Abstract
Maintenance of the drug-addicted state is thought to involve changes in gene expression in different neuronal cell types and neural circuits. Midbrain dopamine (DA) neurons in particular mediate numerous responses to drugs of abuse. Long noncoding RNAs (lncRNAs) regulate CNS gene expression through a variety of mechanisms, but next to nothing is known about their role in drug abuse. The proportion of lncRNAs that are primate-specific provides a strong rationale for their study in human drug abusers. In this study, we determined a profile of dysregulated putative lncRNAs through the analysis of postmortem human midbrain specimens from chronic cocaine abusers and well-matched control subjects (n=11 in each group) using a custom lncRNA microarray. A dataset comprising 32 well-annotated lncRNAs with independent evidence of brain expression and robust differential expression in cocaine abusers is presented. For a subset of these lncRNAs, differential expression was validated by quantitative real-time PCR and cellular localization determined by in situ hybridization histochemistry. Examples of lncRNAs exhibiting DA cell-specific expression, different subcellular distributions, and covariance of expression with known cocaine-regulated protein-coding genes were identified. These findings implicate lncRNAs in the cellular responses of human DA neurons to chronic cocaine abuse.
Keywords: long noncoding RNA, dopamine, cocaine, postmortem, gene expression, drug abuse
Drug addiction is a debilitating chronic disorder characterized by craving, compulsive use of drugs even in the face of adverse consequences, and high incidences of relapse. At a molecular level, long-lived changes in neural gene expression arising through transcriptional and epigenetic mechanisms are thought to constitute a ‘molecular memory’ that contributes to the maintenance of a drug-addicted state (Feng and Nestler 2013). Of the different neural cell types and neural circuits implicated in the effects of drugs of abuse, perhaps none play a more central role than dopamine (DA)-synthesizing neurons of the ventral midbrain which, though few in number, mediate many acute rewarding effects of drugs of abuse, conditioned responses to cues associated with previous drug use, and the emergence of some adverse effects upon cessation of drug use (Koob and Volkow 2010; Volkow et al. 2011). Recent analysis of human postmortem midbrain has revealed a molecular signature of pathophysiological changes in gene expression that are diagnostic for chronic cocaine abuse (Bannon et al. 2014; Bannon et al. 2015), but our understanding of the mediators of these changes remains rudimentary.
Recent transcriptional analyses have revealed that, although only a small fraction of the human genome is translated into proteins, the majority of genomic sequence is transcribed to produce many thousands of noncoding RNAs, a large proportion of which are long noncoding RNAs (lncRNAs), RNAs >200 nucleotides in length but lacking extended open reading frames (Encode Consortium 2012; Derrien et al. 2012; Lipovich et al. 2010). Emergent data suggest that lncRNAs can regulate the expression of protein coding genes through a striking variety of mechanisms, including locus-specific or widespread targeting of epigenetic modifications, nucleating assembly of RNA splicing complexes, or modifying the stability or translation of specific cytoplasmic mRNAs (Clark and Blackshaw 2014; Guttman and Rinn 2012; Mercer and Mattick 2013). In the CNS, some lncRNAs show strong cell-specificity of expression, modulate the developmental specification of individual neuronal subtypes and, most recently, have been implicated in several CNS disorders (Clark and Blackshaw 2014; Modarresi et al. 2012; Ng et al. 2013; Pastori and Wahlestedt 2012; Punzi et al. 2014). In contrast, we know very little about the potential role lncRNAs may play in drug abuse (Michelhaugh et al. 2011; Bu et al. 2012). Because approximately one-third of the thousands of human lncRNAs identified appear to be unique to the primate lineage (Derrien et al. 2012), there is a compelling rationale for studying lncRNAs in the drug-addicted human brain as well as simpler model systems.
To address this significant gap in knowledge, the current study investigated lncRNA expression in the postmortem midbrain of human cocaine abusers and well-matched control subjects. A profile of lncRNAs dysregulated in chronic cocaine abusers was determined. LncRNAs exhibiting DA cell-specific expression, different subcellular distributions, and covariance of expression with known cocaine-regulated protein-coding genes were identified. The findings are consistent with the notion that some lncRNAs may act as mediators of cellular responses to drug abuse.
Materials and Methods
Human Brain Specimens
Human midbrain specimens were obtained by forensic pathologists in the course of the routine autopsy process, and de-identified specimens were subsequently characterized as described previously (Albertson et al. 2004; Albertson et al. 2006; Bannon et al. 2014; Bannon et al. 2015; Bannon and Whitty 1997; Johnson et al. 2012; Michelhaugh et al. 2011; Okvist et al. 2011). Briefly, cause of death was determined by forensic pathologists following medico-legal investigations evaluating the circumstances of death including medical records, police reports, autopsy results, and toxicological data. Case inclusion in the group of cocaine-related fatalities (n=11) was based on a documented history of drug abuse, a toxicology positive for cocaine and/or cocaine metabolites but negative for other drugs of abuse or CNS medications at time of death, and forensic determination of cocaine as a cause of death. Cases in the control group (n=11) had no documented history of drug abuse, and tested negative for cocaine, cocaine metabolites, and other drugs of abuse or CNS medications (other than a single case with a sub-intoxicating ethanol level of 0.06 g/dl). Causes of death for control cases were cardiovascular accidents or gunshot wounds. Cases were not screened for the presence of nicotine or metabolites. Exclusion criteria for either group included a known history of neurological or psychiatric disorder, evidence of neuropathology (e.g. stroke, encephalitis) or chronic illness (e.g. cirrhosis, cancer), death by suicide, or an estimated postmortem interval >20 hr. To reduce variance unrelated to drug abuse, the two groups were matched (Table 1) in terms of gender, race, age, and well-established measures of tissue sample quality (brain pH) and perimortem agonal state (RNA integrity number; RIN) (Schroeder et al. 2006; Stan et al. 2006). The use of de-identified cadaver specimens obtained at autopsy is not defined as human subjects research and therefore exempt from regulation 45 CFR pt 46 (NIH SF424 guide Part II: Human Subjects).
