Abstract
Conjugation of DNAs to defined locations on a protein surface will offer powerful tools for positioning functional groups and molecules in biological and biomedical studies. However, tagging protein with DNA is challenging in physiological environments, which requires a bioorthogonal approach. Here we report a chemical solution to selectively conjugate DNA aptamer with a protein by protein-aptamer template (PAT)-directed reactions. Since protein-aptamer interactions are bioorthogonal, we exploit PAT as a unique platform for specific DNA-protein cross-linking. We develop a series of modified oligonucleotides for PAT-directed reactions and screen out F-carboxyl as a suitable functionality for selective and site-specific conjugation. The functionality is incorporated into aptamers by our F-carboxyl phosphoramidite with easy synthesis. We also demonstrate the necessity of a linker between the reactive functionality and the aptamer sequences.
Introduction
Proteins are essential biomolecules of organisms and participate in every cellular process.1 To study proteins in their native microenvironments, several methods have been developed to equip proteins with reporter tags in vivo for visualization and isolation.2-3 Recently developed bioorthogonal chemistry provides practical methods to tag proteins with small-molecule probes.4-8
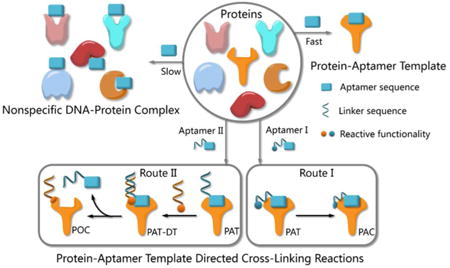
Functionalities of small molecules can be incorporated into oligonucleotides by automated synthesis.9 Furthermore, oligonucleotides with given sequences created by SELEX10-11 can recognize target proteins and provide a unique means for manipulation. Therefore, such DNA-protein conjugates possess many exceptional properties with important applications in biomedical diagnostics, bioanalysis and nanobiology.12-14 Researchers have developed a variety of methods to provide DNA-protein conjugates. In 2012, Famulok et al., for example, employed aptamers modified with reactive functionalities to selectively label target proteins.15 Very recently, Gothelf and coworkers applied DNA-templated synthesis to accomplish site-selective labeling of antibodies, transferrin and metal-binding proteins.16 As our probe molecules of choice, several DNA aptamers that recognize membrane proteins on cancer cells have been developed.17-18 Identification of these membrane proteins is important not only because they are potential biomarkers,19-21 but also because they hold the keys to understanding biological processes in cancer cells.22-23 However, bioorthogonal reactions for aptamer-based biomarker discovery and other applications are challenging when protein substrates cannot be modified in advance. We had prepared modified TD05 aptamers and managed to identify the target proteins on Ramos cells24 by photo-cross-linking,25-27 while this method didn't work for other aptamers selected by our group. Aptamers modified with aldehyde28 were also tried to tag the target membrane proteins. We had tested that the modified aptamers still specifically recognize and bind with the target protein on the cell surface, forming a stable complex named as protein-aptamer template (PAT). PAT-directed cross-linking may provid a platform for bioorthogonal conjugate of aptamers with target proteins. Therefore, we had proposed PAT cross-linking as a one-step bioorthogonal reaction to tag, isolate and identify target membrane protein (Figure 1a) in 2011, and proposed that many factors should be explored for this type of reaction to establish standard and practical cross-linking protocol for aptamer-based biomarker discovery.29 Herein we report a new method for selective conjugation of target proteins at lysine residues through a PAT-directed reaction by aptamers tethered with F-carboxyl group.
Figure 1.
