Abstract
INTRODUCTION
Reporting and sharing pharmacogenetic test results across clinical laboratories and electronic health records is a crucial step toward the implementation of clinical pharmacogenetics, but allele function and phenotype terms are not standardized. Our goal was to develop terms that can be broadly applied to characterize pharmacogenetic allele function and inferred phenotypes.
MATERIALS AND METHODS
Terms currently used by genetic testing laboratories and in the literature were identified. The Clinical Pharmacogenetics Implementation Consortium (CPIC) used the Delphi method to obtain consensus and agree on uniform terms among pharmacogenetic experts.
RESULTS
Experts with diverse involvement in at least one area of pharmacogenetics (clinicians, researchers, genetic testing laboratorians, pharmacogenetics implementers, and clinical informaticians; n=58) participated. After completion of five surveys, consensus (>70%) was reached with 90% of experts agreeing to the final sets of pharmacogenetic terms.
DISCUSSION
The proposed standardized pharmacogenetic terms will improve the understanding and interpretation of pharmacogenetic tests and reduce confusion by maintaining consistent nomenclature. These standard terms can also facilitate pharmacogenetic data sharing across diverse electronic health care record systems with clinical decision support.
Keywords: Pharmacogenetics, Pharmacogenomics, Informatics, CPIC, Nomenclature, LOINC, Logical Observation Identifiers Names and Codes, Terminology, Standard
INTRODUCTION
Many different terms are used to describe a variant allele’s impact on enzyme function and the corresponding inferred phenotypic interpretation of a clinical pharmacogenetic test result. For example, a genetic testing laboratory report could interpret a TPMT *3A allele as leading to “low function,” “low activity,” “null allele,” “no activity” or “undetectable activity.” Moreover, a laboratory might assign a phenotype designation to an individual carrying two non-functional TPMT alleles as being “TPMT homozygous deficient” while another laboratory might use the term “TPMT low activity.” These same laboratories could also use different terminology to describe a similar phenotype for a different gene (e.g., an individual carrying two non-functional DPYD alleles might be described as “DPYD defective”; see Supplemental Tables S1-S2). As a result, the use of inconsistent terms can be confusing to clinicians, laboratory staff and patients. While the actual phenotypes are the same in the TPMT and DPYD examples above (i.e., no function), the terms describing these phenotypes have differed among laboratories and likely have led to confusion in the subsequent interpretation.
The lack of standard vocabularies describing pharmacogenetic results also interferes with the exchange of structured interpretations between laboratories, institutions employing electronic health records (EHRs), and patients’ personal health records. The impact on interoperability may significantly impede the portability of results throughout a patient’s lifetime.1-3 Recently, a joint guideline was developed by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) that standardized the interpretation terms for describing the clinical significance of variants detected in Mendelian disease genes.4 ClinGen has utilized these terms to enable comparison of interpretations from clinical laboratories to identify and potentially resolve differences in variant interpretation,5 a critical step in improving the uniformity of patient care based on genetic information.
The Clinical Pharmacogenetics Implementation Consortium (CPIC) was formed in 2009 as a shared project between PharmGKB (https://www.pharmgkb.org) and the Pharmacogenomics Research Network (PGRN) (http://www.pgrn.org/). CPIC provides clinical guidelines that enable the translation of pharmacogenetic laboratory test results into actionable prescribing decisions for specific drugs,6 which to date has produced 17 clinical guidelines (https://cpicpgx.org/genes-drugs/). Currently, the terms used in CPIC guidelines to describe allele function and phenotype reflect community usage for each gene, and therefore are not standardized across CPIC guidelines (Supplemental Table S3). Ideally, phenotype terms should be easily interpretable by clinicians with basic pharmacogenetic training, and where possible, should be consistent across genes encoding proteins with similar functions (e.g., the use of the term “poor metabolizer” could describe an individual carrying two non-functional alleles for any drug-metabolizing enzyme).
To maximize the utility of pharmacogenetic test results and to facilitate more uniform implementation of CPIC guidelines, it is essential to standardize these terms.7 To achieve this goal, particularly for purposes of clinical pharmacogenetic test reporting, CPIC initiated a project to identify terms that could be used consistently across pharmacogenes by developing consensus among pharmacogenetics experts. A modified Delphi method was employed, which is a structured approach to establish consensus through iterative surveys of an expert panel. Where possible, the goal was to agree upon uniform terms that could apply across pharmacogenes to characterize 1) allele functional status, and 2) inferred phenotypes based on the combined impact of both alleles (i.e., diplotypes).
MATERIALS AND METHODS
The Delphi survey technique is an established approach for seeking expert consensus on a given topic.8-10 The method uses a series of repeated structured questionnaires, or “rounds.” The rounds are usually anonymous and provide written, systematic refinement of expert opinion, where feedback of group opinion is provided after each round.11 Delphi survey technique guidelines proposed by Hasson et al. were consulted in the design of the project.12 The St. Jude Children’s Research Hospital’s Institutional Review Board (IRB) determined that this project does not meet the definition of research and was exempt for IRB purview.