Table 1.
Characteristics of Study Subjects
| Control subjects
|
Cocaine subjects
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | Race/Sex | Cause of Death | pH | RIN | Age | Race/Sex | Cause of Death | pH | RIN | Cocaine μg/mL | BE μg/mL | |
| 66 | BM | MGSW | 6.7 | 6.9 | 64 | BM | cocaine intoxication | 6.7 | 7.6 | 0.07 | 0.40 | |
|
| ||||||||||||
| 50 | BM | GSW | 6.7 | 6.9 | 51 | BM | cocaine abuse | 6.7 | 5.7 | ND | 0.13 | |
|
| ||||||||||||
| 51 | BF | ASCVD | 6.5 | 6.9 | 52 | BF | cocaine abuse, intracerebral | 6.3 | 6.6 | ND | 0.08 | |
|
| ||||||||||||
| 40 | BF | dilated cardiomyopathy | 6.4 | 6.8 | 34 | BF | cocaine abuse | 6.4 | 6.2 | ND | 0.66 | |
|
| ||||||||||||
| 35 | BM | GSW, ASCVD | 6.4 | 6.4 | 35 | BM | cocaine intoxication | 6.7 | 7.7 | 0.28 | 5.3 | |
|
| ||||||||||||
| 47 | WM | ASCVD | 6.2 | 6.1 | 46 | WM | cocaine intoxication | 6.4 | 7.3 | 0.80 | 5.7 | |
|
| ||||||||||||
| 45 | BM | MGSW | 6.3 | 7.5 | 49 | BM | cocaine abuse, aortic aneurysm | 6.4 | 6.7 | ND | 0.42 | |
|
| ||||||||||||
| 49 | BM | ASCVD | 6.8 | 7.0 | 59 | BM | cocaine abuse | 6.6 | 7.3 | ND | 0.07 | |
|
| ||||||||||||
| 52 | BM | hypertensive cardiomyopathy | 6.3 | 7.0 | 52 | BM | cocaine abuse | 6.6 | 5.6 | 0.08 | 0.33 | |
|
| ||||||||||||
| 53 | BM | ASCVD | 6.3 | 7.8 | 52 | BM | cocaine abuse | 6.5 | 6.5 | 0.29 | 3.0 | |
|
| ||||||||||||
| 51 | BM | aortic dissection, hypertension | 6.7 | 6.0 | 52 | BM | cocaine abuse, aortic dissection | 6.3 | 7.2 | ND | 0.58 | |
|
| ||||||||||||
| Mean (SEM) | 49 (7.5) | 6.5 (0.2) | 6.9 (0.5) | 50 (8.5) | 6.5 (0.1) | 6.8 (0.7) | ||||||
Abbreviations: ASCVD, arteriosclerotic cardiovascular disease; BE, major cocaine metabolite benzoylecgonine; BF, black female; BM, black male; GSW, gunshot wound; MGSW, multiple gunshot wounds; ND, not detected; RIN, RNA integrity number; SEM, standard error of the mean; WM, white male.
Sample Processing and Microarray Analysis
All methodologies have been previously described in detail (Albertson et al. 2004; Albertson et al. 2006; Bannon et al. 2014; Bannon et al. 2015; Bannon and Whitty 1997; Johnson et al. 2012; Michelhaugh et al. 2011; Okvist et al. 2011). Briefly, postmortem samples encompassing the entire ventral midbrain (encompassing approximately plates 51–56 of DeArmond et al. 1989) were fresh-frozen upon collection at autopsy, cryosectioned, and DA cell-enriched regions finely dissected and pooled for each subject. RNA was isolated, quantified, and assessed for integrity (by RIN) using an Agilent Bioanalyzer (Santa Clara, CA).
LncRNAs represented on our custom human lncRNA microarray were chosen through a process of genome-wide computational identification and manual annotation of putative lncRNAs, as previously described (Jia et al. 2010). In addition, some protein-coding genes previously shown to be affected by cocaine abuse (Bannon et al. 2014) were included on the microarray as positive controls. Microarray experiments were executed as previously described (Lipovich et al. 2012). Briefly, cRNAs were generated from each case and hybridized to a custom Agilent 4 × 44,000-feature high-density oligonucleotide microarray platform designed to interrogate 5586 unique putative lncRNAs (plus an additional 120 protein-coding and housekeeping genes serving as controls), with seven 60-mer probes assigned to each gene (Jia et al. 2010; Lipovich et al. 2012). Microarray experiments were performed with cocaine-control matched pairs in a dye-flip two-color design, meaning each sample was run in quadruplicate, twice with each dye (Alexa-647 and Alexa-555; Invitrogen, Carlsbad, CA). Microarray slides were scanned with the default Agilent protocol and the intensity of fluorescence between dyes was normalized using a Loess correction. Data across all cases and quadruplicates were quantile-normalized and validated using MA plot density and distribution analysis (Lipovich et al. 2012). Approximately one-half of all probes were detected above background in the majority of subject pairs. The entire microarray dataset is available in the Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo; GSE67281).
Bioinformatics and Statistics
The criteria used for selection of lncRNA transcripts for further analysis are graphically represented in Figure S1. Briefly, for this study a putative lncRNA transcript was classified as differentially expressed in cocaine abusers versus control subjects only if the signal from all 7 non-identical microarray probes sequences was significantly changed, as determined by a two-step mixed model ANOVA, which utilizes both within pairs and between-groups comparisons. A false discovery rate (FDR) of 0.05 was applied to obtain final corrected p-values. Of the 428 putative lncRNA transcripts meeting these criteria, 91 exhibited an average fold change of >1.3. The subset of these transcripts most strongly supported by EST data in the UCSC Genome browser (Dec 2009 hg19 assembly), as well as expression data from the Burge Brain RNA-Seq (Wang et al. 2008), Sestan Brain microarray (Johnson et al. 2009), Allen Brain (Hawrylycz et al. 2012), or FANTOM5 (Andersson et al. 2014; FANTOM Consortium et al. 2014) datasets served as the basis for further study. The UCSC Genome browser (Dec 2009 hg19 assembly) was also used to examine the presence of polyadenylation and pre-RNA splicing consensus sequences in human lncRNA genes and their conservation across species.
