error.
The use of probabilistic methods has become increasingly popular for segmentation procedures to account for differences in the underlying neuroanatomy. Instead of a single template, several templates are manually labeled to account for the variability of any given population. This type of methodology has been popular in the segmentation of sulci in the human cortex (given their inherent variability). One of the earliest uses of this technique was presented by Fischl et al. [2002], where the prior probability of the occurrence of each structure at a specific voxel is used to determine the a priori spatial arrangement of neuroanatomical structures. This arrangement is then refined using a Markov random field. These ideas have also been extended for the segmentation of mouse brain MRI data. In Ali et al. [2005], probabilistic information is obtained about the location and the spatial relationships between structures using multispectral (T1, T2, proton density, and diffusion weighted) MRI acquisitions and then refined using Markov random field theory. This segmentation technique has recently been improved further by introducing a support vector machine into the Markovian random field classification stage [Bae et al., 2009].
Recently, a new class of segmentation technique has emerged which has been referred to as multi-atlas or label fusion based segmentation. In principal, this methodology is well suited for dealing with the limitations of regular model-based segmentation procedures. In this methodology, a library of numerous manually defined templates is generated by a single or multiple manual raters, and each subject undergoes the subject-to-template nonlinear registration procedure numerous times (where the subject is matched to all of or some of the templates in the library). Once all of the nonlinear transformations have been estimated, the labels are warped back onto the subject's anatomy. This is often referred to as the multi-atlas or template library step. However, once this stage is complete a label fusion step must be employed to combine labels in an optimal fashion that can boost the labeling accuracy. Here, many strategies have been employed such as the STAPLE algorithm that computes a probabilistic segmentation based on an ensemble of competing segmentations; others where a subset of templates are selected from the library based on some sort of similarity criteria that attempts to evaluate the homology between the regions of interest to be segmented using metrics such as the sum of squared-differences [Coupe et al., 2011b] or normalized mutual information [Collins and Pruessner, 2010]; or an optimized combination of weights based on the covariance of region of interest intensities [Wang et al., 2011].
One of the first implementations of this was proposed by Rolf Heckemann et al. [2006] where each brain is matched to 30 manually prelabeled templates using a spline-based nonlinear registration technique [Rueckert et al., 1999]. The final segmentation is then implemented using label fusion rules [Rohlfing et al., 2004a,b]. These techniques have since been improved to reduce the computational complexity by preselecting the best N templates which are most similar to the subject's input MRI data based on the normalized mutual information metric [Aljabar et al., 2007, 2009]. Similarly, Collins and Pruessner [2010] implemented a multi-atlas based segmentation strategy for the automatic identification of the human hippocampus and amygdala by determining the N most similar labels based on the estimation of the normalized mutual information [Studholme et al., 2001] in a region-of-interest defined in the temporal lobe. Others have also chosen to use a similarity criterion to select the “best” template from a library [Barnes et al., 2008], thereby reducing the label fusion problem to a template selection problem followed by a traditional atlas-based segmentation procedure.
An open topic in label-fusion based segmentation is how to best generate a template library. To do so manually requires trained personal and a manual segmentation strategy that does not suffer from inter- and intra-rater subjectivity [Pruessner et al., 2002]. A strategy that generates some or all of the templates in a library is far more desirable if it is able to successfully mimic the work of a manual rater. One method that generates a template library from a limited set of manually derived segmentations is the LEAP algorithm [Wolz et al., 2010]. In their method, the pair-wise similarity between all images in a dataset is first estimated and used to define the edge-weights is a graph representation. The segmentation procedure is then implemented as a three-step process. First, the initial atlases are used to segment a predefined number of images from the dataset. The choice of images to be segmented is defined by their proximity in the graph representation. In the second step, these newly segmented images become atlases as well, where they are then used for the segmentation of the other unlabeled images within the dataset. This can lead to a number of segmentations for each subject; thus, the final segmentation is defined using a decision rule at each voxel.
The Wolz technique [2010] is the closest to the work that we have presented here. Unlike Wolz et al, we demonstrate that segmentations can be improved in the special case where there can only be a single or very few input atlases. While the label fusion techniques described above are robust, they are difficult to implement in cases where the initial atlas is derived from a unique dataset. For example, atlases based on the reconstruction of manually segmented serial histological data [Chakravarty et al., 2006; Yelnik et al., 2007] are difficult to generate due to the availability of donated histological data, limitations concerning processing the brain for different histological and immunohistochemical techniques, and time required to manually identify structures on such a high-resolution datasets. Despite the other improved techniques available for the visualization of subcortical structures [Behrens et al., 2003; Deoni et al., 2005], these atlases provide great utility in defining discrete hard-to-visualize structures. In the mouse imaging community, there is no shortage of possible mice with which to create a population-based atlas. However given the resolution of such an atlas and the number of structures which can be resolved with high-fidelity, comprehensive and accurate manual segmentation would typically take months per atlas, making the creation of multiple such segmentations prohibitive. In this manuscript we extend the multi-atlas based segmentation techniques through the development of multiple automatically generated templates from a single labeled brain (MAGeT Brain). We hypothesized that a template library could be automatically generated directly within the dataset being analyzed; specifically in structures (or brains) where anatomical mislabeling should arise from misregistration or resampling errors. To this end, we validate this technique on human brain structures (the striatum, the globus pallidus, and the thalamus) and mouse brains (which are lissencephallic). In other words, we focus on structures and brains where the morphological homology should be maximal in comparison to anatomical structures in the human cerebral cortex. Segmentations were performed using two different registration algorithms to demonstrate that our technique can work using different validated nonlinear warping methods. The segmentation results are validated against manually derived gold-standards using two different “goodness-of-fit” metrics. The results demonstrate that the MAGeT Brain technique improves segmentation over standard model-based segmentation and group-wise segmentation/registration procedures and may be equivalent to multi-atlas label-fusion segmentation techniques in cases where three input templates can be used for the segmentation. We also verify if weighting the votes using local similarity constraints in the template library selection process can further improve segmentation.
METHODS
MAGeT Brain
The MAGeT Brain procedure is a three-step process. Consider any input dataset consisting of n structural MRIs. A template library can be generated through the segmentation of each or a subset of the n subjects in the input dataset using a regular model-based segmentation procedure. Similarly, an input atlas can be created a priori from existing MRI data independent of the dataset being analyzed. The goal is to distribute possible registration and resampling errors (and therefore segmentation errors) across the input data.
The next step is to generate multiple segmentations for each subject from the newly derived template library. This is accomplished through pair-wise registration of each subject to all the templates in the automatically generated template library. For each subject, this process yields as many possible segmentations as there are templates and allows for the averaging of some of the errors present in the initial segmentations used to create the template library. This will serve to filter any spurious errors that arise out of the initial segmentation process.
The final step consists of a voxel voting procedure, where the most frequently occurring label at each voxel is retained for the final segmentation [as in Collins and Pruessner, 2010]. While other voxel voting procedures exist [Heckemann et al., 2006], the evaluation of the best voxel-decision method is outside of the scope of this manuscript. A schematic view of this procedure is given in Figure 1.
Figure 1.
Illustration of the MAGeT Brain Method. Step 1: A template library can be created automatically using the data that one has on one hand. This requires the estimation of a nonlinear transformation matching an input template to each subject in the dataset (A) and then applying that transformation to the label set so that each subject is now labeled (B). Step 2: Multi-atlas segmentation is then performed by estimating all pairwise inter- subject nonlinear transformations (i.e., using the newly labeled subjects as a template library; (2A). This creates a set of different labels for each subject for each of the anatomical structures. Step 3. A label voting technique is applied at each voxel where the most frequently occurring label is retained for the final segmentation output.
Nonlinear Registration Algorithms
In the remainder of this manuscript we evaluate the performance of two different algorithms in the context of MAGeT Brain and all other techniques that are tested (these techniques will be described later in the Methods section):
Automatic nonlinear image matching and anatomical labeling (ANIMAL) [Collins and Evans, 1997; Collins et al., 1995]. The ANIMAL algorithm is an iterative procedure that estimates a 3D deformation field that matches a source volume to a target volume. The algorithm is divided into two steps. The first is the outer loop, where large deformations are estimated using blurred versions of the input data. These larger deformations are then input to subsequent steps where the fit is refined by estimating smaller deformations on data blurred with a Gaussian kernel with a smaller full-width at half maximum. The final transformation is a set of local translations defined on a bed of equally spaced nodes that were estimated through the optimization of the correlation coefficient (Robbins et al., 2004). The ANIMAL algorithm is part of the mni_autoreg package and is freely available (http://www.bic.mni.mcgill.ca/ServicesSoftware/HomePage).
Automatic normalization tools (ANTs) [Avants et al., 2011; Avants et al., 2008]. ANTs is a diffeomorphic registration algorithm which provides great flexibility over the choice of transformation model, objective function, and the consistency of the final transformation. The transformation is estimated in a hierarchical fashion where the MRI data is subsampled, allowing large deformations to be estimated and successively refined at later hierarchical stages (where the data is subsampled to a finer grid). The deformation field and the objective function are regularized with a Gaussian kernel at each level of the hierarchy. The ANTs algorithm is also freely available (http://www.picsl.upenn.edu/ANTS/).
For both choices the parameter setting will be crucial to the accuracy of the transformations estimated. These parameter choices will be further explained below.
Experiments
The experiments were performed using MRI data from both mouse and human brains to demonstrate the general reliability of MAGeT Brain on diverse datasets. The input templates, validation datasets, manually derived gold-standards, and registration parameters are described for the three different validation experiments performed.
