Abstract
Objectives
To report aberrant myeloblasts detected by flow cytometry immunophenotypic studies in an asymptomatic patient with familial platelet disorder with propensity to myeloid malignancy, a rare autosomal dominant disease caused by germline heterozygous mutations in Runt-related transcription factor 1.
Methods
Morphologic evaluation, flow cytometry immunophenotypic studies, nanofluidics-based qualitative multiplex reverse transcriptase polymerase chain reaction, Sanger sequencing, and next-generation sequencing-based mutational hotspot analysis of 53 genes were performed on bone marrow biopsy and aspirate samples.
Results
Flow cytometry immunophenotypic analysis showed 0.6% CD34+ blasts with an abnormal immunophenotype: CD13 increased, CD33+, CD38 decreased, CD117 increased, and CD123 increased.
Conclusions
The acquisition of new phenotypic aberrancies in myeloblasts as detected by flow cytometry immunophenotypic studies might be a harbinger of impending myelodysplastic syndrome or acute myeloid leukemia in a patient with familial platelet disorder with propensity to myeloid malignancy.
Keywords: Familial platelet disorder, Myeloid malignancy, RUNX1
Familial platelet disorder with propensity to myeloid malignancy (FPD/MM; OMIM 601399) is a rare autosomal dominant disease caused by germline heterozygous mutations in Runt-related transcription factor 1 (RUNX1).1 Approximately 40 family pedigrees with FPD/MM have been described to date, but the frequency of this disorder is most likely underestimated. FPD/MM is characterized by inherited thrombocytopenia, normal platelet size, platelet functional defects, and an increased risk of myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML). Thrombocytopenia is usually mild to moderate, but bleeding tends to be more severe than expected (based on the degree of thrombocytopenia) due to platelet dysfunction. Only a proportion of affected individuals develops MDS or AML, and as of today, it is impossible to predict who will develop MDS or AML and when. Here, we describe a population of aberrant myeloid precursors detected by flow cytometry immunophenotypic analysis in an asymptomatic man with FPD/MM.
Case Report
An asymptomatic 54-year-old man was referred to the University of Texas MD Anderson Cancer Center for consultation after he was found to be thrombocytopenic. The patient did not have any bleeding tendency in the past, and a CBC was performed as a part of family screening that was triggered by detection of a germline heterozygous RUNX1 mutation (c.836G>A p.W279X) in his 15-year-old daughter. The daughter had a persistent history of moderate to severe thrombocytopenia (~30 × 109/L) with a bleeding tendency since her premature birth at 24 weeks of gestation; she required platelet transfusions infrequently, mostly for surgical procedures. The CBC of the patient we are reporting showed the following: hemoglobin, 140 g/L (normal range, 140–180 g/L); mean corpuscular volume, 89 fL (normal range, 82–98 fL); WBC count, 6.5 × 109/L (normal range, 4–11 × 109/L); platelet count, 92 × 109/L (normal range, 140–440 × 109/L); and mean platelet volume, 9.5 fL (normal range, 4–10.4 fL). A peripheral blood smear showed decreased numbers of platelets with adequate granularity. Erythrocytes and WBCs were morphologically unremarkable, and no blasts were identified. Twenty-one months later, at last clinical follow-up, the patient had a platelet count of 104 × 109/L, and he remained asymptomatic.
Materials and Methods
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissue blocks were cut in 4-μm-thick sections and processed with heat-induced epitope retrieval. 3,3′-Diaminobenzidine was used as a chromogen. Staining was performed in an automated immunostainer (Ventana Medical Systems, Tucson, AZ). Assessed antibodies against the following antigens were CD34 (MY10, 1:40; BD Biosciences, Franklin Lakes, NJ) and CD61 (2F2, 1:100; Cell Marque, Rocklin, CA).
Flow Cytometry Immunophenotypic Studies
Bone marrow aspirate samples were collected in EDTA anticoagulant and processed within 24 hours of collection. After incubation with monoclonal antibodies for 10 minutes at 4°C, erythrocytes were lysed with ammonium chloride (PharmLyse; BD Biosciences, San Diego, CA) at room temperature for 10 minutes using a standard lyse/wash technique. Samples were acquired on FACSCanto II instruments (BD Biosciences). Antibody panels are shown in Table 1. A total of 200,000 events were acquired.
Table 1.