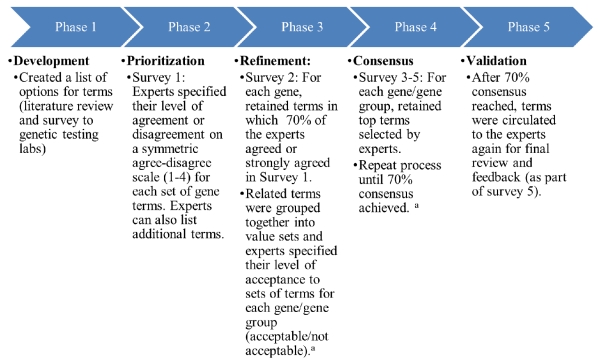
For the Delphi method used (Figure 1), CPIC solicited pharmacogenetic experts by email invitation to members of CPIC, PGRN, pharmacogenetic-related working groups for the Clinical Genome Resource (ClinGen; https://www.clinicalgenome.org), Institute of Medicine (IOM) DIGITizE Action Collaborative (http://iom.nationalacademies.org/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/EHR.aspx), Centers for Disease Control and Prevention PGx nomenclature workgroup,13 Global Alliance for Genomics and Health (GA4GH; http://ga4gh.org), ACMG (https://www.acmg.net), Electronic Medical Records and Genomics (eMERGE; https://emerge.mc.vanderbilt.edu/), the CHAMP online resource for AMP members (http://champ.amp.org), and the College of American Pathologists (CAP). In addition, experts not included in the above groups were solicited by posting a description of the project on the PharmGKB website. All individuals who volunteered were included in survey 1
Figure 1.
Modified Delphi process
aResults from each prior survey were made available to the experts.
Individuals were invited to participate in a series of surveys using an internet-based survey tool (SurveyMonkey Inc, Palo Alto, CA; http://www.surveymonkey.com), supplemented with multiple live webinars that were used to explain the survey and solicit feedback. The webinars were designed to facilitate understanding of the survey to encourage completion; however, towards the end of the process an additional webinar was used to assist in developing consensus. Each survey also included questions regarding the expert’s workplace setting and degree of pharmacogenetic expertise (i.e., role in clinical pharmacogenetics, time devoted to pharmacogenetics). Responses were included in the analysis if the respondent provided their name and contact information, which were necessary to enable follow up with the respondent for the subsequent round (trainees were not excluded). Responses were tabulated as numeric counts and frequencies for each phase to determine whether consensus was reached. Analyses were also performed to determine if there were differences in responses based on the expert’s role in clinical pharmacogenetics. These analyses tested clinician versus non-clinician responses using Chi squared tests with an alpha of 0.05 to ensure the final set of terms would be likely to be adopted by clinicians as well as laboratory based researchers. All analyses were conducted in R version 3.0.1 (R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org).
The goal of this project was to standardize terms used to characterize: 1) allele functional status (i.e. allele descriptive terms), and 2) inferred phenotypes based on the combined impact of both alleles (i.e., diplotypes). The terms used in the initial survey were identified by querying genetic testing laboratories and reviewing literature for currently used terms for CPIC Level A genes (https://cpicpgx.org/genes-drugs/). This was informed by a literature review of references in the CPIC guidelines’ evidence tables and the terms used in these papers to describe allele function and clinical phenotypes for genes with current CPIC guidelines (i.e., CYP2D6, CYP2C19, CYP3A5, CYP2C9, TPMT, DPYD, HLA-B, UGT1A1, SLCO1B1, and VKORC1) (Supplemental Figures S1-S4). We also queried genetic testing laboratories listed at https://www.genetests.org/laboratories/ and translational software companies and created a list of terms currently being used in laboratory reports.
For the first two survey rounds (survey 1 and survey 2), terms that were found acceptable by at least 70% of the experts were retained for use in the next round. To improve semantic consistency, terms that were retained after survey 1 were assembled into value sets, which together described the range of possible descriptors of alleles or phenotypes. These value sets were evaluated in surveys 2 through 4 and the top value sets were retained until 70% consensus was reached. For survey 1 and 2, genes that encode enzymes with similar metabolic function were combined where appropriate (e.g., DPYD and TPMT were combined as were all the CYP enzymes excluding CYP3A5) and experts were given the opportunity to suggest alternative terms. In survey 1, experts were also asked the number of categories of function/phenotype they felt were needed (e.g., three major categories for TPMT - high/normal, medium/some, no activity - versus five major categories for CYP enzymes). To promote consensus, a summary of comments from previous surveys was provided and experts were asked to read the comments prior to answering the questions (https://cpicpgx.org/resources/term-standardization/). These comments were emphasized during the webinars to promote thoughtful discussion. Experts also had access to the full survey results. Of note, experts from surveys 1 and 2 commented in the survey and during webinar discussions that the standardized terms should be consistent across all pharmacogenes if possible. Based on this feedback and feedback from CPIC members, three categories of value sets were proposed and grouped together in survey 3: 1) drug metabolizing enzymes (all CYP enzymes, UGT1A1, DPYD, and TPMT), 2) drug transporters (e.g., SLCO1B1) and non-drug metabolizing enzymes (e.g., VKORC1), and 3) high-risk genotypes (e.g., HLA-B). These groupings were used for the remainder of the surveys. Because consensus was not reached after survey 4, experts were invited to a conference call to discuss and recommend final terms, which included weighing the potential disruptive impact of adopting a new term for clinical laboratories versus any anticipated benefit of adopting a new term. These recommended terms were included in survey 5.