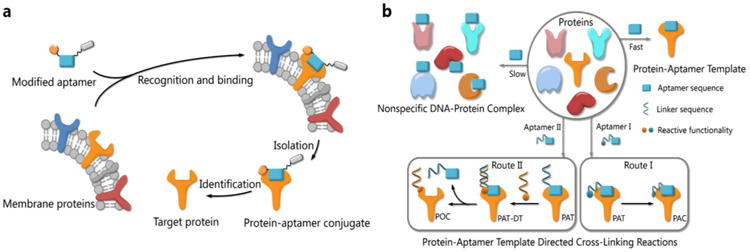
(a) The process of one-step bioorthogonal reaction for biomarker discovery; (b) General approaches to the preparation of protein-aptamer conjugates (PAC, route I) or protein-oligonucleotide conjugates (POC, route II) by PAT-directed reactions.
Results and Discussion
Compared with a DNA template, PAT is more complicated, and it is more difficult to predict the structure; nonetheless, PAT is expected to be a suitable template for directing reactions in a controllable way, as long as the aptamer is well designed. In both physiological environments and buffer solutions, protein-aptamer interactions are bioorthogonal, and aptamers can recognize and bind to their target proteins with high affinity, providing PAT with the requisite orthogonality (Figure 1b). (On the other hand, a nonspecific DNA-protein complex may be formed, albeit more slowly, at low concentration in the same system.) The resultant PAT promotes a cross-linking reaction to give a protein-aptamer conjugate (PAC, route 1, Fig. 1b) when a reactive functionality (aquamarine circle) is incorporated into the aptamer's 3′- or 5′-end via a linker (aquamarine ribbon). PAT may also facilitate the bioorthogonal reaction of the protein with a second oligonucleotide through a PAT-DT complex to give a protein-oligonucleotide conjugate (POC, route II, Figure 1b). Here we report the development of PAT-directed reactions as a practical strategy to selectively modify proteins with aptamers at a specific site. We evolve a series of modified oligonucleotides that react with proteins. By using aptamer incorporated with the α,α-gem-difluoromethyl carboxyl group (F-carboxyl), we successfully label target protein at lysine residues. We demonstrate that both reactive functionality and linker are important factors for PAT-directed reactions.
When a functionalized aptamer binds with its target protein to form a PAT, the aptamer's 3′- or 5′-end may be kept distant from the aptamer's surface in accordance with some crystallographic structures of the protein-aptamer complex.30-32 In addition, the reactive functional group tethered to an aptamer without linker may be kept distant from lysine residues. Therefore, a linker between the reactive functional group and the aptamer sequence is necessary for site-specific conjugation, allowing the functional group to reach out to and cross-link with the site on the protein surface. To verify our hypothesis and explore a proper linker for PAT- directed reactions, we have synthesized a series of aptamers tethered with an aldehyde via different linkers (see Table S1 in Supporting Information for detailed sequences). Aldehyde functionality can be readily incorporated into oligonucleotides by using a RNA cpg33 or commercially available 5-aldehyde-modifier phosphoramidite.34
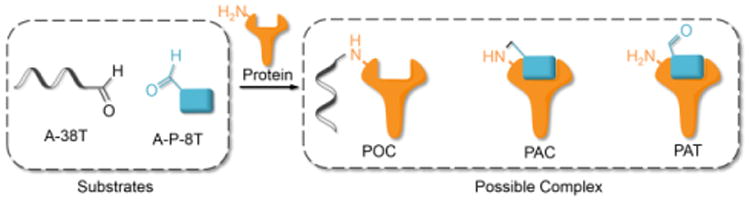
Using RNA-CPG, e.g., U-CPG, a series of thrombin aptamers35 were prepared with a vicinal diol group at the 3′-end. Oxidation of the vicinal diol with NaIO4 led to aldehyde-modified aptamers (Figure 2a). PAT-directed reactions of aldehyde-modified aptamers were initially carried out in 10-20 μL buffer (pH 7) in the presence of NaCNBH3 at 4 °C. The reactions of the aptamers with different linkers were compared, and the results were determined by SDS PAGE gel (Figure 2). Based on the gel results, no reaction was detected between thrombin and aptamer A-1 (with no linker, Figure 2b, entry 2), A-2 (with a PEG linker, Figure 2b, entry 3), or A-3 (with a 3T linker, Figure 2b, entry 4). Fortunately, however, the reaction of aptamer A-4 (with an 8T linker, Figure 2b, entry 5) with thrombin did give the cross-linked product in moderate yield. These results showed the importance of a linker for PAT-directed reactions. To verify if the 8T linker would be applicable to other PAT-directed reactions, another protein aptamer tethered with aldehyde via 8T linker (PDGF-BB aptamer,36 see Table S1 in Supporting Information for detailed sequences) was prepared. The reaction of PDGF-BB with A-5 also gave the cross-linked conjugate in moderate yield, as expected (Figure 2c, entry 2), and the yield was improved when the concentration of aptamer was increased (Figure 2c, entry3).