Panel of Antibodies Used for Flow Cytometry Immunophenotypic Studies
| Tube | FITC | PE | APC | PE-Cy7 | PerCP-CY5.5 | V450 | V500 |
|---|---|---|---|---|---|---|---|
| 1 | CD16 | CD11b | CD13 | CD33 | CD34 | CD19 | CD45 |
| 2 | CD7 | CD64 | CD2 | CD10 | CD34 | CD14 | CD45 |
| 3 | HLA-DR | CD56 | CD123 | CD10 | CD34 | CD4 | CD45 |
| 4 | CD5 | CD117 | CD38 | CD10 | CD34 | CD15 | CD45 |
Data were analyzed using FCS Express software (De Novo Software, Los Angeles, CA). Major cell populations were defined by CD45/SSC (side scatter) characteristics. Aberrancies of myeloblasts were assessed on CD34+CD10−/CD19− myeloid precursors. The normal ranges of intensity of antigen expressions and the percentage of expressions were established on 30 healthy controls (with no hematopoietic neoplasm and with a normal CBC) in our previous study, and the flow cytometric immunophenotypic assay was validated in cytopenic patients without MDS and those with chronic myelomonocytic leukemia.2 For antigens normally not expressed or only partially expressed, the expressions were measured as a percentage, whereas for antigens normally highly expressed, such as CD38, CD117, CD123, CD34, CD45, and CD13, the levels of antigen expressions were measured by mean fluorescence intensity. “Alterations” were two standard deviations from normal controls, also confirmed by at least one-third of log scale changes.3
Conventional Cytogenetics
Conventional G-band karyotype analysis was performed on bone marrow aspirate specimens as described previously.4 Karyotype was written following the 2013 International System for Human Cytogenetics Nomenclature.5
Next-Generation Sequencing
Next-generation sequencing to assess mutational hotspots in 53 genes (ABL1, AKT1, ALK, APC, ATM, BRAF, CDKN2A, CHD1, CSF1R, CTNNB1, DNMT3A, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KLHL6, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP, VHL, and 53 XPO1) was performed on bone marrow aspirate specimens as described previously using a MiSeq sequencer (Illumina, San Diego, CA).6
Nanofluidics
Nanofluidics-based qualitative multiplex reverse transcriptase polymerase chain reaction (RT-PCR) was performed on bone marrow aspirate specimens (Fluidigm, San Francisco, CA). It is designed to detect t(8;21)(q22;q22); RUNX1-RUNX1T1, inv(16)(p13.1q22) or t(16;16)(p13.1q22); CBFβ-MYH11, t(15;17)(q22;q12); PML-RARA, t(9;22)(q34;q11.2); BCR-ABL1, t(12;21)(p13;q22); ETV6-RUNX1, t(1;19)(q23; p13.3); E2A-PBX1, t(4;11)(q21;q23); MLL-AF4, and t(6;9) (p23;q34); DEK-NUP214.
Sanger Sequencing
Sanger sequencing was performed at Baylor College of Medicine (Houston, TX).
Results
Morphologic Findings
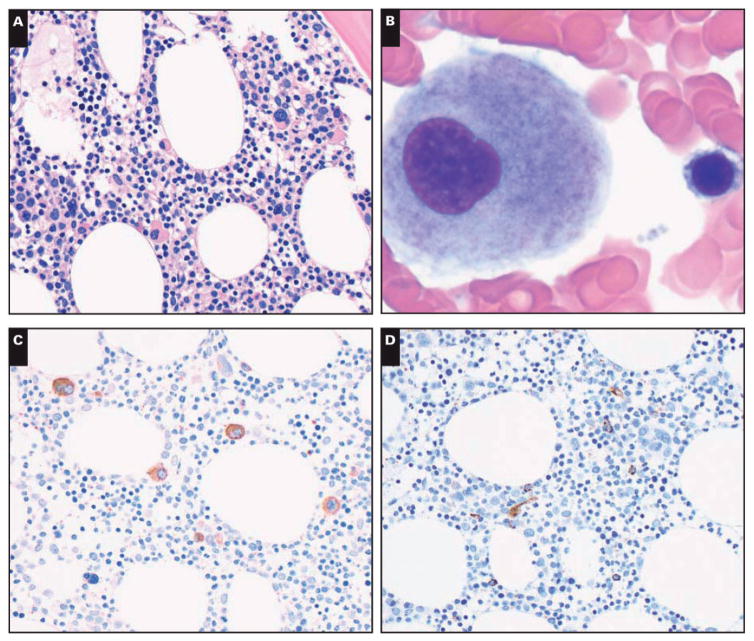
Bone marrow biopsy specimens showed a cellular (40%–50%) bone marrow. Megakaryocytes were increased (10/high-power field) with small monolobated forms Image 1A. Granulocytic and erythroid precursors showed complete maturation. Reticulin and trichrome stains demonstrated slightly increased reticulin fibrosis (MF-1 by European Consensus on grading bone marrow fibrosis).7 Bone marrow aspirate smears showed rare dysplastic megakaryocytes with separated nuclear lobes Image 1B. The granulocytic and precursors showed complete maturation without dysplasia. Blasts were not increased. Iron stain showed decreased storage iron (1+ of 4).
