Abstract
Background and Purpose
To apply automated quantitative volumetric MRI analyses to patients diagnosed with Rasmussen’s encephalitis (RE), to determine the predictive value of lobar volumetric measures, and to assess regional atrophy difference and monitor disease progression using these measures.
Materials and Methods
Nineteen patients (42 scans) with diagnosed RE were studied. Two control groups were used: one with 42 age- and gender-matched normal subjects; the other with 42 non-RE epilepsy patients with the same disease duration as RE patients. Volumetric analysis was performed on T1-weighted images using BrainSuite. Ratios of volumes from the affected hemisphere divided by those from the unaffected hemisphere were used as input to a logistic regression classifier, which was trained to discriminate patients from controls. Using the classifier, we compared the predictive accuracy of all the volumetric measures. These ratios were further used to assess regional atrophy difference and to correlate with epilepsy duration.
Results
Interhemispheric and frontal lobe ratios had the best prediction accuracy to separate RE patients from normal and non-RE epilepsy controls. The insula showed significantly more atrophy compared to all the other cortical regions. Patients with longitudinal scans showed progressive volume loss of the affected hemisphere. Atrophy of the frontal lobe and insula correlated significantly with epilepsy duration.
Conclusions
Automated quantitative volumetric analysis provides accurate separation of RE patients from normal controls and non-RE epilepsy patients, and thus may assist diagnosis of RE. Volumetric analysis could also be included as part of followup for RE patients to assess disease progression.
Keywords: Rasmussen’s Encephalitis, Rasmussen’s Syndrome, epilepsy, MRI, presurgical evaluation, image processing, volumetric analysis, disease progression
Introduction
Rasmussen’s syndrome, also known as Rasmussen’s encephalitis (RE), is a chronic inflammatory disease of the brain, which typically affects only one hemisphere. It is usually associated with a progressive course of focal seizures, characterized by epilepsia partialis continua (EPC), as well as neurologic deficits (most frequently hemiparesis). It is generally believed that RE is driven by a T-cell mediated inflammation that leads to neuronal and astrocytic cell death in one hemisphere.1, 2 The disease is characterized by three stages: (1) the predromal stage of mild hemiparesis and infrequent seizures; (2) an acute stage with frequent seizures from one hemisphere characterized by EPC, with deterioration of neurologic functions (histopathology in this stage shows the highest inflammatory intensity) and (3) a residual stage with a severe, fixed neurological deficit and drug-resistant seizures (histopathology in this stage shows a decrease in inflammation).
MRI characteristics of RE include early cortical swelling followed by cortical and subcortical hyperintensity on fluid-attenuated inversion recovery (FLAIR) and T2-weighted images with progressive atrophy of the affected hemisphere.1 The perisylvian region has been observed to be the predominant site for signal abnormality and atrophy.3 Volume loss of the ipsilateral caudate head is also frequently observed.3
Over the past decade, MRI has become an increasingly important tool in the diagnosis of RE, as well as in assessing disease progression and therapeutic effectiveness. The degree of hemispheric atrophy in RE can be obtained from the MRI based on manual planimetry1 and manual volumetry,4 both of which can be labor intensive, time consuming and rater dependent. Fully automated volumetric methods have been proposed that showed a high concordance with planimetric methods and clinical parameters.5
To date, no studies have examined how volumetric measures can help predict RE, i.e., when a new patient presents with suspected RE, can the extent of atrophy on the MRI be quantified to predict the probability that the patient truly has RE? In the early stage of the disease, atrophy can be too subtle for visual inspection to detect and therefore diagnosis is often uncertain and delayed. Can fully automated volumetric methods be used to complement visual analysis? Furthermore, progression of RE can be slow especially at the residual stage. Can fully automated volumetric methods be used to reveal slow changes over the years? In an attempt to answer these questions, in this study we examined 15 volumetric measures in a cohort of patients with RE and two control groups: one group consisted of 42 age- and gender-matched normal controls; the other group consisted of 42 non-RE epilepsy patients with matched disease duration. Once the volumetric measures were obtained, we used them to form a statistical model to classify patients and controls, in order to determine the measure with the highest predictive accuracy. The same volumetric measures were then used to examine regional atrophy difference and correlated with disease progression.
Materials and Methods
Patients
This retrospective study was approved by the Cleveland Clinic institutional review board. Patients evaluated at the Cleveland Clinic Epilepsy Center were included using the following criteria: 1) clinical diagnosis of RE following guidelines specified in literature,3 and 2) MRI available with T1-weighed Magnetization Prepared Rapid Acquisition with Gradient Echo (MPRAGE) sequence.
Controls
Forty-two normal controls (34 provided by the Pediatric Imaging, Neurocognition and Genetics Study, and 8 from the investigators’ centers) were chosen to be age- and gender-matched to the 42 RE patient scans. The control subjects were free of any neurological diseases.
An additional group of 42 non-RE epilepsy controls (all from the investigators’ center) were chosen so that they have matching epilepsy duration with the 42 RE scans. All these epilepsy patients had negative (nonlesional) MRI scans and had clearly lateralizing epilepsy as documented by their video-EEG monitoring.