MAGeT Brain for the mouse
Input template
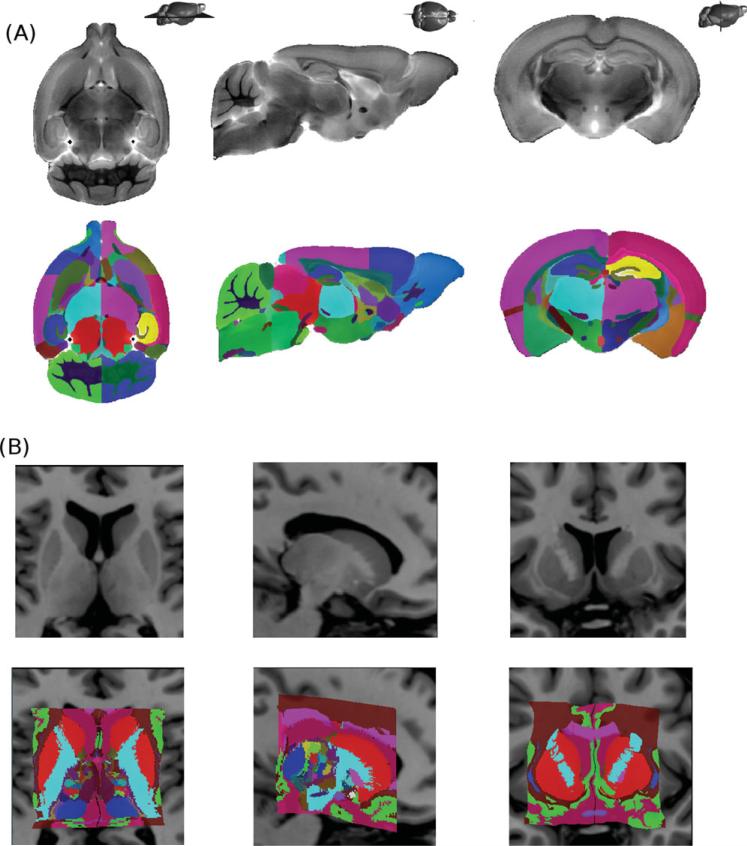
We use a template previously derived from the nonlinear average of 40 T2-weighted MRI volumes of the C57Bl/6 mouse brain as described in Dorr et al., [2008]. The final average image volume contains 32 lm isotropic voxels. The atlas contains 62 different structures that were manually defined by a trained rater. This atlas has been previously used for model-based segmentation in several studies from our group to obtain structure-wise volume values [Ellegood et al., 2010; Lerch et al., 2008, 2011b]. See Figure 2a for an example of this template.
Figure 2.
Input atlases used. (A) MRI mouse brain atlas derived from the nonlinear average of 40 different mouse brain MRIs and contains 62 structures [Dorr et al., 2008]. (B) Atlas of the human basal ganglia and thalamus derived from the reconstruction of 82 serial histological sections which has been warped to a template brain. The final atlas contains 108 subcortical structures [Chakravarty et al., 2006].
Dataset evaluated
MAGeT Brain was evaluated on a dataset of T2-weighted MRI data from 25 C57Bl/6 fixed mice brains. Images were gadolinium-enhanced to increase the contrast within the image. The specimen preparation for postmortem sample preparation has been described elsewhere [Dorr et al., 2008; Lerch et al., 2011b]. Briefly, a multichannel 7.0 T MRI scanner (Varian, Palo Alto, CA) with a 6-cm inner bore diameter insert gradient set was used to acquire anatomical images of brains within skulls. Prior to imaging, the samples were placed into 13-mm-diameter plastic tubes filled with a proton-free susceptibility-matching fluid (Fluorinert FC-77, 3M Corp., St. Paul, MN). Three custom-built, 14-mm-diameter solenoid coils with a length of 18.3 mm and over wound ends were used to image three brains in parallel. Parameters used in the scans were optimized for gray/white matter contrast: a T2-weighted, 3D fast spin-echo sequence, with TR/TE = 325/32 ms, four averages, field-of-view 14 × 14 × 25 mm3 and matrix size = 432 × 432 × 780 giving an image with 32 μm isotropic voxels. Total imaging time was 11.3 h. Geometric distortion due to position of the three coils inside the magnet was corrected using an MR phantom.
Nonlinear registration parameters for murine data
Nonlinear transformations estimated with ANIMAL used parameters previously employed for model-based segmentation and by our group. The transformations draw on a combination of intensity and gradient features for the estimation of the final output transformation. The final nonlinear transformation has a step size of 100 μm and is estimated in the hierarchical iterative fashion described above. The parameters used are summarized in Table I.
TABLE I.
Parameters used for the ANIMAL algorithm for mouse brain nonlinear transformation estimation
| Step size (mm) | Iterations | Gaussian blur (mm) | Feature type |
|---|---|---|---|
| 1 | 60 | 0.25 | Intensity |
| 0.5 | 60 | 0.25 | Intensity |
| 0.5 | 60 | 0.25 | Gradient |
| 0.2 | 10 | 0.25 | Intensity |
| 0.2 | 10 | 0.25 | Gradient |
| 0.1 | 4 | None | Intensity |
All values for lattice diameter were set to 1.5 × Step_size. Regularization parameters of stiffness, weight, and similarity were set to 0.98, 0.8, and 0.8, respectively.
ANTs registration algorithm parameters were determined empirically on a separate dataset by one of the authors (MvE). The optimization performed is beyond the scope of the current manuscript. A cross-correlation objective function was used for all transformations estimated with a symmetric normalization transformation model. Both intensity and gradient information (derived using convolution with a 120 μm Gaussian kernel) were used as input features. The parameters used are summarized in Table II.
TABLE II.
Parameters used for the ANTs algorithm for mouse brain registration
| Parameters | Values |
|---|---|
| Step size (intensity and gradient features) | 4 |
| Gaussian normalization (objective function) | 3 |
| Gaussian normalization (deformation field) | 3 |
| Symmetric normalization | 0.5 |
| Iterations | 100 × 100 × 100 × 40 |
Gold-standard for evaluation and segmentation methods evaluated
Manually derived gold standards were generated by one of the authors of this manuscript (VG) for both the left hippocampus and the entire anterior commissure for 10 of the 25 MRI volumes used in this experiment. In addition, five of the mice were relabeled to determine the intra-rater reliability.
MAGeT Brain segmentations were derived using both nonlinear registration algorithms were compared to the ground truth provided by the manual gold-standards. For the MAGeT Brain segmentations, the template library was created from all 25 of the input MRI volumes. These segmentations were compared to two other segmentation methods. The first is the standard template-based registration procedure, where the labels previously defined on a template are warped based on a template-to-subject nonlinear transformation. Because the purpose of MAGeT Brain is to filter out nonlinear registration and possible resampling errors, we use segmentations derived by first creating a model of the mean neuroanatomy as previously described in several articles from our group [Lerch et al., 2011a,b; Spring et al., 2007]. For both nonlinear registration algorithms, the MRI volumes are first rigidly registered to a common space [Dorr et al., 2008] followed by the estimation of all pairwise 12 parameter registrations (three scales, shears, rotations, and translations) and an average transformation for each mouse. After these average transformations have been applied, all MRI volumes were effectively normalized to the population average of the group and can be represented using a voxel-by-voxel average. An iterative multi-generation nonlinear alignment procedure is then initialized where all subjects are registered to the population average. Subsequently each subject is registered toward the voxel-by-voxel average of the previous generation. The registration schedules for both the ANIMAL and ANTs based group-wise averaging are given in Tables III and IV. The final segmentation for each subject was obtained by first concatenating the transformation mapping the original atlas to the new group-wise average and the inverse of the transformation mapping each subject to the final group-wise average and then applying it to the atlas labels.
TABLE III.
Parameters used for the ANIMAL algorithm for mouse brain group-wise nonlinear transformation estimation
| Generation | Step size (mm) | Iterations | Gaussian blur (mm) |
|---|---|---|---|
| 1 | 0.7 | 20 | 0.3 |
| 2 | 0.6 | 8 | 0.2 |
| 3 | 0.5 | 8 | 0.2 |
| 4 | 0.24 | 8 | 0.2 |
| 5 | 0.12 | 8 | 0.1 |
| 6 | 0.06 | 8 | 0.06 |
All values for lattice diameter were set to 1.5 × Step_size. Regularization parameters of stiffness, weight, and similarity were set to 0.98, 0.8, and 0.8, respectively. In all cases both intensity and gradient features extracted from the blur were used.
TABLE IV.
Parameters used for the ANTs algorithm for mouse brain group-wise nonlinear transformation estimation
| Generation | Iterations | Gaussian normalization (gradient fields) | Gaussian normalization (deformation field) |
|---|---|---|---|
| 1 | 100 × 100 × 100 × 0 | 5 | 1 |
| 2 | 100 × 100 × 100 × 20 | 5 | 1 |
| 3 | 100 × 100 × 100 × 50 | 5 | 1 |
In all cases both the cross correlation object function was used in conjunction with a symmetric normalization transformation model SyN=0.4, weight = 1, radius = 3.
All segmentations derived from model-based, group-wise, and MAGeT Brain techniques (derived for both nonlinear registration algorithms) were compared to the ground truth provided by the manual gold-standards. For the MAGeT Brain segmentations, the template library was created from all 25 of the input MRI volumes. In tables and figures, model-based segmentation using ANIMAL and ANTs will be referred to as Model ANIMAL and Model ANTs, respectively. Segmentations derived from the group-wise model building using the ANIMAL and ANTs algorithms will be referred to as Group-wise ANIMAL and Group-wise ANTs respectively. Similarly, MAGeT Brain segmentation using ANIMAL and ANTs will be referred to as MAGeT ANIMAL and MAGeT ANTs, respectively.