Correlations between lncRNA abundances and cocaine metabolite levels (Table S2), or between lncRNA and protein-coding gene transcript levels (Table 3), were calculated using Pearson’s correlations. The LncRNA2 Function database (http://mlg.hit.edu.cn/lncrna2function) was used for a computational investigation of the potential functionality of lncRNAs based on patterns of co-expression with protein-coding genes in 19 human tissues (Table S3).
Quantitative PCR and In Situ Hybridization Histochemistry
Differential expression of 6 (3 up-regulated and 3 down-regulated) lncRNAs from Table 2 was validated by quantitative real-time (qPCR), as previously described (Albertson et al. 2004; Albertson et al. 2006; Bannon et al. 2014; Bannon et al. 2015; Bannon and Whitty 1997, Johnson et al. 2012; Michelhaugh et al. 2011; Okvist et al. 2011). Primers sequences are provided in Table S1. Pearson’s correlations between the microarray data and qPCR data for these transcripts were determined (Bannon et al. 2014; Michelhaugh et al. 2011).
Using in situ hybridization histochemistry (ISHH), the cellular and subcellular localization of several transcripts were examined in 14μm sections of human midbrain using previously published methods (Bannon and Whitty, 1997; Okvist et al. 2011). Digoxigenin-labeled antisense or sense (control) riboprobes were transcribed (DIG RNA labeling reagents; Roche, Indianapolis, IN) from cloned DNA sequences derived using the same parameters as qPCR validation experiments (Table S1). The signal was developed using anti-digoxigenin-alkaline phosphatase conjugated Fab fragment with NBT/BCIP as substrate. Images were captured using an Olympus BX53 microscope and 60X immersion objective and CellSens software with image deconvolution and brightness adjustment.
Results
Cocaine-related fatalities in this study were closely matched with drug-free control subjects in terms of race, sex, and age (Table 1) in an effort to minimize potential variance in gene expression data unrelated to cocaine abuse. There were also no differences between groups in terms of well-established measures of tissue sample quality (i.e. brain pH) or perimortem agonal state (i.e. RIN values)(Table 1). Postmortem specimens of human ventral midbrain enriched for DA neurons (Bannon et al. 2014) were processed in parallel through all experimental procedures and hybridized in quadruplicate to custom lncRNA microarrays as described (Lipovich et al. 2012) in order to maximize the accuracy of gene expression profiles. The dataset has been deposited in its entirety in the Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo; GSE67281).
LncRNA transcripts were selected for further analysis based on a series of criteria (Figure S1). Briefly, for the purposes of this study, a putative lncRNA transcript was initially classified as differentially expressed in cocaine abusers versus control subjects only if the signal intensity of all 7 non-identical microarray probes sequences was significantly different, as determined by ANOVA with an FDR of 5%. Of 5586 putative lncRNAs represented on the microarray, 428 met this criterion. In order to restrict subsequent analysis to the most robustly changed and well-annotated of these transcripts, the dataset was further pared down using both a magnitude of difference threshold (≥ 1.3 average fold-difference) and the requirement of independent evidence of expression in brain (see Materials and Methods). Application of this stringent set of criteria yielded a final list of 32 well-annotated lncRNAs exhibiting robust differential expression (14 were up-regulated and 18 were down-regulated) in the midbrains of human cocaine abusers (Table 2).
Table 2.
Differentially regulated lncRNAs in the ventral midbrain of cocaine abusers.
| lncRNA | ENSEMBL Gene ID | Gene Location | Average Fold Difference | Relative Abundance | Polyadenylation Consensus Sequence and Conservation | Splice Site Consensus Sequence and Conservation | Detected Brain Expression | Remarks |
|---|---|---|---|---|---|---|---|---|
|
Upregulated lncRNAs
| ||||||||
| RP11-309G3.3 | ENSG00000272198 | chr1q25.3 | 2.35 | 16.2 | no | no | B, S | Intergenic. Adjacent to IER5 (same strand) |
| RP1-212P9.2 | ENSG00000226852 | chr1p35.3 | 2.03 | 74.1 | yes: conserved | yes: conserved | B, F, S | Intergenic. Adjacent to OPRD1 (same strand) |
| LOC101929176 | ENSG00000250579 | chr5p15.32 | 1.77 | 30.1 | yes: primate | yes: mixed | S | Intergenic. Adjacent to ADAMTS16 (same strand) |