MRI Protocol
Out of the 42 scans from patients with RE, 18 MRIs were performed using 1.5T Siemens Avanto scanner (Erlangen, Germany); 22 were scanned using a 3T Siemens Skyra scanner (Erlangen, Germany); 2 were scanned using a 3T Siemens Trio scanner (Erlangen, Germany) at our institute. The 3D volumetric T1-weighted MPRAGE sequence was used for the volumetric processing. Sequence parameters at 1.5T were: repetition time (TR) = 11 milliseconds (ms), echo time (TE) = 4.6 ms, no inversion, flip angle (FA) = 20 degrees, bandwidth (BW) = 130 kHz, slice thickness = 1.25 mm, no gap, and in-plane resolution = 0.9 mm. Sequence parameters at 3T(Trio/Skyra) were TR = 1860/1800 ms, TE = 3.4/2.56 ms, inversion time (TI) = 1100/900 ms, FA = 10 degrees, BW = 130/220 kHz, slice thickness = 0.94/1 mm, no gap, and in-plane resolution of 0.94/0.41 mm.
Out of the 42 normal control scans, 24 were performed with a 3T GE Signa scanner (Milwaukee, WI, USA) using the SPGR (Spoiled Gradient Recalled Echo) sequence. Sequence parameters were: TR = 8.132 ms, TE = 3.452 ms, TI = 640 ms, FA = 8 degrees, BW = 244 kHz, slice thickness = 1.2 mm, no gap, and in-plane resolution of 0.94 mm. Eighteen patients were with a 3T Siemens Trio scanner with the MPRAGE sequence (with the same parameters as reported in the previous section). Out of the 42 non-RE epilepsy control scans, 15 MRIs were performed using 1.5T Siemens Avanto scanner, 25 MRIs were scanned using a 3T Siemens Skyra/Trio scanner, and 2 MRIs were scanned using a 3T Siemens Trio scanner at our institute, with the same parameters as reported in the previous section.
Volumetric Analysis
All MRI processing was performed using standard procedures of the freely available BrainSuite software (Version 15b; http://brainsuite.org/). BrainSuite provides an automatic sequence to extract surface models of the cerebral cortex. The procedure uses anatomical information from both the surface models and volume of the brain images for accurate co-registration between the subject and an atlas6–8. The final parcellation computes volumes of 140 brain regions (70 from each hemisphere). White matter (WM), gray matter (GM) and cerebral spinal fluid (CSF) volumes are generated separately by surface and volume registration (SVReg) for each of the brain regions. Details can be found in Supplementary Material 1.
-
The following volumes on each side of the brain are calculated in milliliters (ml) using built-in statistical tools of BrainSuite:
Hemispheric Volume = GM+WM without brainstem and cerebellum
Lobar volume=GM+WM of a particular lobe
Mesial and subcortical structure volume = volume of amygdala+hippocampus, putamen, caudate nucleus, thalamus, globus pallidus, brainstem, and cerebellum
-
Ratios of the volumes were calculated:
HRvol=volume of the affected hemisphere (AH)/volume of the unaffected hemisphere (UH)
Lobar/GM/WM/mesial/subcortical structure ratios: volume of Lobe/GM/WM/mesial/subcortical ROI from the AH/volume of Lobe/GM/WM/mesial/subcortical ROI from the UH
In total, 15 volumetric ratio measures were generated: interhemispheric (HRvol), insular, frontal, temporal, parietal, occipital, GM, WM, amygdala+hippocampus, putamen, caudate nucleus, thalamus, globus pallidus, brainstem, and cerebellum.
Visualization of the parcellated brain regions (the final output of BrainSuite processing) in one patient is shown in Figure 1 as an example. This patient had two MRIs, separated by 7 years, the second scan showing progressive atrophy around the perisylvian area.
Figure 1.
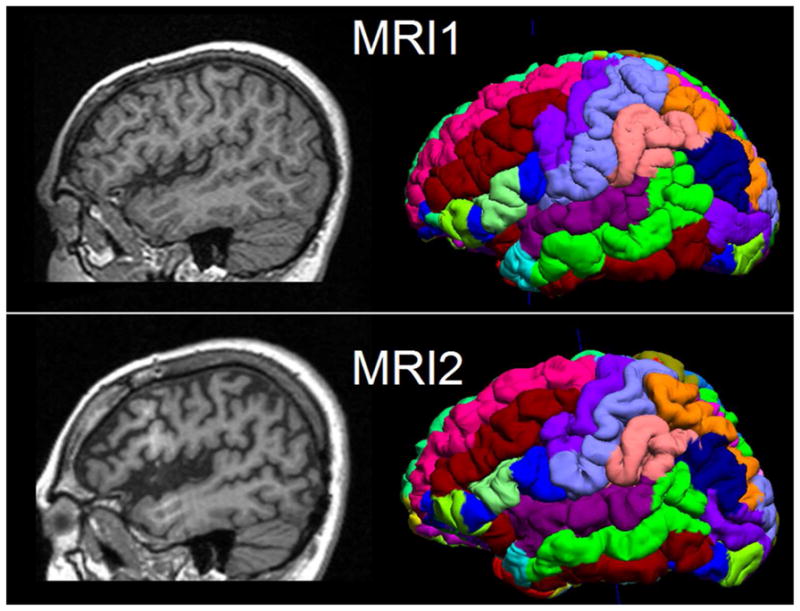
Illustration of two serial MRIs from the same patient (P6) and the surface and volume registration (SVReg) output of BrainSuite. First row: MRI at age 10. Second row: MRI at age 17. Shown in the left column are the sagittal T1-weighted MPRAGE images. The right column is the cortical rendering of the SVReg labels, with different colors denoting different anatomical areas of the brain. Pronounced atrophy can be observed at the perisylvian area, and the interhemispheric ratio (HRvol) showed decrease from 0.79 to 0.70.
