Abstract
As the frequency and prevalence of zoonotic diseases increase worldwide, investigating how mammal host distributions determine patterns of human disease and predicting which regions are at greatest risk for future zoonotic disease emergence are two goals which both require better understanding the current distributions of zoonotic hosts and pathogens. Here we review the existing data about mammalian host species, comparing and contrasting these patterns against global maps of zoonotic hosts from all 27 orders of terrestrial mammals. We discuss the zoonotic potential of host species from the top six most species-rich mammal groups, and review the literature to identify analytical and conceptual gaps that must be addressed to improve our ability to generate testable predictions about zoonotic diseases originating from wild mammals.
Keywords: macroecology, infectious disease, biogeography, hotspot, risk, prediction
Where will future zoonoses come from?
Current understanding about the global distribution of most infectious diseases is surprisingly limited. Even for human infectious diseases, the spatial distributions of the vast majority remain little known [1]. Yet, the frequency with which new infectious diseases are emerging [2], especially zoonoses (see Glossary), underscores the necessity of shifting from a reactionary to a pre-emptive approach to mitigating infectious disease.
Assessing future disease risk requires baseline data – information about where infectious diseases are distributed geographically, taxonomically (with respect to animal reservoirs), and in relation to human populations. Such information is most abundant for records of human infectious disease. Whether looking across multiple diseases to glean generalizable epidemiological insight, or at specific diseases to identify important covariates predicting particular human outbreaks, previous studies have combined detailed data on human infectious disease events and environmental factors to quantify current and to predict future disease hotspots (e.g., [2–6]). Such baseline data provide important starting points for making projections of human disease risk, and can be effectively applied to predict the spread of particular infectious diseases to new areas that are in close proximity, or located in environments similar to historical outbreak locations (e.g., [6,7]). As one example, data describing the ecology of bat reservoirs of Nipah virus can help make projections about the kinds of environments expected to support cases of human disease [5]. Such baseline data can then be applied to identify and manage similar locations where future Nipah outbreaks might be predicted to occur. However, data from past outbreaks may offer little towards efforts to predict outbreaks of completely novel diseases that punctuate the status quo – for example, the emergence of new zoonotic pathogens, such as the Middle East respiratory syndrome coronavirus (MERS-CoV) [8], or outbreaks of known zoonoses in unexpected areas, such as Ebola virus disease in West Africa [9]. Because of their often surprising departures from previous outbreak patterns, some argue that disease events may be inherently unpredictable (e.g., [10]). Predicting outbreaks, caused either by novel pathogens or known pathogens in novel places, remains one of the biggest scientific challenges of our time. We agree that this is a hard problem, but disagree that it is impossible. This causes us to ask, what kind of data are available that may facilitate more effective prediction?
Since most human infectious diseases have animal origins [2,3,11,12], and the majority of emerging human diseases originate from mammals [13,14], better understanding the global distributions of mammal zoonotic hosts could provide a first order prediction of future hotspots for zoonotic disease emergence. Recognizing that a parasite or pathogen is unlikely to persist in all populations of its definitive host(s), we think of the collective geographic ranges of known host species as the maximum potential current geographic range of a zoonosis. Visualizing this potential range offers a baseline from which we can ask basic comparative questions about realized and unrealized risk of zoonotic diseases and offers a launch point for building predictive models of future zoonotic disease events.
Here we review what is known about the geographical distribution of zoonoses carried by wild mammals [15]. We describe global biogeographic patterns of zoonotic hosts across all 27 orders of terrestrial mammals (as confirmed to the species level by the Global Infectious Disease and Epidemiology Network (GIDEON) database, [16]), which provides real-time updates of infectious diseases of zoonotic relevance to humans and reports animal hosts to the species level. For zoonotic hosts in each of the six most speciose mammal groups, we review the geographic ranges recorded by the International Union for the Conservation of Nature [http://www.iucnredlist.org/technical-documents/spatial-data], and address five outstanding research questions about mammal-borne zoonoses.
1. What causes high zoonotic disease risk?
Human zoonotic disease risk can be defined as a function of several factors, including the probability of successful transmission of a zoonotic pathogen from an animal host into human hosts (transmission of infection) and the probability of an infection transitioning to a state of disease in human hosts (transition to disease) [18,19]. These components of disease risk rely on several factors that are external to the host-pathogen system. Extrinsic factors, such as urbanization, agriculture and socioeconomic standing, control host and human population dynamics underlying the frequency of transmissible contacts at the human-wildlife and the wildlife-livestock interface [5,18,20–22]. Intrinsic factors (of hosts, pathogens, and vectors) combine with extrinsic factors to contribute to disease risk in humans. Intrinsic factors include life history [23–25], behavior [26,27], competence [28–30], and rapid evolutionary changes in animal hosts and pathogens [31–33]; transmission modes and host breadth in pathogens [13,34,35]; and differences in host susceptibility, often conferred by prosperity or poverty in human populations [36], and pristine or degraded communities in wildlife hosts [37]. Thus, regions can have high zoonotic risk for multiple reasons – people living in regions with inherently high zoonotic potential may be considered at high risk, but so too can those living in regions with low host and pathogen diversity but increasing external pressures (such as warming or urbanization) that may facilitate the transmission of some zoonoses through a cascade of environmental changes [38–41].
Notwithstanding these complexities, areas that are currently experiencing zoonotic outbreaks are places where a high zoonotic risk has been realized as observable disease events. Investigating the features shared in common among regions with high realized disease risk (in the form of recurring or new observed outbreaks) is a first step to understanding what triggers these events (e.g., [2,4,5,42]). But comparing regions with high realized disease risk offers limited utility for forecasting unexpected disease events, which requires quantifying unrealized disease risk. Disruptive extrinsic pressures in regions of high zoonotic potential where host or pathogen richness is already particularly high could trigger spillover to human hosts. Identifying locations with high zoonotic potential but without overt signs of disease emergence could signal regions where pathogen transmission is high but transition to disease is low (Ebola virus outside Africa, [43]) or areas where transition to disease is low because pathogen transmission is currently low, but once transmission occurs, disease will increase as a matter of course (e.g., Ebola virus disease within Africa, [6]).