| LOC100507534 | ENSG00000261325 | chr16q12.1 | 1.67 | 117.2 | yes: primate | yes: conserved | B, F, S | Intergenic. Within a cluster of lncRNAs (both strands) |
| RP11-109G23.3 | ENSG00000260278 | chr4q21.21 | 1.61 | 8.9 | yes: conserved | yes: mixed | S | Intergenic. Adjacent to BMP2K (opposite strand) |
| HOTAIRM1 | ENSG00000233429 | chr7p15.2 | 1.53 | 293.5 | yes: conserved | yes: conserved | F, S | Antisense. Regulates transcription of HOXA1 and HOXA2 genes. |
| RPPH1 | ENSG00000259001 | chr14q11.2 | 1.49 | 22.4 | no | no | B, F, S | Antisense. Component of RNAse P involved in tRNA maturation |
| TRAF3IP2-AS1 | ENSG00000231889 | chr6q21 | 1.45 | 477.4 | yes: conserved | yes: conserved | A, B, F, S | Antisense to TRAF3IP2 |
| RP11-552F3.9 | ENSG00000267801 | chr17q25.1 | 1.40 | 5.9 | no | no | F, S | Antisense to TRIM47 |
| RP11-49I11.1 | ENSG00000260552 | chr18q12.2 | 1.37 | 4.4 | yes: primate | yes: conserved | F, S | Antisense to MOCOS |
| RP4-809F18.2 | ENSG00000257061 | chr12q24.32 | 1.37 | 8.0 | no | yes: mixed | B, S | Intergenic. Adjacent to LINC RP4-809F18.1 (same strand) |
| AC083843.1 | ENSG00000259820 | chr8q24.22 | 1.33 | 60.4 | yes: conserved | no | B, S | Intergenic. Adjacent to MIR30B and MIR30D (same strand) |
| WDR11-AS1 | ENSG00000227165 | chr10q26.12 | 1.33 | 36.9 | no | yes: conserved | B, F, S | Antisense to WDR11 |
| RP11-521016.2 | ENSG00000260163 | chr2q14.3 | 1.31 | 56.9 | yes: primate | yes: mixed | B, F, S | Antisense to LINC RP11-521016.1 |
|
| ||||||||
|
Down-regulated lncRNAs
| ||||||||
| RNF219-AS1 | ENSG00000234377 | chr13q22.3 | −1.31 | 87.8 | yes: conserved | yes: mixed | B, F, S | Antisense to RNF219 |
| RP11-553L6.5 | ENSG00000259976 | chr3q13.31 | −1.31 | 3471.8 | yes: conserved | no | B | Sense overlapping with ZBTB20 |
| PRKCQ-AS1 | ENSG00000237943 | chr10p14 | −1.33 | 149.2 | yes: primate | yes: conserved | B, F, S | Antisense to (shared 5′ region with) PRKCQ |
| RP11-388C12.1 | ENSG00000263063 | chr17q25.3 | −1.33 | 34.4 | no | no | S | Intergenic. Adjacent to FN3KRP (opposite strand) |
| STX18-AS1 | ENSG00000247708 | chr4p16.2 | −1.35 | 5.1 | yes: conserved | no | B, F, S | Antisense to STX18 |
| LINC00540 | NA | chr13q12.11 | −1.36 | 10.1 | no | yes: conserved | S | Intergenic. Nearest neighbors are clusters of lncRNAs |
| LOC100507140 | ENSG00000237166 | chr2q33.1 | −1.37 | 196.1 | no | yes: conserved | B, F, S | Antisense to AOX2P |
| LINC00403 | ENSG00000224243 | chr13q34 | −1.39 | 865.9 | no | yes: mixed | A, B, F, S | Sense overlapping with SOX1, SNORD441, and RP11-450H6.3 |
| RP11-539L10.3 | ENSG00000251580 | chr4p16.1 | −1.42 | 985.1 | yes: conserved | yes: primate | B, F, S | Antisense to lncRNAs LOC93622 and AC093323.3 |
| AP001505.9 | ENSG00000261706 | chr21q22.3 | −1.43 | 47.3 | yes: conserved | no | F, S | Intergenic. Adjacent to lncRNAs LINC00163 and LINC00162 |
| LOC400548 | ENSG00000278214 | chr16q24.1 | −1.44 | 17.0 | no | yes: mixed | B, F, S | Intergenic. Nearest genes FAM92B and CTC- 786C10.1 |
| LOC643763 | ENSG00000274956 | chr8q12.3 | −1.46 | 1569.0 | yes: conserved | no | A, B, F | Sense intronic within single exon of NKAIN3 |
| LINC01314 | ENSG00000259417 | chr15q25.1 | −1.49 | 543.8 | yes: primate | yes:conserved | A, B, F, S | Intergenic. Adjacent to LINC00927 and FAH (opposite strand) |
| RP11-23P13.6 | ENSG00000174171 | chr15q15.1 | −1.50 | 6.1 | no | no | B, F, S | Antisense to SPTBN5 |
| AFAP1-AS1 | ENSG00000272620 | chr4p16.1 | −1.62 | 7.1 | yes: conserved | yes: mixed | A, B, F, S | Antisense AFAP1 |
| LINC00645 | ENSG00000258548 | chr14q12 | −1.69 | 87.1 | yes: conserved | yes: mixed | S | Antisense intronic to lncRNA BC148262 |
| LINC00162 | ENSG00000224930 | chr21q22.3 | −1.74 | 65.1 | no | yes: mixed | A, B, F, S | Intergenic. Adjacent to LINC000163, and lncRNA AP001505.9 |
| LINC01010 | ENSG00000236700 | chr6q23.2 | −1.99 | 36.9 | yes: conserved | yes: mixed | B, F, S | Antisense to lncRNAs LH15.4OC101928231 and RP11-557H15.3 |
Relative abundance is overall fluorescence signal/background. In addition to expression data from UCSC Genome Browser, Dec 2009 hg19 chr assembly, transcript detection: A, in substantia nigra by Allen Brain Atlas microarray (Hawrylycz et al, 2012); B, in Burge brain RNA-Seq (Wang et al, 2008); S, in Sestan brain microarray (Johnson et al, 2013); F in substantia nigra by FANTOM5 CAGE database (Andersson et al, 2014).A gene feature found in primates as well as at least one non-primate species is denoted as conserved. Primate-specific gene features, and mixtures of conserved and primate-specific features, are also indicated.