Scans from controls were processed with the same methodology as patients. In normal controls (who did not have an affected hemisphere), all the ratios were calculated by randomly dividing the two sides. In non-RE epilepsy controls, all ratios were determined by dividing the epilepsy side by the non-epileptic side.
Statistics
Statistical analyses were performed using the Statistics Toolbox in MATLAB 2013b (MathWorks, Natick, Massachusetts). The Mann-Whitney U-test was performed to compare the degree of atrophy between different brain regions (multiple comparison corrected by FDR9). The two-tailed Pearson correlation coefficient test was used to assess the correlation between lobar ratio and epilepsy duration. The significance level was set at p<0.05. Methodology of classification is described below.
Overall Workflow
As shown in Supplementary Figure 1, the 15 volumetric ratio measures from both patient and controls were first normalized by subtracting the pooled mean and dividing by the pooled standard deviation. The logistic regression classifier was trained to discriminate the patients and controls using each of the volumetric ratio measures. The binary logistic regression classifier can be described as follows:
| (1) |
where fi(n) is the nth observation from the ith feature, β0, β1, are the regression parameters and is the probability that the feature fi(n) belong to the patient class. For example, a value of π(fi(n)) − 0.8 denotes that there is 80% probability that the feature fi(n) belong to the patient class. The decision boundary for the classifier was set at 0.5, i.e., a value of π(fi(n)) > 0.5 denotes a patient class.
Cross-Validation
The performance of the classifier was assessed via 5-fold cross-validation. For the cross-validation, the original features were randomly partitioned into 5 equal-sized subgroups (G1, G2 … G5) containing equal numbers of patients and controls. From the five subgroups, a single subgroup was used as validation data set for testing the model and the remaining four subgroups were used as training data. The process was repeated five times so that each of the subgroups was used once as the testing data set. The performance of the classifier was then quantified by estimating the accuracy of the predictor on the validation data set (or test data) as follows
| (2) |
where TP, TN, FP, FN denote the number of True Positives, True Negatives, False Positives and False Negatives respectively. The mean values of accuracy across the five trials were used to quantify the performance of the classifier.
Classifier Design
Based on the results of the cross-validation analysis described in the previous section, the feature with the highest accuracy was selected for constructing the optimal classifier. A leave-one-out cross validation strategy was used, i.e., the classifier was constructed using N-1 samples and was tested on the left-out sample. The process was repeated N times so that each of the samples was used once as the testing data. The sensitivity and specificity of the classifier were estimated and the Receiver Operating Characteristic (ROC) curve was constructed. The performance of the classifier was evaluated by estimating the Area under the Curve (AUC).
Probability Curve
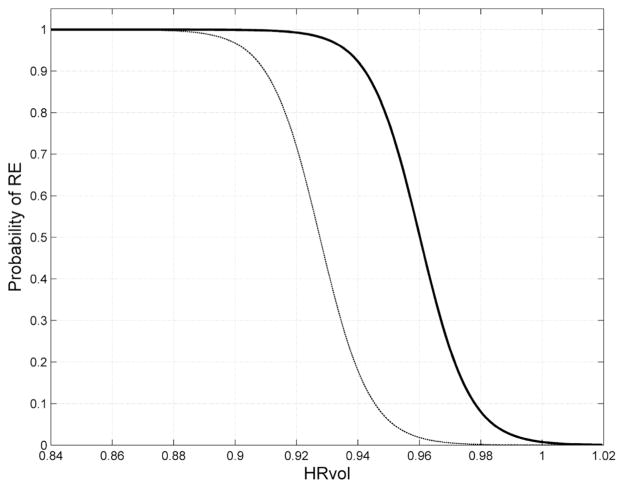
Equation (1) was used to generate a probability curve to describe how the chosen feature predicts the probability of the patient belonging to the RE group. The probability curve was generated from the 42 RE scans and 42 non-RE epilepsy scans from this study (assuming pre-test probability of 50%); another probability curve was additionally generated to correct for the difference in incidence for RE and non-RE epilepsy (1 in 1,000,000 vs. 1 in 100) with methods established by Whittemore et al.10
Results
Patient and Control Demographics
Table 1 describes the 19 RE patients included in this study. All patients met the diagnostic criteria as previously published.3 In 15 patients the diagnosis was confirmed by biopsy and/or surgical pathology. In the remaining 4 patients, RE diagnosis was based on the presence of the following features: (1) focal seizures and unilateral cortical deficits; (2) EEG with unihemispheric slowing with or without epileptiform activity and unilateral seizure onset; and (3) MRI with unihemispheric atrophy and T2/FLAIR hyperintense signal in GM/WM or caudate. A total of 42 MRIs from these 19 RE patients were included in this study.
Table 1.