Image 1.
Representative images of bone marrow biopsy and aspiration specimens. A, Bone marrow biopsy specimen shows adequate cellularity for age and increased number of small, hypolobated megakaryocytes (H&E, ×400). B, Bone marrow aspiration smear shows a small, hypolobated megakaryocyte (Giemsa, ×1,000). C, Anti-CD61 stain on bone marrow biopsy section highlights increased number of small, hypolobated megakaryocytes (×400). D, Anti-CD34 stain on bone marrow biopsy section shows rare CD34+ immature cells (×400).
Immunohistochemistry and Flow Cytometry Immunophenotypic Studies
Immunohistochemical analysis was performed on the bone marrow biopsy specimens. The anti-CD61 antibody showed dysplastic small monolobated megakaryocytes Image 1C. Anti-CD34 stain showed 1% of CD34+ blasts Image 1D. Flow cytometry immunophenotypic studies performed using bone marrow aspirate material showed 0.6% CD34+ blasts with an abnormal immunophenotype: CD13 increased, CD33+, CD38 decreased, CD117 increased, and CD123 increased Image 2. No monoclonal B cells or aberrant T cells were detected.
Image 2.
A, Flow cytometric analysis of bone marrow aspirate material shows a small number of a CD34+ cell population (0.6%). Of the CD34+ cells, there are markedly reduced stage I hematogones (normal precursor B cells) (arrowheads). The CD34 myeloid precursors (arrows) show increased expression of CD117, CD123, and CD13 and decreased expression of CD38, as expressed by the mean fluorescence intensity (MFI). The median and two standard deviation of normal MFI are 4,100 (1,120–5,920) for CD117, 25,020 (19,100–39,400) for CD38, 649 (240–1,080) for CD123, and 1,870 (920–3,300) for CD13. B, A normal bone marrow of an age-matched individual is shown for comparison.
Cytogenetic and Molecular Studies
Conventional cytogenetic analysis detected a normal male karyotype in 20 metaphases. Next-generation sequencing did not detect any mutations. Nanofluidics-based qualitative multiplex RT-PCR was negative. Sanger sequencing detected a germline heterozygous c.836G>A (p.W279X) mutation in the RUNX1 gene, which was identical to the mutation identified previously in his daughter.
Discussion
RUNX1, formerly known as AML1 and CBFA2, is located at chromosome locus 21q22.12. RUNX1 protein has a highly conserved Runt homology domain (RHD) at the N-terminal and a transactivation domain at the C-terminal. The RHD domain mediates DNA binding and heterodimerization with CBFβ, which enhances the affinity of RUNX1 to DNA.8 RUNX1 is essential for terminal differentiation of the megakaryocytic and T-lymphoid lineages. Several mis-sense, nonsense, and frameshift mutations; intragenic deletions; and intragenic duplication have been reported in patients with FDP/MM.1,8–14
FPD/MM was first reported in 1978 by Luddy et al,15 who described a family of three siblings with lifelong history of a bleeding disorder and thrombocytopenia. The first well-studied pedigree was reported by Dowton et al16 in 1985, suggesting autosomal dominant inheritance. In 1996, the causal region was found at 22q22.1–22.2 by linkage analysis, and RUNX1 was implicated as one of the candidate genes.17 RUNX1 mutations were identified by Song et al1 in 1999. Since that time, approximately 40 families with FPD/MM have been reported. The disease appears to have complete penetrance, but the clinical presentation is variable.18 Of note, thrombocytopenia is not a requisite for diagnosis, and many patients do not have a history of bleeding.10,19 However, when a patient has a bleeding tendency, the degree of bleeding can be disproportionate to the level of thrombocytopenia because of qualitative defects in platelets such as decreased adenosine diphosphate (ADP) and serotonin and a decreased number of dense granules.20 Platelet aggregation tests often show abnormal results in response to arachidonic acid, collagen, and epinephrine, but platelet aggregation in response to ADP and ristocetin is normal.17,20 The underlying mechanism of qualitative and quantitative platelet defects in FDP/MM is incompletely understood. Available data demonstrate that RUNX1 mutation is associated with impaired agonist-induced platelet myosin light chain phosphorylation and receptor-mediated GPIIb-IIIa activation.21 Myosin light chain phosphorylation takes part in platelet shape change and platelet granule secretion upon activation. In one patient reported, there was a 77-fold downregulation of one of the myosin light chain genes, MYL9, which is a target of RUNX1.22,23 Quantitative platelet defects in patients with FPD/MM can be partially explained by decreased expression of the MPL receptor, impaired thrombopoietin-induced signaling, and impaired megakaryocyte maturation with proplatelet formation.24,25
The bone marrow of patients with FDP/MM generally shows mildly decreased megakaryocytes with a predominance of immature forms. The immature megakaryocytes usually show a high nuclear-to-cytoplasm ratio, strongly basophilic cytoplasm, and poorly lobulated nuclei. In contrast, erythropoiesis and granulopoiesis are completely normal with no bone marrow fibrosis unless MDS or AML develops.