While there is not a universal definition of consensus for the Delphi method, 70% has been recommended and it was considered a reasonable threshold given our diverse group of experts.14,15 Several new terms were added to survey 3 based on the feedback from rounds 1 and 2; these terms were built from existing terms and were included to improve semantic uniformity within a value set (Supplemental Figures S1-S4). The final survey (survey 5) measured the level of acceptance of the final sets of terms. Results from each round were posted on PharmGKB (https://cpicpgx.org/resources/term-standardization/) and were available to respondents throughout the process.
RESULTS
Expert Panel Composition
A total of 222 individuals and approximately 2,000 subscribers to the CHAMP discussion board of AMP were invited to participate in the surveys, and 58 completed Survey 1, 54 completed Survey 2, 46 completed survey 46, and 36 completed survey 5. The response group represented diverse involvement in at least one area of pharmacogenetics: 43% identified as clinicians, 67% as pharmacogenetics researchers, 19% as genetic testing laboratory staff, 43% as pharmacogenetics implementers, and 12% as clinical informaticians. 86% of the participants were from the United States, 10% from Europe and 3% from other (i.e. Brazil and Egypt). Individuals were permitted to self-identify in more than one area. 48% of survey 1 respondents indicated that they spend >75% of their time devoted to pharmacogenetics. 57% of the experts were CPIC members and 93% indicated they were involved in other pharmacogenetic-related groups (Table 1). See Table 1 for additional demographics and number of experts for subsequent surveys.
Table 1. Demographics of experts.
| No. (%) respondents | |||||
|---|---|---|---|---|---|
| Survey 1 (n=58) N (%) |
Survey 2 (n=54) N (%) |
Survey 3 (n=47) N (%) |
Survey 4 (n=46) N (%) |
Survey 5 (n=36) N (%) |
|
| Clinical pharmacogenetics role* | |||||
| Clinician (physician, pharmacist, nurse, etc.) | 25 (43) | 26 (48) | 22 (47) | 22 (48) | 18 (50) |
| Pharmacogenetics Researcher | 39 (67) | 37 (69) | 33 (70) | 32 (70) | 25 (69) |
| Genetic Testing Laboratory Staff | 11 (19) | 11 (20) | 11 (23) | 11 (24) | 7 (19) |
| Pharmacogenetics implementer | 25 (43) | 26 (48) | 25 (53) | 25 (54) | 23 (64) |
| Clinical Informatics | 7 (12) | 7 (13) | 7 (15) | 7 (15) | 7 (19) |
| Other (please specify) | 3 (5) | 2 (4) | 1 (2) | 1 (2) | 1 (3) |
| Workplace setting* | |||||
| For profit hospital or clinic | 1 (2) | 1 (2) | 1 (2) | 1 (2) | 1 (3) |
| Nonprofit or academic hospital or clinic | 29 (50) | 28 (52) | 25 (53) | 25 (54) | 22 (61) |
| Reference/clinical laboratory | 14 (24) | 12 (22) | 10 (21) | 10 (22) | 6 (17) |
| Educational or research resource | 9 (16) | 7 (13) | 6 (13) | 6 (13) | 5 (14) |
| University | 24 (41) | 21 (39) | 19 (40) | 19 (41) | 15 (42) |
| Research or clinical institute | 11 (19) | 10 (19) | 10 (22) | 9 (20) | 8 (22) |
| laboratory test interpretation service | 6 (10) | 4 (7) | 3 (6) | 3 (7) | 3 (8) |
| Other (please specify) | 2 (3) | 1 (2) | 1 (2) | 1 (2) | 1 (3) |
| Percentage of time devote to pharmacogenetics | |||||
| 0%-5% | 3 (5) | 1 (2) | 1 (2) | 1 (2) | 1 (3) |
| 6%-26% | 8 (14) | 7 (13) | 7 (15) | 7 (15) | 4 (11) |
| 26%-50% | 8 (14) | 6 (11) | 5 (11) | 5 (11) | 7 (19) |
| 51%-75% | 11 (19) | 12 (22) | 11 (23) | 11 (24) | 4 (11) |
| 76%-100% | 28 (48) | 28 (52) | 23 (49) | 22 (48) | 20 (56) |
| Pharmacogenetics group membership* | |||||
| CPIC | 33 (57) | 33 (61) | 31 (66) | 31 (67) | 26 (72) |
| ClinVar | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| PGRN | 14 (24) | 14 (26) | 13 (28) | 13 (28) | 10 (28) |
| IOM’s Roundtable on Translating Genomic-Based Research for Health |
7 (12) | 5 (9) | 4 (9) | 4 (9) | 4 (11) |
| ClinGen's PGx working group | 2 (3) | 3 (6) | 3 (6) | 3 (7) | 3 (8) |
| IOM’s EHR Action Collaborative | 2 (3) | 3 (6) | 2 (4) | 2 (4) | 2 (6) |
| CDC Pgx nomenclature group | 11 (19) | 11 (20) | 10 (21) | 10 (22) | 9 (25) |
| GA4GH’s Clinical Working Group | 2 (3) | 2 (4) | 2 (4) | 2 (4) | 2 (6) |
| ACMG Laboratory Standards and Guidelines Committee |
3 (5) | 3 (6) | 3 (6) | 3 (7) | 2 (6) |
| CAP Pharmacogenetics Working Group | 1 (2) | 1 (2) | 1 (2) | 1 (2) | 1 (3) |
| AMP | 2 (3) | 2 (4) | 1 (2) | 1 (2) | 0 (0) |
| Other (please specify) | 17 (29) | 14 (26) | 14 (30) | 13 (28) | 11 (31) |
Participants could appear in multiple groups
Phase 1: Development
A total of seven clinical testing laboratories submitted terms and results can be found in Supplemental Tables S1 and S2. Terms identified in the literature review can be found in Supplemental Table S3.