Detailed demographics and clinical data of the 19 patients with Rasmussens Encephalitis (RE) and the two control groups. SF=seizure-free with >12 months postoperative followup.
| RE patients (N=19) | |
| Mean age at epilepsy onset | 7.3 (SD±5.3, median 8, range 1.5, 22) |
| Mean disease duration at first MRI | 4.0 (SD±4.8, median 2.9, range 0.1, 20) |
| Gender | |
| Female | 11 |
| Male | 8 |
| Handedness | |
| Right | 13 |
| Left | 5 |
| Ambidextrous | 1 |
| Surgery Location | |
| Hemispherectomy | 12 (10 SF) |
| Frontal | 1 (1 SF) |
| Insular/opercular | 1 |
| Temporal | 1 |
| No surgery | 4 |
| Number of MRI scans | |
| Single scan | 10 |
| Multiple scans | 9 (range 2,7) |
| Mean age at MRI (N=42) | |
| RE scans | 14.2 (SD±8.0, median 14.8, range 3, 43) |
| Normal controls | 14.3 (SD±8.0, median 14.0, range 3.6, 43) |
| Non-RE epilepsy controls | 16.9 (SD±7.6, median15.5, range 5, 31) |
| Mean disease duration at MRI (N=42) | |
| RE scans | 7.0 (SD±5.6, median 6.3, range 0.8, 21.4) |
| Non-RE epilepsy controls | 7.6 (SD±5.1, median 6.5, range 1, 22) |
Predictive Values
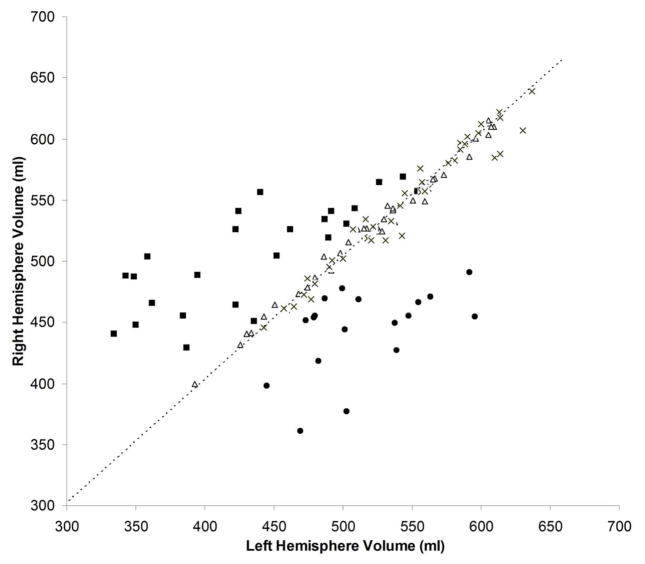
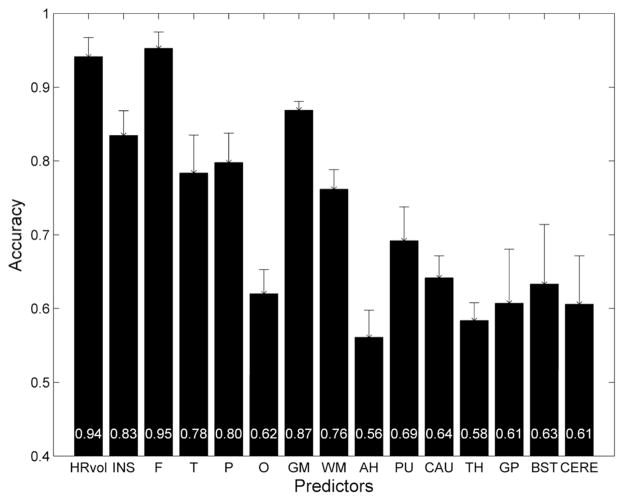
As shown in Figure 2, hemispheric asymmetry of total brain volume produced clearly defined separation of RE patients from controls (both normal and epilepsy). Both control groups clustered tightly along the diagonal line that represents hemispheres with equal volumes, whereas the patients (regardless of left or right sided) showed markedly more dispersion in the hemispheric volumes. In all RE patients, HRvol were <1.0 indicating a smaller volume of the affected hemisphere. Figure 3 shows the mean accuracy (across the five cross-validation runs) of the logistic regression classifier for all volumetric measure ratios, when compared to the non-RE epilepsy controls. The highest prediction accuracy was for HRvol and frontal lobe ratio (0.94 and 0.95), the two of which were not significantly different.
Figure 2.
Absolute volume of the right hemisphere plotted versus the left hemisphere. Triangles = normal controls; crosses = non-RE epilepsy controls; squares = patients with left RE; circles = patients with right RE. The dashed line is the diagonal line representing hemispheres with equal volumes.
Figure 3.
Mean accuracy across the five cross-validation runs of the logistic regression classifier for 15 volumetric ratio measures. RE patients were compared with non-RE epilepsy controls with matching disease duration. Error bars denote standard deviation. The mean accuracy values for each measure were plotted at the bottom of the bars. HRvol=hemispheric volume, INS=insula, F=frontal, T=temporal, P=parietal, O=occipital, GM=gray matter, WM=white matter, AH=amygdala+hippocampus, PU=putamen, CAU=caudate nucleus, TH=thalamus, GP=globus pallidus, BST=brainstem, and CERE=cerebellum.