2. Where are global hotspots of zoonotic mammal hosts?
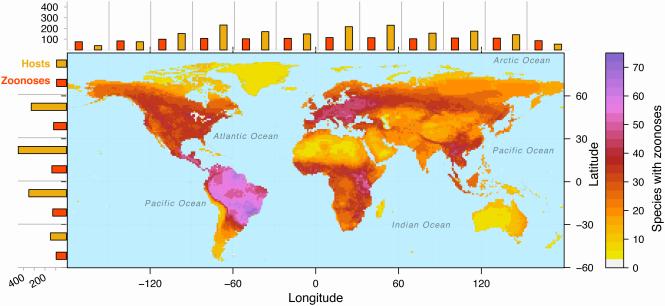
In general, we expect more zoonotic host species to occur in regions with high biodiversity [44]. Indeed, mapping the overlapping geographic ranges of zoonotic mammal hosts (Figure 1) reveals that regions with the greatest host richness include Central and South America (southeast of the Andes); Central East Africa; Europe (Eastern Europe, some of Western Europe), and mainland Southeast Asia (Figure 1). In general, these regions align with global geographic patterns of mammal biodiversity (Figure S5; [45]) with the exception of the hotspot in the north temperate zone (Europe), which contains a higher diversity of mammal hosts than expected from global biodiversity patterns. We postulate that this pattern may be driven in part by the high richness of rodents and insectivores found in this region (see Question 3, below).
Figure 1. Geographic ranges of zoonotic mammal hosts.
Mammal reservoirs of zoonotic diseases are globally distributed, with noteworthy hotspots in Amazonia and Eurasia. Overlapping geographic ranges of mammal species recognized to carry one or more zoonotic diseases, with counts of unique host species (gold bars) and unique zoonotic pathogens (red bars) found within 30° latitudinal and longitudinal bands. This map depicts 5007 total wild mammal species from 27 orders.
To what degree do these zoonotic host hotspots align with global patterns of human zoonotic disease? While the diversity of host species corresponds well to global biodiversity patterns, the biogeography of human outbreaks is less straightforwardly explained by biodiversity. For rodents, Han et al.[23] found a greater diversity of zoonotic hosts and diseases in regions with low overall mammal biodiversity. They also found that the number of human outbreaks caused by rodent-borne zoonoses (since 1990) is concentrated in northerly latitudes, especially Europe [23]. In terms of emergence, the majority of human infectious diseases emerged for the first time in northerly latitudes, with a hotspot in Europe [2]. Unsurprisingly, zoonoses with wildlife (as opposed to domesticated species) origins were found to emerge more frequently in humans populations occurring in biodiverse areas (as measured by wildlife species richness, which includes, but is not restricted to, mammals) [2]. In contrast, the emergence frequency of zoonoses from non-sylvatic (e.g., domesticated species) origins is best explained by human population density and by latitude [2], with no identifiable significance of biodiversity. These patterns provide additional evidence for the general expectation that zoonoses pose greater risks with increasing human contact with animal sources [18,46,47].
Many of the zoonotic host hotspots in higher latitudes also overlap with centers of high human population density (e.g., Europe, Southeast Asia; Figure 1), which suggests an important role of reporting or study biases in determining larger biogeographical patterns of zoonotic disease [48]. As more resources are dedicated to disease-related research and treatment in the northern hemisphere [2,34,36,44], the number of hosts and pathogens discovered in these countries tends to increase (e.g., [49,50]). However, if such biases were the sole factor driving this pattern, hotspots of similar magnitude would be expected in the United States and Canada, since these countries span similar latitudes, exhibit similar overall species richness [45], and apportion the greatest expenditures towards biological and health-related research [http://apps.who.int/nha/database/World_Map/Index/en]. An open question is whether human population density alone is driving these geographic patterns, as human populations are spread over larger areas in North America compared to Europe.
An underexplored possibility is that there are intrinsic differences between species comprising north temperate communities. These communities display different patterns of functional diversity compared to those in the tropics, with each temperate species contributing disproportionately to ecosystem processes compared to tropical species [52]. Studies that move beyond occurrence-based metrics (e.g., richness) to consider the functional trait diversity of host and pathogen species may reveal another dimension to what is driving geographic patterns of zoonotic diversity. Whether temperate hosts carry proportionately more zoonoses or whether zoonotic pathogens in temperate zones exhibit proportionately greater host breadth compared to tropical systems is an open question [53]. Comparing the topologies of host-pathogen networks in temperate vs. tropical regions is one approach to answering this question (e.g.,[54,55]).
3. Which mammal groups pose the greatest zoonotic disease risk?
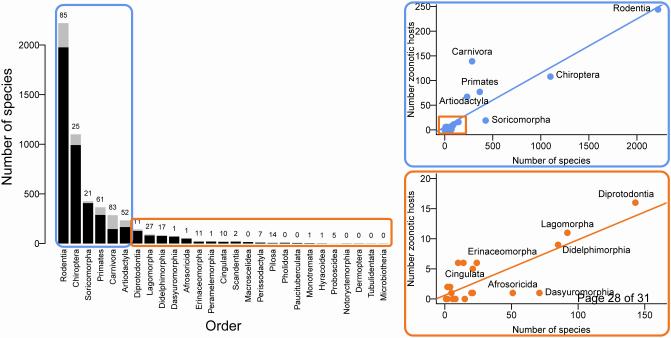
Across the 27 terrestrial mammal orders, there is wide variation in both the total number of species, and the fraction of species in each order that are zoonotic hosts (Figure 2; Figures S1-S3). In the most speciose orders (blue boxes, Figure 2), the number of zoonotic hosts increases with total species richness of the order, a pattern which may be explained in part by accumulating evidence that parasitism drives host diversification (reviewed in [56]). But further examination of the variation around this general trend is warranted. For example, groups with more zoonotic host species than expected for the richness of the clade (e.g., orders that fall above a regression line through points in Figure 2) may share suites of similar intrinsic or extrinsic factors enabling more successful pathogen transmission (e.g., [57]), or more frequent human contacts that, over time, facilitate the transition of a novel infection to a disease state in humans [18,19,58] (e.g., the ungulates, a paraphyletic group that includes the Artiodactyla and Perissodactyla; ungulates comprise the majority of domesticated mammal species). Conversely, orders with fewer zoonotic hosts than expected for the richness of the clade (those below a regression line) may reflect species that have been poorly sampled for zoonoses, or species that carry fewer zoonoses due to unique combinations of intrinsic and extrinsic features. For example, the marsupial carnivores (Dasyuromorpha), much like placental carnivores (Carnivora), have less direct contact with humans compared to other clades (e.g., Rodents, Ungulates) [59], which may reduce the risk of zoonotic transmission to humans. However, many dasyuromorphs have regular contact with domesticated species (for example, domesticated dogs, especially when Tasmanian devils and quolls are attracted to livestock on farms [60]), providing opportunity for human exposure through farm animals on human-modified environments [46].
Figure 2. The number of zoonotic hosts increases with total species richness of the order.