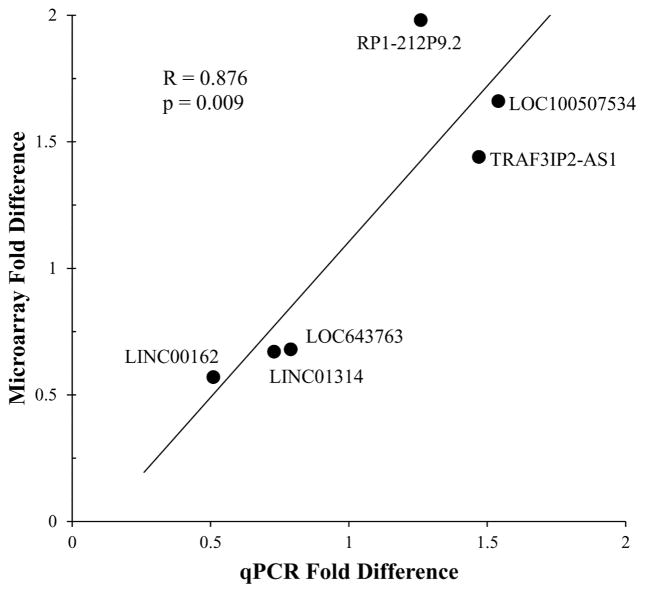
Of these 32 lncRNAs, 3 up-regulated and 3 down-regulated transcripts, representing a range of fold-differences and abundances, were further analyzed by qPCR. In each instance examined, qPCR data validated the custom microarray data (p=0.009; Fig. 1), supporting the robust nature of the findings. Importantly, the abundances of differentially expressed lncRNAs were not correlated with subjects’ levels of cocaine metabolite (Table S2), providing evidence that the recency of cocaine use was likely not a major determinant of the differential expression seen in cocaine abusers.
Fig 1.
Validation of differential lncRNA expression detected by microarray. Six lncRNAs spanning a range of transcript abundances and magnitude differences were selected from Table 2 for validation by quantitative real-time PCR. Pearson’s correlation between microarray data and qPCR data is shown. Primer sequences can be found in Table S1.
The features of the lncRNAs listed in Table 2 were consistent with those described in global analyses of lncRNAs (Derrien et al. 2012; Lipovich et al. 2010; Wight and Werner 2013). Approximately two-thirds of the these differentially expressed lncRNAs included a polyadenylation consensus sequence within 100 bases of the transcript 3’ end; of these, one-third contained primate lineage-specific sequence, whereas two-thirds showed sequence conservation beyond the primate lineage (Table 2). Canonical consensus sequences for pre-RNA splicing were also found in over two-thirds of the corresponding lncRNA genes; these were equally divided between genes with a mix of conserved and primate-specific splice sites and genes with no primate-specific consensus sequences (only a single gene showed primate-specific splice site sequences exclusively)(Table 2). In terms of genomic localization, approximately one-half of the lncRNA genes were antisense (opposite strand) to protein-coding or other lncRNA genes; the bulk of the remaining lncRNA genes were intergenic (i.e., not overlapping with known genes) (Table 2).
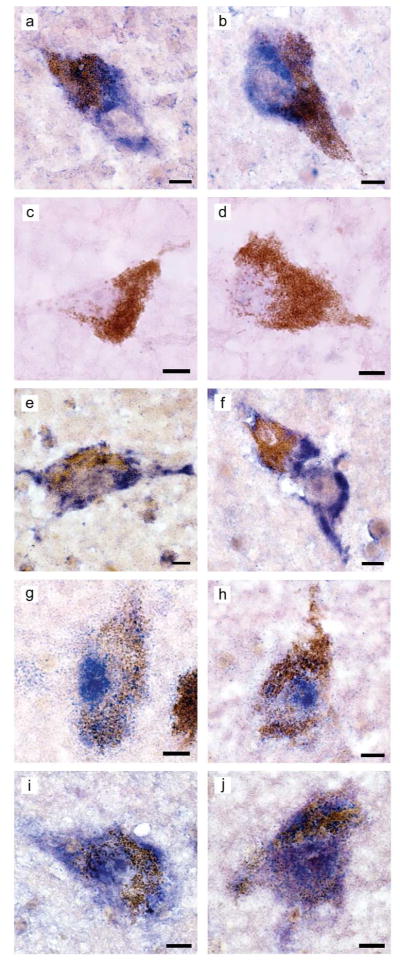
From among the 6 lncRNAs validated by qPCR, the cellular and subcellular localization of a down-regulated transcript (long intergenic noncoding RNA 00162; LINC00162) and an up-regulated transcript (tumor necrosis factor receptor-associated factor 3-interacting protein 2-antisense 1; TRAF3IP2-AS1) was examined in human ventral midbrain by means of ISHH (Fig. 2). As a positive control, the robust expression of DA transporter-encoding transcript was visualized within the processes and soma of DA neurons (readily identifiable by their characteristic large nuclei and high intracellular neuromelanin content)(Fig. 2a and 2b). Specificity of the ISHH procedure was further demonstrated by the absence of signaling using a negative control riboprobe (directed against bacterial neomycin gene sequence; Fig. 2c and 2d). Qualitative analysis indicated that both LINC00162 and TRAF3IP2-AS1 lncRNA transcripts were visualized nearly exclusively in DA neurons (Fig. 2e–2h). Similar to DA transporter transcript, LINC00162 transcript was robustly expressed within the processes and soma of DA cells, with nuclear exclusion (Fig. 2e and 2f).
Fig 2.
Cellular localization and subcellular distribution of selected transcripts determined by in situ hybridization histochemistry. (a,b) Robust expression of DA transporter mRNA within the processes and soma of DA neurons (also readily identifiable by their characteristic large nuclei and high intracellular neuromelanin content). (c,d) Specificity was demonstrated by the absence of signaling using a riboprobe derived from bacterial neomycin gene sequence as a negative control. (e,f) Similar to DA transporter mRNA localization, LINC00162 transcript was robustly expressed within the processes and soma of DA cells, with nuclear exclusion. (g,h) TRAF3IP2-AS1 transcript exhibited a strong nuclear localization in DA cells. (i,j) TRAF3IP2 protein-coding transcript distribution was distinctly different from TRAF3IP2-AS1 transcript, and was found throughout the nucleus, cytoplasm and processes of DA neurons. Probe sequences can be found in Table S1. Images captured with a 60X objective. Scale bars equal 10 microns.