MAGeT Brain for subcortical labeling in the human
Input template
The input template used for segmentation of subcortical structures is an atlas of the basal ganglia and thalamus derived from a set of 84 serial histological slices [Chakravarty et al., 2006] where 108 different anatomical regions were defined. These regions include the Hirai and Jones [1989] and the Hassler [Schaltenbrand and Wahren, 1977] subdivisions for the thalamus and other subcortical nuclei and the Gloor [1997] definitions for temporal lobe structures. As in previous work [Chakravarty et al., 2008, 2009a,b], we collapse sublabels into three different anatomical structures for the striatum, globus pallidus, and thalamus. In previous work, the histology was reconstructed to minimize inconsistencies in slice-to-slice morphology and intensity inhomogeneities. All voxels within the reconstructed volume were assigned a unique voxel label. These voxel labels were then modified to match the intensity and contrast of the colin27 [Holmes et al., 1998] template to create a “pseudo-mri.” Using this pseudo-mri, a nonlinear atlas-to-template nonlinear transformation was estimated to match the atlas to the template. The template, and as a result the atlas labels, can be customized to the anatomy of a new subject using standard model based segmentation procedures. See Figure 2b for an example of the input data used.
Dataset evaluated
Twenty MRI volumes acquired from human adolescents aged 5–17 (10 male and 10 female) were used for this experiment. Half of the subjects suffer from attention deficit hyperactivity disorder (ADHD) while the other half are healthy normal controls [Shaw et al., 2007, 2011]. Because there are known thalamic abnormalities in subcortical structures in subjects that suffer from ADHD [Ivanov et al., 2010], the heterogeneity of the subjects used serves to add some anatomical variance to this study and has been used in other segmentation procedures [Fischl et al., 2002]. All images were acquired from a GE Signa 1.5T MR system (General Electric, Milwaukee, WI) using a coronal three-dimension spoiled gradient recalled sequence (TR/TE = 14/3 ms, 256 × 192 acquisition matrix, 124-mm slices; voxel size: 0.94 × 0.94 × 1.5 mm3).
Nonlinear registration parameters for human data
For segmentation with the ANIMAL algorithm we use the nonlinear registration parameters previously validated [Chakravarty, 2009a,b] and evaluated against other nonlinear registration methods [Chakravarty, 2009b]. Briefly, this method involves a linear registration followed by a nonlinear registration focused on a region-of-interest limited predominantly to the subcortical structures and other noncortical anatomical structures (such as the corpus callosum and lateral ventricles) to help guide the nonlinear transformation estimation process. See Tables V and VI for a summary of the parameters used in the nonlinear transformation process for ANIMAL and ANTs respectively. As in the mouse segmentation experiments, we also checked to see if we could improve segmentations by mapping the template to a group-wise average created from the population under study [Borghammer et al., 2010; Grabner et al., 2006]. The nonlinear registration parameters for each generation are given in Tables VII and VIII for ANIMAL and ANTs, respectively.
TABLE V.
Parameters used for the ANIMAL algorithm for human nonlinear transformations estimated for subcortical segmentation
| Step size (mm) | Iterations | Gaussian blur (mm) | Feature type |
|---|---|---|---|
| 4 | 20 | None | Intensity |
| 2 | 20 | None | Intensity |
| 1 | 15 | None | Intensity |
All values for lattice diameter were set to 3 × Step_size. Regularization parameters of stiffness, weight, and similarity were set to 1, 1, and 0.3, respectively as per the optimization of (Robbins et al., 2004).
TABLE VI.
Parameters used for the ANTs algorithm for registration of human subcortical structures
| Parameters | Values |
|---|---|
| Step size (Intensity and gradient features) | 5 |
| Gaussian normalization (objective function) | 5 |
| Gaussian normalization (deformation field) | 0 |
| Symmetric normalization | 0.5 |
| Iterations | 20 × 20 × 20 |
TABLE VII.
Parameters used for the ANIMAL algorithm for human subcortical group-wise nonlinear transformation estimation
| Generation | Step size (mm) | Iterations | Gaussian blur (mm) |
|---|---|---|---|
| 1 | 8 | 50 | 16 |
| 2 | 8 | 20 | 8 |
| 3 | 4 | 20 | 8 |
| 4 | 4 | 20 | 4 |
| 5 | 2 | 20 | 4 |
| 6 | 2 | 20 | 2 |
All values for lattice diameter were set to 1.5 × Step_size. Regularization parameters of stiffness, weight, and similarity were set to 1, 1, and 0.3, respectively (as per {Robbins et al., 2004, Med Image Anal, 8, 311-23}). In all cases intensity features extracted from the blur were used.
TABLE VIII.
Parameters used for the ANTs algorithm for mouse brain group-wise nonlinear transformation estimation
| Generation | Iterations | Gaussian normalization (gradient fields) | Gaussian normalization (deformation field) |
|---|---|---|---|
| 1 | 60 × 5 × 0 | 3 | 0 |
| 2 | 60 × 30 × 0 | 3 | 0 |
| 3 | 60 × 30 × 10 | 3 | 0 |
In all cases both the cross correlation object function was used in conjunction with a symmetric normalization transformation model SyN = 0.5, weight = 1, radius = 4.
Parameters for the ANTs algorithm were determined by one of the authors of this manuscript (PS) and are similar to those previously reported by the group who developed the algorithm [Avants et al., 2008]. Unlike the mouse brain registration using only intensity features was shown to produce acceptable segmentation results. The probabilistic registration objective function was a symmetric normalization transformation model.
Gold-standard for evaluation
Gold standards of the striatum, globus pallidus, and thalamus were manually derived, for both hemispheres, by one of the authors of this manuscript (RDC) for all 20 subjects using the manual segmentation rules previously described in [Chakravarty, 2009b]. In addition, the subcortical structures from the left hemisphere were relabeled on five randomly chosen subjects to verify intra-rater reliability. Model-based segmentation and the MAGeT Brain procedure were compared to the gold standards. As in the murine experiment, MRI data from all 20 of the input subjects were used in the creation of the automatically generated template library. The same convention will be used as those defined in the murine experiments for reporting results in the figures and tables (i.e., Model ANIMAL, Model ANTs, MAGeT ANIMAL, and MAGeT ANTs).
Using more than one input template
In certain cases, there may be a possibility for the development of more than a single “hard-to-define” atlas. Similarly, one may be confronted with the analysis of a subject population with highly atrophic or enlarged brain structures. In these scenarios, it may be possible to devote resources to the manual segmentation of certain structures in multiple MRI volumes. We used the manual segmentations described above for the validation of the subcortical segmentations. Unlike the MAGeT Brain method, we randomly chose three subjects from the 20 manually labeled subjects as input templates. In this way, three segmentations are available for each subject in the template library. As a result the voxel voting now occurs with 3 × (n - 1) possible segmentations. We chose three unique atlases three times to verify the impact of the atlas choice on the quality of the final segmentations. MAGeT Brain and traditional “multi-atlas” label fusion based segmentation results were compared to the gold-standard. We consider the traditional multi-atlas based segmentation result as the upper bound on segmentation quality as other publications have demonstrated its increased accuracy over traditional model-based segmentation procedures [Aljabar et al., 2007, 2009; Collins and Pruessner, 2010]. We also evaluate multi-atlas segmentation in the case where there are only three templates in the library (instead of a larger template library) to evaluate the level of improvement that MAGeT Brain provides. Here we use the same templates as in the MAGeT Brain experiments. In the figures and tables mutliatlas segmentation with using a template library of three templates will be referred to as 3 Template Multi-Atlas; MAGeT Brain results using three input templates will be referred to as 3 Template MAGeT Brain; and finally multi-atlas label fusion using all 20 templates in the library will simply be referred to as Multi-Atlas.
Checking for optimal templates from the template library
To determine if there is some optimal subset of templates from the automatically generated template library that can yield more accurate segmentation we employed a strategy similar to the work in [Collins and Pruessner, 2010]. In their work on hippocampal and amygdala segmentation using a template library and label fusion, each subject is linearly transformed to match all templates in a template library of 80 manually segmented subjects. The labels that are then chosen for label fusion based on a rank order of the normalized mutual information estimated in a region of interest around the medial temporal lobe. Similarly, others have demonstrated that a label-fusion strategy weighted towards an optimal combination of subjects in the template library will improve segmentation [Aljabar et al., 2007, 2009; Heckemann et al., 2006; Rohlfing et al., 2004a,b]. We have previously demonstrated that nonlinear registration focused on an ROI that define the extents of the structures to be segmented can improve accuracy [Chakravarty, 2009b], and have chosen to combine these two ideas here.
In our implementation, we estimated the cross-correlation between an input subject and each of the subjects within the automatically generated template library. All cross-correlations were estimated using only those voxel intensities in the region of interest as defined after linear registration of the input subject to each of the templates in the template library. For each subject, the templates are rank-ordered by their cross-correlation value and only the top “n” subjects are retained. The labels are then fused via majority vote (i.e., the mode label at each voxel location) using only these top “n” subjects.
Our aim was to determine if there is an optimal number of subjects that should be retained from the template library and if this number differed for different structures. Optimality was determined using the mean kappa and Jaccard values for the group after varying “n” from 3 to 19.
Goodness-of-Fit Metrics
Segmentations were evaluated against the manual gold standards using two different goodness-of-fit metrics.
- The Dice Kappa (κ) metric was used to measure the quality of the overlap with the manually derived gold-standard:
Where a is common to the automatic segmentation and the gold-standard, and b + c is the sum of the voxels uniquely identified by the segmentation and the gold standard. The κ metric can take on values between 0 and 1.0, where 1.0 indicates perfect agreement [see Chakravarty et al., 2008] for a demonstration of the sensitivity of the κ metric. - The Jaccard similarity (J) was also used as another overlap measure:
Where a, b, and c are defined the same as above. l are always lower (and perhaps more sensitive) than κ values and range from 0 to 1.0 (where 1.0) represents perfect overlap.
Both of the methods described above have been previously used to evaluate the quality of automated segmentation procedures [Chakravarty, 2009b; Collins and Pruessner, 2010; Klein et al., 2009].
RESULTS
Intra-rater Reliability
Before using the manually derived segmentations to evaluate the quality of the automated segmentation procedures, we verified the intra-rater reliability to determine if the rater was able to recreate the initial segmentations with a high level of accuracy. The results for both metrics for both the mouse and the human datasets are given in Table IX. For the mouse segmentations the manual rater showed high labeling consistency for both the hippocampus (mean Kappa (κ) and Jaccard [J] = 0.883) and the anterior commissure (mean κ = 0.904, J = 0.826) and all values were within a very small range.