Figure 2.
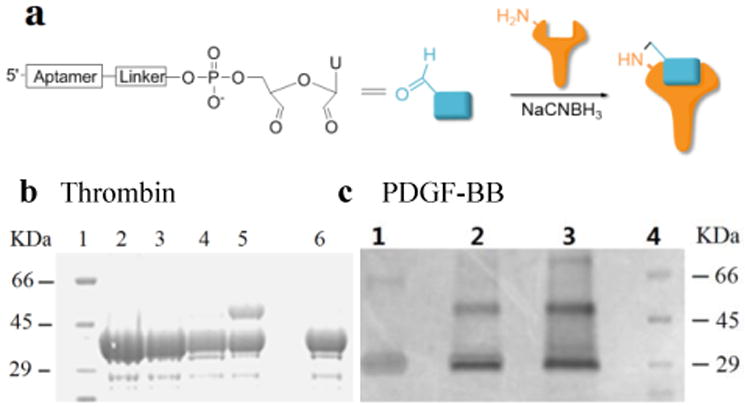
Cross-linking of proteins with aldehyde-modified aptamers. (a) Structure of aldehyde-modified aptamer and its reaction with protein. (b) SDS PAGE analysis of Thrombin reacted with aptamers (5×) with different linkers: Lane 1: molecular weight marker; Lane 2: Thrombin+A-1; Lane 3: Thrombin+A-2; Lane 4: Thrombin+A-3; Lane 5: Thrombin+A-4; Lane 6: Thrombin. (c) SDS PAGE analysis of PDGF-BB reacted with aptamers with 8T linker (5×) at different concentrations: Lane 1: PDGF-BB; Lane 2: PDGF+A-5 (5×); Lane 3: PDGF-BB+A-5 (20×); Lane 4: molecular weight marker.
The interaction of an aptamer with its target protein is highly selective, and high-affinity binding results in the rapid formation of a PAT (Figure 1b). In contrast, nonspecific interactions of aptamers with other proteins may slowly lead to formation of an unstable protein-oligonucleotide complex (POC, Figure 1b) at low percentages. PAT promotes bimolecular reaction by pulling two reactants close to each other in a manner similar to that of an intramolecular reaction. We propose that PAT-directed conjugation is a bioorthogonal reaction based on the hypothesis that protein-aptamer cross-linking proceeds mainly within the template. However, nonspecific conjugation may also proceed effectively to provide a protein-oligonucleotide conjugate (POC, Figure 3) without the promotion by PAT, when the aptamer is modified with a functionality that is highly reactive with proteins. Therefore, the reactivity of modified aptamers is another critical component ensuring the selectivity of PAT-directed conjugation.
Figure 3.
Interactions of aldehyde-modified aptamers with protein.
The aldehyde group represents a highly reactive functionality with protein and has been incorporated into oligonucleotides for the preparation of nonspecific DNA-protein conjugates. Hence, the interactions of aldehyde-modified oligonucleotides with proteins may result in the formation of three different complexes, as shown in Figure 3: protein-oligonucleotide conjugate (POC, nonspecific), protein-aptamer conjugate (PAC, specific), and protein-aptamer template (PAT, specific). The reduction reaction of PAT with NaCNBH3 provides PAC as the product if the linker tethering aldehyde with aptamer is proper (as we demonstrate in Figure 2a).
