Abstract
Cells communicate with one another to create microenvironments and share resources. One avenue by which cells communicate is through the action of exosomes. Exosomes are extracellular vesicles that are released by one cell and taken up by neighbouring cells. But how exosomes instigate communication between cells has remained largely unknown. We present evidence here that particular long non-coding RNA molecules are preferentially packaged into exosomes. We also find that a specific class of these exosome associated non-coding RNAs functionally modulate cell viability by direct interactions with L-lactate dehydrogenase B (LDHB), high-mobility group protein 17 (HMG-17), and CSF2RB, proteins involved in metabolism, nucleosomal architecture and cell signalling respectively. Knowledge of this endogenous cell to cell pathway, those proteins interacting with exosome associated non-coding transcripts and their interacting domains, could lead to a better understanding of not only cell to cell interactions but also the development of exosome targeted approaches in patient specific cell-based therapies.
Keywords: non-coding RNA, extracellular RNA, exosomes, retroelement, pseudogene
INTRODUCTION
Exosomes are 40–100 nm extracellular vesicles secreted by mammalian cells, the result of multivesicular endosome fusion to the cells plasma membrane. These extracellular particles are involved in intercellular communication by acting as transport vehicles for proteins and RNA, both mRNA and non-coding RNAs (van der Pol et al., 2012). Small non-coding RNAs, such as miRNAs, as well as long non-coding RNAs (lncRNAs) have also been observed packaged into exosomes (Kogure et al., 2011) and found to act as messengers in cell-to-cell communication (Valadi et al., 2007). Non-coding RNAs exhibit a plethora of functions, ranging from chromatin and enhancer modifiers to scaffolding and transcript sponge functions (Morris and Mattick, 2014). Some lncRNAs such as MALAT1 and NEAT1 have also been found to function in cancer cell signalling (Clemson et al., 2009; Guffanti et al., 2009; Gutschner et al., 2013; Kogure et al., 2013; Gezer et al., 2014; Chen et al., 2015; Guo et al., 2015; Y. Li et al., 2015; Pan et al., 2015), with MALAT1 being previously observed to be packaged into exosomes (Gezer et al., 2014).
One possible mode of action for these particular lncRNAs might be to act distally in the creation of a pre-metastatic niche for metastatic cells (Bard et al., 2004; Hood et al., 2011). We set out to explore this notion and find a set of exosome-associated lncRNAs that functionally modulate cell viability by interactions with several proteins involved in cellular metabolism and signalling. Previous studies have identified that RNA content can be dependent on the cell type from which it originated from. This has led to an increase in the investigation of exosomal RNAs as possible biomarkers for a variety of cancers (Skog et al., 2008; Nilsson et al., 2009; Ogata-Kawata et al., 2014). It has also been observed that Viruses are capable of affecting RNA content within exosomes, which can repress the target genes of the virus (Pegtel et al., 2010; El Andaloussi et al., 2013). In light of these discoveries we set out to determine whether a cervical cancer affected by the HPV virus (in this case Hela) affect the RNA content in the exosome when compared to a cervical cell line without HPV (C33A).
MATERIALS AND METHODS
Cell Culture
Hela and C33A cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin solution and 2 mM L-glutamine. The cells were grown in T-175 flasks at 37°C and 5% CO2. Cultures were passaged when they were ~90% confluent. The media was removed from the cells and they were washed with 8 mL of 1X PBS. Trypan blue exclusion was used to assess cell viability and estimate cell numbers.
Calcium Phosphate Transfection of Hela Cells
Hela cells were transfected with 5-Bromouridine 5′-Triphosphate (BrUTP) using the calcium phosphate transfection method (Kingston et al., 2003). When cells take up the BrUTP, it is utilised in transcription and incorporated into RNA transcripts causing them to be labelled. The BrUTP-labelled transcripts are packaged into exosomes, which are isolated from the cell culture media, described below. One day prior to transfection, 100 mm cell culture plates were seeded with approximately 2 million Hela cells and the volume of each dish was made up to 10 mL with fresh media. For each plate, a transfection mix was made by adding 500 μL of 1X HBS and 0.5 μL of 10 mM BrUTP to a sterile microcentrifuge tube and vortexing to mix. To each transfection mix 30 μL of 2.5 M calcium chloride was added. The tubes were mixed by vortexing and left to sit at room temperature for 20 minutes. The calcium phosphate transfection mix was added to each plate drop-wise and mixed by rocking. The final BrUTP concentration in each plate was 0.5 μM. Exosomes from the cell culture media were collected over a 3 day period post-transfection.
