Abstract
Metformin is the first-line antidiabetic drug with over 100 million users worldwide, yet its mechanism of action remains unclear1. Here the Metformin Genetics (MetGen) Consortium reports a three-stage genome-wide association study (GWAS), consisting of 13,123 participants of different ancestries. The C allele of rs8192675 in the intron of SLC2A2, which encodes the facilitated glucose transporter GLUT2, was associated with a 0.17% (p=6.6×10−14) greater metformin-induced in haemoglobin A1c (HbA1c) in 10,577 participants of European ancestry. rs8192675 is the top cis expression quantitative trait locus (cis-eQTL) for SLC2A2 in 1,226 human liver samples, suggesting a key role for hepatic GLUT2 in regulation of metformin action. Among obese individuals, C-allele homozygotes at rs8192675 had a 0.33% (3.6 mmol/mol) greater absolute HbA1c reduction than T-allele homozygotes. This was about half the effect seen with the addition of a DPP-4 inhibitor, and equated to a dose difference of 550mg of metformin, suggesting rs8192675 as a potential biomarker for stratified medicine.
Metformin was commercialized before the modern era of target-based drug discovery. It typically reduces HbA1c by 1-1.5% (11-16mmol/mol) and has an excellent safety record, but considerable variation exists in how well patients respond to metformin2,3. We have recently established that genetic factors influence glycaemic response to metformin, with many common variants across the genome together explaining a substantial proportion of the variation, ranging from 21% to 34%, depending on how glycaemic response was measured4. Hypothesis-driven studies of pharmacokinetic variants have shown no consistent results5-10. The only GWAS published to date showed an association with rs11212617 near the ATM locus, which has been further replicated11,12.
Here we extended the previous GWAS by an additional 345 samples to a screening set of 1,373 participants. As in our previous report12, rs11212617 remained the top signal with no other genome-wide significant hit (Supplementary Figure 1). We undertook a systematic three-stage replication, with the work flow shown in Supplementary Figure 2. Only rs8192675 in the intron of SLC2A2 was replicated through the first two stages with a combined p=1×10−7 derived from a linear regression meta-analysis of 3,456 participants (Supplementary Data and Supplementary Table 1).
The MetGen Consortium performed the final replication of rs8192675 as a meta-analysis. Measures of glycaemic response to metformin were aligned across the cohorts as the absolute HbA1c reduction (expressed as reduction in percentage of HbA1c). Within each cohort, we tested associations with rs8192675 using two multiple linear models with or without the adjustment of baseline HbA1c, in addition to other available clinical covariates (Supplementary Table 2). In the meta-analysis of 10,557 participants of European ancestry (Figure 1), each copy of the C-allele was associated with a greater HbA1c reduction of 0.07% (p=2×10−8, P value (Phet) =0.35) when adjusting for baseline HbA1c; without adjustment the allelic effect of C-allele was 0.17% (p=6.6×10−14, Phet =0.52). There was no effect of rs8192675 on the efficacy of metformin in delaying progression to diabetes, or on metformin efficacy in a small insulin treated cohort (Supplementary Table 3).
Figure 1.
Pharmacogenetic impact of rs8192675 on metformin response in participants of European ancestry. The forest plots show meta-analyses of association test results for metformin-induced change in HbA1c in a total of 10,557 participants from 10 MetGen cohorts. Results from linear regression models with (left) and without (right) adjustment for baseline HbA1c are presented. The x axis represents the impact on metformin-induced HbA1c reduction of each copy of the C allele. HbA1c was measured in percentage.
We tested the pharmacogenetic effect of rs8192675 in 2,566 participants of non-European ancestries (Supplementary Table 4). The meta-analysis showed the C-allele was associated with a 0.08% greater HbA1c reduction (p=0.006, Phet =0.63) when adjusting for baseline HbA1c, and without the baseline adjustment the allelic effect of the C allele was 0.15% (p=0.005, Phet =0.95) without the baseline adjustment. In the meta-analysis of 13,123 participants of any ancestry (data not shown), we observed no genetic heterogeneity (Phet >0.29) between different ethnic groups despite the frequency of the C allele ranging from 24% in Latino populations to around 70% in African American populations.