TABLE IX.
Intra-rater reliability given as the mean (range)
| Structure | Kappa | Jaccard |
|---|---|---|
| Mouse | ||
| Hippocampus | 0.938 (0.924–0.956) | 0.883 (0.858–0.915) |
| Anterior Commissure | 0.904 (0.883–0.928) | 0.826 (0.790–0.865) |
| Human | ||
| Striatum | 0.910 (0.892–0.92) | 0.834 (0.805–0.852) |
| Globus Pallidus | 0.793 (0.658–0.839) | 0.662 (0.500–0.723) |
| Thalamus | 0.861 (0.829–0.910) | 0.758 (0.707–0.834) |
The intra-rater reliability results for the human subcortical structures were consistent as well. The rater's most consistent performance was in the labeling of the striatum (mean κ = 0.910, J = 0.834). The rater also achieved good consistency in the labeling of the thalamus (mean κ = 0.861, J = 0.758). The labeling of the globus pallidus was more variable than the other structures (range κ = 0.658–0.839, J = 0.500–0.723). Nonetheless, the mean overlap values for the pallidal segmentations were acceptable (mean κ = 0.793, J = 0.662).
Mouse Brain Experiment Results
Hippocampal and anterior commissure model-based segmentations using both the ANIMAL and ANTs algorithms demonstrate good overlap with manually derived gold-standards see (Table X). Qualitatively, both ANIMAL and ANTs-based segmentations demonstrate labeling that is consistent with manual segmentations (see Fig. 3). In addition, some mislabeling caused by either resampling or registration error is accounted for when using the MAGeT Brain technique. However, the ANTs-based segmentations appear to be prone to systematic segmentation errors that cannot be accounted for. In the case of the hippocampus, very little improvement in overlap is gained through the use of the MAGeT Brain technique (see Table X) for either the ANIMAL (model-based mean κ = 0.863, J = 0.768; MAGeT Brain mean κ = 0.869, J = 0.768) or the ANTs (model-based mean κ = 0.847, J = 0.735; MAGeT Brain mean κ = 0.850, J = 0.740) nonlinear registration algorithms. However, MAGeT Brain does provide significant improvement (P < 0.001) over the group-wise segmentation method tested for both the ANIMAL and ANTs algorithms. ANIMAL MAGeT Brain segmentations furthermore show increased segmentation accuracy over model-based segmentations of the anterior commissure (P < 0.001; model-based mean κ = 0.752, J = 0.603; MAGeT Brain mean κ = 0.801, J = 0.661). Like in the segmentation of the hippocampus, the ANTs MAGeT Brain does not greatly improve the overall accuracy of the segmentations (model-based mean κ = 0.751, J = 0.601; MAGeT Brain mean κ = 0.759, J = 0.611); although a tighter confidence interval is observed for the MAGeT Brain case in comparison to the model-based segmentation (model-based κ confidence interval: 0.732–0.772; MAGeT Brain κ confidence interval: 0.742–0.775) suggesting an improvement in the precision of the MAGeT Brain procedure. Similar to the hippocampal results, both and ANIMAL and ANTs based MAGeT Brain results perform significantly better (P < 0.001) than the homologous Group-wise segmentation procedures). All ANIMAL-based techniques provide greater segmentation accuracy for the mouse brain experiments.
TABLE X.
Results for model-based, group-wise and MAGeT Brain segmentations using atlases given as mean (confidence interval)
| Model-based |
Group-wise |
MAGeT Brain |
||||
|---|---|---|---|---|---|---|
| Structure | ANIMAL | ANTs | ANIMAL | ANTs | ANIMAL | ANTs |
| Kappa | ||||||
| Mouse | ||||||
| Hippocampus | 0.863 (0.850–0.881) | 0.847 (0.829–0.864) | 0.816 (0.797–0.836) | 0.833 (0.813–0.849) | 0.869## (0.853–0.884) | 0.850### (0.832–0.867) |
| Anterior commissure | 0.752 (0.732–0.772) | 0.751 (0.734–0.766) | 0.692 (0.672–0.710) | 0.728 (0.708–0.750) | 0.801***,### (0.790–0.811) | 0.759### (0.742–0.775) |
| Human | ||||||
| Striatum | 0.818 (0.808–0.827) | 0.816 (0.809–0.823) | 0.796 (0.787–0.804) | 0.807 (0.799–0.815) | 0.833**,### (0.825–0.837) | 0.845***,### (0.833–0.843) |
| Globus pallidus | 0.735 (0.712–0.752) | 0.740 (0.722–0.759) | 0.722 (0.697–0.746) | 0.728 (0.702–0.753) | 0.743 (0.723–0.762) | 0.752# (0.733–0.771) |
| Thalamus | 0.850 (0.838–0.856) | 0.848 (0.840–0.856) | 0.843 (0.833–0.854) | 0.849 (0.837–0.860) | 0.861# (0.881–0.900) | 0.856 (0.850–0.864) |
| Jaccard | ||||||
| Mouse | ||||||
| Hippocampus | 0.768 (0.740–0.789) | 0.735 (0.708–0.762) | 0.691 (0.671–0.710) | 0.712 (0.686–0.738) | 0.768## (0.745–0.792) | 0.740### (0.713–0.767) |
| Anterior commissure | 0.603 (0.578–0.629) | 0.601 (0.581–0.623) | 0.529 (0.507–0.550) | 0.574 (0.546–0.599) | 0.661***,### (0.653–0.683) | 0.611### (0.591–0.632) |
| Human | ||||||
| Striatum | 0.692 (0.683–0.702) | 0.690 (0.680–0.700) | 0.661 (0.649–0.673) | 0.740 (0.732–0.748) | 0.710**,### (0.702–0.719) | 0.740***,### (0.732–0.748) |
| Globus pallidus | 0.581 (0.557–0.606) | 0.591 (0.569–0.614) | 0.568 (0.538–0.597) | 0.610 (0.582–0.631) | 0.585 (0.570–0.620) | 0.610# (0.582–0.631) |
| Thalamus | 0.735 (0.723–0.749) | 0.738 (0.725–0.748) | 0.730 (0.714–0.745) | 0.731 (0.718–0.740) | 0.751# (0.740–0.766) | 0.741 (0.738–0.760) |
* represent significance levels of P < 0.05, P < 0.01, and P < 0.001 for MAGeT Brain as compared to its Model-based counterpart.
represent significance levels of P < 0.05, P < 0.01, and P < 0.001 for MAGeT Brain as compared to its Model-based counterpart.
represent significance levels of P < 0.05, P < 0.01, and P < 0.001 for MAGeT Brain as compared to its Model-based counterpart.
represent significance levels of P < 0.05, P < 0.01, and P < 0.001 for MAGeT Brain as compared to its group-wise counterpart.
represent significance levels of P < 0.05, P < 0.01, and P < 0.001 for MAGeT Brain as compared to its group-wise counterpart.
represent significance levels of P < 0.05, P < 0.01, and P < 0.001 for MAGeT Brain as compared to its group-wise counterpart.
Only segmentations using the same nonlinear registration algorithms were compared to one another (e.g., only the model-based segmentation using ANIMAL) was compared to the MAGeT Brain segmentation using ANIMAL.
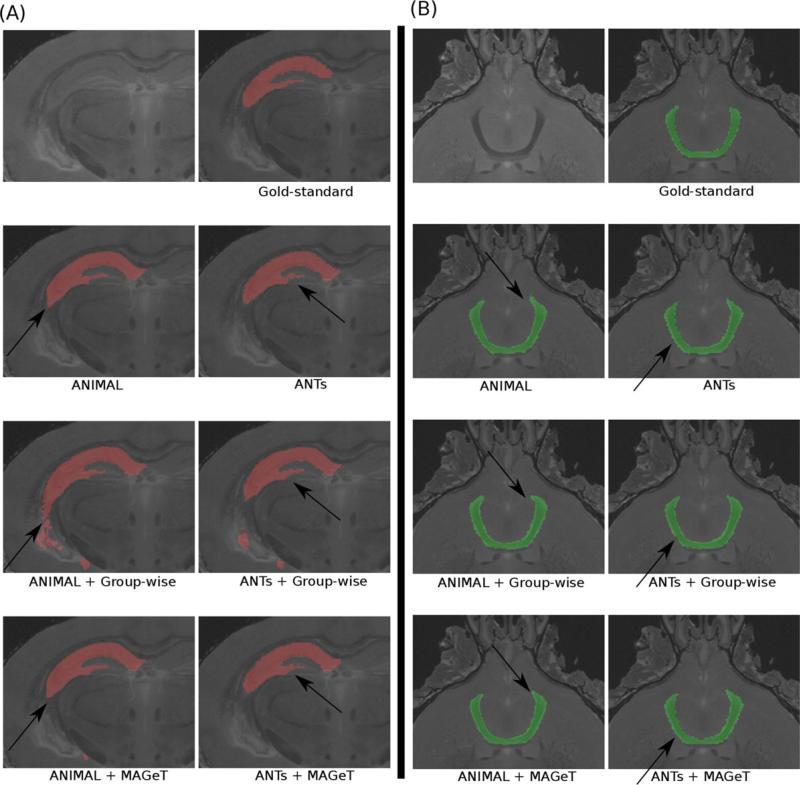
Figure 3.
Results from segmentations for the (A) hippocampus and (B) anterior commissure of the mouse brain. Black arrows indicate areas where the MAGET Brain method is able to provide an improvement in segmentation accuracy. Note how the tail of the hippocampus has improved definition in the MAGeT Brain results.