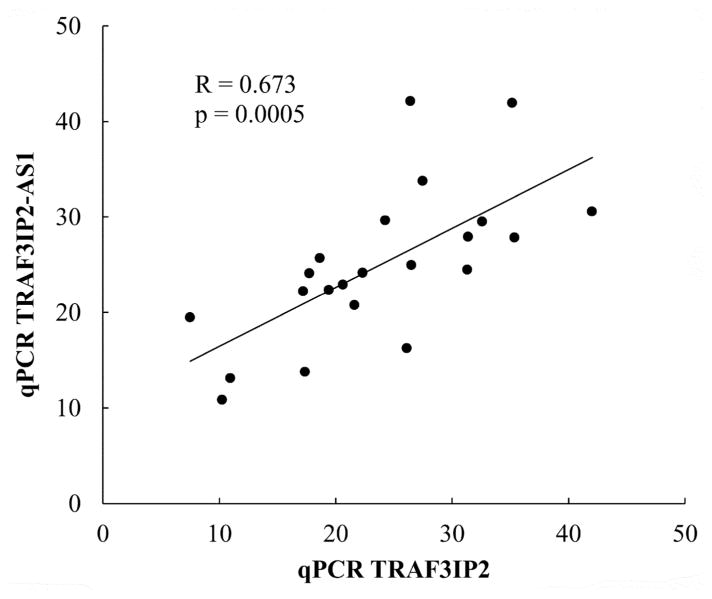
In contrast to LINC00162, TRAF3IP2-AS1 transcript showed a surprisingly strong nuclear localization in DA cells (Fig. 2g and 2h). Interestingly, the subcellular distribution of TRAF3IP2 protein-coding transcript (from the strand opposite TRAF3IP2-AS1) was quite distinct from that of TRAF3IP2-AS1 transcript, being found throughout the nucleus, cytoplasm and processes of DA neurons (Fig. 2i and 2j). As shown by qPCR, TRAF3IP2-AS1 transcript abundance correlated significantly with the levels of TRAF3IP2 protein-coding transcript (Fig. 3), consistent with a potential effect of TRAF3IP2-AS1 on the expression of its cognate protein-coding gene.
Fig 3.
The expression of a protein-coding transcript and lncRNA transcript from the TRAF3IP2 locus are significantly correlated. Pearson’s correlation between TRAF3IP2 protein-coding transcript and TRAF3IP2-AS1 transcript abundances (as determined by qPCR) is shown. Primer sequences can be found in Table S1.
Given the dearth of functional data for most lncRNAs, potential functionality has been inferred through computational investigations of the co-expression patterns of lncRNAs and protein-coding genes across different conditions or tissues (Jiang et al. 2015). Our custom lncRNA microarray (Lipovich et al. 2012) included (as positive controls) probes for a number of protein-coding neuroplasticity, stress-response, and immediate early genes known to be up-regulated in cocaine abusers’ midbrains (Bannon et al. 2014). As shown in Table 3, the expression of these known cocaine-responsive genes correlated significantly with expression of a subset of the differentially expressed lncRNAs, including RP11-309G3.3, the lncRNA most up-regulated in our dataset. It was perhaps noteworthy that the RP11-309G3.3 gene lies immediately adjacent to an immediate early gene (IER5). In addition, our microarray also included probes for several DA cell phenotypic genes that are down-regulated in cocaine abusers’ midbrains (Bannon et al., 2014). In the current study, the abundance of HOX antisense intergenic RNA myeloid 1(HOTAIRM1), an lncRNA implicated in neurogenesis and brain development (Lin et al. 2011), was negatively correlated with the expression of transcripts encoding the DA transporter (−.443; p<0.05), the DA biosynthetic enzyme tyrosine hydroxylase (−.503; p<0.05), and the DA-specifying transcription factor nurr1 (−.469; p<0.05). These correlative data suggest potential functional relationships between the expression of numerous cocaine-responsive protein-coding genes and some specific lncRNAs identified in this study. Furthermore, a global examination of all 32 lncRNAs (Table 2) for patterns of co-expression with known protein-coding genes across 19 human tissues (using the lncRNA2Function database; see Materials and Methods), revealed a highly significant association with gene ontology terms related to synapse and neuron (cellular component), and transporter and channel activities (molecular function) (Table S4), further implicating these lncRNAs in the regulation of neural function.
Table 3.
Correlations between differentially expressed lncRNAs and protein-coding stress, neuroplasticity, and immediate early gene transcripts.
| RP11-309G3.3 | RP11-109G23.3 | RPPH1 | AC083843.1 | RP11-552F3.9 | LINC00540 | |
|---|---|---|---|---|---|---|
| FOS | .699*** | .606** | .797*** | .775*** | .430* | −.454* |
| FOSB | .714*** | .622** | .802*** | .778*** | .448* | −.465* |
| JUN | .804*** | .730*** | .848*** | .760*** | .483* | −.485* |
| CEBPD | .720*** | .717*** | .758*** | .559** | .512* | −.438* |
| ATF3 | .969*** | .917*** | .919*** | .611** | .416 | −.531* |
| HSPA1A | .959*** | .947*** | .886*** | .602** | .358 | −.483* |
| BAG3 | .955*** | .966*** | .864*** | .519* | .377 | −.413 |
| GADD45B | .710*** | .634** | .828*** | .793*** | .541** | −.456* |
| CDKN1A | .848*** | .848*** | .775*** | .346 | .478* | −.392 |
| GADPH | −.298 | −.379 | −.312 | −.363 | −.264 | −.019 |
Significantly correlated
p < .001,
p < .01,
p < .05 (2-tailed).
GAPDH is a housekeeping gene included as a negative control.