We examined whether rs8192675 had an impact on baseline HbA1c, because the effect sizes of its association with glycaemic response to metformin differed depending on whether there was adjustment for the baseline HbA1c. In the 10,557 participants of European ancestry, the C allele was associated with a 0.13% (p=2.6×10−8) higher baseline HbA1c but a 0.04% (p=0.007) lower on-treatment HbA1c, which together contributed to the observed 0.17% (p=6.6×10−14) pharmacogenetic impact on HbA1c reduction in the model without baseline adjustment (Supplementary Figure 3).
Given the association of rs8192675 with HbA1c before treatment with metformin, we assessed whether this variant was marking a general ability to respond to any antihyperglycaemic treatment. Therefore we studied the pharmacogenetic impact of rs8192675 in 2,654 participants treated with sulfonylureas (Supplementary Table 5), another commonly used class of antidiabetic drug13,14. As in metformin users, the C allele was also associated with a higher baseline HbA1c in these users of sulfonylureas (beta=0.15%, P = 3.1×10−4). However, in contrast to the case for users of metformin, the C allele remained associated with a higher on-treatment HbA1c (beta=0.09%, p=0.006) in the users of sulfonylureas, which resulted in no net pharmacogenetic impact (beta=0.04%, p=0.44) on sulfonylurea-induced HbA1c reduction. These data suggest that rs8192675 is marking a genetic defect in glucose metabolism in type 2 diabetes that is ameliorated by metformin treatment but not by sulfonylurea treatment. The fact that rs8192675 is not associated with sulfonylurea response strongly supports a specific role for this variant on glycaemic response to metformin, rather than simply reflecting the higher pretreatment (baseline) HbA1c seen within carriers of this C-allele. In addition, the association with metformin-induced HbA1c reduction remained significant (P = 2×10−8; Figure 1) after adjustment for baseline HbA1c, corroborating a specific effect on response beyond its effect on baseline glycaemia.
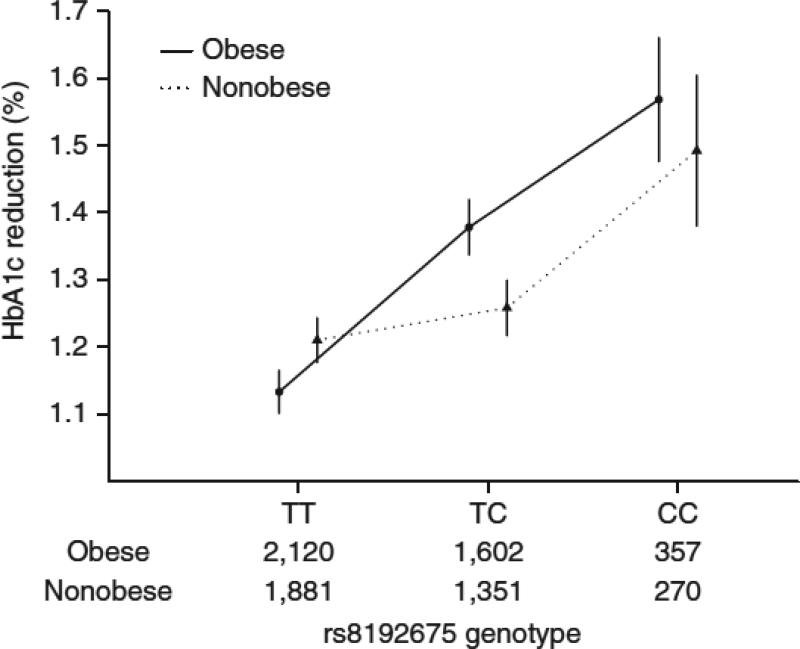
Metformin is particularly recommended for the treatment of diabetes in obese individuals owing to its beneficial effect on body weight15-17. Therefore, we explored whether the pharmacogenetic impact of rs8192675 varied by body mass index (BMI) in the MetGen cohorts (n=7,581 participants). BMI was associated with HbA1c reduction (beta = −0.01%; P = 1.7×10−4) but not rs8192675 genotype (p=0.52). Adjusting for BMI did not attenuate the observed pharmacogenetic effect of rs8192675 (Supplementary Table 6). When we stratified participants into nonobese (BMI<30 kg/m2) and obese groups (BMI≥30kg/m2) groups, there was a significant (P = 0.02) gene by BMI group interaction (Figure 2). The pharmacogenetic effect size of the C allele was 0.13% (s.e.m. = 0.04%, P = 0.001) in the non-obese participants as compared to that of 0.24% (s.e.m. =0.04%, P =5.0×10−11) in the obese participants.