Segmentation Results for Subcortical Labeling in the Human
The use of MAGeT Brain with both the ANIMAL and ANTs registration algorithms improves segmentations when using the atlas derived from serial histological data as the input template (Table X and Fig. 4). Segmentations of the striatum and thalamus show greater increases in accuracy through the use of the MAGeT Brain technique. ANTs-based MAGeT Brain shows the greatest improvement in striatal segmentation accuracy (P < 0.001) over the model-based segmentation technique (model-based mean κ = 0.816, J = 0.690; MAGeT Brain mean κ = 0.845, J = 0.740); however ANIMAL-based MAGeT Brain results provide comparable levels of improvement (P < 0.01) in segmentation accuracy (model-based mean κ = 0.818, J = 0.692; MAGeT Brain mean κ = 0.833, J = 0.710). ANTs-based MAGeT also improves the confidence interval of the mean (e.g., model-based κ confidence interval: 0.809–0.823; MAGeT Brain κ confidence interval: 0.833–0.843). Both ANIMAL- and ANTs-based MAGeT Brain provide significant improvement over their group-wise counter-parts (P < 0.001 in both cases).
Figure 4.
Subcortical segmentation results for regular model-based segmentation, group-wise segmentation and MAGeT Brain (using ANIMAL and ANTs). Black arrows demonstrate improvement of segmentation accuracy using MAGeT Brain at the posterior end of the globus pallidus and in the medial edge of the thalamus and improvement in identification of the medial edge of globus pallidus and the lateral edge of the thalamus.
ANIMAL-based MAGeT shows the greatest improvement in thalamic segmentation accuracy (model-based mean κ = 0.850, J = 0.735; MAGeT Brain mean κ = 0.861, J = 0.751; see Table X). Although this method is not significantly better than the ANIMAL model-based segmentation, it did demonstrate improvement at a trend level (P = 0.11). ANTs-based segmentations also provide reasonable increases in accuracy (model-based mean κ = 0.848, J = 0.738; MAGeT Brain mean κ = 0.856, J = 0.740). Trend levels of significant improvement were also observed in comparison to the ANTs template-based segmentation. Only ANIMAL-based MAGeT Brain shows a significant improvement over its group-wise counterpart (P < 0.05).
Mean pallidal overlap values both for ANIMAL (model-based mean κ = 0.735, J = 0.581; MAGeT Brain mean κ = 0.743, J = 0.581; and ANTs-based (model-based mean κ = 0.740, J = 0.591; MAGeT Brain mean κ = 0.752, J = 0.610) MAGeT Brain techniques demonstrate improvement However, MAGeT Brain does little to improve the confidence intervals of overlap values irrespective of the algorithm used.
Segmentation Results for Subcortical Labeling Using More Than One Input Template
Using MAGeT Brain with three templates improved segmentation for all structures in comparison to multi-atlas label fusion with three templates (see Table XI and Fig. 5). For the ANIMAL-based results of the striatum, MAGeT Brain improves (P < 0.05) the overlap with the gold standard (3 template multi-atlas mean κ = 0.885, J = 0.784; 3 template MAGeT Brain mean κ = 0.891, J = 0.810). These results also hold for the ANTs-based segmentations (P < 0.05; 3 template multi-atlas mean κ = 0.888, J = 0.795; 3 template MAGeT Brain mean κ = 0.894, J = 0.806).
TABLE XI.
Results for model-based and MAGeT Brain segmentations using manually generated template library using atlases given as mean (confidence interval)
| 3 Template Multi-Atlas |
3 Template MAGeT |
Multi-Atlas |
||||
|---|---|---|---|---|---|---|
| Structure | ANIMAL | ANTs | ANIMAL | ANTs | ANIMAL | ANTs |
| Kappa | ||||||
| Striatum | 0.885 (0.880–0.889) | 0.888 (0.882–0.891) | 0.891* (0.887–0.896) | 0.894* (0.890–0.898) | 0.892 (0.887–0.895) | 0.900# (0.896–0.904) |
| Globus pallidus | 0.790 (0.772–0.808) | 0.797 (0.781–0.812) | 0.815* (0.800–0.827) | 0.805 (0.789–0.820) | 0.811 (0.797–0.824) | 0.819 (0.804–0.834) |
| Thalamus | 0.883 (0.874–0.891) | 0.887 (0.878–0.895) | 0.895 (0.883–0.901) | 0.895 (0.884–0.902) | 0.891 (0.882–0.900) | 0.902 (0.893–0.911) |
| Jaccard | ||||||
| Striatum | 0.784 (0.774–0.793) | 0.795 (0.790–0.803) | 0.810* (0.797–0.812) | 0.806* (0.799–0.813) | 0.803 (0.797–0.811) | 0.820# (0.814–0.826) |
| Globus pallidus | 0.645 (0.622–0.670) | 0.662 (0.643–0.685) | 0.687* (0.667–0.707) | 0.676 (0.654–0.697) | 0.684 (0.664–0.703) | 0.696 (0.675–0.717) |
| Thalamus | 0.794 (0.781–0.811) | 0.794 (0.784–0.811) | 0.806 (0.792–0.822) | 0.807 (0.792–0.821) | 0.804 (0.790–0.819) | 0.821 (0.809–0.837) |
Results are given for multi-atlas and MAGeT Brain segmentation with 3 templates (3 Template Multi-Atlas and 3 Template MAGeT, respectively) and full multi-atlas segmentation.
Represents significance levels of P < 0.05 for 3 Template MAGeT Brain as compared to its 3 Template Multi-Atlas counterpart.
Represents significance levels of P < 0.05 for Multi-Atlas segmentation compared to 3 Template Multi-Atlas counterpart.
Only segmentations using the same nonlinear registration algorithms were compared to one another (e.g., only the model-based segmentation using ANIMAL was compared to the MAGeT Brain segmentation using ANIMAL).
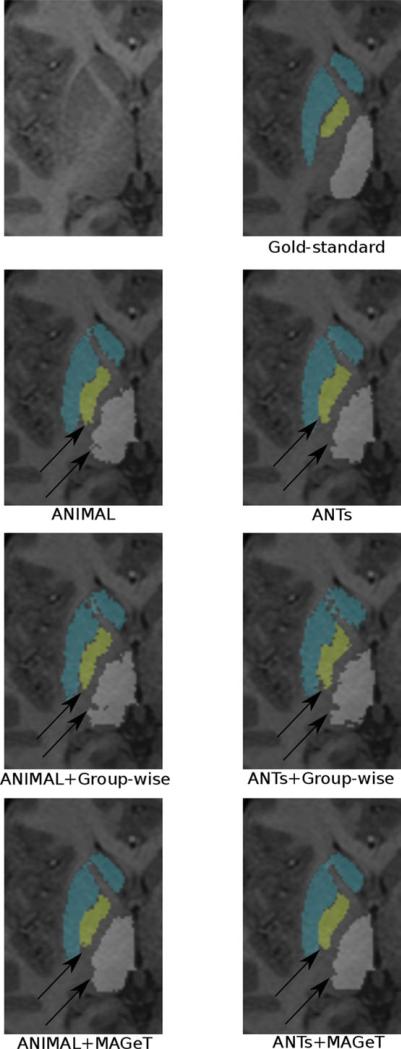
Figure 5.
Using multiple templates for segmentation. Results for multi-atlas and MAGeT Brain segmentations using three input templates. Black arrows pointing to region of visible improvement. In the ANIMAL case, the MAGeT Brain technique corrects the inaccurate segmentation on the medial wall of the globus pallidus. In the ANTs case, label ambiguity at the striatal-pallidal edge is corrected. For both ANIMAL and ANTs, the full multi-atlas-based segmentation yields results most similar to the gold-standard segmentation.
Similar to the single template case (from the previous section), using three manually generated input templates in the MAGeT Brain technique also improves the thalamic segmentation for both the ANIMAL (3 template multi-atlas mean κ = 0.883, J = 0.794; 3 template MAGeT Brain mean κ = 0.895, J = 0.806) and ANTs-based (3 template multi-atlas mean κ = 0.887, J = 0.794; 3 template MAGeT Brain mean κ = 0.895, J = 0.807) techniques. Also as in the single template case, these results showed improvement at trend levels (P = 0.12 and P = 0.09 for the ANIMAL and ANTs algorithms, respectively). ANIMAL-based MAGeT Brain with three input templates may be approaching or even exceeding the segmentation accuracy of true multi-atlas segmentation (multi-atlas mean κ = 0.891, J = 0.804). However, the best segmentation results are achieved using ANTs multi-atlas segmentation (multi-atlas mean κ = 0.902, J = 0.821).
Mean overlap values for the segmentation of the globus pallidus improve via the use of the MAGeT Brain with three templates in comparison to the three-template multi-atlas for both the ANIMAL (P < 0.05; 3 template multi-atlas mean κ = 0.790, J = 0.645; 3 template MAGeT Brain mean κ = 0.815, J = 0.687) and ANTs-based (3 template multi-atlas mean κ = 0.797, J = 0.662; 3 template MAGeT Brain mean κ = 0.805, J = 0.676) techniques. The ANIMAL-based MAGeT Brain results demonstrate equivalent accuracy to the regular multi-atlas segmentations (multi-atlas mean κ = 0.811, J = 0.684); however the ANTs-based multi-atlas segmentation show the greatest mean overlap with the manually derived gold standards (multi-atlas mean κ = 0.819, J = 0.696).