To explore the specificity of aldehyde-modified aptamers, we compared the reaction of A-5 with the control reaction of A-6 (see Table S1 in Supporting Information for detailed sequences and reaction conditions). As shown in Figure 4, the reaction of A-6 (20×) gave POC in moderate yield (Lane 4, Figure 4), comparable to that of A-5 (5×, Figure 3c, Lane 2). Since nonspecific aldehyde-modified oligonucleotides still react with proteins without the promotion of PAT, the results confirm the hypothesis that the reactivity of modified aptamer is critical for the selectivity of a PAT-directed reaction. Interestingly, this nonspecific reaction can be inhibited by the addition of aptamer A-7 (without aldehyde-modification, Lane 3, Figure 4). We infer that A-7 prevents the nonspecific interaction of A-6 from PDGF-BB and their cross-linking when the protein is binding with A-7. In the presence of A-7, the reaction of A-5 and PDGF-BB still gave the cross-linked PAC at moderate yield (Lane 5, Figure 4) because protein-aptamer binding is a reversible interaction and A-5 competes equally with A-7 to form the template. Not surprisingly, no PAC was observed for the reaction of A-5 with PDGF-BB without the addition of NaCNBH3 (Lane 5, Figure 4). The results indicate that aldehyde-aptamer may still be suitable for bioorthogonal labeling of membrane proteins on cell surfaces because aptamers nonspecifically binding with other proteins can be washed away before the addition of NaCNBH3, eliminating POC formation.
Figure 4.
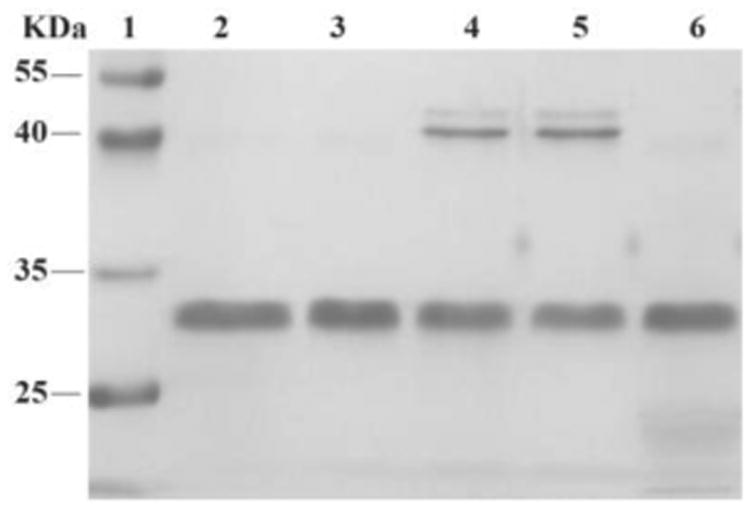
SDS-PAGE analysis of control reactions. Lane 1: molecular weight marker; Lane 2: PDGF-BB; Lane 3: PDGF-BB, A-6, A-7; Lane 4: PDGF-BB, A-6 (20×); Lane 5: PDGF-BB, A-5, A-7 (5×); Lane 6: PDGF-BB, A-5, without addition of NaCNBH3.
On the observation of the partial selectivity of aldehyde-modified aptamer, we had been looking for a proper functional group to realize both bioorthogonal and site-specific modification of proteins for general PAT-directed reactions. A carboxyl group is coupled with an amine group directly to form peptide bonding as a result of enzyme catalysis in the biosynthesis of proteins, but the carboxyl has to be activated for efficient coupling in peptide synthesis.37 The reactivity of the carboxyl group is tunable, and the introduction of an adjacent electron-withdrawing group (EWG) can enhance reactivity with the nucleophilic amine.