Recently, Owen and colleagues26 suggested that FPD/MM may be underdiagnosed based on their detection of RUNX1 mutations in 50% of families with more than one first-degree relatives with MDS and/or AML. Individual mutations might cause different degrees of functional loss of RUNX1 protein and partially explain variable clinical presentations in FPD/MM. Missense and nonsense mutations in the RHD cause aberrant RUNX1 proteins with impairment of DNA binding and with dominant negative effect. Large deletions or frame-shift mutations induce RUNX1 haploinsufficiency.25 Mutations causing haploinsufficiency are more commonly reported than ones with a dominant-negative effect.1,27,28 Interestingly, RUNX1 mutations with a dominant-negative effect are more frequently associated with subsequent leukemia than mutations associated with haploinsufficiency.28
Patients with FPD/MM have an increased risk of myeloid neoplasms, including acute myeloid leukemia, myelodysplastic syndromes, and chronic myelomonocytic leukemia.29 The morphologic types of acute myeloid leukemia, as designated using the French-American-British group terminology, include M1, M2, M4, and M5. The frequency of myeloid malignancies in patients with FDP/MM has been reported to range from 20% to 65%.17,18 Rare patients who developed T-lymphoblastic leukemia and diffuse large B-cell lymphoma also have been reported rarely.14,18,26,30 The median patient age when they develop leukemia is 37 years, but children as young as 6 years are reported.26 Germline RUNX1 mutation alone is insufficient for development of leukemia; a second hit is required.31 Bluteau et al25 showed that RUNX1 mutations increase the clonogenic potential of immature CD34+CD38− progenitors, hence increasing the pool of cells susceptible to a second hit. A high frequency of RUNX1 biallelic alterations has been shown in cases of AML that arose in patients with FPD/MM,14 but other genetic aberrations might be sufficient for leukemogenesis. Cytogenetic abnormalities, including del(5q), −7, +8, del(11q), and t(1;7)(p34.1;q22), have been reported at the time of onset of leukemia.8,16,17,30
There are no established guidelines for the management of patients with FDP/MM. When bleeding tendency is severe, patients are treated similarly to patients with other platelet function disorders. When AML or MDS develops, patients are treated with chemotherapy or stem cell transplantation. Prior studies have shown that the same mutation can be detected in siblings who are being evaluated as potential transplant donors.9,26 Although most of the reported RUNX1 mutations are located in exons 3 to 8, whole or partial hemizygous RUNX1 deletion and intragenic gene duplications have also been described. These latter alterations can be missed by Sanger sequencing and require further genetic assessment of copy number abnormalities using a single-nucleotide polymorphism array or array comparative genomic hybridization.8 It is recommended that screening for germline RUNX1 mutation be performed when there is a bleeding tendency in a family, unexplained mild thrombocytopenia with normal platelet size in combination with abnormal platelet aggregation studies, or isolated dysplastic changes in megakaryocytes.