Exosome Isolation
Exosomes were isolated from conditioned media as they are secreted by cells into the culture media. During cell passaging, the conditioned media was collected and centrifuged at 700 x g for 5 minutes to remove any live cells. The conditioned media was stored at −20°C. To isolate exosomes from the conditioned media, ultracentrifugation on a 30% sucrose density cushion was used. The conditioned media was centrifuged at 10,000 x g for 30 minutes with a Hitachi P28S rotor to remove any dead cells or cellular debris. The supernatants were then transferred to clean 40PA tubes and centrifuged at 100,000 x g for 90 minutes in the P28S rotor to pellet the exosomes and contaminating proteins. The supernatant was discarded and the pellet was resuspended with 1 mL of 1X PBS. The resuspended exosomes were pooled together and the total volume was made up to 9 mL with 1X PBS. In a 13PA tube, 1 mL of the 30% sucrose density cushion (30 g sucrose, 2.4 g Tris base, 100 mL deuterium oxide, pH 7.4) was added and the resuspended exosome solution was placed above the sucrose cushion. Great care was taken not to disturb the interface between the two solutions. Using a Hitachi P40S rotor, the sucrose cushion and exosome solution was centrifuged at 100,000 x g for 90 minutes. During this ultracentrifugation step contaminating proteins are pelleted while the exosomes float in the sucrose cushion. The sucrose cushion was transferred to a clean 13PA; 9 mL of 1X PBS was added and the exosomes were washed by centrifuging at 100,000 x g for 90 minutes. The supernatant was removed and final exosome pellet was resuspended in 100 μL of 1X PBS. All centrifugation steps were performed at 4°C using a Hitachi HiMac CP100WX ultracentrifuge. A Bradford protein assay (Bio-Rad) was performed to estimate the exosome concentration.
Western Blot of Exosomal Markers
Western blots of exosomal markers CD9, CD63 and flotillin-1 were performed to confirm the isolation of exosomes. The cellular protein calnexin was also analysed as a control, as it is not present in exosomes. The anti-CD9 and anti-CD63 antibodies required non-reducing conditions while reducing conditions were required for the anti-flotillin-1 and anti-calnexin antibodies required. Hela exosomes (10 μg) and cells (105) were mixed with either non-reducing sample buffer (4X Laemmli sample buffer) or reducing sample buffer (4X Laemmli sample buffer with 10% 2-mercaptoethanol) depending on the condition required by the specific antibody. The samples in the reducing buffer were heated at 95°C for 10 minutes. The samples and Precision Plus Protein Kaleidoscope Standard were loaded onto a 4–20% gradient Mini-PROTEAN TGX precast gel. Electrophoresis was performed at 150 V for 45 minutes in 1X Tris/Glycine/SDS buffer. The proteins in the gel were transferred onto to a PVDF membrane using a Bio-Rad Mini-Trans Blot Electrophoretic Transfer Cell at 90 V for 1 hour in 1X Tris/glycine buffer with 20% methanol. The PVDF membranes were blocked overnight at 4°C in 15 mL of 1X PBS/5% skim milk/0.1% Tween-20 blocking solution. The anti-CD9 (ab2215), anti-CD63 (ab59479) and anti-calnexin (ab21290) primary antibodies were diluted 1:1000 in blocking solution and the anti-flotillin-1 antibody (ab41927) was diluted 1:500. The membranes were incubated with 3 mL of a specific primary antibody solution for 4 hours at room temperature on a shaking platform. They were then washed three times for 10 minutes in 50 mL of 1X PBS/0.1% Tween-20 at room temperature with shaking. The anti-mouse IgG horseradish peroxidase conjugate secondary antibody (Bio-Rad) was diluted 1:3000 times in blocking solution. The membranes were incubated in 3 mL of secondary antibody solution for 1 hour at room temperature with shaking before being washed again three times. Bound secondary antibodies were colourimetrically detected using an Opti-4CN Substrate Kit.
Electron Microscopy of Exosomes
Electron microscopy of exosomes was performed by Rebekka Williams of the Children’s Cancer Institute Australia (as a fee for service) to confirm that exosomes were isolated from conditioned cell culture media. A 10 μL suspension of Hela exosomes was fixed with 2% glutaraldehyde for 30 minutes. The fixed exosomes (5 μL) were dropped onto 300 mesh carbon coated formvar-copper TEM grids and left to settle for 15 minutes. Excess sample was removed by wicking with filter paper and the grids were washed with water droplets. The exosomes were negatively stained by placing the grid on a droplet of 2% uranyl acetate for 2 minutes and any excess was wicked off using filter paper. A Joel JEM-1400 transmission electron microscope was used to image the grids. Negative control grids containing no exosomes (only glutaraldehyde and PBS) were also prepared.
Sequencing of Exosomal RNA
The Ramaciotti Centre for Gene Function Analysis performed library preparation and sequencing of Hela and C33A exosomal RNA on an Illumina HiSeq 2000 as a fee for service. The 100 bp paired end reads generated by RNA sequencing were analysed by first quality control performed using FastQC v0.10.1 available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Reads were quality filtered, trimmed and adapter sequences were removed using Trimmomatic v0.3.0 (Lohse et al., 2012). Reads were then aligned to the hg19 Homo sapiens genome with tophat2 (v2.0.8b) using the default settings except for the following (--b2-sensitive) (Kim et al., 2013). Transcripts were assembled using Cufflinks v2.1.1 (Trapnell et al., 2010). All transcripts were merged using cuffmerge and FPKM values were determined using cuffdiff.