Results From Cross-Correlation Based Weighting and Label Fusion
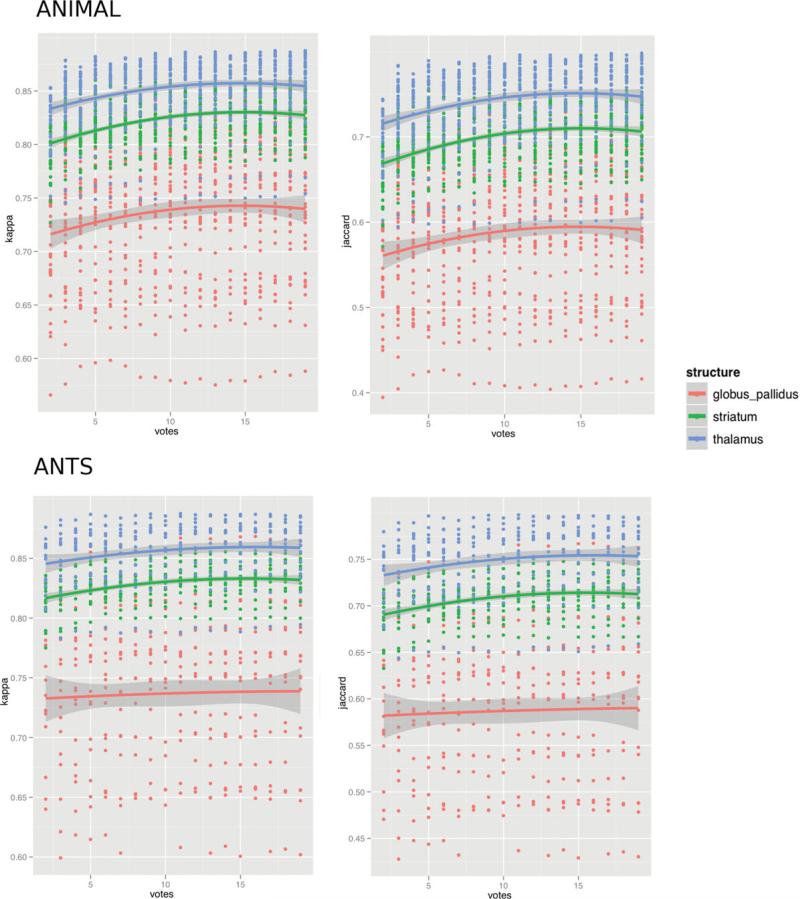
The result from the weighted voting technique demonstrate that at least 15 templates must be selected for registration to achieve optimal results when using ANIMAL-based MAGeT with a single input template for the globus pallidus, striatum, and thalamus. Small decreases in accuracy are observed when more than 15 templates are selected (see Fig. 6). Similarly, optimal accuracy is shown when 15 templates are selected using the ANTs algorithm for the both the striatum and thalamus. However, for the globus pallidus shows very little improvement regardless of the number of templates/labels that are included in the label fusion step.
Figure 6.
Cross-correlation-based label fusion with the subcortical ROI. Kappa and Jaccard statistics are given for voting based on the rank order of correlations between the subcortical ROI in the subject to be segmented and each of the templates in the template library. A majority of voxels are kept for the top “n” hits, where “n” was varied from 3 to 19. Results are shown for both ANIMAL- and ANTs-based MAGeT Brain for the globus pallidus, striatum, and thalamus. Except for the case of the pallidum segmentations using ANTs-based MAGeT Brain, an optimal segmentation is reached when using the top 15 templates based on the cross-correlation metric.
DISCUSSION
In this manuscript we have presented a methodology that addresses the need for a multi-atlas based segmentation approach that can be used with a single input template; particularly in cases when the input template is an atlas which may be difficult and time-consuming to create [Chakravarty et al., 2006; Dorr et al., 2008]. The MAGeT Brain technique begins by creating a template library from a subset or all of the input data through model-based segmentation techniques. The multi-atlas procedure is then used to minimize the effects of spurious nonlinear registration and resampling errors. In this case all of the data to be segmented is matched to each template in the library using a nonlinear transformation. This then yields as many different segmentations as there are unique templates. The final segmentation is achieved through a voxel voting procedure where the most frequently occurring label at each voxel is retained. Two different nonlinear registration algorithms, ANIMAL [Collins et al., 1995] and ANTs [Avants et al., 2008], were tested against manually derived gold standards using two different overlap metrics. The technique was tested using a variety of data types. First, the quality of the MAGeT Brain technique was evaluated on high-resolution mouse brain MRI data (for segmentations of the hippocampus and the anterior commissure). In the second experiment, MAGeT Brain was tested against model-based segmentation of the striatum, globus pallidus, and the thalamus in a dataset of 20 human adolescent MRI volumes. Finally, the possibility of using three input templates was tested against regular multi-atlas label fusion based segmentation and multi-atlas segmentation using a template library of three templates.
In most cases the results demonstrated improvement in the segmentation by using the MAGeT Brain technique. One of the exceptions to this was the result for the segmentation of the mouse hippocampus. Because very little improvement was gained through the MAGeT Brain procedure, we suspect that segmentations were running into a ceiling effect for this particular structure. The mouse hippocampus is morphologically noncomplex, takes up a large proportion of the entire mouse brain, and is well defined by high contrast borders which identify changes in tissue type (particularly through the use of the techniques previously described by our group [Lerch et al., 2011a; Nieman et al., 2007]). In addition, the ANTs algorithm, in our implemented, may not be well suited for the nonlinear registration of the mouse brain (see below in the Discussion section). This is further supported by the mild improvement derived from the ANTs-based MAGeT Brain segmentation of the anterior commissure—especially given the remarkable improvement in segmentation quality in the ANIMAL-based MAGeT Brain segmentations. Although it is difficult to compare segmentation results across studies, gold-standards and algorithms, the technique of Ali and colleagues [Ali et al., 2005] demonstrate a voxel overlap percentage of about 85 and 57% for the mouse hippocampus and anterior commissure, respectively (based on Fig. 7 in [Ali et al., 2005]). Extending their technique through the use of a support vector machine to represent probabilistic information improves the accuracy of their segmentations for the hippocampus, but not the anterior commissure (86.2 and 50.76%, respectively, from Table II in [Bae et al., 2009]). Because the Jaccard metric is comparable to the percentage voxel overlap metric that they used for the evaluation of segmentation quality, we surmise that MAGeT Brain may be providing improved segmentation quality.
All other results show that MAGeT Brain improves the quality of the overlap of model-based segmentations and multi-atlas segmentations using three templates. This is particularly evident when considering previous work [Chakravarty et al., 2008, 2009a,b] presented using the sub-cortical atlas [Chakravarty et al., 2006] used here. Previous comparisons of different model-based segmentation procedures demonstrated maximum Kappa values of 0.573, 0.754, and 0.818 for striatum, globus pallidus, and thalamus, respectively. One should make note that these previous experiments were performed using MRI used for presurgical planning that had lower signal- and contrast-to-noise ratios than the data presented here. Our results when using three input templates for MAGeT Brain are also comparable to those demonstrated in the recent release of the FIRST algorithm [Patenaude et al., 2011] which reports Kappas of ~0.898, 0.871, 0.821, and 0.894 for the caudate, putamen, globus pallidus, and thalamus respectively (see Table 3 of their paper; dependent on number of modes of variation used). Similarly, our results match or exceed the overlap values presented in other subcortical segmentation techniques [Babalola et al., 2008; Fischl et al., 2002].
In some instances we have reported results that exceed the intra-rater reliability. Specifically, this was observed in the accuracy of the thalamic segmentations when using MAGeT Brain with a single template (see Table XI). Using MAGeT Brain with three input templates also exceeds the intra-rater reliability for both the globus pallidus and the thalamus despite the nonlinear registration algorithm used (see Table XI). These results underscore the necessity of having a reliable segmentation technique that is robust and consistent. It also underscores the limitation inherent to the evaluation of all segmentation algorithms. Specifically, how does one best evaluate the correspondence in neuroanatomy as estimated through a nonlinear transformation? The lack of an actual gold standard is one of the primary limitations in these studies.
Unlike some of the other segmentation procedures primary advantages of using MAGeT Brain would be the flexibility of implementation. Many other label-fusion techniques require more than just the sharing of scripts and computer code, but also the sharing of a template library. This can be cumbersome, especially as some groups report the usage of 30 [Heckemann et al., 2010], 80 [Collins and Pruessner, 2010], or even 160 [Eskildsen et al., 2011] subjects within a single template library. One of the main advantages of our work is that it can be easily shared given it requires only a single input template. Although two different nonlinear registration algorithms were presented in this article, our technique could be implemented using any well-validated nonlinear registration algorithm [Klein et al., 2009] or input template [Yelnik et al., 2007]. One possible implementation would be using an input atlas of the hippocampal subfields derived from high-field, high-resolution MRI imaging of postmortem specimens [Van Leemput et al., 2008; Yushkevich et al., 2008, 2009]. However, our results from the mouse brain experiment demonstrate the importance of the choice of the nonlinear registration algorithm used. While this is fairly intuitive, it demonstrates how multi-atlas techniques are incapable of overcoming consistent errors in the registration algorithm. Our choice of nonlinear registration algorithms, in the case of ANIMAL, was based on our familiarity and depth of knowledge of the technique. In addition, ANIMAL has previously been extensively validated in experiments for segmentation of the mouse brain and subcortical structures [Chakravarty et al., 2009a,b; Lau et al., 2008]. ANTs was used based on the quality of the nonlinear registration previously reported [Avants et al., 2011; Klein et al., 2009]. To the best of our knowledge, ANTs has yet to be used extensively in the mouse imaging literature, but we felt that it would be appropriate to attempt to use it in the segmentation experiments reported here. ANTs’ performance on the mouse brain experiments may be caused by the interplay between the morphometry of the mouse brain and the considerable elastic constraints placed on the nonlinear transformation estimation process [Avants et al., 2008, 2011]. These constraints may be ideal to account for the considerable variable morphometry in the human, but may be overconstraining the nonlinear transformation estimation process in the mouse. Nonetheless, ANTs does perform favorably in the segmentation of subcortical structures. It should be noted here, however, that the goal of the paper was not to explicitly evaluate the accuracy of nonlinear registration algorithms, but rather to demonstrate the improvement in segmentation gained by implementing MAGeT Brain.