Phase 2: Prioritization
Terms identified in Phase 1 were used to create the first Delphi survey (survey 1) (see Supplemental Tables S1-S3 and Figures S1-S4 for complete list of terms). The prioritization phase was utilized to eliminate terms that experts found not appropriate. See https://cpicpgx.org/resources/term-standardization/ and Supplemental Figures S1 and S2 for results.
Phase 3, 4, and 5: Refinement and Consensus
After survey 3, consensus was reached (77%) for high-risk genotype genes but not for the other gene categories. Experts participating in survey 3 indicated that terms used to describe transporter function may not be suitable for all non-drug metabolizing enzymes such as VKORC1 or genes encoding drug receptors. Thus, VKORC1 was excluded from future surveys (see Discussion for further explanation). Notably, assessing response rates between clinicians and non-clinicians did not reveal any significant differences (Supplemental Figure S5).
At the conclusion of survey 4, one phenotype designation had not reached the targeted 70% consensus level. Although the phenotype designation of “intermediate metabolizer” was widely used in the literature to designate individuals in between “normal metabolizer” and “poor metabolizer,” that term had not gained 70% consensus. After a conference call to discuss and recommend final terms to include in survey 5, and following completion of the final survey, 100% of experts agreed to terms for allele functional status for drug metabolizing enzymes and transporters, 91.7% for drug metabolizing enzyme phenotypes, and 91.7% for transporter phenotypes (Supplemental Figure S6). The final terms and definitions are summarized in Table 2.
Table 2. Final consensus terms for allele functional status and phenotype.
| Term/Gene Category |
Final Term* | Functional Definition | Genetic Definition | Example diplotypes/alleles |
|---|---|---|---|---|
| Allele Functional Status-all genes |
Increased Function | Function greater than normal function | N/A | CYP2C19*17 |
| Normal Function | Fully functional/wild-type | N/A | CYP2C19*1 | |
| Decreased Function | Function less than normal function | N/A | CYP2C19*9 | |
| No Function | Non-functional | N/A | CYP2C19*2 | |
| Unknown Function | No literature describing function or the allele is novel |
N/A | CYP2C19*29 | |
| Uncertain Function | Literature supporting function is conflicting or weak |
N/A | CYP2C19*12 | |
| Phenotype- Drug Metabolizing Enzymes (CYP2C19, CYP2D6, CYP3A5, CYP2C9, TPMT, DPYD, UGT1A1) |
Ultra-rapid Metabolizer |
Increased enzyme activity compared to rapid metabolizers. |
Two increased function alleles, or more than 2 normal function alleles |
CYP2C19*17/*17
CYP2D6*1/*1XN |
| Rapid Metabolizer | Increased enzyme activity compared to normal metabolizers but less than ultra- rapid metabolizers. |
Combinations of normal function and increased function alleles |
CYP2C19*1/*17 | |
| Normal Metabolizer | Fully functional enzyme activity | Combinations of normal function and decreased function alleles |
CYP2C19*1/*1 | |
| Intermediate Metabolizer |
Decreased enzyme activity (activity between normal and poor metabolizer) |
Combinations of normal function, decreased function, and/or no function alleles |
CYP2C19*1/*2 | |
| Poor Metabolizer | Little to no enzyme activity | Combination of no function alleles and/or decreased function alleles |
CYP2C19*2/*2 | |
| Phenotype- Transporters (SLCO1B1) |
Increased Function | Increased transporter function compared to normal function. |
One or more increased function alleles |
SLCO1B1*1/*14 |
| Normal Function | Fully functional transporter function | Combinations of normal function and/or decreased function alleles |
SLCO1B1*1/*1 | |
| Decreased Function | Decreased transporter function (function between normal and poor function) |
Combinations of normal function, decreased function, and/or no function alleles |
SLCO1B1*1/*5 | |
| Poor Function | Little to no transporter function | Combination of no function alleles and/or decreased function alleles |
SLCO1B1*5/*5 | |
| Phenotype- High risk genotype status (HLA-B) |
Positive | Detection of high-risk allele | Homozygous or heterozygous for high-risk allele |
HLA-B*15:02 |
| Negative | High risk-allele not detected | No copies of high-risk allele |
All terms should begin with the gene name (e.g., CYP2D6 Poor metabolizer, TPMT Normal metabolizer, SLCO1B1 Decreased Function)
Discussion
We successfully engaged a diverse group of experts to establish standard terms through consensus for both pharmacogenetic allele function and inferred phenotypes. The final terms presented in Table 2 will be used in all new and updated CPIC guidelines, and we recommend these terms be considered as standard terminology across all areas of clinical pharmacogenetics, including clinical genetic testing laboratory reporting. Moreover, these terms can be used for clinical decision support (CDS) to guide drug use and dosing (Table 3) using the suggested alerts in CPIC guidelines.16-19
Table 3. Examples of phenotype terms that trigger clinical decision support (CDS).