Fluorescence-Activated Cell Sorting (FACS)
FACS was used to demonstrate the transfer of RNA from exosomes into recipient cells. This was achieved by exposing C33A cells to Hela exosomes (20μl) containing either normal RNA or BrUTP-labelled RNA for 24 or 48 hours. The exosome exposed cells were collected, permeabilised and stained with an anti-BrdU primary antibody (B8434) and an IgG-FITC secondary antibody (F0257) (Sigma-Aldrich). The C33A exosome exposed cells were analysed with a BD Biosciences FACSCanto II flow cytometer at the Biological Resources Imaging Laboratory at UNSW as a fee for service.
Chromatin Immunoprecipitation of Biotin linked RNAs
Chromatin immunoprecipitation (ChIP) was performed on T7 expressed biotin dUTP containing lncNRAs (Table S1). The biotin RNAs were generated as described in (Johnsson et al., 2013; Saayman et al., 2014). The Biotin containing RNAs were transfected into HEK293 cells and CHIP carried out 48 hours later (as described in (Johnsson et al., 2013)). The enrichment of the various RNAs at predicted target loci (Figure S2) was determined by qPCR with various primers (Table S2).
Immunoprecipitation and Mass Spectrometry of RNA Associated Proteins
Proteins associated with exosomal RNAs were determined by transfection of biotin labelled RNAs (Table S1) into 293HEK cells and immunoprecipitation carried out 48hrs later from either the cells or exosomes collected from the transfected cells (as described in (Hawkins and Morris, 2010; Saayman et al., 2014)). The resultant elutes were then analysed by mass spectrometry using the fee for service Bioanalytical Mass Spectrometry Facility at UNSW.
RESULTS
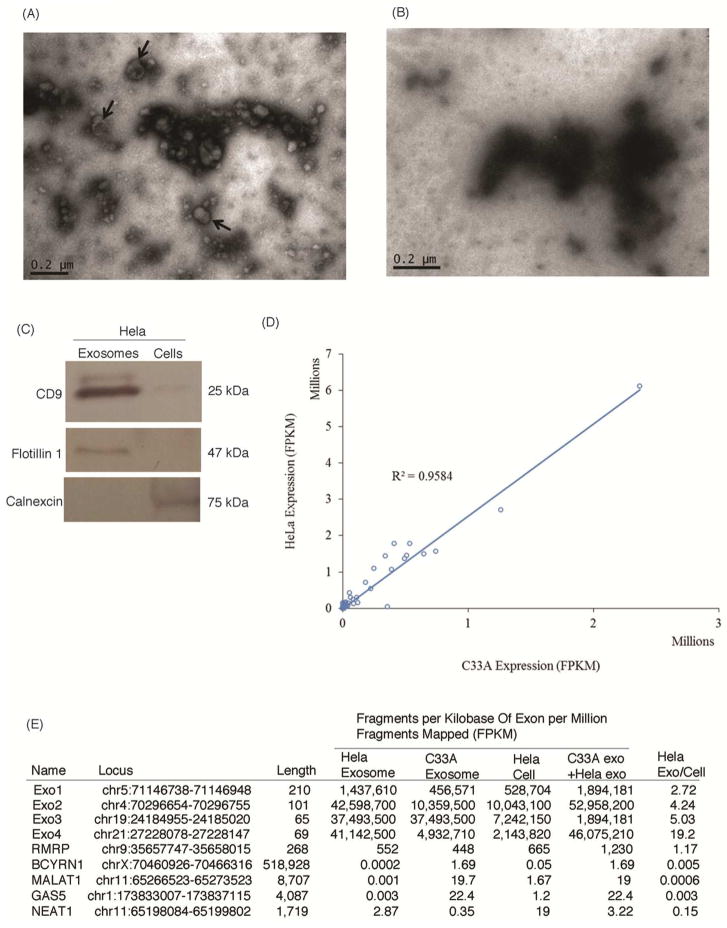
To explore the notion that particular lncRNAs are preferentially packaged into exosomes we collected extracellular particles containing exosomes from cultures grown in pre-cleared conditions (Figure 1A–B). Western blot analysis for exosomal markers CD9 and Flotillin-1 contrasted with the cellular protein Calnexin (Taylor and Gercel-Taylor, 2011) confirmed that the predominant isolate from the extracellular particles utilized here contained known exosomal associated proteins (Figure 1C). CD9 is a tetraspanin found in cells but also recognized as an exosomal marker as they are found highly enriched on the surface of exosomes and Flotillin-1 is a lipid raft associated protein found in exosomes while Calnexin is an endoplasmic reticulum protein predominately localized in the cell (Thery et al., 2006). Collectively, these data suggest that our protocol, based on (Thery et al., 2006; Graça Raposo and Stoorvogel, 2013b) results in the isolation of exosome associated proteins.