Zoonotic diseases are found in the majority of terrestrial mammal orders (21/27), with the most species rich orders containing the greatest diversity of zoonoses. This split bar plot shows the total number of host species (black and gray) and the fraction of species that are confirmed zoonotic hosts for one or more zoonotic diseases (gray). The number above each bar represents a tally of the total unique zoonoses per order. Mammal orders are arranged in descending order of species richness. The number of zoonotic host species in each order is represented in scatterplots, with the most speciose orders contained in the blue boxes (top right; regression R2=0.81), and all other orders in the orange boxes (bottom right; regression R2=0.63).
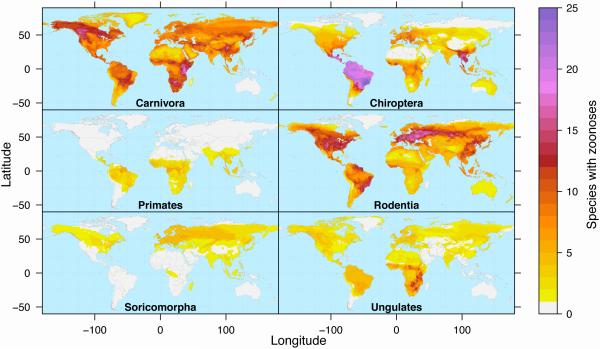
Maps depicting geographic range distributions of zoonotic hosts across the most speciose orders (Figure 3, Key Figure) show that global hotspots (Figure 1) are driven in part by striking differences in the distribution of zoonotic hosts from specific clades, as will be explained below. Maps depicting geographic range distributions of zoonotic hosts from mammal orders endemic to Australia can be found in Figure S2, and maps for all other orders can be found in Figure S3.
Figure 3. Key Figure. Global hotspots of zoonoses are driven by differences in the distribution of zoonotic hosts from specific clades.
Mapping overlapping geographic ranges of mammal species recognized to carry one or more zoonotic diseases highlights regions of high and low zoonotic host diversity arising from particular clades. Mammal zoonotic host richness is depicted by color for carnivores, bats (Chiroptera), primates, rodents, shrews and moles (Soricomorpha), and the hoofed mammals (Ungulates, which combine the orders Perissodactyla and Artiodactyla and exclude domesticated species).
Rodentia
Among mammals, rodents are the most abundant and most species-rich and include a greater number of zoonotic hosts than any other order: approximately 10.7% of rodents are hosts (244/2220 species, updated from proportions reported by [23]), carrying 85 unique zoonotic diseases. Rodent reservoirs of zoonotic diseases are distinguished by features that support a fast life history profile, reproducing earlier in life and more frequently compared to other rodent species [23]. Figure 3, Key Figure, shows that north temperate areas in North America and Europe and the tropical Atlantic forest region of Brazil contain the most rodent host species. We note that the larger global zoonotic host hotspot observed in Europe and Russia (Figure 1) may be driven in part by the diversity of rodent and small-bodied insectivore hosts (Soricomorpha, see below), as well as their predators (Carnivora, see below) (Figure 3, Key Figure).
Chiroptera
Because they are also relatively small-bodied, speciose, and associated with numerous EID events in humans, bats are often compared to rodents with respect to their risk of carrying zoonotic pathogens [61]. There are about half as many zoonotic bat hosts compared to rodents (108/1100 bat species are hosts, approximately 9.8%), and they carry about a third of the number of unique zoonoses (27) compared to rodents.
The mammal host hotspot in the Neotropics is likely to be driven in part by the high diversity of bat hosts in this region (Figure S5, [45]). Figure 3, Key Figure, shows hotspots of zoonotic bat hosts in Central and South America (wet regions east of Chile, north of Paraguay and Uruguay), as well as in Southeast Asia. These patterns are generally consistent with bat biodiversity patterns [45], except for the following departures: 1) within South America, bat species richness is highest in the Andean countries and in northern Brazil whereas the hotspot of bat hosts is in southern Brazil; 2) Southeast Asia and equatorial Africa display similar patterns of bat species richness, but Southeast Asia has many more zoonotic host species even though it is a much smaller land mass. If, as ecological theory suggests, we expect the number of hosts to be proportional to overall species richness [54,62,63], these maps suggest either that Africa is understudied or that Southeast Asia has more zoonotic hosts than expected for its mammal species richness and for its area.
Soricomorpha
Among the insectivoran mammals, relatively few shrews and moles are known to be zoonotic hosts, with only about 4% (19/426 species) carrying 19 unique zoonoses. This small percentage could be due to this group being understudied compared to other, similarly species rich mammal groups. A Web of Science search on the Latin binomials of all extant mole and shrew species returned a total of 4600 citations, which is 1-2 orders of magnitude fewer studies than for other speciose mammal orders (blue box, Figure 2). Zoonotic hosts in this order are distributed widely across north temperate latitudes, with the greatest number of host species overlapping across Europe and across the Atlantic coast of the United States (Figure 3, Key Figure). Given their species richness, wide geographical distribution, and degree of biological and ecological similarity to rodent and bat hosts (e.g., small body size, high metabolic rates, and plasticity in reproductive traits), zoonotic disease ecology in insectivorous mammals stands out as an important area for further research.
Carnivora
While ungulates were previously thought to share the most pathogens with humans [13], we find that carnivoran hosts nearly tie with the rodents to harbor more unique zoonoses than other terrestrial mammal clades. Approximately 49% (139/285) of all carnivore species – the highest proportion of any mammal order – carry one or more of 83 unique zoonotic pathogens. Carnivoran hosts are among the most widely distributed in terms of spatial extent (Figure 3, Key Figure), with hotspots of host diversity in Southern and East Africa, Southeast Asia, and the subarctic region of North America. This contrasts patterns of carnivoran species richness, which is concentrated in the southern hemisphere (particularly in Africa and Southeast Asia; Figure S5). Previous studies showed that pathogen richness (zoonotic and non-zoonotic) closely tracked carnivore species richness [64], and that the range of carnivoran host species infected by a pathogen depends primarily on host phylogenetic relatedness [65]. In addition, research exploring the degree to which carnivorans accumulate the infectious agents of their prey (which can themselves be zoonotic hosts) will contribute to understanding whether the distribution of carnivoran hosts are tracking the diversity of prey items.
Ungulates
Ungulate reservoirs of zoonotic disease have been of particular interest because of high human contact rates through hunting, and the degree of contact and relatedness between wild and domesticated species (livestock) [13,20,40,66]. Recent work also shows that the time since domestication correlates positively with the number of zoonotic infections shared between ungulates and humans, and that species with the longest history of domestication do not only carry more zoonotic pathogens, but may also transmit infection to a greater diversity of alternative host species [67]. For wild ungulates (excluding domesticated species), we find that approximately 32% of species were zoonotic hosts (73/247 species), carrying 68 unique zoonoses. Ungulates cover a greater spatial extent than bats, primates and insectivores, and the majority of host species overlap in East and Southern Africa (Figure 3, Key Figure).