To our knowledge, the detection of aberrant myeloblasts by flow cytometry immunophenotypic analysis has not been reported previously in asymptomatic patients with FPD/MM. Our group has demonstrated high sensitivity and specificity of flow cytometry immunophenotypic studies for the detection of aberrant myeloblasts in patients with MDS.2 Since only a proportion of patients with FPD/MM develops MDS or AML, and it can take many years to do so, there is a desperate need for better tools to predict which patients with FDP/MM may develop MDS or AML. It is conceivable that a new, second clone emerging after a “second genetic hit” and slowly progressing into overt MDS or AML also could be detected by flow cytometry immunophenotypic studies at an asymptomatic or oligosymptomatic phase, well in advance of the onset of clinically relevant MDS or AML. If it is shown that the presence of clonal progression indicates that patients are doomed to develop AML or MDS in the near future, these patients might benefit from aggressive therapeutic intervention. However, prospective studies are needed to address this issue.
Footnotes
For permissions, please e-mail: journals.permissions@oup.com
References
- 1.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 2.Tang G, Jorgensen LJ, Zhou Y, et al. Multi-color CD34(+) progenitor-focused flow cytometric assay in evaluation of myelodysplastic syndromes in patients with post cancer therapy cytopenia. Leuk Res. 2012;36:974–981. doi: 10.1016/j.leukres.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Shen Q, Ouyang J, Tang G, et al. Flow cytometry immunophenotypic findings in chronic myelomonocytic leukemia and its utility in monitoring treatment response. Eur J Haematol. 2015;95:168–176. doi: 10.1111/ejh.12477. [DOI] [PubMed] [Google Scholar]
- 4.Khoury JD, Sen F, Abruzzo LV, et al. Cytogenetic findings in blastoid mantle cell lymphoma. Hum Pathol. 2003;34:1022–1029. doi: 10.1053/s0046-8177(03)00412-x. [DOI] [PubMed] [Google Scholar]
- 5.Shaffer LG, McGowan-Jordan JMS. An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: Karger; 2013. [Google Scholar]
- 6.Luthra R, Patel KP, Reddy NG, et al. Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: applicability for diagnostics and disease monitoring. Haematologica. 2014;99:465–473. doi: 10.3324/haematol.2013.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiele J, Kvasnicka HM, Facchetti F, et al. European consensus on grading bone marrow fibrosis and assessment of cellularity. Haematologica. 2005;90:1128–1132. [PubMed] [Google Scholar]
- 8.Jongmans MC, Kuiper RP, Carmichael CL, et al. Novel RUNX1 mutations in familial platelet disorder with enhanced risk for acute myeloid leukemia: clues for improved identification of the FPD/AML syndrome. Leukemia. 2010;24:242–246. doi: 10.1038/leu.2009.210. [DOI] [PubMed] [Google Scholar]
- 9.Buijs A, Poddighe P, van Wijk R, et al. A novel CBFA2 single-nucleotide mutation in familial platelet disorder with propensity to develop myeloid malignancies. Blood. 2001;98:2856–2858. doi: 10.1182/blood.v98.9.2856. [DOI] [PubMed] [Google Scholar]
- 10.Walker LC, Stevens J, Campbell H, et al. A novel inherited mutation of the transcription factor RUNX1 causes thrombocytopenia and may predispose to acute myeloid leukaemia. Br J Haematol. 2002;117:878–881. doi: 10.1046/j.1365-2141.2002.03512.x. [DOI] [PubMed] [Google Scholar]
- 11.Kirito K, Sakoe K, Shinoda D, et al. A novel RUNX1 mutation in familial platelet disorder with propensity to develop myeloid malignancies. Haematologica. 2008;93:155–156. doi: 10.3324/haematol.12050. [DOI] [PubMed] [Google Scholar]
- 12.Ripperger T, Steinemann D, Gohring G, et al. A novel pedigree with heterozygous germline RUNX1 mutation causing familial MDS-related AML: can these families serve as a multistep model for leukemic transformation? Leukemia. 2009;23:1364–1366. doi: 10.1038/leu.2009.87. [DOI] [PubMed] [Google Scholar]
- 13.Beri-Dexheimer M, Latger-Cannard V, Philippe C, et al. Clinical phenotype of germline RUNX1 haploinsufficiency: from point mutations to large genomic deletions. Eur J Hum Genet. 2008;16:1014–1018. doi: 10.1038/ejhg.2008.89. [DOI] [PubMed] [Google Scholar]