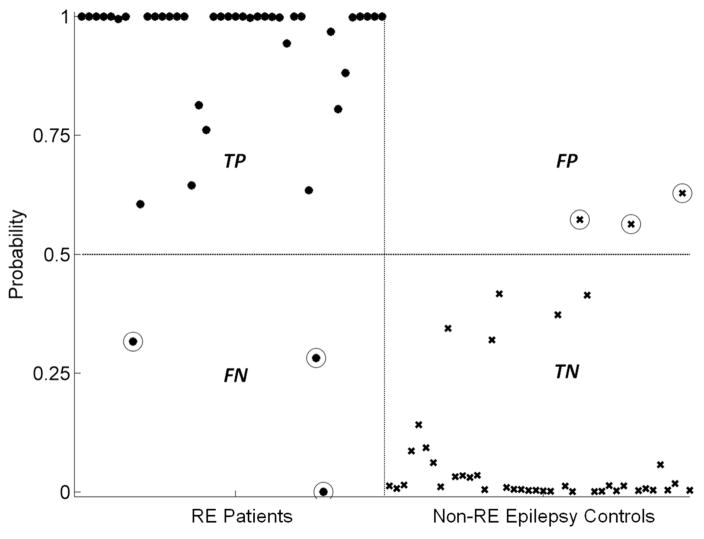
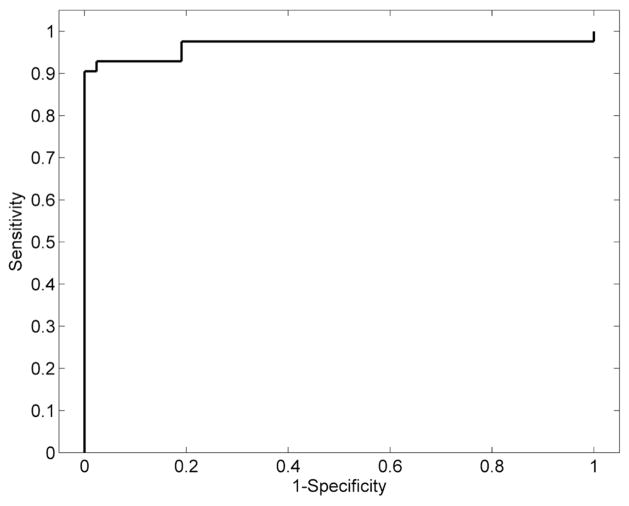
We used HRvol as the feature for the classifier and evaluated the performance of the classifier to separate RE patients from the non-RE epilepsy controls. Using this classifier, the majority of the patients and controls were correctly classified. As shown in Figure 4, of 42 RE patient scans, only 3 were misclassified; of 42 non-RE epilepsy controls, only 3 were misclassified. The classifier attained a sensitivity of 0.93 and a specificity of 0.93. The ROC curve of the detector is shown in Figure 5. The AUC was 0.97, which denotes a highly discriminative classifier. Figure 6 shows the probability curves that depict the relationship between HRvol and the probability of the patient belonging to the RE group. The classifier using frontal lobe ratio had similar performance (sensitivity = 0.95, specificity = 0.97, AUC = 0.97).
Figure 4.
Performance of the classifier constructed using HRvol. RE patients were denoted with dots, and non-RE epilepsy controls were denoted with crosses. True positives (TP) are defined as patients being correctly identified as patients by the classifier. True negatives (TN) are defined as normals being correctly identified as normals. False positives (FP) are defined as normals being incorrectly identified as patients. False negatives (FN) are defined as patients being incorrectly identified as normals. Circled dots/crosses denote the subjects that were misclassified (3 FP and 3 FN).
Figure 5.
Receiver operating characteristic analyses showing a highly discriminative classifier using HRvol.
Figure 6.
Probability curves depicting the relationship between HRvol and the probability of RE. The solid curve was estimated based on comparison of 42 RE patient scans and 42 non-RE epilepsy control scans with the same disease duration. The dashed curve was additionally generated to correct for the difference in incidence for RE and non-RE epilepsy (1 in 1,000,000 vs. 1 in 100).
Degree of Atrophy
As shown by Table 2, among all the lobar regions, insula exhibited the most atrophy at the most recent MRI (p<0.05). Among all the basal ganglia and mesial temporal structures, the regional atrophy differences were not statistically significant. Nine patients showed ipsilateral brainstem atrophy, and 3 patients showed contralateral cerebellar atrophy, with ratio < 0.95 at the most recent MRI.
Table 2.
Regional atrophy difference in all the lobar/basal ganglia and mesial temporal structures regions.