Figure 2.
HbA1c reduction by BMI group and rs8192675 genotype. Participants were stratified into obese (BMI ≥ 30 kg/m2) and nonobese (BMI < 30 kg/m2) groups. The number of obese and nonobese individuals in each genotype group is noted along the x axis. Error bars, s.e.m.
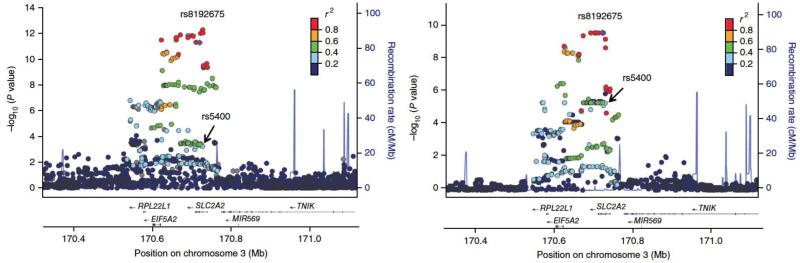
We performed a locus-wise meta-analysis to narrow down the candidate causal gene and variant list. Variant rs8192675 and its proxies showed the strongest association with HbA1c reduction (Figure 3). The linkage disequilibrium (LD) block covered three genes, of which SLC2A2 encodes the facilitated glucose transporter GLUT2, and EIF5A2 and RPL22L1 have little known functionality. Previous GWAS showed the nonsynonymous rs5400 in SLC2A2 is the main variant associated with glycaemic traits such as fasting glucose and HbA1c18,19. Because rs8192675 and rs5400 are in partial LD (D'=1; r2=0.35), here rs5400 was also associated with metformin response (beta=0.13%, p=5.2×10−4). However, when conditioning on rs5400, rs8192675 remains strongly associated with metformin response (beta=0.21%, s.e.m. =0.04%, P = 2.3×10−9); when conditioning on rs8192675, rs5400 is non-significant (P = 0.29). These results suggest the pharmacogenetic impact of rs8192675 is unlikely to be via the amino acid change of GLUT2 at rs5400.
Figure 3.
Regional plots of the SLC2A2 locus. SNPs are plotted by position on chromosome 3 against association with meta-analysis of HbA1c reduction without baseline adjustment (−log10 P) in 7,223 participants (left) and meta-analysis of SLC2A2 expression (−log10 P) in 1,226 liver samples (right). In both plots rs8192675 (purple circle) and its proxies are the top signals. The nonsynonymous SNP rs5400 (arrow) was also nominally associated with HbA1c reduction. Estimated recombination rates (cM/Mb) are plotted in blue to reflect the local LD structure. The SNPs surrounding the most significant SNP, rs8192675, are color coded to reflect their LD with this SNP. This LD was taken from pairwise r2 values from the HapMap CEU data. Genes, the position of exons and the direction of transcription from the UCSC Genome Browser are noted.
Given that liver is the most established site of metformin action, we examined whether rs8192675 is an eQTL in 1,226 liver samples of European ancestry. In Figure 3 we show rs8192675 as the top cis-eQTL for SLC2A2, with the C allele associated with decreased (P = 4.2×10−12) expression. In the 48 tissues examined by Genotype-Tissue Expression (GTEx) Project, SLC2A2 was sufficiently expressed in seven tissues (Supplementary Table 7). rs8192675 showed a significant (p=5.7 ×10−4) impact on SLC2A2 expression in the 271 samples of transformed fibroblasts, but no other significant associations20. Beyond GTEx, we sought additional eQTL evidence for other tissues that have been implicated in metformin action or glucose homeostasis. We found directionally consistent and supportive evidence of rs8192675 or its proxies being SLC2A2 cis-eQTLs in 118 islets (rs8192675, p = 0.0025)21, 173 intestinal samples (rs5398, p = 0.007)22, and 44 kidney samples (rs1905505, p = 0.04) (Supplementary Table 7).