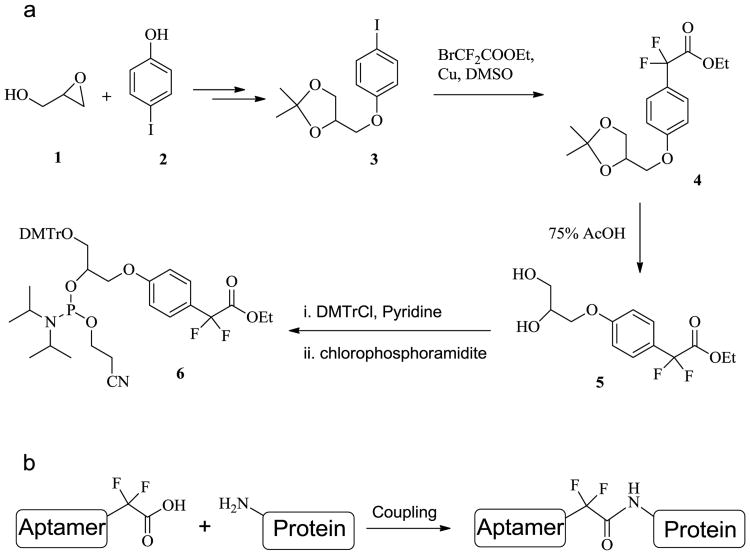
Perfluoroalkyl (F-alkyl) groups are EWGs with chemically inert properties. α,α-gem-Difluoromethyl (F-Carboxyl) is the smallest perfluoroalkyl group in size, and it has been verified that F-carboxyl is a moderate amine-reactive electrophile.38 We extrapolated that nonspecific reactions of F-carboxyl-functionalized aptamers with the amine of lysine residues in aqueous solution might be negligible, but that intramolecular reactions within the PAT could be efficient in view of the fact that the F-carboxyl group is assembled close to the amine group. Thus, the cross-linking reaction of an F-carboxyl aptamer with target protein could efficiently give site-specific PAC with selectivity. Moreover, the linker between the aptamer and protein is an amide bond (Figure 5b), which can be enzymatically dissociated. Therefore we have designed and synthesized an F-carboxyl phosphoramidite for convenient preparation of F-carboxyl aptamer.
Figure 5.
(a) The synthetic route to F-carboxyl phosphoramidite 6. (b) The coupling of F-alkyl aptamer with protein.
As shown in Figure 5a, the synthesis of F-alkyl carboxyl phosphoramidite 6 starts with the coupling of commercially available glycidol 1 and 4-iodophenol 2.39 Cross-coupling of ethyl bromodifluoroacetate with acetonide-protected precursor 3 provides α,α-gem-difluoromethyl derivative 4.40 Removal of the acetonide group gives diol 5 in high yield, where two hydroxyl groups are protected by Dmitri and the phosphoramidite group, respectively,41 giving phosphoramidite 6 (see Supporting Information for details).
From phosphoramidite 6, we prepared F-carboxyl aptamer F-1 and oligonucleotide F-2 as control (see Table S1 in Supporting Information for detailed sequences). PAT-directed reaction of F-carboxyl aptamer F-1 with PDGF-BB gave a cross-linked PAC product with a yield (Lane 3, Figure 6) comparable to that of aldehyde-modified aptamers. While nonspecific reaction of F-carboxyl oligonucleotide F-2 was carried out in similar conditions, no cross-linked product was detected by SDS-PAGE (Lane 4, Figure 6). The PAC band in Lane 3 was cut and sent for mass analysis, and the results confirmed the structure of the conjugate.
Figure 6.