Because our technique effectively increases accuracy through distributing the randomly distributed error (due to anatomical differences, registration error, and resampling error) across a template library, we also compared MAGeT Brain to the group-wise averaging paradigms that our groups have used in studies of both humans [Borghammer et al., 2010] and mice [Lau et al., 2008; Lerch et al., 2008]. We were surprised to find that this methodology did not even improve segmentation accuracy over the template-based procedures that we tested in this manuscript. This lack of accuracy can probably be attributed to the step in which the initial input atlas is matched to the population average. There may be errors present in this stage that cannot easily be accounted for, and thus these errors get propagated when following the transformation chain back to each individual subject.
Our technique also presents an interesting design choice. While many multi-atlas segmentation techniques rely on the use of an extensive manually defined template library and weighted label-fusion techniques [Aljabar et al., 2007, 2009; Collins and Pruessner, 2010] our results demonstrates that some of these issues can be dealt with by using our multiple registration strategy. This, in effect provides, a trade-off between finding and investing time and resources in a trained manual-rater who can create an extensive template library versus increasing the computational complexity of the segmentation procedure. However, given the availability of modern high-performance computing technology most segmentations can be estimated in parallel, thereby reducing the quantity of time required to perform the final labeling. However, our experiment using the weighted voxel-voting demonstrates that marginal gains in accuracy can be achieved by constraining the label-fusion step to only those templates in the library that are most like the subject to be segmented. This is useful computationally as it can effectively reduce the number of nonlinear registrations required between the subject and template library. In the hippocamapal segmentation presented in [Collins and Pruessner, 2010], they demonstrate that there are diminishing returns in accuracy after 11 templates are chosen from their manually labeled template library. Although it is difficult to compare segmentation techniques across methods and different neuroanatomical structures there is an interesting comparison to be made here. One of the goals of the work presented here is to minimize and potentially obviate the need for an exhaustive manually labeled template library. As such we demonstrate that through the creation of an automatically generated template library that we can reach a ceiling using almost the same number subject-to-template library segmentations. It bears mentioning that results could possibly be improved further in the template library creation stage, where a similar selection strategy could be used. This is a future line of investigation of our group. It is also reasonable to assume that our results could further benefit from a more sophisticated voxel-voting strategy [Aljabar et al., 2007, 2009; Coupe et al., 2011a,b; Eskildsen et al., 2011].
A question that was raised was whether the improvement that we are observing by using the MAGeT Brain technique may simply be a product of smoothing. We tested this hypothesis using both the mice and human data as input. For the mice data we blurred each of the hippocampal and anterior commissure labels using 60, 90, and 120 lm kernels. The human data were blurred with 2, 4, and 6 mm kernels. A threshold was then applied to each of the blurred structures (from 0.1 to 0.9 in steps of 0.1). Overlap with the gold standards was then evaluated using the Dice Kappa. Graphs for the overlaps over all structures and both nonlinear registration algorithms are given below in Supporting Information Figs. S1 and S2 (for mouse and human data, respectively). The results demonstrate that the improvement in accuracy is not simply a function of smoothing. In both experiments similar trends are observed: namely that there is some combination of blurring and thresholding that yields a high overlap. However, this combination is not the same for all combination of structures and blurring kernels. Further, the accuracy is heavily dependent on the blurring threshold and therefore it would be extremely difficult to ensure accurate segmentation if this were to be used with in any technique.
We chose to validate our results using two measures of overlap with manually derived gold standards. Using these metrics, our raters showed consistency in labeling of all structures tested. Although there are other metrics which can be used to evaluate quality (Chakravarty et al., 2009a,b; Hellier et al., 2003; Klein et al., 2009; Robbins et al., 2004) we chose to focus on the general applicability of the MAGeT Brain method. One segmentation that may have benefited from a more probabilistic evaluation was the segmentation the globus pallidus. It is unclear whether the larger uncertainty in the results was due to the variability of the manual rater or as a result of the segmentation techniques. Work by Warfield et al. [2004] has addressed this issue by developing an expectation maximization algorithm that estimates an optimal segmentation from the different methods being evaluated. Using this technique (referred to as STAPLE), each method can then be weighted depending upon its estimated performance level with respect to the other methods being tested. The specificity and sensitivity parameters used in STAPLE could add valuable information to the evaluation of the data presented in this article. Furthermore, the probabilistic ground truth of the manual rater data generated by STAPLE could also be used for evaluation instead of the gold-standard generated through consensus used in the work presented here.
In conclusion, we have presented a multi-atlas technique that can be implemented for the segmentation of structures where morphological homology exists. This can be used to improve segmentation when using hard to define atlases or when segmenting populations with unique anatomical properties. Further, the work demonstrated here may allow for improved segmentation accuracy instead of creating extensive manually defined template libraries [Collins and Pruessner, 2010; Shattuck et al., 2008]. The trade-off in using this technique is the increased computational burden of the technique that should be mitigated by modern super computing infrastructure.
Supplementary Material
ACKNOWLEDGMENTS
Computations were performed on the SciNet supercomputer at the SciNet HPC Consortium. SciNet is funded by the Canada Foundation for Innovation under the auspices of Compute Canada, the Government of Ontario, Ontario Research Fund — Research Excellence and the University of Toronto. MMC would also like to thank G. Clinton, E. Hazel, and B. Worrell for inspiring this work.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Ali AA, Dale AM, Badea A, Johnson GA. Automated segmentation of neuroanatomical structures in multispectral MR microscopy of the mouse brain. Neuroimage. 2005;27:425–435. doi: 10.1016/j.neuroimage.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Aljabar P, Heckemann R, Hammers A, Hajnal JV, Rueckert D. Classifier selection strategies for label fusion using large atlas databases. Med Image Comput Comput Assist Interv. 2007;10(Part 1):523–531. doi: 10.1007/978-3-540-75757-3_64. [DOI] [PubMed] [Google Scholar]
- Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage. 2009;46:726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babalola KO, Cootes TF, Twining CJ, Petrovic V, Taylor CJ. 3D brain segmentation using active appearance models and local regressors. Med Image Comput Comput Assist Interv. 2008;11(Part 1):401–408. doi: 10.1007/978-3-540-85988-8_48. [DOI] [PubMed] [Google Scholar]
- Bae MH, Pan R, Wu T, Badea A. Automated segmentation of mouse brain images using extended MRF. Neuroimage. 2009;46:717–725. doi: 10.1016/j.neuroimage.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajcsy R, Lieberson R, Reivich M. A computerized system for the elastic matching of deformed radiographic images to idealized atlas images. J Comput Assist Tomogr. 1983;7:618–625. doi: 10.1097/00004728-198308000-00008. [DOI] [PubMed] [Google Scholar]
- Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, Frost C, Scahill RI, Fox NC. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. Neuroimage. 2008;40:1655–1671. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Behrens TE, Johansen-Berg H, Woolrich MW, Smith SM, Wheeler-Kingshott CA, Boulby PA, Barker GJ, Sillery EL, Sheehan K, Ciccarelli O, Thompson AJ, Brady JM, Matthews PM. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nat Neurosci. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- Bock NA, Kovacevic N, Lipina TV, Roder JC, Ackerman SL, Henkelman RM. In vivo magnetic resonance imaging and semiautomated image analysis extend the brain phenotype for cdf/cdf mice. J Neurosci. 2006;26:4455–4459. doi: 10.1523/JNEUROSCI.5438-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghammer P, Østergaard K, Cumming P, Gjedde A, Rodell A, Hall N, Chakravarty MM. A deformation-based morphometry study of patients with early-stage Parkinson's disease. Eur J Neurol. 2010;17:314–320. doi: 10.1111/j.1468-1331.2009.02807.x. [DOI] [PubMed] [Google Scholar]
- Burk K, Globas C, Wahl T, Bühring U, Dietz K, Zuhlke C, Luft A, Schulz JB, Voigt K, Dichgans J. MRI-based volumetric differentiation of sporadic cerebellar ataxia. Brain. 2004;127(Part 1):175–181. doi: 10.1093/brain/awh013. [DOI] [PubMed] [Google Scholar]
- Chakravarty MM, Bertrand G, Hodge CP, Sadikot AF, Collins DL. The creation of a brain atlas for image guided neurosurgery using serial histological data. Neuroimage. 2006;30:359–376. doi: 10.1016/j.neuroimage.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Chakravarty MM, Sadikot AF, Germann J, Bertrand G, Collins DL. Towards a validation of atlas warping techniques. Med Image Anal. 2008;12:713–726. doi: 10.1016/j.media.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Chakravarty MM, Broadbent S, Rosa-Neto P, Lambert CM, Collins DL. Design, construction, and validation of an MRI-compatible vibrotactile stimulator intended for clinical use. J Neurosci Methods. 2009a;184:129–135. doi: 10.1016/j.jneumeth.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Chakravarty MM, Sadikot AF, Germann J, Hellier P, Bertrand G, Collins DL. Comparison of piece-wise linear, linear, and nonlinear atlas-to-patient warping techniques: Analysis of the labeling of subcortical nuclei for functional neurosurgical applications. Hum Brain Mapp. 2009b;30:3574–3595. doi: 10.1002/hbm.20780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DL, Evans AC. ANIMAL: Validation and applications of non-linear registration-based segmentation. Int J Pattern Recogn Artif Intell. 1997;11:1271–1294. [Google Scholar]