Figure 1. Analysis of Hela and C33A associated exosomes.
(A–B) Detection of exosomes by electron microscope. (A) a negative control grid with no exosomes and (B) a grid containing Hela exosomes. The Hela exosomes are approximately 40 nm to 100 nm in size. The three arrows mark exosomes that display the commonly observed cup-shaped morphology. (C) Western blot of exosomal markers. Western blots for the exosomal markers CD9, Flotillin-1 and Calnexin was performed on Hela exosomal and cellular proteins to confirm that exosomes were isolated from conditioned cell culture media using ultracentrifugation and a 30% sucrose density cushion. (D) Correlation in the RNA content of Hela and C33A exosomes. Hela and C33A exosomal RNA transcripts greater than 4 FPKM were plotted against each other and a R2 value of 0.9584 was calculated. (E) Candidate transcripts found in exosomes from both C33A and Hela cells along with the enrichment of MALAT1, BCYRN1, GAS5 and NEAT1, previously reported in cancer exosomes and the enzymatic RNA RMRP. The top candidate RNAs Exo1-4 and RMRP and were investigated.
Next, we sought to determine the predominant species of lncRNAs associated with these exosomes. Hela and C33A exosomes were isolated and their RNA content sequenced using Illumina RNA-sequencing. A strong correlation in the RNA content of Hela and C33A exosomes was observed as determined and those RNA transcripts greater than 4 fragments per kilobase of exon per million fragments mapped (FPKM) plotted against each other (Figure 1D). The observed R2 value of 0.9584 suggested that the RNA content of Hela and C33A exosomes is approximately 96% similar (Figure 1D). Notably, some of the previously observed cancer-exosome associated lncRNAs BCYRN1(Hu and Lu, 2015), MALAT1 (Gutschner et al., 2013), GAS5 (Smith and Steitz, 1998; Mourtada-Maarabouni et al., 2008; Mourtada-Maarabouni et al., 2009; Kino et al., 2010) and NEAT1(Souquere et al., 2010; Chen et al., 2015; Guo et al., 2015; Y. Li et al., 2015; Pan et al., 2015; Wang et al., 2016) were not observed present above cellular RNAs in the Hela and C33A exosomes assessed here (Figure 1E and Table S3). Interestingly, the top 4 most abundant transcript candidates observed associated with both Hela and C33A exosomes were un-annotated transcripts emanating from genomic deserts that contained DNase hypersensitive regions of high histone acetylation as well as RNAPII and CTCF binding sites. Mapping to Human Feb. 2009 (GRCh37/hg19) assembly suggested that these transcripts are ribosomal RNAs (Figure S1A–D). These transcripts were designated Exo1-4 (Figures 1E, S1A–D and Tables S1 and S3) and were contrasted with the enzymatic RNA, RMRP, a mitochondrial RNA-processing endoribonuclease that is imported into the mitochondria to cleave mitochondrial RNA (Hsieh et al., 1990), which was also observed to be lowly associated with the Hela and C33A exosomes (Figure 1E, Tables S1 and S2)
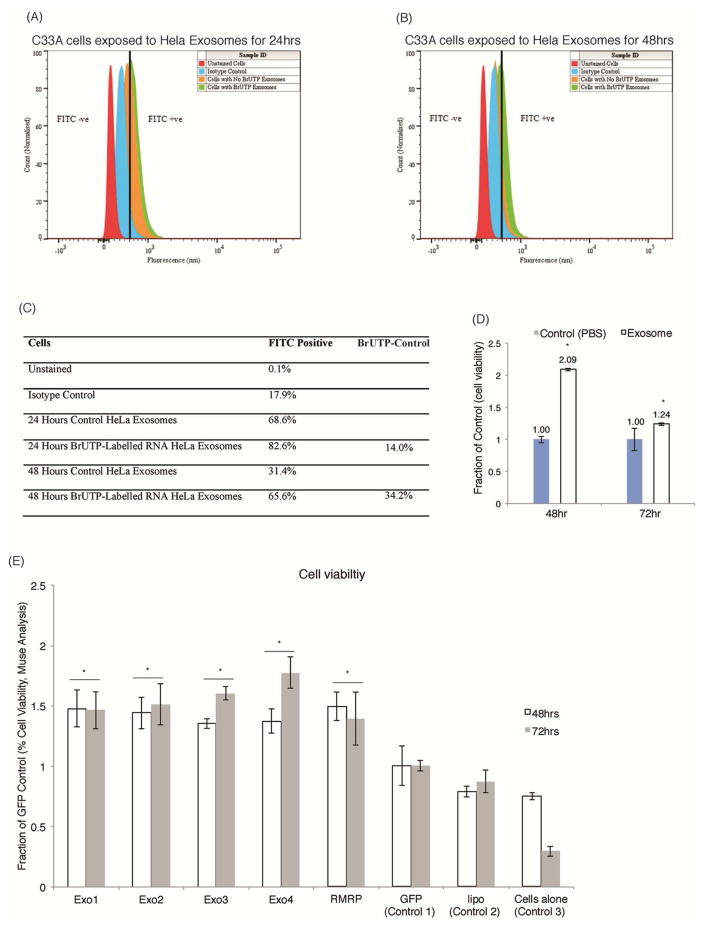