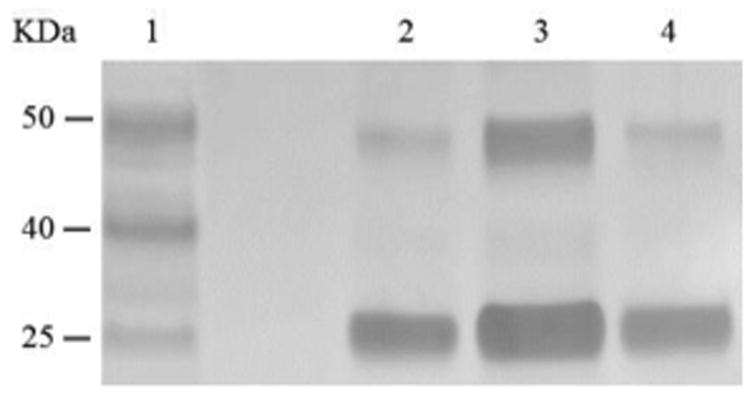
SDS-PAGE analysis of PDGF-BB reacted with F-carboxyl oligonucleotides. Lane 1: molecular weight marker; Lane 2: PDGF-BB; Lane 3: PDGF-BB, F-1; Lane 4: PDGF-BB, F-2.
When reactive functionalities are tethered to the 3′- or 5′-end of the cell-aptamer sequence, they may affect binding affinity. Accordingly, we evaluated the specific recognition and binding ability of biotinylated aptamers KDED2a-3 and KCHA10a42 modified with an aldehyde at the 3′-end (Biotin-KDED2a-3-Aldehyde and Biotin-KCHA10-Aldehyde; see Table S1 in Supporting Information for detailed sequences). For visualization, each complex was combined with a streptavidin-PE-Cy5.5 (Phycoerythrin) dye for the cell binding assay. In this assay, flow cytometry was used to monitor the fluorescence intensities of cells, with aptamer Biotin-KDED2a-3 and Biotin-KCHA10a as positive control and random sequences (Biotin Library and Biotin-Library-aldehyde) as negative control. As shown in Figure 7, the fluorescence intensities of cells incubated with aldehyde-modified control (Biotin-Library-aldehyde) are as weak as those bound with Biotin Library, excluding the possibility of nonspecific binding caused by aldehyde. The fluorescence intensities of DLD-1 cells and HCT 116 cells bound with respective aldehyde-modified aptamers were comparable to those bound with positive controls, indicating that these modified aptamers still bound specifically with their target proteins.
Figure 7.
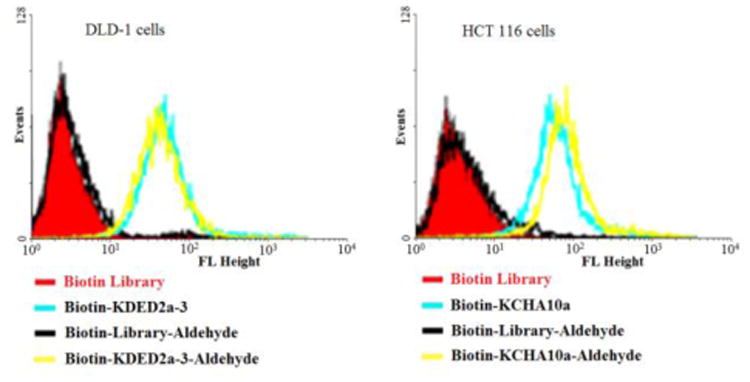
Aptamers modified with aldehyde at 3′ and biotin at 5′ could still bind specifically to the target cells.
Similarly, retention of selectivity and binding affinity were also observed for F-carboxyl aptamer (F-3). The binding affinity assay indicates the formation of PAT.