Discussion
The major goal of this study was to identify lncRNAs that are significantly dysregulated in the ventral midbrain of human cocaine abusers. Since they do not encode protein products, lncRNA transcripts constitute the final mediators of lncRNA gene function. Using an experimental design that incorporated parallel processing and quadruplicate hybridization of specimens from well-matched subject pairs of cocaine fatalities and drug-free control subjects (Table 1), followed by the application of various statistical, magnitude difference, and expression data filters (Lipovich et al. 2012), we identified 32 well-annotated lncRNAs with clear differential expression in the midbrains of human cocaine abusers (Table 2). The robustness of the dataset obtained was confirmed by the successful validation (by qPCR) of differential lncRNA expression in every instance examined (Fig. 1).
A number of limitations associated with this study warrant mention. The application of stringent subject inclusion and exclusion criteria, and the careful matching of the cocaine-abusing and control cohorts in terms of numerous demographic and sample quality parameters, limited the number of subjects available for study. The list of differentially expressed lncRNAs we identified by microarray is, in all likelihood, far from exhaustive; future RNA-seq experiments involving larger cohorts and encompassing ongoing advances in lncRNA annotation, will undoubtedly extend the findings of this preliminary analysis. In addition, as the current experiments involved only cocaine abusers, other studies are needed to determine the extent to which these differentially expressed lncRNAs reflect changes common to all drug abusers versus cocaine-specific effects. Previous studies of nucleus accumbens have identified both commonalities and differences in profiles of gene expression between cocaine and heroin abusers (Albertson et al. 2004; Albertson et al. 2006; Michelhaugh et al. 2011). Furthermore, genomic studies in human and/or animal models are required to address the possibility that some differentially expressed lncRNAs might be associated with a predisposition to, rather than a response to, drug abuse. Finally, although the two lncRNAs selected for ISHH were subsequently shown to be expressed nearly exclusively within DA neurons (Fig. 2), the cellular locus of expression of the remaining lncRNAs was not examined; it is quite plausible that glia or non-DA neurons contribute to the pattern of differential lncRNA expression observed in our microarray and qPCR experiments. Additional studies are clearly needed to advance our understanding of these issues.
As is the case for nearly all lncRNAs (Jiang et al. 2015), the biological functions of the cocaine-responsive lncRNAs identified in this study are not currently understood. Computational investigations were therefore used to provide some preliminary insights into their potential functionality. As discussed, the lncRNA dataset as a whole (Table 2), based on the pattern of co-expression with protein-coding genes across human tissues, was very strongly associated with gene ontology terms related to neuronal function (Table S3). Further, inclusion in our custom lncRNA microarray of probes for numerous protein-coding genes that are up-regulated (i.e., neuroplasticity, stress-response, and immediate early genes) or down-regulated (i.e., DA cell phenotypic genes) in cocaine abusers’ midbrains (Bannon et al. 2014; Bannon et al. 2015) allowed us to identify a specific subset of lncRNAs (Table 3) whose expression was significantly correlated with these known cocaine-responsive genes. The potential functional relationship between these cocaine-responsive lncRNAs and protein-coding genes warrants further investigation.
Another interesting finding was the up-regulation in cocaine abusers of the lncRNA TRAF3IP2-AS1 (Table 2), and its positive correlation with the opposite strand protein-coding transcript TRAF3IP2 (Fig. 3), despite their distinct subcellular localizations (Fig. 2). The exclusively nuclear localization of TRAF3IP2-AS1 transcript and its lack of complementarity with TRAF3IP2 protein-coding transcript sequence suggest a possible epigenetic effect of TRAF3IP2-AS1 transcript on TRAF3IP2 gene expression through alterations of chromatin state at this locus, as has been shown for some other antisense lncRNAs (Khorkova et al. 2014). Another lncRNA gene we found dysregulated in cocaine abusers, PRKCQ-AS1 (protein kinase C, theta-antisense 1) (Table 2) is antisense to the protein-coding PRKCQ (protein kinase C, theta) gene with which it shares a common promoter region. It is noteworthy that both TRAF3IP2 and PRKCQ proteins interact with other signaling molecules to activate the transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) (Chuang et al. 2011; Valente et al. 2013). As we have previously identified, dysregulation of several NF-kB-associated genes in cocaine abusers’ midbrains (Bannon et al. 2014) and NF-kB signaling has been shown to regulate cocaine reward (Russo et al. 2009). TRAF3IP2-AS1 and PRKCQ-AS1 lncRNAs represent potential mediators of a disruption of NF-kB signaling seen in cocaine abuse.
In summary, the current experiments represent, to our knowledge, the first profile of lncRNA dysregulation associated with human drug abuse. A small dataset of well-annotated lncRNAs exhibiting robust differential expression in cocaine abusers’ midbrains was identified. Examples of lncRNAs with DA cell-specific expression, differential subcellular distribution, or covariance with known cocaine-responsive protein-coding genes were identified. In keeping with the emerging myriad roles of lncRNAs in brain development and some other CNS disorders (Clark and Blackshaw, 2014; Michelhaugh et al. 2011; Modarresi et al. 2012; Ng et al. 2013; Pastori and Wahlstedt, 2012; Punzi et al. 2014), we hypothesize that a number of the lncRNAs identified in this study mediate broader downstream changes in gene expression arising within the DA neurons of chronic drug abusers. Delineating the contributions of specific lncRNAs to the molecular processes underlying drug addiction will require experimental interventions in animal models, but could ultimately lead to the development of novel therapeutic approaches for the treatment of addiction.
Supplementary Material
Figure S1. Criteria used for selection of lncRNAs for further analyses.
Table S1. Sequences of primers used for qPCR validation and of riboprobes used for in situ hybridization histochemistry.
Table S2. Absence of correlation between cocaine metabolite benzoylecgonine and differentially expressed lncRNAs from Table 2.
Table S3. Gene ontology terms associated with the lncRNA dataset in Table 2.