| Lobar Regions | Ratio of Atrophy |
|---|---|
| Insula | 0.77 (SD±0.04) |
| Frontal | 0.84 (SD±0.03) |
| Temporal | 0.88 (SD±0.03) |
| Parietal | 0.87 (SD±0.03) |
| Occipital | 0.90 (SD±0.05) |
| GM | 0.85 (SD±0.02) |
| WM | 0.88 (SD±0.02) |
| Basal Ganglia and Mesial Structures | Ratio of Atrophy |
|---|---|
| AH | 0.90 (SD±0.02) |
| PU | 0.88 (SD±0.03) |
| CAU | 0.89 (SD±0.06) |
| TH | 0.95 (SD±0.04) |
| GP | 0.94 (SD±0.02) |
Longitudinal Analyses
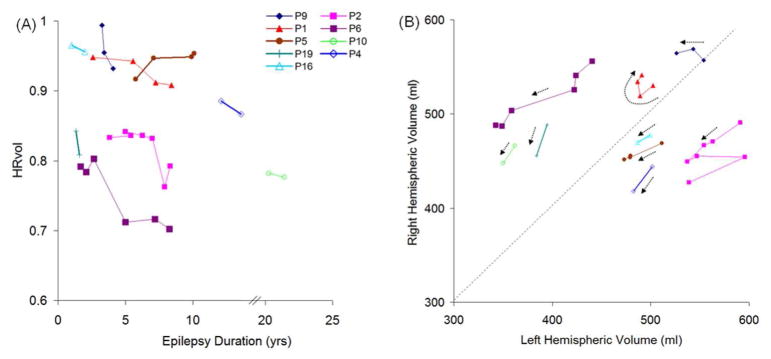
Figure 7 (A) shows the disease progression in the 9 patients with multiple scans. The median change of HRvol per year was −1.4%. All patients had decreasing HRvol indicating disease progression, with the exception of one patient (P5) who showed an increase of HRvol over time. Seven out of these 9 patients had intravenous immunoglobulin (IVIG) treatment with varying responses. P9 responded well with a reduction of seizures; P1 and P6 had only transient response in terms of seizure control after each IVIG cycle; P2, 4, 5 and 16 had no clear benefit. In P2, a significant drop of HRvol was observed at the sixth MRI, at which time the patient was admitted for status epilepticus. The seventh MRI in the same patient was taken after seizures were controlled (IVIG + steroids + tacrolimus) and a slight increase of HRvol was observed at that time, although not back to the baseline level of the previous 5 MRIs.
Figure 7.
(A) HRvol plotted over epilepsy duration in the 9 patients with serial MRI. All but P5 showed a decrease in HRvol over the observed period of time. The axis was broken from 14 to 20 years since there are no data points for these durations. (B) Absolute hemispheric volume (right side plotted vs. left side) of the same 9 patients. Dotted arrows for the data of each patient show progression of time; beginning of arrow denotes earlier scans. Panel A and panel B share the same symbol for each patient for direct comparison.
Additional information can be obtained by analyzing absolute volumes of the hemispheres as shown in Figure 7 (B). The majority of patients moved downward (towards origin) over time, as indicated by the arrows. P2 (who had a significant drop of HRvol at the sixth MRI due to status epilepticus) had abrupt changes in the absolute volumes, and review of the MRI showed bilateral swelling, more on the unaffected side. P1 and P9 did not show a clear downward trend of the arrows.
Correlation of Volume Loss with Disease Duration
The ratios of frontal lobe and insular volumes were significantly negatively correlated with disease duration (p=0.018, p<0.01, respectively, two tailed Pearson correlation test). The correlation is not significant for the ratio of the other lobes, mesial or deep brain structures, or interhemispheric ratio.
Discussion
We present here automated volumetric analyses of a large series of patients diagnosed with Rasmussen’s Syndrome. Independently using a different platform of processing routines, our results confirmed the volumetric findings reported in a previous study by Wagner et al.;5 we further expand the previous study by evaluating the predictive values of volumetric findings, with the addition of normal and epilepsy controls. Our study provides further evidence that automatic volumetric analysis can be useful to the diagnosis of patients with RE on an individual basis.
The most important finding from our study is that volumetric measures, particularly interhemispheric and frontal lobe ratios, had a high degree of accuracy to separate patients from non-RE epilepsy patients with the same disease duration. Used in the relevant clinical settings, such as initial and follow-up investigations in epilepsy patients with suspected RE, the probability curve that was estimated using HRvol can potentially provide an objective measure to solidify the confidence of the diagnosis of RE. Additionally, with the methodology established in this paper, such patients can be studied prospectively with volumetric findings compared with surgical pathology/biopsy.
In terms of lobar atrophy, we found the insula to be significantly more atrophic than all other lobes. In terms of predictive value, the frontal lobe ratio is found to be the most predictive measure for RE among all lobes, i.e. frontal lobe ratio separates patients from controls with the highest accuracy. One should not confuse prediction accuracy with the size of an effect (atrophy), as having a large effect does not necessarily imply higher accuracy. This was seen in our data, where frontal lobe ratio had the best accuracy but insula had the most atrophy. Segmentation of insula cortex can be difficult and thickness can be over-estimated, causing more noise in the measured data, which can explain the lower accuracy. Both frontal and insular ratios, however, correlated significantly with disease duration. These findings support that frontal lobe and insula were preferentially involved in the atrophic process, as compared to temporal, parietal and occipital lobes. This finding is consistent with those from previous studies.1, 5, 11–13
Our results showed that the predictive accuracy of GM is greater than WM, which may suggest that GM is preferentially affected in RE as compared to WM. In terms of the severity of atrophy, both GM and WM were affected and lost volume over the years. Although GM had a lower mean ratio than WM, the difference was not statistically significant. Overall, this finding is consistent with the previous study by Wagner et al., in which they found a preferential effect of RE on GM, and others studies that documented additional WM involvement (especially at the advanced stages of RE) by pathology and imaging methods.14, 15
Three patients (P8, P9, P16) were misclassified as normals by the HRvol classifier and the frontal lobe ratio classifier. All three patients had pathologic confirmation of RE based on biopsy and/or surgical pathology. In P9 and P16, the first MRIs that were misclassified were at 3 and 1 years of epilepsy onset, respectively; later MRIs at (3.5 and 2 years, respectively) were correctly classified as RE. This data indicates a need for improving sensitivity of our methodology especially for detecting subtle changes at early stage of the disease; alternatively, this data could suggest that volume loss in select patients at the initial stage of RE may not be sufficient to differentiate them from normal volume asymmetry. On the other hand, the three patients in our cohort who had the shortest disease duration at the first MRI (0.1, 0.83, 0.91 years respectively) were all correctly classified, demonstrating the effectiveness of our methodology in these patients.