Summary
In conclusion, we have developed a new method for selective conjugation of target proteins at lysine residues through a PAT-directed reaction. This was achieved by an aptamer tethered with F-carboxyl group via an 8T linker. PAT promotes the cross-linking of protein and aptamer by pulling together the F-carboxyl group and the amine of a lysine residue. We evolved a series of oligonucleotides tethered with amine-reactive functionalities via different linkers. Comparing the reactions of aldehyde-modified aptamers with different linkers, we optimized 8T as a proper linker between the aptamer sequence and the reactive functionality. On the observation of partial selectivity of the aldehyde-modified aptamer, we designed and developed a novel phosphoramidite to incorporate the F-carboxyl group as a proper functional group for PAT-directed reactions. We found that nonspecific reaction of F-carboxyl oligonucleotide with protein is negligible, while, on the other hand, PAT-directed reaction of F-carboxyl aptamer provides the cross-linked product efficiently. Flow cytometry studies confirmed that aptamers retain their specific recognition and binding abilities, even when they are modified with reactive functionalities. It has been illustrated that PAT is a suitable template to direct reactions in a controllable way. We believe that this method will be especially valuable for cell membrane protein imaging and isolation and other related biological studies.
Experimental details
All experimental details are written as a separate section in the SI materials. These include the synthetic procedures and testing protocols etc. They are written in detail and complete.
Supplementary Material
Acknowledgments
This work is supported by NIH grants (GM079359, CA133086), the National Key Scientific Program of China (2011CB911000), the Foundation for Innovative Research Groups of the NSFC (21221003), the China National Instrumentation Program (2011YQ03012412), and the Project Sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry. The authors thank Dr. Kwame Sefah at University of Florida, and Feng Zhang and Prof. Xingang Zhang at Shanghai Institute of Organic Chemistry for the help with synthesis of compounds.
Footnotes
Notes: The authors declare no competing financial interests.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Contributor Information
Ruowen Wang, Email: ruowenwang@ufl.edu.
Weihong Tan, Email: tan@chem.ufl.edu.
References
- 1.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 3.Lippincott-Schwartz J, Patterson GH. Science. 2003;300:87–97. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 4.Prescher JA, Bertozzi CR. Nat Chemical Biology. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 5.Scheck RA, Schepartz A. Acc Chem Res. 2011;44:654–665. doi: 10.1021/ar2001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sletten EM, Bertozzi CR. Acc Chem Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Best MD, Rowland MM, Bostic HE. Acc Chem Res. 2011;44:686–698. doi: 10.1021/ar200060y. [DOI] [PubMed] [Google Scholar]
- 8.Hao Z, Hong S, Chen X, Chen PR. Acc Chem Res. 2011;44:742–751. doi: 10.1021/ar200067r. [DOI] [PubMed] [Google Scholar]
- 9.Verma S, Echstein F. Annu Rev Biochem. 1998;67:99–134. doi: 10.1146/annurev.biochem.67.1.99. [DOI] [PubMed] [Google Scholar]
- 10.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Sano T, Smith CL, Cantor CR. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 13.Vestweber D, Schatz G. Nature. 1989;338:170–172. doi: 10.1038/338170a0. [DOI] [PubMed] [Google Scholar]
- 14.Niemeyer CM. Angew Chem Int Ed. 2010;49:1200–1216. doi: 10.1002/anie.200904930. [DOI] [PubMed] [Google Scholar]
- 15.Vinkenborg JL, Mayer G, Famulok M. Angew Chem Int Ed. 2012;51:9176–9180. doi: 10.1002/anie.201204174. [DOI] [PubMed] [Google Scholar]
