Abstract
We report the extraction of a bed bug mitogenome from high-throughput sequencing projects originally focused on the nuclear genome of Cimex lectularius. The assembled mitogenome has a similar AT nucleotide composition bias found in other insects. Phylogenetic analysis of all protein-coding genes indicates that C. lectularius is clearly a member of a paraphyletic Cimicomorpha clade within the Order Hemiptera.
Keywords: Hemiptera, pest, true bugs, blood feeding, arthropods
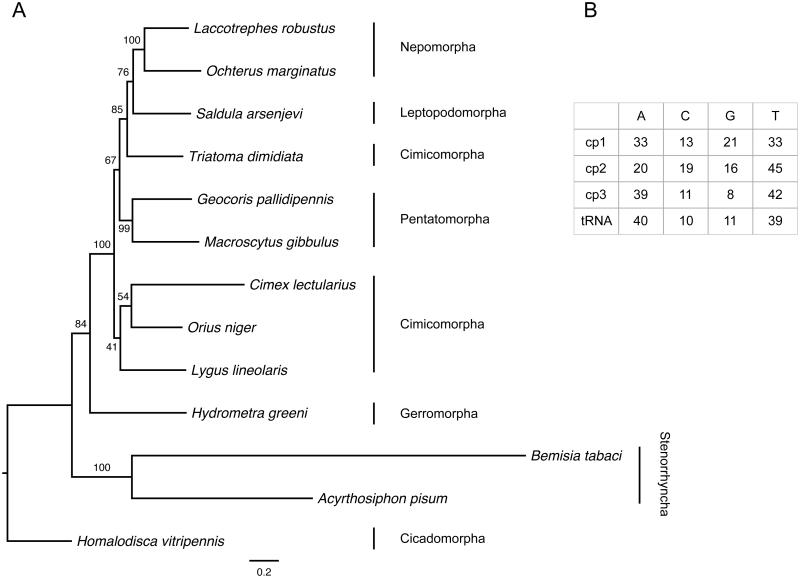
The common bed bug, Cimex lectularius Linnaeus, 1758 (Hemiptera: Cimicidae), has been intimately associated with humans for thousands of years (Panagiotakopulu & Buckland 1999; Booth et al. 2015). It is an obligate ectoparasite that primarily feeds on human blood in a hematophagic lifestyle, but will readily feed on many bird and mammalian species as well (Usinger 1966). The insecticide-susceptible laboratory strain Har-73 (= Harlan) of C. lectularius was used for whole-genome shotgun sequencing and sequence assembly de novo, performed by two independent research groups: one in New York City, based primarily at the American Museum of Natural History (AMNH) and Weill Cornell Medicine (WCM) (Rosenfeld et al. 2016), and another one at Baylor College of Medicine (BCM) as part of the i5k genome sequencing initiative (http://www.arthropodgenomes.org/wiki/i5K) (Benoit et al. 2016). Specimens are kept in the AMNH Invertebrate Zoology collection and stored in liquid nitrogen in the Ambrose Monell Cryo Collection (AMCC). Living colonies are maintained by L.N. Sorkin and fed on human blood and an inbred line is maintained by C. Schal and fed on rabbit blood. Purified DNA and RNA samples are also stored in the AMCC and at WCM. Sequence data for the original genome projects can be accessed on GenBank/EMBL/DDBJ under BioProject PRJNA259363 and PRJNA167477. In order to identify orthologous loci and assemble the bed bug mitogenome, we first queried complete mitogenome sequences from related species Adelphocoris fasciaticollis (GenBank accession NC_023796), Empoasca vitis (NC_024838), Orius sauteri (NC_024583), Peirates arcuatus (NC_024264), and Triatoma dimidiata (NC_002609) against the genome assemblies of both genome projects. This did not yield significant hits; therefore, we subsequently queried the abovementioned mitogenome sequences against the high-quality Illumina reads, which yielded bona fide matches. The mitogenome assembly was annotated using MITOS (Bernt et al. 2013), resulting in 13 protein-coding genes, two rRNA loci and 21 tRNA loci (tRNA-Leu was missing), and a mitogenome size of 15,217 bp. The mitogenome sequence is deposited in GenBank under accession code KU350871. We compared our mitogenome sequence to a subset of related mitogenome sequences (Li et al. 2011) in a maximum likelihood phylogenetic analysis using the 13 encoded proteins in RAxML 8.2.4 (Stamatakis 2014) with the MtArt replacement matrix (Abascal et al. 2007) and empirical residue frequencies, along with among-site rate heterogeneity modeled with the Γ distribution and four discrete rate categories (Yang 1994), through ten searches starting from random-addition maximum parsimony trees (Figure 1A). C. lectularius was placed as sister to Orius niger and Lygus lineolaris within the Cimicomorpha clade. This is in agreement with a larger phylogenetic analysis of hemipteran mitochondrial genomes that displayed a comparable lack of branch support for the backbone of the phylogenetic tree (Song et al. 2012). In terms of nucleotide composition, the C. lectularius mitogenome was slightly purine-rich (52%) with a high AT content (71%). Compositional bias was pronounced (Figure 1B), as is common in insects (Cameron 2014) with high AT bias across tRNA loci and 3rd codon positions – a characteristic also shared by other Hemiptera (Liu & Liang 2013). This sequence will serve as a resource for evolutionary and comparative genomics studies of true bugs and other animals, as well as the basis of microevolutionary studies of bed bug colonization, infestation, and heteroplasmy (Robison et al. 2015).