Previous studies have demonstrated that lncRNAs can be transmitted from one cell to another by the action of the exosome (van der Pol et al., 2012). To determine to what extent the exosome associated transcripts are transmitted between Hela and C33A cells, a BrUTP cellular RNA labelling assay was performed. In this assay 5-Bromouridine 5′-Triphosphate UTPs (BrUTP) are transfected into Hela cells and the resultant cellular exosomes collected 24–48hrs post-transfection. The exposure of C33A cells to these BrUTP labelled RNA containing exosomes demonstrated that Hela exosomes can transfect C33A cells and disseminate BrUTP transcripts to ~34% of the treated cells over a 48hr time course (Figures 2A–C). To determine to what extent these Hela derived exosomes can affect HEK293 cell function, HEK293 cells were exposed to Hela cell derived exosomes. Notably, Hela exosome treatment of HEK293 cells results in significant changes in cell viability, suggesting that these exosomes functionally modulate the recipient target cells (Figure 2D). This observed effect on cell viability was dosage dependent, suggesting that the lncRNAs exhibit a bona fide and quantitative effect on the recipient cells (Figure S2). To determine to what extent the candidate non-coding RNAs, Exo1-4 and RMRP are functionally involved in modulating recipient cells, cultures were transfected and the effects on cell viability determined. All of the top candidate exosome associated lncRNAs, Exo1-4 and RMRP functionally enhanced cell viability relative to controls (Figure 2E). Collectively these observations suggest that exosomes contain a subset of lncRNAs that modulate recipient cell function resulting in enhanced recipient cell viability.
Figure 2. Exosomal transfer enhances recipient cell viability.
(A–C) Exosomal-mediated RNA transfer into recipient cells was determined. C33A cells exposed to Hela exosomes containing normal RNA or BrUTP-labelled RNA were analysed by FACS to determine if RNA can transfer into cells from exosomes. RNA transfer was determined by FITC fluorescence in cells exposed to exosomes for (A) 24 hours or (B) 48 hours and (C) the percentage transfer of BrUTP-labelled exsomal RNA determined. (D) Exosomal cell viability of HEK293 cells treated in triplicate with Hela exosomes or a PBS control. Cell viability was the analysed at 48 and 72 hours using the Millipore Muse cell analyser. The effects of the top candidate exosome associated RNAs expression on cell viability (E) HEK293 cells were transfected in triplicate to overexpress candidate lncRNAs (Exo1-4, and RMRP) along with a GFP control. Lipofectamine only and untransfected cells served as the controls. Cell viability was measured using the Millipore Muse cell analyzer at 48 and 72 hours post-transfection. For (D–E) Error bars represent standard error of the mean, * represents P<0.05 relative to GFP Control 1.
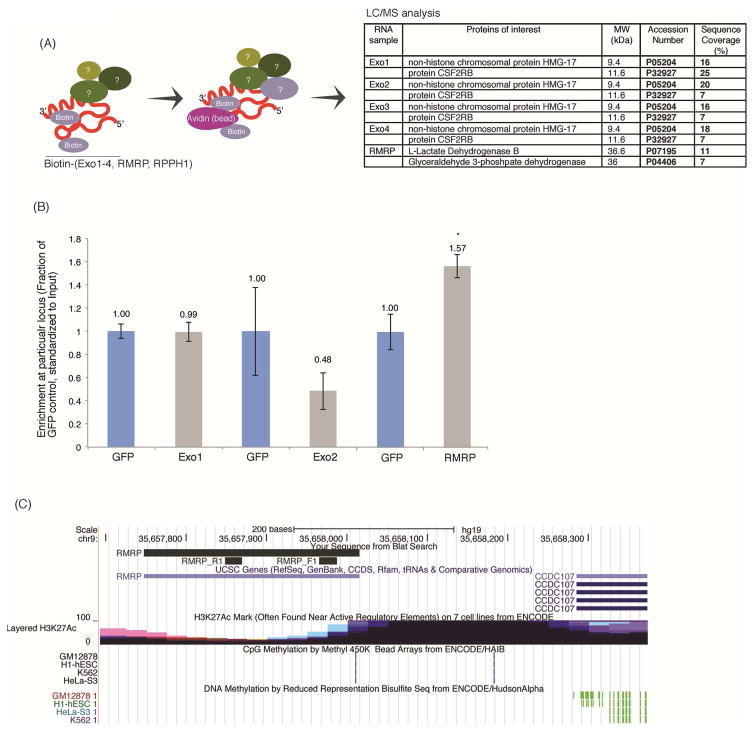
The list of functions of non-coding RNAs continues to expand (Morris and Mattick, 2014). To determine how the exosome-associated lncRNAs Exo1-4 and RMRP might function in the cell, biotin-labelled lncRNAs were generated, transfected into cells and the resultant complexes eluted and bound protein partners determined by mass-spectrophotometry analysis (MS)(Figure 3A). Interestingly, relative to the biotin-GFP control transcripts, Exo1-4 and RMRP precipitated with L-lactate dehydrogenase B (LDHB), high-mobility group protein 17 (HMG-17), and CSF2RB; proteins