| Gene | High Risk Phenotype | High Risk Drug |
Example CDS provided in CPIC guideline | Reference |
|---|---|---|---|---|
| CYP2C19 | CYP2C19 Ultrarapid Metabolizer |
Citalopram | This patient is predicted to be a CYP2C19 ultrarapid metabolizer and may be at an increased risk of a poor response due to low plasma concentrations of citalopram. Consider selecting an alternative SSRI not extensively metabolized by CYP2C19. |
Hicks, et al. (2015)17 |
| CYP3A5 | CYP3A5 Normal Metabolizer |
Tacrolimus | Based on the genotype result, this patient is predicted to have lower tacrolimus serum drug levels if initiated on a standard tacrolimus starting dose. Consider increasing the starting dose to 1.5 times to 2 times the standard dose. Total starting dose should not exceed 0.3mg/kg/day. Further dose adjustments or selection of alternative therapy may be necessary due to other clinical factors (e.g., medication interactions, or hepatic function). Use therapeutic drug monitoring to guide dose adjustments. |
Birdwell, et al. (2015)18 |
| CYP2C9 | CYP2C9 Intermediate Metabolizer |
Phenytoin | Based on the genotype result, this patient is predicted to be a CYP2C9 Intermediate Metabolizer and is at increased risk for developing phenytoin-induced toxicities. Consider a 25% reduction of recommended starting maintenance dose. Subsequent maintenance doses should be adjusted according to therapeutic drug monitoring and response. |
Caudle, et al. (2014)40 |
| HLA-B*57:01 | HLA-B*57:01 positive |
Abacavir | The HLA-B*57:01 allele has been detected in this patient. This allele is associated with high risk of severe hypersensitivity to abacavir. DO NOT prescribe abacavir per the FDA’s black box warning. Please choose an alternate antiretroviral. |
Martin, et al. (2014)19 |
In surveys 1 and 2 and during survey discussions experts indicated that terms should be consistent across all genes if possible. Thus, terms describing phenotype were grouped together for subsequent surveys based on related enzyme functions. Final consensus terms included one set of terms to describe allele functional status and three sets of terms describing inferred phenotype depending on the type of pharmacogene: 1) drug metabolizing enzymes (e.g., CYP2D6, DYPD, TPMT, etc.), 2) transporters (e.g., SLCO1B1) and 3) high-risk genotypes (e.g., HLA-B) (Table 2). These terms are suitable for use in most CPIC level A and B genes (https://cpicpgx.org/genes-drugs/).
Many experts felt that the historical term “extensive metabolizer” was too confusing for clinicians, often requiring clarification that it reflects “normal.” Therefore, the final consensus term “normal metabolizer” was selected and the widely used term “extensive metabolizer” will no longer be used in the CPIC guidelines. Furthermore, applying these standardized terms across all drug metabolizing enzymes means that terms like “normal metabolizer” will also be used for genes such as TPMT and DPYD that historically used other designations (i.e., TPMT wild-type activity).
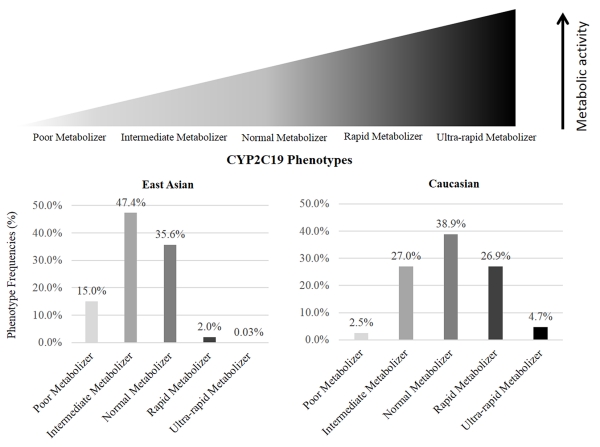
The speed with which we achieved consensus was based on the complexity of the gene and historical use of the term. Because of their simplicity and some level of standardization prior to this project, we quickly achieved consensus for the high-risk genotype genes (e.g. HLA-B). However, the phenotype terms describing drug metabolizing enzymes were the most challenging to standardize due to the different terms that have been used in research and clinical settings. Specifically, defining the term to distinguish the metabolizer status between “normal” and “poor” generated significant discussion. The panel eventually reached consensus on the commonly used term “intermediate metabolizer” after an additional review of the literature and after considering the difficulty of changing this specific term. Drug metabolism terms often need to be interpreted considering the relative nature of the phenotypes to each other on a scale, going from very low function to very high function, which is more complex than expressing high-risk genotype genes as positive or negative for a specific variant allele. Visual depiction of such a scale (Figure 2) may be a helpful addition to interpretive reports.