Our findings also demonstrated the feasibility that progression of RE can be measured by volumetric analysis. The majority of patients with serial MRIs showed a decrease in HRvol, indicating disease progression despite IVIG and anti-epileptic drug treatments. Additionally, by analyzing the absolute hemispheric volumes, we found that RE patients also have atrophy in the unaffected hemisphere and may suffer bilateral brain volume loss, i.e., the majority of patients moved downward (towards origin) over time as shown on the absolute hemisphere volume plot. The patients who showed unusual time courses of longitudinal hemispheric volumes and ratios could be explained by several reasons: (1) natural progression of the disease (acute and chronic), e.g., P2 (who had a significant drop of HRvol at the sixth MRI due to status epilepticus) had abrupt changes in the absolute volumes, and review of the MRI showed bilateral swelling, more on the unaffected side, which might have contributed to the volume change. (2) Transient response to treatment, e.g. P1 had had transient response to IVIG which may have caused corresponding volume changes. (3) Natural brain development, i.e., P1 and P9, who did not show a clear downward trend of the arrows, were 8 and 3 years of age respectively. It is conceivable that between the scans the brains were still developing and growing; therefore absolute volume may not be a good measure of disease progression. In fact, HRvol of both patients showed clear downward trend over time. (4) The possibility of bilateral RE, e.g., P5 was the only patient whose HRvol increased over time, indicating a higher volume of atrophy in the unaffected hemisphere than the affected hemisphere. This finding raises the possibility of bilateral RE, despite the fact that the seizures were found to be in the affected hemisphere only by EEG. However, the presence of bilateral RE is debated and likely very rare; only 2 cases out of the reported 200–300 cases in the literature showed histopathological proof for bilateral RE.16, 17 No biopsy was obtained from the unaffected hemisphere to confirm or disprove this hypothesis.
The performance of the classifier constructed in our study had similar performance when comparing RE patients to either the normal controls or the non-RE epilepsy controls. Taken into the account of the fact that the epilepsy controls had the same disease duration as the RE patients, our data provides quantitative evidence that brain volume decrease (likely caused by extensive neuronal loss1–3) is a characteristic of RE, which as we show in this study, can be utilized to accurately separate RE patients from non-RE epilepsy patients.
Several limitations should be considered when interpreting the results of our study:
There are three patients in this study with an adolescent or adult life onset age (13, 13, 22 years of age, respectively). These patients are considered less common presentation of RE,18 and could bring heterogeneity in the study cohort. Additionally, there are 4 patients who did not have pathology/biopsy to confirm RE. Although we followed the commonly accepted diagnostic criteria,3 the lack of pathological confirmation could still potentially contribute to inaccurate diagnosis of RE.
Due to the retrospective design of the study, there was a relatively long interval between disease onset and earliest MRI available for volumetric analysis in our cohort. In only a small subset of RE patients (3 of 19), we had access to the MRI within one year of their initial disease onset. For the other patients, the initial MRI was either performed outside of Cleveland Clinic, or was not performed with volumetric T1 sequences and could not be used for analysis. Therefore, based on the current data, it was not possible to conclude the effectiveness of our methodology for early diagnosis. Additionally, many of the RE patients were referred to our center for surgery; in these patients, only one pre-surgical MRI was available for analysis and there is no data for the later stages of the disease. Overall, further prospective studies are warranted to confirm validity of the findings from our current study.
The 42 MRIs from the 19 RE patients are treated as 42 different cases and matched to 42 different control subjects, introducing potential bias to the comparison.
This study was not set up to evaluate the effectiveness of immune therapy, i.e. we could not assess whether IVIG was an effective treatment to alleviate the disease impact as there is no proper control group (patients without IVIG). A more rigorously study design is needed to make any definite conclusions on treatment effectiveness.
As future work, it is conceivable to use the established methodology to (1) prospectively analyze patients with suspected RE and compare with biopsy and surgical pathology; and (2) monitor disease progression in patients with confirmed RE by calculating volumetric changes in hemispheric and lobar regions on their repeat MRIs. In terms of methodological improvement, in addition to volumetric measures, we will also extract other important features of the T1 dataset (such as signal change) to be utilized as the input to the classifier, in order to improve classification of patients at the early stage of the disease.
Conclusion
In summary, our study highlights the usefulness of volumetric analysis to assist diagnosis of patients with Rasmussen’s Syndrome. We demonstrate that interhemispheric and frontal lobe ratios can accurately classify individual RE patients from normal controls and non-RE epilepsy patients with the same disease duration. The probability curves generated in our study can be used in appropriate clinical settings to solidify the confidence of the diagnosis of RE. We also demonstrate that progression of hemispheric atrophy can be measured reliably by volumetric analysis. Such analysis, when used in conjunction with other clinical data, can provide insight into disease progression and treatment effectiveness, and could be consider as part of the followup process for patients suspected to have RE.