involved in metabolism, nucleosomal architecture and cell signalling respectively (Figure 3A). Previous studies have found some lncRNAs can associate with homologous containing loci in the genome. To determine to what extent the exosome-associated lncRNAs Exo1-4 and RMRP might be targeting the genome a chromatin immunoprecipiation analysis was carried out with the biotin-labelled lncRNAs. Only RMRP exhibited any binding to homologous containing genomic loci relative to the controls (Figures 3B). Notably, the RMRP binding locus is on the shoulder of histone acetylation for CCDC107, a gene expressing a coiled-coil containing membrane protein (Figure 3C) which also contains Rho guanine nucleotide exchange factor 39 (ARHGEF39), a gene known to be involved in cell migration, in overlapping and antisense orientation (Figure S3). Collectively, these data suggest that the exosome-associated lncRNAs interact with particular cellular proteins to affect cell viability and the majority of these interactions, with the exception of RMRP, do not appear to be chromatin based.
Figure 3. Mechanism of exosome associated RNA regulation of cell viability and associated proteins.
(A) Biotin containing transcripts Exo1-4, and RMRP were transfected into HEK293 cells and elutes from the ChIP sent for mass spec analysis at the UNSW BMFS facility in triplicate. Proteins present in at least two of the samples and not in the GFP controls were flagged as candidates of interest. (B) Relative enrichment of the candidate exosome associated RNAs at homology containing loci. Biotin tagged transcripts (Exo1, Exo2 and RMRP were transfected into HEK293 cells in triplicate and forty-eight hours later immunoprecipitated with steptavidin beads. Enrichment of each biotin transcript is shown at homology containing loci relative to the GFP control. Error bars represent standard error of the mean,* represents p<0.05. (C) A UCSC snapshot of the RMRP target locus found enriched by CHIP (in B above) is shown with the relative CHIP primer binding sites and associated epigenetic state of the locus
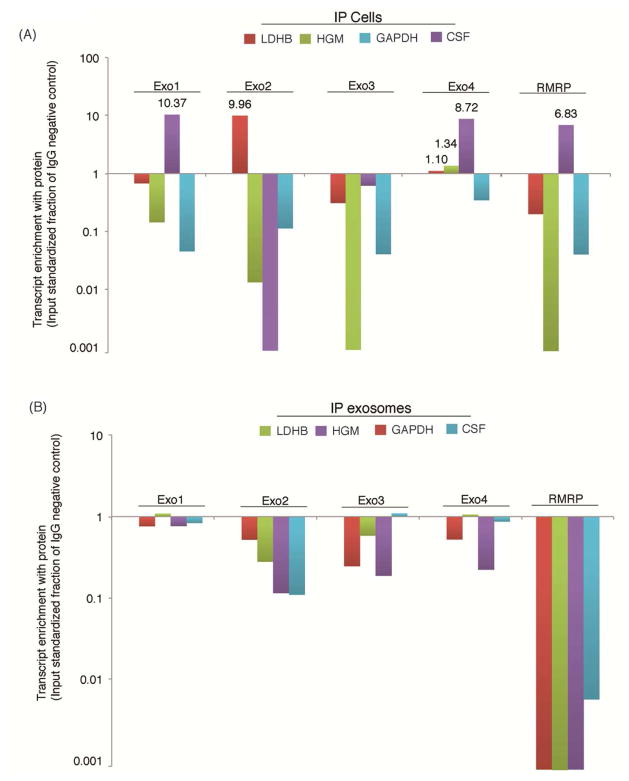
The observations that the exosome associated lncRNAs bound to particular proteins (Figure 3B), suggested that either the candidate lncRNAs bind particular proteins and are packaged into exosomes or functionally interact with the candidate proteins upon being introduced into the cells via exosome uptake in recipient cells. To determine to what extent the exosome associated lncRNAs are interacting with the candidate proteins in either exosomes or recipient cells an immunoprecipitation of the candidate proteins LDH, HGM, CSF and GAPDH (Figures 3A and 3B) was carried out in both cells and exosomes and qRTPCR carried out on the elutes for detection of the candidate transcripts in association with the proteins of interest. Interestingly, the exosome associated lncRNAs only associated with CSF, LDH, and HGM in the context of the recipient cells (Figure 4A) and not within the context of the exosome (Figure 4B). These observations suggest that the exosome-associated lncRNAs interact with particular cellular proteins; CSF, LDH and HGM, upon exosomal entry into the target cells to affect cellular viability.