Primates
The high degree of phylogenetic relatedness between human and non-human primates is thought to contribute to greater risk of pathogen spillover [68]. For example, species that are closely related and share habitat show the most similar parasite communities [69], suggesting that spatial overlap and phylogenetic relatedness are likely to be important for understanding transmission in humans and in wild host species. Primates are generally found in the global equatorial zone, with greatest species diversity in the rainforests of Africa, the Neotropics, and Asia [70]. Primate zoonotic host richness is greatest in equatorial Africa (in central Africa in the Congo Basin, and West Africa), in Southeast Asia, and in the tropical/mixed forest regions of northern Brazil and the Guyana Shield. Over 20% of primate species are zoonotic hosts (21%; 77/365 species) for at least one of 63 unique zoonoses. Thus, while there are fewer species of primates overall, a greater proportion of primate are zoonotic hosts than either the rodents or the bats.
Zoonotic potential of hosts and human disease risk
Importantly, the hotspots identified here depict regions with the greatest number of overlapping zoonotic host species, excluding other components of disease risk. In general, increasing opportunities for human exposure to animal groups with high zoonotic potential will increase the risk of zoonotic disease in humans. In addition to overall host richness of an area, zoonotic potential depends on the chance that a species is a zoonotic host given the order it belongs to, and breadth of zoonoses harbored by host species in a given order [13]. Thus, based on the results described above, rodents can be considered to have high zoonotic potential because there are more rodent host species than hosts from any other mammal group. And while there are fewer carnivoran species in comparison to rodents, they may also be considered to have high zoonotic potential because a greater fraction of them are zoonotic hosts (i.e., given an encounter with a carnivore, the likelihood that it is a zoonotic host is higher than for an equivalent encounter with a rodent from which there are comparatively fewer zoonotic hosts). If the high degree of parasite sharing suggests that the majority of zoonoses has already been transmitted from wild reservoirs to humans, the fact that humans already share a large number of zoonoses with carnivorans may indicate there are few zoonoses left to spillover anew (novel emergence) but many available to seed re-emergence events. Carnivorans also carry a greater diversity of zoonotic pathogens than any other mammal group. While zoonotic potential is considered high for rodents based purely on numerical frequency, there may be something intrinsic about carnivores where, despite relatively low direct contact with humans, they are more permissive to zoonotic infections and may be particularly good at sustaining them even if the risk of transmissible contacts with humans is lower. Similarly, while there are fewer species of ungulates compared to rodents, bats, primates, carnivorans, and insectivorans (Figure 2, Artiodactyla and Perissodactyla), many more wild ungulates are closely related to domesticated livestock species that humans are in regular and increasingly close contact, which could facilitate successful transmission [13,20,67,71]. Bats are suspected to be particularly permissive hosts of zoonotic infection [72,73], but outside of the paleotropics where bats are hunted as bushmeat, direct human contact with bats is likely surpassed by contact with rodents. Thus, for both bats and carnivorans, it may be more likely that spillover is facilitated by indirect transmission (e.g., for bats, contact with partially consumed food, including insects, vertebrates, and fruit; [12,74]; for carnivorans, contact with domesticated pets or livestock, and through parasite shedding in encroaching habitats [75]). These patterns should also be tempered by more explicit quantification of surveillance bias. For example, the surveillance scrutiny applied to bats for particular pathogen types (e.g., viruses [76,77]), may skew the number of species found to be positive for zoonotic viruses. Similar surveillance biases across other clades will impede comparisons of relative zoonotic disease risk posed by particular mammal groups or species.
4. Where are global hotspots of mammal-borne zoonoses?
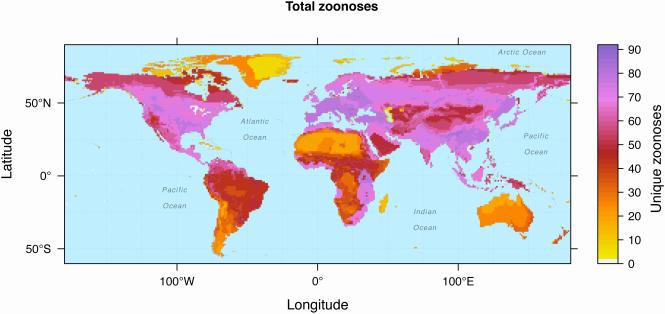
If we make the simplifying assumption that zoonoses are distributed throughout the geographic ranges of the mammals carrying them [64], then we can generate some baseline hypotheses about where zoonotic potential may be greatest by identifying zoonotic pathogen hotspots. Such hotspots occur where many zoonotic hosts overlap in geographic range, and thus their zoonotic pathogens also overlap. The histograms in Figure 1 show that species richness of zoonotic hosts reflects latitudinal gradients reminiscent of well-known biogeographical patterns in free-living organisms (more host species at lower latitudes) [78]. However, the richness of zoonotic diseases does not exhibit this pattern: despite wide variation in the global distribution of land mass and species richness, chi-squared tests showed that the numbers of unique zoonoses found across 30-degree longitudinal bands were not significantly different from each other (=17.571, p-value=0.09; similar analyses across 30-degree bands of latitude and longitude showed expected geographical patterns of higher richness of hosts and zoonoses in the tropics, and in the longitudinal bands capturing greater land mass, Table S1). Compared to zoonotic mammal hosts, zoonoses are distributed more evenly worldwide (Figure 4) suggesting that, compared to wildlife-specific pathogens whose ranges are delimited by ecological interactions regulating their host and vector populations [42,79–81], the distributions of zoonoses are not as tightly coupled to the distribution of their host species. Zoonoses are not as broadly distributed as human-specific diseases [34], but are possibly more labile than wildlife diseases due to the mobility and range expansion of human populations [34,42,82] while adhering to biogeographic grouping patterns reflecting ecological barriers to animal host species establishment and dispersal [42].
Figure 4. Overlapping geographic ranges of zoonotic diseases carried by wild terrestrial mammal host species from 27 orders.