Figure 1.
(A) Phylogenetic relationships among Hemiptera species based on the translated sequences of 13 protein-coding genes. Values at the internode branches denote support drawn from 500 rapid bootstrap pseudoreplicates (Stamatakis et al. 2008) mapped onto the best-known maximum likelihood phylogram (-lnLik=63126.175, α=0.53). Infraorder groupings are indicated. GenBank accessions used: Acyrthosiphon pisum (NC_011594), Bemicia tabaci (NC_006279), Geocoris pallidipennis (NC_012424), Homalodisca vitripennis (NC_006899), Hydrometra greeni (NC_012842), Laccotrephes robustus (NC_012817), Lygus lineolaris (NC_021975), Macroscytus gibbulus (NC_012457), Ochterus marginatus (NC_12820), Orius niger (NC_12429), Saldula arsenjevi (NC_012463), Triatoma dimidiata (NC_002609). (B) Nucleotide percent composition for each codon position and all tRNA loci.
Acknowledgements
The AMNH team thanks the Sackler Institute for Comparative Genomics, the Korein Foundation and The Lewis and Dorothy B. Cullman Molecular Systematics Program at the AMNH for support of this project. C.E. Mason would to like to thank the Alfred P. Sloan Foundation (grant no. 2015-13964), the Bert L. and N. Kuggie Vallee Foundation, the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts, the WorldQuant Foundation, the STARR Consortium (grant no. I9-A9-071), and support from the National Institutes of Health (NIH) (grant no. R25EB020393 and the Ruth L. Kirschstein National Research Service Award). D. Reeves was supported by NIH grant no. F31GM111053. The BCM team acknowledges funding for genome sequencing, assembly and automated annotation from the National Human Genome Research Institute (grant no. U54 HG003273 to Richard A. Gibbs). W. Booth was supported by the Oklahoma Center for the advancement of Science and Technology (grant no. HR13-211).
Footnotes
Declaration of Interest
The authors have no competing interests to declare.
References
- Abascal F, Posada D, Zardoya R. MtArt: a new model of amino acid replacement for Arthropoda. Mol Biol Evol. 2007;24:1–5. doi: 10.1093/molbev/msl136. [DOI] [PubMed] [Google Scholar]
- Benoit JB, Adelman ZN, Reinhardt K, Dolan A, Poelchau M, Jennings EC, Szuter EM, Hagan RW, Gujar H, Shukla JN, et al. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat Commun. 2016;6:10165. doi: 10.1038/ncomms10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Booth W, Balvin O, Vargo EL, Vilimova J, Schal C. Host association drives genetic divergence in the bed bug, Cimex lectularius. Mol Ecol. 2015;24:980–992. doi: 10.1111/mec.13086. [DOI] [PubMed] [Google Scholar]
- Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- Li H, Gao J, Liu H, Liu H, Liang A, Zhou X, Cai W. The architecture and complete sequence of mitochondrial genome of an assassin bug Agriosphodrus dohrni (Hemiptera: Reduviidae) Int J Biol Sci. 2011;7:792–804. doi: 10.7150/ijbs.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liang A. The complete mitochondrial genome of spittlebug Paphnutius ruficeps (Insecta: Hemiptera: Cercopidae) with a fairly short putative control region. Acta Biochim Biophys Sin. 2013;45:309–319. doi: 10.1093/abbs/gmt009. [DOI] [PubMed] [Google Scholar]
- Panagiotakopulu E, Buckland PC. Cimex lectularius L., the common bed bug from Pharaonic Egypt. Antiquity. 1999;73:908–911. [Google Scholar]
- Robison GA, Balvin O, Schal C, Vargo EL, Booth W. Extensive mitochondrial heteroplasmy in natural populations of a resurging human pest, the bed bug (Hemiptera: Cimicidae) J Med Entomol. 2015;52:734–738. doi: 10.1093/jme/tjv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Reeves D, Brugler MR, Narechania A, Simon S, Durrett R, Foox J, Shianna K, Schatz MC, Gandara J, et al. Genome assembly and geospatial phylogenomics of the bed bug Cimex lectularius. Nat Commun. 2016;6:10164. doi: 10.1038/ncomms10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Liang AP, Bu CP. A molecular phylogeny of Hemiptera inferred from mitochondrial genome sequences. PLoS ONE. 2012;7:e48778. doi: 10.1371/journal.pone.0048778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Usinger RL. Monograph of Cimicidae. Entomological Society of America; Washington (DC): 1966. [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]