Figure 2. Example of interpretive scale to visualize a drug metabolism gene’s phenotype.
Phenotype frequencies were estimated using the equation describing Hardy Weinberg equilibrium based on the allele frequencies published in the CPIC guideline.17 For CYP2C19, phenotype frequencies differ substantially by ancestry. “Caucasian” includes those identified as European or North American in primary literature.
Experts also had varying opinions about terms used to differentiate between alleles where there is no literature describing function and alleles where there is conflicting data to support the resulting function. In survey 2, the choices of terms were identical for “no literature describing function” and “conflicting data” and experts chose different terms for each type of variant. While the distinction may not be immediately apparent to clinical providers, we speculate that the experts differentiated these terms to be clear on the level and existence of evidence for a given variant. Distinguishing these concepts may provide value in certain contexts to distinguish lack of evidence from conflicting evidence and this distinction may be emerging as a standard across genomic medicine (e.g., ClinVar)20.
Additional standardization opportunities exist beyond the genes presented here. For example, VKORC1 is the one CPIC level A gene (https://cpicpgx.org/genes-drugs/) where we did not reach consensus. This gene is primarily tested in the context of predicting starting doses of the common anticoagulant warfarin, which is also dependent on CYP2C9. Therefore, many laboratories report a drug-centered phenotype such as “Greatly increased sensitivity to warfarin” (see CPIC guideline for warfarin21), which complicated standardization of VKORC1 terms following the formats used for other genes. In addition, VKORC1 genotype and inferred phenotypes for warfarin dosing are also reported by some laboratories and the CAP proficiency testing surveys according to the CYP2C9 and VKORC1 policy statement published by the ACMG in 2008,22 which further could have added to the difficulty in standardizing VKORC1.
This project and recent work13 have demonstrated that there is great diversity in how genetic test results are reported and interpreted,23 which can lead to confusion among clinicians, patients, and researchers in the exchange and use of clinical genetic data. Clear opportunities exist to develop new terminologies and improve existing standards to represent genetic results and interpretations24. While they do not represent comprehensive solutions, some recent progress has been made. An HL7 standard now exists that outlines how genetic test results could be reported.25 The Logical Observation Identifier Names and Codes (LOINC) terminology, which is a widely used standard for reporting laboratory test results and interpretations,26,27 is one terminology that could be used reporting genetic interpretations and has recently been extended to support genetic data.28 Therefore, to enable precise communication beyond the CPIC guidelines, encourage use of these terms within EHRs, and facilitate the implementation of pharmacogenetic CDS, we obtained LOINC identifiers for pharmacogenetic interpretation codes and answer lists (Supplemental Tables S4 and S5). Our work with LOINC has focused on standardizing pharmacogenetic test interpretation codes, and all of the terms from the CPIC terminology standardization project were registered as LOINC answer lists and were released on December 21, 2015 as part of LOINC 2.54.
The use of standardized vocabularies, such as LOINC, addresses a limitation identified in early implementations of pharmacogenetic CDS.29 Because pharmacogenetic expertise may remain concentrated in specialized health care centers, but patients commonly move to and from a variety of health care providers, the consistent use of standard terms will improve the ability to share patient specific pharmacogenetic knowledge across disparate clinical systems, including those systems with fewer resources for genomic medicine. In addition, the use of standard codes in CPIC guidelines to represent pharmacogenetic interpretation will facilitate further implementation of CDS rules, which are often triggered based on specific pharmacogenetic diagnoses with high-risk phenotypes.29,30
The Action Collaborative on Developing Guiding Principles for Integrating Genomic Information Into the Electronic Health Record Ecosystem (DIGITizE) (http://iom.nationalacademies.org/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/EHR.aspx), an ad hoc activity under the auspices of the IOM Roundtable on Translating Genomic-Based Research for Health, engages key stakeholders from health information technology and management vendors, academic health centers, government agencies, and other organizations to work together to examine how genomic information can be uniformly represented and integrated into EHRs in a standards-based format. As an initial step, DIGITizE developed a CDS implementation guide for two pharmacogenetic use cases, HLA-B*57:01/abacavir and TPMT/azathioprine, based on the aforementioned HL7 standard and published CPIC guidelines. The implementation guide provides examples of HL7 messages for communicating the results of pharmacogenetic testing and CDS logic using the CPIC LOINC codes for HLA-B*57:01 and TPMT. As part of this effort, there was a careful decision to include only interpretations in the guide and not guidance for the genetic data itself. We anticipate that the availability of standard codes for pharmacogenetic interpretations will encourage the incremental development and dissemination of additional implementation resources.
In addition to facilitating LOINC implementation, another goal of CPIC is to have these standardized pharmacogenetic terms adopted broadly by clinical genetic testing laboratories and relevant professional societies and organizations. Importantly, after reviewing the CPIC term standardization project and outcome, the AMP, which is an international society of over 2000 molecular and genomic laboratory medicine professionals, formally endorsed these pharmacogenetic terms on October 26th 2015 (http://www.amp.org/documents/AMPendorsementoftheCPICinitiative2015-10-26.pdf). The terms from this study also may have significant utility for collaborative genomic variation curation and interpretation efforts, including ClinGen and ClinVar.31 PharmGKB is currently working with ClinVar to deposit CPIC Level A gene/drug pairs using these standardized pharmacogenetic terms, and term adoption by other ClinVar submitters in the future would facilitate comparison across submissions. Additionally, these terms may be useful for proficiency testing programs that are designed to improve quality assurance and uniform pharmacogenetic interpretation among clinical genetic testing laboratories (e.g., College of American Pathologists (CAP-PGX).