Though major hotspots of mammal hosts occur in the New and Old World tropics (South America and Eastern Africa, particularly; Figure 1), there are more zoonoses concentrated in northern latitudes, Eastern Africa, and Southeast Asia (Figure 4). This is opposite to the patterns depicted for zoonotic hosts in the tropics, where host richness is expected to match global patterns of high species richness that increase the frequency of consumer-resource interactions overall, including parasitic interactions [83]. In addition to the hotspots of human emerging diseases observed in the tropics and Europe, Figure 4 draws attention to the global subarctic. While this region has lower zoonotic host and species diversity compared to other biogeographic regions, mammals found in the subarctic zone harbor more zoonoses than hosts from other regions. In general, mammal species are predominantly constrained by their abiotic environments, but pathogens contend primarily with the biotic environment presented by their hosts (e.g., [84]). Thus, one possible explanation of this pattern is that while there are fewer host species in the global subarctic compared to other regions of the world, the pathogens causing zoonoses in these species are saturating all available niches, leading to greater evenness (i.e., a larger fraction of species are zoonotic hosts; Figure S3) and higher prevalence (i.e., a larger fraction of the host population that is infected). These broad and opposing patterns call for future studies to investigate more directly what is driving geographic patterns of zoonotic host richness vs. richness of zoonotic diseases, an endeavor which subsumes direct confrontation of pervasive issues of sampling bias (for wildlife species) and reporting bias (for human disease cases) inherent to data on pathogens and disease. Such studies will also benefit from the large literature investigating biogeography of parasitic organisms writ large [85].
5. What is the zoonotic risk posed by different pathogen types?
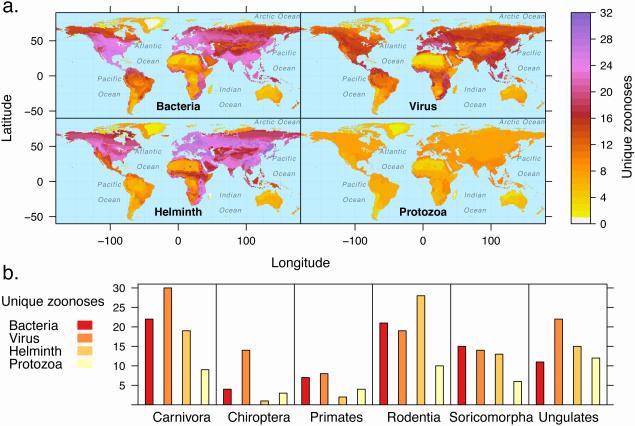
There are more zoonoses caused by bacteria than any other pathogen type, followed by viruses, helminths, protozoa, and fungi (Figure 5, Figure S4). While zoonotic pathogens are geographically widespread (Figure 5a), they are unevenly distributed across mammal groups (Figure 5b), with carnivores carrying the greatest number of viral and bacterial zoonoses, rodents carrying the most zoonotic helminths, and ungulates carrying more zoonotic protozoa compared to other mammal groups (Figure 5b). Discerning the human disease risk posed by various pathogen types will require cross-referencing these broad patterns with zoonotic host patterns (Figure 3, Key Figure). For example, based purely on frequencies (i.e., excluding factors influencing contact rates with humans, etc.), the zoonotic virus hotspot in Europe (Figure 5a) could be driven by the high diversity of rodent hosts (Figure 3d). Mapping zoonoses by pathogen type and mammal clade may better characterize the host and pathogen community contributing to broad patterns. Human disease risk posed by different pathogen types will also depend on their host breadth. In response to the emergence of prominent viral zoonoses in humans (e.g., MERS, SARS) and evidence of frequent host switching [86,87], recent studies have aimed to understand the zoonotic potential of viruses, particularly those arising from bats [86,88]. In addition to characterizing the propensity for host switching among particular host clades and pathogen types, comparative studies of host competence will inform the relative contributions of host species as pathogen sinks or conduits for further transmission.
Figure 5. Zoonoses caused by the four major pathogen types are globally distributed, with notable hotspots for bacteria and helminths in North America and Eurasia.
Richness patterns are depicted by pathogen type in descending order: bacteria, viruses, helminths, protozoa. B) A histogram showing the number of unique zoonoses caused by each pathogen types in the six most species rich mammal groups: the carnivores, bats (Chiroptera), primates, rodents, shrews and moles (Soricomorpha), and the hoofed mammals (Ungulates, which combine the orders Perissodactyla and Artiodactyla and exclude domesticated species).
Concluding Remarks and Future Perspectives
Tallying and mapping zoonotic mammal hosts is an important step to assessing which mammal groups and pathogen types hold the greatest potential to originate new human diseases. Ultimately, there are many factors describing hosts, pathogens, and environments that combine to determine the risk of zoonotic disease emergence from mammal species. Understanding the current geographic and taxonomic distributions of zoonotic mammalian hosts offers an important baseline of empirical data against which observed patterns of human disease can be compared to improve first-order predictions of human disease risk posed by wild mammals. Here, we have summarized the global distributions of mammal hosts and the zoonoses they carry, and discuss them in context of existing research on human zoonoses. These hotspots provide only a partial view of zoonotic disease risk in humans and highlight many outstanding questions for future research (see Outstanding Questions). Moving forward, studies comparing hotspots of mammal hosts or particular pathogen types against the spatial landscape of epidemiological metrics (e.g., prevalence, incidence, frequency of disease events in human populations) and extrinsic drivers (e.g., environmental, sociopolitical, economic) will continue to triangulate those areas where high realized risk of human disease coincides with high zoonotic potential. Such comparisons may also reveal zoonotic coldspots that offer important points of contrast to identifying sources and triggers of unrealized disease risk.
The diversity of zoonotic pathogens and their wild reservoirs may erroneously be considered synonymous with human disease risk, rather than a measure of underlying zoonotic potential, which is itself mediated by many additional interactions in ecological communities (e.g., [54,89– 91]. We argue that human zoonotic disease risk in a geographical region relies on both zoonotic potential and the factors that drive zoonotic potential to be realized as human infection and disease. Many case studies now concur that these drivers can be intrinsic to the hosts [23,72], or result from changing human ecology [5,48,92]. Future studies that partition and quantify the components of zoonotic transmission and the transition of infection to human disease will offer much-needed inroads to assessing the most effective approaches to prevent, manage, or intervene in wild animal systems to achieve the broader goal of mitigating zoonotic disease burden worldwide.
Supplementary Material
Outstanding Questions.
Where will future zoonoses come from? Assessing this question will require the generation of baseline hypotheses about the distribution of animal reservoirs and their pathogens from which future diseases will emerge.
What is the underlying zoonotic potential of regions, where unrealized risk of future disease is particularly high? Analyses to formally quantify the underlying zoonotic potential of a region, a host group, or a pathogen type will be necessary to compare unrealized disease risk across the landscape. Quantifying the components of unrealized zoonotic disease risk will also be critical for developing a predictive approach to managing human and animal populations to minimize or prevent outbreaks.
Are there intrinsic differences in permissiveness and transmissibility between reservoir species or among ecological communities? For example, is a zoonosis more virulent if it arises from a particular animal group (e.g., bats)? Are future zoonoses more likely to arise from regions that have more zoonoses (northerly latitudes) or more reservoir species (the tropics)?