- Collins DL, Pruessner JC. Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. Neuroimage. 2010;52:1355–1366. doi: 10.1016/j.neuroimage.2010.04.193. [DOI] [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Hum Brain Mapp. 1995;3:190–208. [Google Scholar]
- Coupe P, Eskildsen SF, Manjon JV, Fonov V, Collins DL. Simultaneous segmentation and grading of hippocampus for patient classification with Alzheimer's disease. Med Image Comput Comput Assist Interv. 2011a;14(Part 3):149–157. doi: 10.1007/978-3-642-23626-6_19. [DOI] [PubMed] [Google Scholar]
- Coupe P, Eskildsen SF, Manjon JV, Fonov V, Collins DL. Simultaneous segmentation and grading of anatomical structures for patient's classification: Application to Alzheimer's disease. Neuroimage. 2011b;59:3736–3747. doi: 10.1016/j.neuroimage.2011.10.080. [DOI] [PubMed] [Google Scholar]
- Deoni SC, Peters TM, Rutt BK. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn Reson Med. 2005;53:237–241. doi: 10.1002/mrm.20314. [DOI] [PubMed] [Google Scholar]
- Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuro-image. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- Ellegood J, Pacey LK, Hampson DR, Lerch JP, Henkelman RM. Anatomical phenotyping in a mouse model of fragile X syndrome with magnetic resonance imaging. Neuroimage. 2010;53:1023–1029. doi: 10.1016/j.neuroimage.2010.03.038. [DOI] [PubMed] [Google Scholar]
- Eskildsen SF, Coupe P, Fonov V, Manjon JV, Leung KK, Guizard N, et al. BEaST: Brain extraction based on nonlocal segmentation technique. Neuroimage. 2011;59:2362–2373. doi: 10.1016/j.neuroimage.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Frey S, Pandya DN, Chakravarty MM, Bailey L, Petrides M, Collins DL. An MRI based average macaque monkey stereotaxic atlas and space (MNI monkey space). Neuroimage. 2011;55:1435–1442. doi: 10.1016/j.neuroimage.2011.01.040. [DOI] [PubMed] [Google Scholar]
- Gloor P. The Temporal Lobe and Limbic System. Oxford University Press; New York, USA: 1997. [Google Scholar]
- Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL. Symmetric atlasing and model based segmentation: An application to the hippocampus in older adults. Med Image Comput Comput Assist Interv. 2006;9(Part 2):58–66. doi: 10.1007/11866763_8. [DOI] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. Neuroimage. 2010;51:221–227. doi: 10.1016/j.neuroimage.2010.01.072. [DOI] [PubMed] [Google Scholar]
- Hellier P, Barillot C, Corouge I, Gibaud B, Le Goualher G, Collins DL, Evans A, Malandain G, Ayache N, Christensen GE, Johnson HJ. Retrospective evaluation of intersubject brain registration. IEEE Trans Med Imaging. 2003;22:1120–1130. doi: 10.1109/TMI.2003.816961. [DOI] [PubMed] [Google Scholar]
- Hirai T, Jones EG. A new parcellation of the human thalamus on the basis of histochemical staining. Brain Res Brain Res Rev. 1989;14:1–34. doi: 10.1016/0165-0173(89)90007-6. [DOI] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. J Comput Assist Tomogr. 1998;22:324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Ivanov I, Bansal R, Hao X, Zhu H, Kellendonk C, Miller L, Sanchez-Pena J, Miller AM, Chakravarty MM, Klahr K, Durkin K, Greenhill LL, Peterson BS. Morphological abnormalities of the thalamus in youths with attention deficit hyperactivity disorder. Am J Psychiatry. 2010;167:397–408. doi: 10.1176/appi.ajp.2009.09030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JC, Lerch JP, Sled JG, Henkelman RM, Evans AC, Bedell BJ. Longitudinal neuroanatomical changes determined by deformation-based morphometry in a mouse model of Alzheimer's disease. Neuroimage. 2008;42:19–27. doi: 10.1016/j.neuroimage.2008.04.252. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Carroll JB, Spring S, Bertram LN, Schwab C, Hayden MR, et al. Automated deformation analysis in the YAC128 Huntington disease mouse model. Neuroimage. 2008;39:32–39. doi: 10.1016/j.neuroimage.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Sled JG, Henkelman RM. MRI phenotyping of genetically altered mice. Methods Mol Biol. 2011a;711:349–361. doi: 10.1007/978-1-61737-992-5_17. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Yiu AP, Martinez-Canabal A, Pekar T, Bohbot VD, Frankland PW, et al. Maze training in mice induces MRI-detectable brain shape changes specific to the type of learning. Neuroimage. 2011b;54:2086–2095. doi: 10.1016/j.neuroimage.2010.09.086. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hof PR, Grant SC, Blackband SJ, Bennett R, Slatest L, McGuigan MD, Benveniste H. A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience. 2005;135:1203–1215. doi: 10.1016/j.neuroscience.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: Theory and rationale for its development. The International Consortium for Brain Mapping (ICBM). Neuroimage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palo-mero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci. 2001a;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palo-mero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Feidler J, Smith K, Boomsma D, Hulshoff Pol H, Cannon T, Kawashima R, Mazoyer B. A four-dimensional probabilistic atlas of the human brain. J Am Med Inform Assoc. 2001b;8:401–430. doi: 10.1136/jamia.2001.0080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Banerjee A, Christensen G, Joshi S, Khaneja N, Grenander U, Matejic L. Statistical methods in computational anatomy. Stat Methods Med Res. 1997;6:267–299. doi: 10.1177/096228029700600305. [DOI] [PubMed] [Google Scholar]
- Nieman BJ, Bishop J, Dazai J, Bock NA, Lerch JP, Feintuch A, Chen XJ, Sled JG, Henkelman RM. MR technology for biological studies in mice. NMR Biomed. 2007;20:291–303. doi: 10.1002/nbm.1142. [DOI] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausova Z, Paus T, Abrahamowicz M, Almerigi J, Arbour N, Bernard M, Gaudet D, Hanzalek P, Hamet P, Evans AC, Kramer M, Laberge L, Leal SM, Leonard G, Lerner J, Lerner RM, Mathieu J, Perron M, Pike B, Pitiot A, Richer L, Séguin JR, Syme C, Toro R, Tremblay RE, Veillette S, Watkins K. Genes, maternal smoking, and the offspring brain and body during adolescence: Design of the Saguenay Youth Study. Hum Brain Mapp. 2007;28:502–518. doi: 10.1002/hbm.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessner JC, Köhler S, Crane J, Pruessner M, Lord C, Byrne A, Kabani N, Collins DL, Evans AC. Volumetry of temporopolar, perirhinal, entorhinal and parahippocampal cortex from high-resolution MR images: Considering the variability of the collateral sulcus. Cereb Cortex. 2002;12:1342–1353. doi: 10.1093/cercor/12.12.1342. [DOI] [PubMed] [Google Scholar]
- Robbins S, Evans AC, Collins DL, Whitesides S. Tuning and comparing spatial normalization methods. Med Image Anal. 2004;8:311–323. doi: 10.1016/j.media.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Brandt R, Menzel R, Maurer CRJ. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage. 2004a;21:1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Russakoff DB, Maurer CRJ. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. IEEE Trans Med Imaging. 2004b;23:983–994. doi: 10.1109/TMI.2004.830803. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Schaltenbrand G, Wahren W. Atlas for the Stereotaxy of the Human Brain. Georg Thieme Verlag; Stuttgart: 1977. [Google Scholar]
- Schulz JB, Skalej M, Wedekind D, Luft AR, Abele M, Voigt K, Dichgans J, Klockgether T. Magnetic resonance imaging-based volumetry differentiates idiopathic Parkinson's syndrome from multiple system atrophy and progressive supra-nuclear palsy. Ann Neurol. 1999;45:65–74. [PubMed] [Google Scholar]
- Seeck M, Dreifuss S, Lantz G, Jallon P, Foletti G, Despland PA, Delavelle J, Lazeyras F. Subcortical nuclei volumetry in idiopathic generalized epilepsy. Epilepsia. 2005;46:1642–1645. doi: 10.1111/j.1528-1167.2005.00259.x. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Eckstrand K, Sharp W, Blumenthal J, Lerch JP, Green-stein D, Clasen L, Evans A, Giedd J, Rapoport JL. Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci USA. 2007;104:19649–19654. doi: 10.1073/pnas.0707741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Gilliam M, Liverpool M, Weddle C, Malek M, Sharp W, Greenstein D, Evans A, Rapoport J, Giedd J. Cortical development in typically developing children with symptoms of hyperactivity and impulsivity: Support for a dimensional view of attention deficit hyperactivity disorder. Am J Psychiatry. 2011;168:143–151. doi: 10.1176/appi.ajp.2010.10030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring S, Lerch JP, Henkelman RM. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage. 2007;35:1424–1433. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Studholme C, Novotny E, Zubal IG, Duncan JS. Estimating tissue deformation between functional images induced by intracranial electrode implantation using anatomical MRI. Neuroimage. 2001;13:561–576. doi: 10.1006/nimg.2000.0692. [DOI] [PubMed] [Google Scholar]
- Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, Dickerson BC, Golland P, Fischl B. Model-based segmentation of hippocampal subfields in ultra-high resolution in vivo MRI. Med Image Comput Comput Assist Interv. 2008;11(Part 1):235–243. doi: 10.1007/978-3-540-85988-8_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Suh JW, Pluta J, Altinay M, Yushkevich P. Optimal weights for multi-atlas label fusion. Inf Process Med Imaging. 2011;22:73–84. doi: 10.1007/978-3-642-22092-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Aljabar P, Hajnal JV, Hammers A, Rueckert D. LEAP: Learning embeddings for atlas propagation. Neuro-image. 2010;49:1316–1325. doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelnik J, Bardinet E, Dormont D, Malandain G, Ourselin S, Tandé D, Karachi C, Ayache N, Cornu P, Agid Y. A three-dimensional, histological and deformable atlas of the human basal ganglia. I. Atlas construction based on immunohisto-chemical and MRI data. Neuroimage. 2007;34:618–638. doi: 10.1016/j.neuroimage.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Avants BB, Pluta J, Minkoff D, Detre JA, Grossman M, Gee JC. Shape-based alignment of hippocampal subfields: Evaluation in postmortem MRI. Med Image Comput Comput Assist Interv. 2008;11(Part 1):510–517. doi: 10.1007/978-3-540-85988-8_61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Avants BB, Pluta J, Das S, Minkoff D, Mechanic-Hamilton D, Glynn S, Pickup S, Liu W, Gee JC, Grossman M, Detre JA. A high-resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. Neuroimage. 2009;44:385–398. doi: 10.1016/j.neuroimage.2008.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.