Supplementary Material
Acknowledgments
Grant Support
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number R01NS074980 (ZIW, SEJ, DWS, RML, AAJ, NBA). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Normal control data collection and sharing for this project were funded by the Pediatric Imaging, Neurocognition and Genetics Study (PING) via the National Institute of Health Grant RC2DA029475 and the National Institute on Drug Abuse and the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
The authors would like to thank James Bena, Lead Biostatistician, Cleveland Clinic Quantitative Health Sciences, for his assistance on the statistical analyses.
Abbreviations
- RE
Rasmussen’s Encephalitis
- HRvol
interhemispheric ratio (affected side/unaffected side)
Footnotes
Presented in part at ASNR 2015 Annual Meeting.
References
- 1.Bien CG, Widman G, Urbach H, et al. The natural history of Rasmussen’s encephalitis. Brain. 2002;125:1751–1759. doi: 10.1093/brain/awf176. [DOI] [PubMed] [Google Scholar]
- 2.Bauer J, Elger CE, Hans VH, et al. Astrocytes are a specific immunological target in Rasmussen’s encephalitis. Ann Neurol. 2007;62:67–80. doi: 10.1002/ana.21148. [DOI] [PubMed] [Google Scholar]
- 3.Varadkar S, Bien CG, Kruse CA, et al. Rasmussen’s encephalitis: clinical features, pathobiology, and treatment advances. Lancet Neurol. 2014;13:195–205. doi: 10.1016/S1474-4422(13)70260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larionov S, Konig R, Urbach H, et al. MRI brain volumetry in Rasmussen encephalitis: the fate of affected and “unaffected” hemispheres. Neurology. 2005;64:885–887. doi: 10.1212/01.WNL.0000152895.23010.52. [DOI] [PubMed] [Google Scholar]
- 5.Wagner J, Schoene-Bake JC, Bien CG, et al. Automated 3D MRI volumetry reveals regional atrophy differences in Rasmussen encephalitis. Epilepsia. 2012;53:613–621. doi: 10.1111/j.1528-1167.2011.03396.x. [DOI] [PubMed] [Google Scholar]
- 6.Joshi A, Shattuck D, Leahy R. A Method for Automated Cortical Surface Registration and Labeling. In: Dawant B, Christensen G, Fitzpatrick JM, et al., editors. Biomedical Image Registration. Springer; Berlin Heidelberg: 2012. pp. 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shattuck DW, Leahy RM. BrainSuite: An automated cortical surface identification tool. Medical Image Analysis. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- 8.Pantazis D, Joshi A, Jiang J, et al. Comparison of landmark-based and automatic methods for cortical surface registration. Neuroimage. 2010;49:2479–2493. doi: 10.1016/j.neuroimage.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 10.Whittemore AS. Logistic regression of family data from case-control studies. Biometrika. 1995;82:57–67. [Google Scholar]
- 11.Bhatjiwale MG, Polkey C, Cox TC, et al. Rasmussen’s encephalitis: neuroimaging findings in 21 patients with a closer look at the basal ganglia. Pediatr Neurosurg. 1998;29:142–148. doi: 10.1159/000028709. [DOI] [PubMed] [Google Scholar]
- 12.Chiapparini L, Granata T, Farina L, et al. Diagnostic imaging in 13 cases of Rasmussen’s encephalitis: can early MRI suggest the diagnosis? Neuroradiology. 2003;45:171–183. doi: 10.1007/s00234-002-0923-7. [DOI] [PubMed] [Google Scholar]
- 13.Rajesh B, Kesavadas C, Ashalatha R, et al. Putaminal involvement in Rasmussen encephalitis. Pediatr Radiol. 2006;36:816–822. doi: 10.1007/s00247-006-0176-4. [DOI] [PubMed] [Google Scholar]
- 14.Cauley KA, Burbank HN, Filippi CG. Diffusion tensor imaging and tractography of Rasmussen encephalitis. Pediatr Radiol. 2009;39:727–730. doi: 10.1007/s00247-009-1261-2. [DOI] [PubMed] [Google Scholar]
- 15.Pardo CA, Vining EP, Guo L, et al. The pathology of Rasmussen syndrome: stages of cortical involvement and neuropathological studies in 45 hemispherectomies. Epilepsia. 2004;45:516–526. doi: 10.1111/j.0013-9580.2004.33103.x. [DOI] [PubMed] [Google Scholar]
- 16.Chinchilla D, Dulac O, Robain O, et al. Reappraisal of Rasmussen’s syndrome with special emphasis on treatment with high doses of steroids. J Neurol Neurosurg Psychiatry. 1994;57:1325–1333. doi: 10.1136/jnnp.57.11.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobias SM, Robitaille Y, Hickey WF, et al. Bilateral Rasmussen encephalitis: postmortem documentation in a five-year-old. Epilepsia. 2003;44:127–130. doi: 10.1046/j.1528-1157.2003.36602.x. [DOI] [PubMed] [Google Scholar]
- 18.Oguni H, Andermann F, Rasmussen TB. The syndrome of chronic encephalitis and epilepsy. A study based on the MNI series of 48 cases. Adv Neurol. 1992;57:419–433. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.