What is driving geographic patterns of zoonotic host richness vs. richness of zoonotic diseases?
Where does high zoonotic potential align with extrinsic pressures that trigger human disease events, and intrinsic features among hosts and pathogens that facilitate pathogen spillover to humans?
What is the zoonotic risk posed by different pathogen types and transmission modes?
Are there comparative patterns of host breadth across zoonoses or across biogeographic regions?
What are the implications of climate warming for zoonosis patterns in northerly latitudes?
Trends Box.
- Predicting zoonotic disease events remains a prominent scientific challenge.
- In response to increasing frequency of emerging infectious disease events caused by animal-borne (zoonotic) pathogens, recent advances assess the biogeographic patterns of human infectious diseases.
- A disproportionate representation of mammal-borne zoonoses among emerging human disease has sparked research emphasis on mammal reservoirs, as improved understanding of mammal host distributions may lead to improved predictions of future hotspots for zoonotic disease emergence.
- In addition to spatial distributions of animal hosts and human disease, the concept of “disease risk” is a topic of intense analysis, and has been quantified on the basis of hindsight where regions undergoing frequent or intense human disease events are categorized as possessing numerous factors that interact to increase disease risk.
Acknowledgements
Authors thank P. Feinberg, E. O’Dea, and M. Gillespie for assistance with data cleaning, figure generation, and manuscript review. Research reported here was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 5F32GM087811 and U01GM110744. JMD acknowledges funding support from the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health. This work benefited from participation in meetings of a Research Coordination Network on Macroecology of Infectious Diseases (NSF DEB 1316223).
Glossary
- Competence
the degree to which a host can successfully transmit a pathogen to its vector
- Disease hotspots
regions where infectious diseases should increase in incidence or geographic range, or regions most likely to generate novel disease events
- Disease event
a general term referring to a collection of human disease cases, including the emergence of novel zoonoses or resurgence of known zoonoses over any temporal or spatial extent
- Emerging infectious disease (EID)
any infectious disease that is increasing in incidence or geographic range
- Host breadth
the range of host species that a given pathogen is able to successfully infect, also commonly referred to as the host range of a pathogen or parasite
- Outbreak
defined by a group of epidemiologically connected disease cases that exceed historical incidence; generally used in this article to refer to the emergence of a zoonotic disease over a relatively short period of time
- Realized disease risk
the component of the overall risk of zoonotic disease in humans that is apparent from current and ongoing disease in human populations
- Reporting bias
bias that arises from infection or disease cases being reported more frequently due to greater resource allocation; thus reporting bias can be high in rich countries
- Richness
the number of unique species within a particular geographic area; richness is a count-based metric for quantifying diversity, which contrasts with other metrics, such as functional trait diversity (the different types of traits represented within a geographic area) or genetic diversity
- Spillover
occurs when a pathogen or parasite successfully infects a human host
- Study bias
bias that can arise when particular organisms (e.g., hosts and pathogens) or geographic areas are better studied than others
- Unrealized disease risk
the component of overall zoonotic disease risk in humans that is not yet apparent, and is comprised by underlying zoonotic potential (see below); intrinsic features of the host, pathogen, and vector; and extrinsic pressures that affect human contact
- Zoonosis or Zoonotic disease (plural, Zoonoses)
a disease that is caused by an infectious pathogen or parasite that originates in (or is maintained in the wild by) one or more non-human hosts, but can be transmitted to and cause disease in humans.
- Zoonotic coldspots
areas of unrealized disease risk where zoonotic host or pathogen richness is higher than observed human disease
- Zoonotic host
any species confirmed to carry one or more zoonotic diseases either as natural reservoirs or as hosts susceptible to some degree of infection; in this article, hosts are limited to mammals
- Zoonotic potential
the dynamic potential of a particular location or animal clade to contribute zoonotic infection to human hosts; conferred by a high underlying diversity of zoonotic hosts or pathogens from which infection may spill over to humans
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Barbara A. Han, Cary Institute of Ecosystem Studies, Box AB Millbrook, NY. 12545.
Andrew M. Kramer, Odum School of Ecology, University of Georgia 140 E. Green St. Athens, GA. 30602 | kramera3@uga.edu
John M. Drake, Odum School of Ecology, University of Georgia 140 E. Green St. Athens, GA. 30602 | jdrake@uga.edu
Bibliography
- 1.Hay SI, et al. Global mapping of infectious disease. Philos. Trans. R. Soc. B Biol. Sci. 2013;368:20120250. doi: 10.1098/rstb.2012.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morse SS, et al. Prediction and prevention of the next pandemic zoonosis. The Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith KF, et al. Global rise in human infectious disease outbreaks. J. R. Soc. Interface. 2014;11:20140950. doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh MG. Mapping the risk of Nipah virus spillover into human populations in South and Southeast Asia. Trans. R. Soc. Trop. Med. Hyg. 2015;109:563–571. doi: 10.1093/trstmh/trv055. [DOI] [PubMed] [Google Scholar]
- 6.Pigott DM, et al. Mapping the zoonotic niche of Ebola virus disease in Africa. eLife. 2014;3 doi: 10.7554/eLife.04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson AT. Mapping Disease Transmission Risk: Enriching Models Using Biogeography and Ecology. 1 Johns Hopkins University Press; 2014. [Google Scholar]
- 8.Chan JFW, et al. Middle East Respiratory Syndrome Coronavirus: Another Zoonotic Betacoronavirus Causing SARS-Like Disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baize S, et al. Emergence of Zaire Ebola Virus Disease in Guinea. N. Engl. J. Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 10.Murphy FA. Emerging zoonoses. Emerg. Infect. Dis. 1998;4:429–435. doi: 10.3201/eid0403.980324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor LH, et al. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruse H, et al. Wildlife as Source of Zoonotic Infections. Emerg. Infect. Dis. 2004;10:2067–2072. doi: 10.3201/eid1012.040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleaveland S, et al. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woolhouse MEJ, Gowtage-Sequeria S. Host Range and Emerging and Reemerging Pathogens. Emerg. Infect. Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson DE, Reeder DM. Mammal Species of the World: A Taxonomic and Geographic Reference. JHU Press; 2005. [Google Scholar]
- 16.Berger SA. GIDEON: a comprehensive Web-based resource for geographic medicine. Int. J. Health Geogr. 2005;4:10. doi: 10.1186/1476-072X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.IUCN IUCN Terrestrial Mammals [Online]. Available: http://www.iucnredlist.org/technical-documents/spatial-data.