- 14.Preudhomme C, Renneville A, Bourdon V, et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood. 2009;113:5583–5587. doi: 10.1182/blood-2008-07-168260. [DOI] [PubMed] [Google Scholar]
- 15.Luddy RE, Champion LA, Schwartz AD. A fatal myeloproliferative syndrome in a family with thrombocytopenia and platelet dysfunction. Cancer. 1978;41:1959–1963. doi: 10.1002/1097-0142(197805)41:5<1959::aid-cncr2820410540>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Dowton SB, Beardsley D, Jamison D, et al. Studies of a familial platelet disorder. Blood. 1985;65:557–563. [PubMed] [Google Scholar]
- 17.Ho CY, Otterud B, Legare RD, et al. Linkage of a familial platelet disorder with a propensity to develop myeloid malignancies to human chromosome 21q22.1-22.2. Blood. 1996;87:5218–5224. [PubMed] [Google Scholar]
- 18.Liew E, Owen C. Familial myelodysplastic syndromes: a review of the literature. Haematologica. 2011;96:1536–1542. doi: 10.3324/haematol.2011.043422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganly P, Walker LC, Morris CM. Familial mutations of the transcription factor RUNX1 (AML1, CBFA2) predispose to acute myeloid leukemia. Leuk Lymphoma. 2004;45:1–10. doi: 10.1080/1042819031000139611. [DOI] [PubMed] [Google Scholar]
- 20.Gerrard JM, Israels ED, Bishop AJ, et al. Inherited platelet-storage pool deficiency associated with a high incidence of acute myeloid leukaemia. Br J Haematol. 1991;79:246–255. doi: 10.1111/j.1365-2141.1991.tb04529.x. [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Mao G, Rao AK. Association of CBFA2 mutation with decreased platelet PKC-theta and impaired receptor-mediated activation of GPIIb-IIIa and pleckstrin phosphoryl-ation: proteins regulated by CBFA2 play a role in GPIIb-IIIa activation. Blood. 2004;103:948–954. doi: 10.1182/blood-2003-07-2299. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Gorospe JR, Hoffman EP, Rao AK. Decreased platelet expression of myosin regulatory light chain polypeptide (MYL9) and other genes with platelet dysfunction and CBFA2/RUNX1 mutation: insights from platelet expression profiling. J Thromb Haemost. 2007;5:146–154. doi: 10.1111/j.1538-7836.2006.02271.x. [DOI] [PubMed] [Google Scholar]
- 23.Jalagadugula G, Mao G, Kaur G, et al. Regulation of platelet myosin light chain (MYL9) by RUNX1: implications for thrombocytopenia and platelet dysfunction in RUNX1 haplodeficiency. Blood. 2010;116:6037–6045. doi: 10.1182/blood-2010-06-289850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heller PG, Glembotsky AC, Gandhi MJ, et al. Low Mpl receptor expression in a pedigree with familial platelet disorder with predisposition to acute myelogenous leukemia and a novel AML1 mutation. Blood. 2005;105:4664–4670. doi: 10.1182/blood-2005-01-0050. [DOI] [PubMed] [Google Scholar]
- 25.Bluteau D, Glembotsky AC, Raimbault A, et al. Dysmegakaryopoiesis of FPD/AML pedigrees with constitutional RUNX1 mutations is linked to myosin II deregulated expression. Blood. 2012;120:2708–2718. doi: 10.1182/blood-2012-04-422337. [DOI] [PubMed] [Google Scholar]
- 26.Owen CJ, Toze CL, Koochin A, et al. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood. 2008;112:4639–4645. doi: 10.1182/blood-2008-05-156745. [DOI] [PubMed] [Google Scholar]
- 27.Matheny CJ, Speck ME, Cushing PR, et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 2007;26:1163–1175. doi: 10.1038/sj.emboj.7601568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaud J, Wu F, Osato M, et al. in vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99:1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- 29.Arepally G, Rebbeck TR, Song W, et al. Evidence for genetic homogeneity in a familial platelet disorder with predisposition to acute myelogenous leukemia (FPD/AML) Blood. 1998;92:2600–2602. [PubMed] [Google Scholar]
- 30.Nishimoto N, Imai Y, Ueda K, et al. T cell acute lymphoblastic leukemia arising from familial platelet disorder. Int J Hematol. 2010;92:194–197. doi: 10.1007/s12185-010-0612-y. [DOI] [PubMed] [Google Scholar]
- 31.Osato M, Yanagida M, Shigesada K, Ito Y. Point mutations of the RUNx1/AML1 gene in sporadic and familial myeloid leukemias. Int J Hematol. 2001;74:245–251. doi: 10.1007/BF02982056. [DOI] [PubMed] [Google Scholar]