Figure 4. Enrichment of various exosome-associated lncRNAs with mass spec identified proteins.
IP was carried out for the various mass spec identified proteins in triplicate with either (A) cells or (B) exosomes. The resultant elutes were subjected to qRT-PCR and enrichment of each lncRNA with the candidate protein relative to the IgG negative control following input standardization. The averages of triplicate IPs are shown.
DISCUSSION
To date many studies have been carried out looking at proteins, mRNAs and even miRNAs associated with exosomes (reviewed in (G. Raposo and Stoorvogel, 2013a)). However, little work has been carried out with regards to the association and putative role of lncRNAs in exosome biology. In light of the emerging realizations that lncRNAs play an underappreciated role in controlling chromosomal content, in particular transcriptional and epigenetic states, we sought to determine those lncRNAs associated with both Hela and C33A cell exosomes. We find here that exosomes exhibit an abundance of transcripts (Table S3), with the top 4 most abundant exosome associated transcripts in both Hela and C33A cells being Exo1-4 (Figures 1, S1 and Tables S1–S2), that mapped to intergenic gene deserts, several kilobases away from their nearest transcribed coding and/or non-coding neighbour. These transcripts have been considered ribosomal associated transcripts and are found embedded in DNase I hypersensitive clusters with exceedingly high levels of histone H3 at lysine 27 acetylation (H3K27ac), two chromatin marks associated with actively transcribed regions of the genome. Interestingly, we were unable to observe significant enrichment of well-known lncRNAs involved in human cell cancers, BCYRN1, MALAT1, GAS5 and NEAT1, which have been observed previously to be associated with exosomes in human cells and human cell cancers (Souquere et al., 2010; Gutschner et al., 2013; Chen et al., 2015; Guo et al., 2015; Hu and Lu, 2015; Y. Li et al., 2015; Pan et al., 2015; Wang et al., 2016).
The observations presented here indicate that the top candidate assessed exosome associated lncRNAs Exo1-4 and RMRP, when over-expressed in recipient cells, affect cell viability. Mechanistically, we find that the candidate exosome associated lncRNAs interact directly with several proteins. The exosome associated lncRNA Exo2 appears to interact directly with lactate dehydrogenase B (LDHB), while Exo4 and RMRP interact with High mobility group protein 17 (HMG-17), and Type I cytokine high affinity receptor for IL-3, IL-5 and CSF (CSF2RB) in cells (Figure 5). GAPDH is known to bind AU rich transcripts (Nagy and Rigby, 1995) and LDHB is known to bind RNA specifically in the NAD+ RNA binding region (Pioli et al., 2002). Interestingly, LDHB appears to be packaged into exosomes (M. Li et al., 2012), though observations here did not indicate that LDHB was binding directly to any of the exosome associated lncRNAs within the context of the exosome. Regardless, LDHB may be one protein that can bind lncRNAs and recruit them specifically to blebbing exosomes in particular cell types, though the data presented here does not suggest this is the case in Hela and C33A cells. Notably, the proteins LDHB, GAPDH, CSF and HMG-17 found here to be associated with exosome associated lncRNAs have been reported previously to be involved in cellular metabolism, nucleosomal architecture and cell signalling. It is also noteworthy that RMRP could also have significant impacts on cell viability, when introduced to recipient cells from the context of exosomes, by direct cis interactions in the chromatin, possibly affecting CCDC107 and ARHGEF39 expression and cell motility (Figure S3). The exosome-associated lncRNAs described here don’t appear to interact with the cellular proteins in the context of the exosome but rather specifically inside the recipient cells (Figure 5), suggesting that the regulatory effect these exosome associated transcripts play is upon entry into recipient cells and not within the context of the exosome per say. Collectively, the observations presented here suggest that exosome-associated lncRNAs functional modulate cellular protein function and expand on the ever-growing functional characteristics of lncRNAs in the human cell. Knowledge of this molecular pathway may prove useful in controlling cellular micro-environments or in developing designer RNA packaged exosomes, an eventuality that could have profound impact on patient specific disease targeted therapeutics.