- 18.Wolfe ND, et al. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd-Smith JO, et al. Epidemic dynamics at the human-animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiethoelter AK, et al. Global trends in infectious diseases at the wildlife–livestock interface. Proc. Natl. Acad. Sci. 2015;112:9662–9667. doi: 10.1073/pnas.1422741112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones BA, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottdenker NL, et al. Anthropogenic Land Use Change and Infectious Diseases: A Review of the Evidence. Ecohealth. 2014;11:619–632. doi: 10.1007/s10393-014-0941-z. [DOI] [PubMed] [Google Scholar]
- 23.Han BA, et al. Rodent reservoirs of future zoonotic diseases. Proc. Natl. Acad. Sci. 2015;112:7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostfeld RS, et al. Life History and Demographic Drivers of Reservoir Competence for Three Tick-Borne Zoonotic Pathogens. PLoS ONE. 2014;9:e107387. doi: 10.1371/journal.pone.0107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayman DTS. Biannual birth pulses allow filoviruses to persist in bat populations. Proc. R. Soc. Lond. B Biol. Sci. 2015;282:20142591. doi: 10.1098/rspb.2014.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altizer S, et al. Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu. Rev. Ecol. Evol. Syst. 2003;34:517–547. [Google Scholar]
- 27.Dizney L, Dearing MD. Behavioural differences: a link between biodiversity and pathogen transmission. Anim. Behav. 2016;111:341–347. doi: 10.1016/j.anbehav.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gervasi SS, et al. The context of host competence: a role for plasticity in host–parasite dynamics. Trends Parasitol. doi: 10.1016/j.pt.2015.05.002. In Press. DOI: 10.1016/j.pt.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LoGiudice K, et al. The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proc. Natl. Acad. Sci. U. S. A. 2003;100:567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy JL, et al. Intrinsic Factors Affecting Vector Competence of Mosquitoes for Arboviruses. Annu. Rev. Entomol. 1983;28:229–262. doi: 10.1146/annurev.en.28.010183.001305. [DOI] [PubMed] [Google Scholar]
- 31.Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altizer S, et al. Rapid evoluationary dynamics and disease threats to biodiversity. Trends Ecol. Evol. 2003;18:589–596. [Google Scholar]
- 33.Woolhouse MEJ, et al. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 2005;20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith KF, et al. Globalization of human infectious disease. Ecology. 2007;88:1903–19010. doi: 10.1890/06-1052.1. [DOI] [PubMed] [Google Scholar]
- 35.Woolhouse MEJ. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 2002;10:s3–s7. doi: 10.1016/s0966-842x(02)02428-9. [DOI] [PubMed] [Google Scholar]
- 36.Bonds MH, et al. Disease Ecology, Biodiversity, and the Latitudinal Gradient in Income. PLoS Biol. 2012;10:e1001456. doi: 10.1371/journal.pbio.1001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daszak P, et al. Emerging infectious diseases of wildlife--threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 38.Benedict MQ, et al. Spread of The Tiger: Global Risk of Invasion by The Mosquito Aedes albopictus. Vector-Borne Zoonotic Dis. 2007;7:76–85. doi: 10.1089/vbz.2006.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gould EA, Higgs S. Impact of climate change and other factors on emerging arbovirus diseases. Trans. R. Soc. Trop. Med. Hyg. 2009;103:109–121. doi: 10.1016/j.trstmh.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones BA, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. 2013;110:8399–8404. doi: 10.1073/pnas.1208059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patz JA, et al. Disease emergence from global climate and land use change. Med. Clin. North Am. 2008;92:1473–1491. doi: 10.1016/j.mcna.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Murray KA, et al. Global biogeography of human infectious diseases. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1507442112. DOI: 10.1073/pnas.1507442112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhn JH. Filoviruses. [electronic resource] : a compendium of 40 years of epidemiological, clinical, and laboratory studies. Wien; New York: 2008. SpringerWeinNew York, c2008. [PubMed] [Google Scholar]
- 44.Guernier V, et al. Ecology Drives the Worldwide Distribution of Human Diseases. PLoS Biol. 2004;2:e141. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenkins CN, et al. Global patterns of terrestrial vertebrate diversity and conservation. Proc. Natl. Acad. Sci. 2013;110:E2602–E2610. doi: 10.1073/pnas.1302251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFarlane R, et al. Synanthropy of Wild Mammals as a Determinant of Emerging Infectious Diseases in the Asian–Australasian Region. EcoHealth. 2012;9:24–35. doi: 10.1007/s10393-012-0763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karesh WB, et al. Ecology of zoonoses: natural and unnatural histories. The Lancet. 2012;380:1936–1945. doi: 10.1016/S0140-6736(12)61678-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morand S, et al. Biodiversity and Emerging Zoonoses. In: Yamada A, et al., editors. Confronting Emerging Zoonoses. Springer; Japan: 2014. pp. 27–41. [Google Scholar]
- 49.Hopkins ME, Nunn CL. A global gap analysis of infectious agents in wild primates. Divers. Distrib. 2007;13:561–572. [Google Scholar]
- 50.Peterson AT, et al. Mapping transmission risk of Lassa fever in West Africa: the importance of quality control, sampling bias, and error weighting. PloS One. 2014;9:e100711–e100711. doi: 10.1371/journal.pone.0100711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WHO Global Health Expenditure Database. 2014 [Online]. Available: http://apps.who.int/nha/database/World_Map/Index/en. [Accessed: 12-Jan-2016]
- 52.Stuart-Smith RD, et al. Integrating abundance and functional traits reveals new global hotspots of fish diversity. Nature. 2013;501:539–542. doi: 10.1038/nature12529. [DOI] [PubMed] [Google Scholar]
- 53.Guernier V, Guégan J-F. May Rapoport’s Rule Apply to Human Associated Pathogens? EcoHealth. 2010;6:509–521. doi: 10.1007/s10393-010-0290-5. [DOI] [PubMed] [Google Scholar]