We chose to use a modified Delphi technique to build consensus among pharmacogenetic experts as it is an established and powerful tool to develop standards across different disciplines.8,9,11 Key risks to the validity of a Delphi study include overestimating the expertise of participants and attrition across the consensus rounds. Given that each participant had involvement in at least one area of pharmacogenetics and 48% of survey one respondents indicated that they spend >75% of their time devoted to pharmacogenetics and 93% indicated they are involved with pharmacogenetic-related groups, we feel this is adequate support of the pharmacogenetic expertise among our survey participants. Participant attrition did occur across consensus rounds during our study; however, it was relatively low (Table 1) and determined to be non-systematic. Although only 60% of the experts participated in survey 5, relative to other Delphi panels and the recommended minimum panel size, our final consensus panel was quite large, which reinforces the validity of our results.32 To reduce bias, especially the authority or reputation of specific individuals, Delphi panel participants are often kept anonymous throughout the process. Although survey creators and analysts were not blinded to participants, identifying information was not shared among survey participants. The only points of participant identification were in between surveys when non-blinded email invitations were sent to participate in conference calls and webinars during which interim results were discussed.
Because these terms were established by experts, an opportunity for further research is to formally assess the terms in end-user usability studies to understand their comprehension among clinicians and patients without formal training or experience in pharmacogenetics. The clinicians’ specific practice site may influence their view of these terms. While surveys of general populations of physicians have indicated limited knowledge and experience with pharmacogenetics33,34 and genome-guided prescribing through CDS,35 a more recent study conducted in a setting with a pre-emptive pharmacogenetics testing program revealed that their physicians were supportive of this type of program and that pharmacogenetic-guided therapy, particularly for cardiovascular medications, has clinical utility.36 Although our consensus terms were generated by experts, nearly 50% of our participants identified as clinicians, and the use of terms by non-expert clinicians and patients was considered throughout the process and most of our experts practice in clinical settings with non-experts.
We aimed to achieve consensus on acceptable terms for multiple pharmacogenes. On their own these terms may not always be an adequate interpretation to guide clinicians, and additional interpretation information can be provided to set the observed phenotype in context of other possible phenotypes. For example, with CYP enzymes, a normal metabolizer status would not typically trigger a dose that is different than the standard recommendation. However, in the case of tacrolimus, a CYP3A5 normal metabolizer (i.e., a CYP3A5 expresser) would require a higher recommended starting dose than the CYP3A5 poor metabolizer (a phenotype that is actually more common among those of European ancestry).18 In practice, it will be necessary to provide the patient’s phenotypic designation in combination with other interpretive information designed for clinicians and patients, and various models of this approach already exist (Table 3).37-39
In conclusion, we anticipate that broad adoption of these proposed standardized pharmacogenetic terms will improve the understanding and interpretation of pharmacogenetic tests by clinicians and patients and reduce confusion by maintaining nomenclature consistency among pharmacogenes. Furthermore, these uniform references will reduce the complexity of the underlying coded vocabulary needed to transmit pharmacogenetic phenotypes between independent laboratories and sites of care, and to trigger CDS.
Supplementary Material
Acknowledgments
The members of CPIC and the CPIC Informatics Working Group are acknowledged for their support in this project. All members are listed here https://www.pharmgkb.org/page/cpicMembers. We specifically thank J. Kevin Hicks, Pharm.D., Ph.D., Andrea Gaedigk, Ph.D., Gillian C. Bell, Pharm.D., Cyrine E. Haidar, Pharm.D., Roseann S. Gammal, Pharm.D., and Katrin Sangkuhl, Ph.D. This work was funded by the National Institutes of Health (NIH) CPIC (R24GM115264), PharmGKB (R24 GM61374), S.A.S. (K23 GM104401), R.R.F. and PGRN (U19 GM61388), and ALSAC.