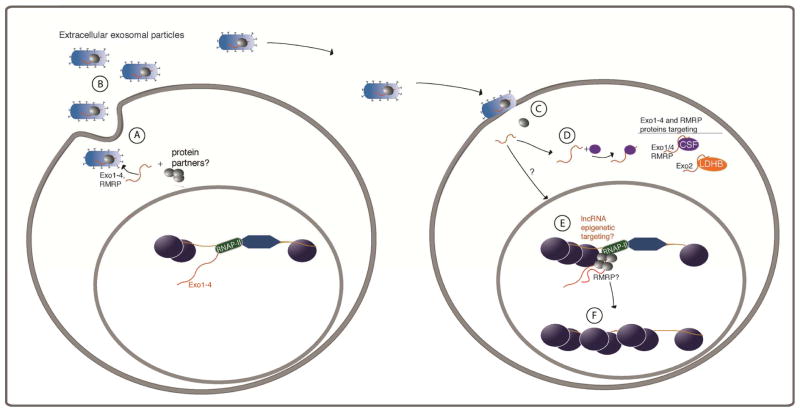
Figure 5.
Model for exosomal mediated spread of lncRNAs. (A) RNAs Exo1-4 and RMRP may interact with host proteins or may be free of interactions and be packaged into exosomes by a specific, yet to be determined set of exosomal directed proteins. (B) The lncRNA containing exosomes are blebbed from the cell and can then (C) interact with recipient target cells. Once inside the recipient cells the lncRNAs can (D) bind proteins, such as Exo1, Exo4 and RMRP binding to CSF or Exo2 binding to LDHB or Exo4 binding HGM-17 (HGM) to affect protein function and cellular states. (E) Some yet to be characterized exosomal-associated lncRNAs such as RMRP may interact with epigenetic regulatory mechanisms to control particular gene expression states by (F) targeting stable epigenetic marks that may lead to targeted heterochromatin.
Supplementary Material
Acknowledgments
We acknowledge the helpful bioinformatics discussions with Shingo Miyauchi, INRA, UMR1163 Biodiversite et Biotechnologie Fongiques, Aix Marseille Universite, UMR1163 Biodiversite et Biotechnologie Fongiques, F-13288 Marseille, France.
FUNDING INFORMATION This project was supported by NIAID P01 AI099783-01, CA151574-01 and ARC Future Fellow FT130100572 to KVM.
Footnotes
The authors declare no conflict of interest from this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Bard MP, Hegmans JP, Hemmes A, et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- Chen X, Kong J, Ma Z, et al. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res. 2015;5:2808–2815. [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S, Lakhal S, Mager I, et al. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65:391–397. doi: 10.1016/j.addr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Gezer U, Ozgur E, Cetinkaya M, et al. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell biology international. 2014;38:1076–1079. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- Guffanti A, Iacono M, Pelucchi P, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Chen W, Luo Y, et al. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8:5395–5402. [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcr. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- Hsieh CL, Donlon TA, Darras BT, et al. The gene for the RNA component of the mitochondrial RNA-processing endoribonuclease is located on human chromosome 9p and on mouse chromosome 4. Genomics. 1990;6:540–544. doi: 10.1016/0888-7543(90)90483-b. [DOI] [PubMed] [Google Scholar]
- Hu T, Lu YR. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015;15:36. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson P, Ackley A, Vidarsdottir L, et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Okayama H. Calcium phosphate transfection. Curr Protoc Cell Biol. 2003;Chapter 20(Unit 20):23. doi: 10.1002/0471143030.cb2003s19. [DOI] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, et al. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science signaling. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure T, Lin WL, Yan IK, et al. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure T, Yan IK, Lin WL, et al. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes & cancer. 2013;4:261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Aliotta JM, Asara JM, et al. Quantitative proteomic analysis of exosomes from HIV-1-infected lymphocytic cells. Proteomics. 2012;12:2203–2211. doi: 10.1002/pmic.201100376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li Y, Chen W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6:27641–27650. doi: 10.18632/oncotarget.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic acids research. 2012;40:W622–627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014 doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Hedge VL, Kirkham L, et al. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J Cell Sci. 2008;121:939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. British journal of cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan LJ, Zhong TF, Tang RX, et al. Upregulation and clinicopathological significance of long non-coding NEAT1 RNA in NSCLC tissues. Asian Pac J Cancer Prev. 2015;16:2851–2855. doi: 10.7314/apjcp.2015.16.7.2851. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioli PA, Hamilton BJ, Connolly JE, et al. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J Biol Chem. 2002;277:35738–35745. doi: 10.1074/jbc.M204002200. [DOI] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology. 2013a;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology. 2013b;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S, Ackley A, Turner AM, et al. An HIV-Encoded Antisense Long Noncoding RNA Epigenetically Regulates Viral Transcription. Mol Ther. 2014;22:1164–1175. doi: 10.1038/mt.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S, Beauclair G, Harper F, et al. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Seminars in immunopathology. 2011;33:441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. In: Bonifacino Juan S, et al., editors. Current protocols in cell biology. Unit 3. Chapter 3. 2006. p. 22. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van der Pol E, Boing AN, Harrison P, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacological reviews. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- Wang P, Wu T, Zhou H, et al. Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. Journal of experimental & clinical cancer research : CR. 2016;35:22. doi: 10.1186/s13046-016-0297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.