- 16.Rosen CB, Kodal ALB, Nielsen JS, Schaffert DH, Scavenius C, Okholm AH, Voigt NV, Enghild JJ, Kjems J, Torring T, Gothelf KV. Nature Chem. 2014;6:804–809. doi: 10.1038/nchem.2003. [DOI] [PubMed] [Google Scholar]
- 17.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sefah K, Yang Z, Bradley KM, Hoshika S, Jimenez E, Zhang L, Zhu G, Shanker S, Yu F, Turek D, Tan W. Proc Natl Acad Sci USA. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berezovski MV, Lechmann M, Mucheev MU, Mak TW, Sergey NK. J Am Chem Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 20.Shangguan D, Cao Z, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan W. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simaeys DV, Turek D, Champanhac C, Vaizer J, Sefah K, Zhen J, Sutphen R, Tan W. Anal Chem. 2014;86:4521–4527. doi: 10.1021/ac500466x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabindran S, Ross DD, Doyle LA, Yang W, Greenberger LM. Cancer Res. 2000;60:47–50. [PubMed] [Google Scholar]
- 23.Varsano S, Rashkovsky L, Shapiro H, Ophir D, Mark-Bentankur T. Clin Exp Immunol. 1998;113:173–182. doi: 10.1046/j.1365-2249.1998.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallikaratchy P, Tang Z, Kwame S, Meng L, Shangguan D, Tan W. Mol Cell Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Golden MC, Resing KA, Collins BD, Willis MC, Koch TA. Protein Sci. 1999;8:2806–2812. doi: 10.1110/ps.8.12.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golden MC, Collins BD, Willis MC, Koch TH. J Biotechnology. 2000;81:167–178. doi: 10.1016/s0168-1656(00)00290-x. [DOI] [PubMed] [Google Scholar]
- 27.Smith D, Collins BD, Hell J, Koch TH. Mol Cell Proteomics. 2003;2:11–18. doi: 10.1074/mcp.m200059-mcp200. [DOI] [PubMed] [Google Scholar]
- 28.Eaton BE, Gold L, Hiche BJ, Janjic N, Jucker FM, Sebesta DP, Tarasow TM, Willis MC, Zichi DA. Bioorg Med Chem. 1997;5:1087–1096. doi: 10.1016/s0968-0896(97)00044-8. [DOI] [PubMed] [Google Scholar]
- 29.Wang R, Sefah K, Simaeys DV, Xiong X, You M, Gulbakan B, Williams KR, Tan W. Protein-aptamer template cross-linking (PATCL): a bioorthogonal reaction for biomarker discovery. Pittcon Conference and Expo; Atlanta, Georgia, USA. 2011. [Google Scholar]
- 30.Huang R, Fremont DH, Diener JL, Schaub RG, Sadler JE. Structure. 2009;17:1476–1484. doi: 10.1016/j.str.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, III, Ghosh G. Proc Natl Acad Sci USA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chayen NE, Saridakis E. Nature Methods. 2008;5:147–153. doi: 10.1038/nmeth.f.203. [DOI] [PubMed] [Google Scholar]
- 33.Misra HK, Khan NN, Agrawal S, Wright GE. Nucl Acids Res. 1992;20:4547–4551. doi: 10.1093/nar/20.17.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podyminogin MA, Lukhtanov EA, Reed MW. Nucl Acids Res. 2001;29:5090–5098. doi: 10.1093/nar/29.24.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 36.Green LS, Jellinek D, Jenison R, Ostman A, Heldin CH, Janjic N. Biochemistry. 1996;35:14413–14424. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 37.Merrifield RB. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 38.Middleton WJ, Bingham EM. J Org Chem. 1980;45:2883–2887. [Google Scholar]
- 39.Bredikhina ZA, Savelev DV, Bredikhin AA. Russian J Org Chem. 2002;38:213–219. [Google Scholar]
- 40.Taguchi T, Kitagawa O, Morikawa T, Nishiwaki T, Uehara H, Endo H, Kobayashi Y. Tetrahedron Lett. 1986;50:6103–6106. [Google Scholar]
- 41.Wang R, Zhu G, Mei L, Xie Y, Ma H, Ye M, Qing FL, Tan W. J Am Chem Soc. 2014;136:2731–2734. doi: 10.1021/ja4117395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sefah K, Meng L, Lopez-Colon D, Jimenez E, Liu C, Tan W. PLoS One. 2010;5:e14269. doi: 10.1371/journal.pone.0014269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.