Acknowledgments
This work was supported by NIH grants DA006470 (MJB) and DA026021 (LL). The authors thank Ms. Becky Cai for technical assistance with preliminary PCR experiments, Mr. Zachary Hartley and Mr. Syed Mahmood for technical assistance with cloning of some riboprobe constructs, Ms. Donghong Ju and Ms. Amanda Goldstone for assistance in the analysis of lncRNA gene features, and Drs. Sokol Todi and Wei-Ling Tsou for their advice regarding cloning and microscopy procedures.
Abbreviations used
- DA
dopamine
- FDR
false discovery rate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HOTAIRM1
HOX antisense intergenic RNA myeloid 1
- ISHH
in situ hybridization histochemistry
- LINC00162
long intergenic noncoding RNA 00162
- lncRNA
long noncoding RNA
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- MIAT
myocardial infarction associated transcript
- NEAT1
nuclear enriched abundant transcript 1
- NF-kB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PRKCQ
protein kinase C theta
- PRKCQ-AS1
protein kinase C theta-antisense 1
- qPCR
quantitative real-time PCR
- RIN
RNA integrity number
- TRAF3IP2
tumor necrosis factor receptor-associated factor 3-interacting protein 2
- TRAF3IP2-AS1
tumor necrosis factor receptor-associated factor 3-interacting protein 2-antisense 1
Footnotes
Conflict of interest disclosure
The authors report no biomedical financial interests or potential conflicts of interest.
References
- Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem. 2004;88:1211–1219. doi: 10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31:2304–2312. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon MJ, Johnson MM, Michelhaugh SK, Hartley ZJ, Halter SD, David JA, Kapatos G, Schmidt CJ. A molecular profile of cocaine abuse includes the differential expression of genes that regulate transcription, chromatin, and dopamine cell phenotype. Neuropsychopharmacology. 2014;39:2191–2199. doi: 10.1038/npp.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon MJ, Savonen CL, Hartley ZJ, Johnson MM, Schmidt CJ. Investigating the potential influence of cause of death and cocaine levels on the differential expression of genes associated with cocaine abuse. PloS One. 2015;10:e0117580. doi: 10.1371/journal.pone.0117580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon MJ, Whitty CJ. Age-related and regional differences in dopamine transporter mRNA expression in human midbrain. Neurology. 1997;48:969–977. doi: 10.1212/wnl.48.4.969. [DOI] [PubMed] [Google Scholar]
- Bu Q, Hu Z, Chen F, et al. Transcriptome analysis of long non-coding RNAs of the nucleus accumbens in cocaine-conditioned mice. J Neurochem. 2012;123:790–799. doi: 10.1111/jnc.12006. [DOI] [PubMed] [Google Scholar]
- Chuang HC, Lan JL, Chen DY, et al. The kinase GLK controls autoimmunity and NF-kappaB signaling by activating the kinase PKC-theta in T cells. Nat Immunol. 2011;12:1113–1118. doi: 10.1038/ni.2121. [DOI] [PubMed] [Google Scholar]
- Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest AR, Kawai H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeArmond SJ, Fusco MM, Dewey MM. Structure of the Human Brain: A Photographic Atlas. 3. Oxford University Press; New York: 1989. [Google Scholar]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Nestler EJ. Epigenetic mechanisms of drug addiction. Curr Opin Neurobiol. 2013;23:521–528. doi: 10.1016/j.conb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Dev Growth Differ. 2012;54:44–54. doi: 10.1111/j.1440-169X.2011.01303.x. [DOI] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Ma R, Wang J, et al. LncRNA2Function: A comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics. 2015;16(Suppl 3):S2. doi: 10.1186/1471-2164-16-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MM, David JA, Michelhaugh SK, Schmidt CJ, Bannon MJ. Increased heat shock protein 70 gene expression in the brains of cocaine-related fatalities may be reflective of postdrug survival and intervention rather than excited delirium. J Forensic Sci. 2012;57:1519–1523. doi: 10.1111/j.1556-4029.2012.02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokova O, Myers AJ, Hsiao J, Wahlestedt C. Natural antisense transcripts. Human Mol Genet. 2014;23:R54–63. doi: 10.1093/hmg/ddu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, Lachman HM. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PloS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H, Loeb JA. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics. 2012;192:1133–1148. doi: 10.1534/genetics.112.145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim et Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. 2011;116:459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Okvist A, Fagergren P, Whittard J, et al. Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biol Psychiatry. 2011;69:245–252. doi: 10.1016/j.biopsych.2010.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Wahlestedt C. Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA Biol. 2012;9:860–870. doi: 10.4161/rna.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzi G, Ursini G, Shin JH, Kleinman JE, Hyde TM, Weinberger DR. Increased expression of MARCKS in post-mortem brain of violent suicide completers is related to transcription of a long, noncoding, antisense RNA. Mol Psychiatry. 2014;19:1057–1059. doi: 10.1038/mp.2014.41. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Wilkinson MB, Mazei-Robison MS, et al. Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. Journal Neurosci. 2009;29:3529–3537. doi: 10.1523/JNEUROSCI.6173-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, Tamminga CA. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente AJ, Sakamuri SS, Siddesha JM, Yoshida T, Gardner JD, Prabhu R, Siebenlist U, Chandrasekar B. TRAF3IP2 mediates interleukin-18-induced cardiac fibroblast migration and differentiation. Cell Signal. 2013;25:2176–2184. doi: 10.1016/j.cellsig.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Tomasi D, Telang F. Addiction: beyond dopamine reward circuitry. Proc Natl Acad Sci. 2011;108:15037–15042. doi: 10.1073/pnas.1010654108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight M, Werner A. The functions of natural antisense transcripts. Essays Biochem. 2013;54:91–101. doi: 10.1042/bse0540091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Criteria used for selection of lncRNAs for further analyses.
Table S1. Sequences of primers used for qPCR validation and of riboprobes used for in situ hybridization histochemistry.
Table S2. Absence of correlation between cocaine metabolite benzoylecgonine and differentially expressed lncRNAs from Table 2.
Table S3. Gene ontology terms associated with the lncRNA dataset in Table 2.