References
- 1.Payne TH, Corley S, Cullen TA, et al. Report of the AMIA EHR-2020 Task Force on the status and future direction of EHRs. Journal of the American Medical Informatics Association : JAMIA. 2015;22(5):1102–1110. doi: 10.1093/jamia/ocv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei WQ, Leibson CL, Ransom JE, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. Journal of the American Medical Informatics Association : JAMIA. 2012;19(2):219–224. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526(7573):343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rehm HL, Berg JS, Brooks LD, et al. ClinGen--the Clinical Genome Resource. The New England journal of medicine. 2015;372(23):2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Current drug metabolism. 2014;15(2):209–217. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirts BH, Salama JS, Aronson SJ, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. Journal of the American Medical Informatics Association : JAMIA. 2015;22(6):1231–1242. doi: 10.1093/jamia/ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalkey N, Helmer O. An experimental application of the Delphi method to the use of experts. Management Science. 1963;9(3):458–467. [Google Scholar]
- 9.Beretta R. A critical review of the Delphi technique. Nurse Researcher. 1996;3:79–89. doi: 10.7748/nr.3.4.79.s8. [DOI] [PubMed] [Google Scholar]
- 10.Green B, Jones M, Hughes D, Williams A. Applying the Delphi technique in a study of GPs’ information requirements. Health & social care in the community. 1999;7(3):198–205. doi: 10.1046/j.1365-2524.1999.00176.x. [DOI] [PubMed] [Google Scholar]
- 11.von der Gracht HA. Consensus measurement in Delphi studies Review and implications for future quality assurance. Technological Forecasting & Social Change. 2012;79:125–1536. [Google Scholar]
- 12.Hasson F, Keeney S, McKenna H. Research guidelines for the Delphi survey technique. Journal of advanced nursing. 2000;32(4):1008–1015. [PubMed] [Google Scholar]
- 13.Kalman LV, Agundez JA, Appell ML, et al. Pharmacogenetic Allele Nomenclature: International Workgroup Recommendations for Test Result Reporting. Clinical pharmacology and therapeutics. 2015 doi: 10.1002/cpt.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu CC, Sandford BA. The Delphi technique: making sense of consensus. Practical Assess Res Eval. 2007;12:1–8. [Google Scholar]
- 15.Sumsion T. The Delphi technique: an adaptive research tool. British Journal of Occupational Therapy. 1998;61(4):153–156. [Google Scholar]
- 16.Gammal RS, Court MH, Haidar CE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing. Clinical pharmacology and therapeutics. 2015 doi: 10.1002/cpt.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clinical pharmacology and therapeutics. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birdwell KA, Decker B, Barbarino JM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clinical pharmacology and therapeutics. 2015;98(1):19–24. doi: 10.1002/cpt.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin MA, Hoffman JM, Freimuth RR, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA-B Genotype and Abacavir Dosing: 2014 update. Clinical pharmacology and therapeutics. 2014;95(5):499–500. doi: 10.1038/clpt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. [Accessed April 14, 2016];Clinical significance on ClinVar aggregate records. http://www.ncbi.nlm.nih.gov/clinvar/docs/clinsig/#clinsig_agg.
- 21.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clinical pharmacology and therapeutics. 2011;90(4):625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flockhart DA, O’Kane D, Williams MS, et al. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genetics in medicine : official journal of the American College of Medical Genetics. 2008;10(2):139–150. doi: 10.1097/GIM.0b013e318163c35f. [DOI] [PubMed] [Google Scholar]
- 23.Tsuchiya KD, Shaffer LG, Aradhya S, et al. Variability in interpreting and reporting copy number changes detected by array-based technology in clinical laboratories. Genetics in medicine : official journal of the American College of Medical Genetics. 2009;11(12):866–873. doi: 10.1097/GIM.0b013e3181c0c3b0. [DOI] [PubMed] [Google Scholar]
- 24.Wiley LK, Tarczy-Hornoch P, Denny JC, et al. Harnessing next-generation informatics for personalizing medicine: a report from AMIA’s 2014 Health Policy Invitational Meeting. Journal of the American Medical Informatics Association : JAMIA. 2016;23(2):413–419. doi: 10.1093/jamia/ocv111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.HL7 V2 Implementation Guide: Clinical Genomics; Fully LOINC-Qualified Genetic Variation Model, Release 2. 2013 Mar;
- 26.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clinical chemistry. 2003;49(4):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 27.Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. Journal of the American Medical Informatics Association : JAMIA. 1998;5(3):276–292. doi: 10.1136/jamia.1998.0050276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deckard J, McDonald CJ, Vreeman DJ. Supporting interoperability of genetic data with LOINC. Journal of the American Medical Informatics Association : JAMIA. 2015;22(3):621–627. doi: 10.1093/jamia/ocu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. Journal of the American Medical Informatics Association : JAMIA. 2014;21(e1):e93–99. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldspiel BR, Flegel WA, DiPatrizio G, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. Journal of the American Medical Informatics Association : JAMIA. 2014;21(3):522–528. doi: 10.1136/amiajnl-2013-001873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic acids research. 2014;42(Database issue):D980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okoli C, Pawlowski S. The Delphi method as a research tool: an example, design considerations and applications. Information and management. 2004;42(1):15–29. [Google Scholar]
- 33.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clinical pharmacology and therapeutics. 2012;91(3):450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 34.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clinical genetics. 2012;82(4):388–394. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overby CL, Erwin AL, Abul-Husn NS, et al. Physician Attitudes toward Adopting Genome-Guided Prescribing through Clinical Decision Support. Journal of personalized medicine. 2014;4(1):35–49. doi: 10.3390/jpm4010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson JF, Field JR, Shi Y, et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. The pharmacogenomics journal. 2015 doi: 10.1038/tpj.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clinical pharmacology and therapeutics. 2012;92(5):563–566. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson JF, Bowton E, Field JR, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genetics in medicine : official journal of the American College of Medical Genetics. 2013;15(10):833–841. doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. American journal of medical genetics. Part C, Seminars in medical genetics. 2014;166C(1):45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caudle KE, Rettie AE, Whirl-Carrillo M, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clinical pharmacology and therapeutics. 2014;96(5):542–548. doi: 10.1038/clpt.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.