- 54.Morand S, et al. Infectious Diseases and Their Outbreaks in Asia-Pacific: Biodiversity and Its Regulation Loss Matter. PLoS ONE. 2014;9:e90032. doi: 10.1371/journal.pone.0090032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris RJ, et al. Antagonistic interaction networks are structured independently of latitude and host guild. Ecol. Lett. 2014;17:340–349. doi: 10.1111/ele.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morand S. (macro-) Evolutionary ecology of parasite diversity: From determinants of parasite species richness to host diversification. Int. J. Parasitol. Parasites Wildl. 2015;4:80–87. doi: 10.1016/j.ijppaw.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roche B, et al. Does host receptivity or host exposure drives dynamics of infectious diseases? The case of West Nile Virus in wild birds. Infect. Genet. Evol. 2015;33:11–19. doi: 10.1016/j.meegid.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Cleaveland S, et al. Diseases of Humans and Their Domestic Mammals: Pathogen Characteristics, Host Range and the Risk of Emergence. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ripple WJ, et al. Status and Ecological Effects of the World’s Largest Carnivores. Science. 2014;343:1241484–1241484. doi: 10.1126/science.1241484. [DOI] [PubMed] [Google Scholar]
- 60.Jones M, et al. Predators with Pouches: The Biology of Carnivorous Marsupials. Csiro Publishing; 2003. [Google Scholar]
- 61.Luis AD, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. R. Soc. B Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krasnov BR, et al. Flea species richness and parameters of host body, host geography and host “milieu.”. J. Anim. Ecol. 2004;73:1121–1128. [Google Scholar]
- 63.Thieltges DW, et al. Host diversity and latitude drive trematode diversity patterns in the European freshwater fauna: Trematode diversity patterns. Glob. Ecol. Biogeogr. 2011;20:675–682. [Google Scholar]
- 64.Harris NC, Dunn RR. Using host associations to predict spatial patterns in the species richness of the parasites of North American carnivores. Ecol. Lett. 2010;13:1411–1418. doi: 10.1111/j.1461-0248.2010.01527.x. [DOI] [PubMed] [Google Scholar]
- 65.Huang S, et al. Phylogenetically related and ecologically similar carnivores harbour similar parasite assemblages. J. Anim. Ecol. 2014;83:671–680. doi: 10.1111/1365-2656.12160. [DOI] [PubMed] [Google Scholar]
- 66.Cleaveland S, et al. Overviews of Pathogen Emergence: Which Pathogens Emerge, When and Why? In: Childs SRSJE, et al., editors. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission. Springer; Berlin Heidelberg: 2007. pp. 85–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morand S, et al. Domesticated animals and human infectious diseases of zoonotic origins: Domestication time matters. Infect. Genet. Evol. 2014;24:76–81. doi: 10.1016/j.meegid.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 68.Brack M. Agents Transmissible from Simians to Man. Springer; Berlin Heidelberg: 1987. [Google Scholar]
- 69.Pedersen AB, Davies TJ. Cross-Species Pathogen Transmission and Disease Emergence in Primates. Ecohealth. 2009;6:496–508. doi: 10.1007/s10393-010-0284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lehman SM, Fleagle JG. Primate Biogeography. Springer; US: 2006. Biogeography and Primates: A Review; pp. 1–58. [Google Scholar]
- 71.Robertson LJ, et al. Impacts of globalisation on foodborne parasites. Trends Parasitol. 2014;30:37–52. doi: 10.1016/j.pt.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Brook CE, Dobson AP. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. doi: 10.1016/j.tim.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plowright RK, et al. Ecological dynamics of emerging bat virus spillover. Proc. Biol. Sci. 2015;282:20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobson AP. What Links Bats to Emerging Infectious Diseases? Science. 2005;310:628–629. doi: 10.1126/science.1120872. [DOI] [PubMed] [Google Scholar]
- 75.Stieger C, et al. Spatial and temporal aspects of urban transmission of Echinococcus multilocularis. Parasitology. 2002;124:631–640. doi: 10.1017/s0031182002001749. [DOI] [PubMed] [Google Scholar]
- 76.Leendertz SAJ, et al. Assessing the Evidence Supporting Fruit Bats as the Primary Reservoirs for Ebola Viruses. EcoHealth. 2015 doi: 10.1007/s10393-015-1053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young CCW, Olival KJ. Optimizing Viral Discovery in Bats. PLOS ONE. 2016;11:e0149237. doi: 10.1371/journal.pone.0149237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lawton JH. Are there general laws in ecology? Oikos. 1999;84:177–192. [Google Scholar]
- 79.Jean K, et al. An equilibrium theory signature in the island biogeography of human parasites and pathogens. Glob. Ecol. Biogeogr. 2015 DOI: 10.1111/geb.12393. [Google Scholar]
- 80.Orrock JL, et al. Biogeographic and Ecological Regulation of Disease: Prevalence of Sin Nombre Virus in Island Mice Is Related to Island Area, Precipitation, and Predator Richness. Am. Nat. 2011;177:691–697. doi: 10.1086/659632. [DOI] [PubMed] [Google Scholar]
- 81.Streicker DG, et al. Host Phylogeny Constrains Cross-Species Emergence and Establishment of Rabies Virus in Bats. Science. 2010;329:676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- 82.Smith KF, Guégan J-F. Changing Geographic Distributions of Human Pathogens. Annu. Rev. Ecol. Evol. Syst. 2010;41:231–250. [Google Scholar]
- 83.Holt RD. The community context of disease emergence: could changes in predation be a key driver? In: Ostfeld RS, et al., editors. Infectious Disease Ecology: Effects of Ecosystems on Disease and of Disease on Ecosystems. Princeton University Press; 2008. pp. 324–346. [Google Scholar]
- 84.Wasik BR, et al. Generalized Selection to Overcome Innate Immunity Selects for Host Breadth in an RNA Virus. Evolution. 2016 doi: 10.1111/evo.12845. DOI: 10.1111/evo.12845. [DOI] [PubMed] [Google Scholar]
- 85.Kamiya T, et al. What determines species richness of parasitic organisms? A meta-analysis across animal, plant and fungal hosts. Biol. Rev. 2014;89:123–134. doi: 10.1111/brv.12046. [DOI] [PubMed] [Google Scholar]
- 86.Drexler JF, et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luis AD, et al. Network analysis of host–virus communities in bats and rodents reveals determinants of cross-species transmission. Ecol. Lett. 2015 doi: 10.1111/ele.12491. DOI: 10.1111/ele.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brierley L, et al. Quantifying Global Drivers of Zoonotic Bat Viruses: A Process-Based Perspective. Am. Nat. 2015 doi: 10.1086/684391. DOI: 10.1086/684391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Civitello DJ, et al. Biodiversity inhibits parasites: Broad evidence for the dilution effect. Proc. Natl. Acad. Sci. 2015;112:8667–8671. doi: 10.1073/pnas.1506279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson PTJ, et al. Why infectious disease research needs community ecology. Science. 2015;349:1259504. doi: 10.1126/science.1259504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keesing F, Ostfeld RS. Ecology. Is biodiversity good for your health? Science. 2015;349:235–236. doi: 10.1126/science.aac7892. [DOI] [PubMed] [Google Scholar]
- 92.Myers SS, et al. Human health impacts of ecosystem alteration. Proc. Natl. Acad. Sci. 2013;110:18753–18760. doi: 10.1073/pnas.1218656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.