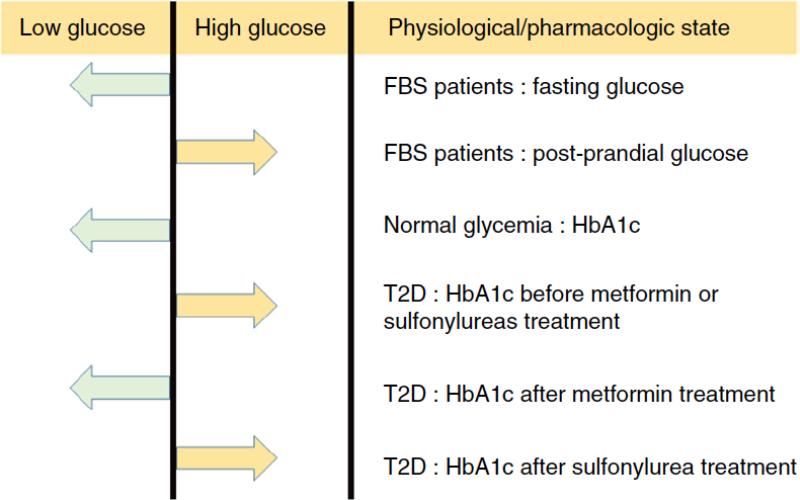
Patients with Fanconi-Bickel Syndrome (Online Mendelian Inheritance in Man (OMIM), 227810), who carry rare loss-of-function variants of GLUT2, can provide useful insight into the role of GLUT2 in glucose homeostasis and into the differing impact of common GLUT2 variants in various physiological states (Figure 4). Patients with Fanconi-Bickel syndrome exhibit low fasting glucose but high postprandial glucose23,24. In parallel, the C allele of rs8192675 that is associated with reduced SLC2A2 expression is associated with lower fasting glucose and HbA1c among individuals of normal glycemia18,19. We found that, in patients with type 2 diabetes, the expression-decreasing C allele of rs8192675 was associated with a higher HbA1c before treatment with either metformin or sulfonylureas. This deleterious genetic effect of rs8192675 on HbA1c was reversed with metformin treatment (C allele associated with lower on-treatment HbA1c and therefore better response to metformin), but not by sulfonylurea treatment.
Figure 4.
Genetic impact of GLUT2 variants on glucose homeostasis in different physiological and pharmacologic states. In patients with the monogenic Fanconi–Bickel syndrome (FBS), the loss-of-function variants led to lower fasting glucose but higher post-prandial glucose; the expression-reducing C allele at rs8192675 was associated with lower HbA1c in normal glycemia state but higher HbA1c in hyperglycemia state (before pharmacological treatment was indicated in patients with type 2 diabetes (T2D)); metformin but not sulfonylurea treatment reversed the genetic impact on HbA1c.
In humans, GLUT2 is a facilitative glucose transporter that is highly produced in the liver, kidney, small intestine and islets, and to a lesser extent in certain brain regions and other tissues. Defects in GLUT2-encoding gene could potentially alter glucose homeostasis at any or all of these sites4. Metformin's main site of action is widely believed to be the liver, where it primarily act to suppress hepatic glucose production1,25-27. In mice with Glut2 inactivation, glucose and glucose-6-phosphate accumulated in the cytoplasm owing to reduced glucose efflux, resulting in increased expression of nuclear ChREBP, L-pyruvate kinase and lipogenic genes29. Our eQTL data in liver samples (Figure 3) and corresponding reporter assays (Supplementary Figure 4) showed that the C allele at rs8192675 was associated with lower expression of SLC2A2. This suggests that the variant may lead to similar effects on hepatic gene expression in humans, which will be potentially modulated by metformin's well-described effect on hepatic glucose production and lipogenesis29,30. An alternative explanation could be that reduced SLC2A2 expression owing to rs8192675 is associated with reduced glucose-mediated glucose clearance (glucose effectiveness) owing to a decreased ability for glucose to enter the liver. This is seen in mice lacking Glut2 in the liver, and is an effect that is improved by metformin treatment31, although the mechanism for this is not understood.
Metformin is also increasingly believed to exert some of its beneficial effects by acting on the intestines to increase gut glucose uptake and non-oxidative glucose disposal, as well as increasing bile acid reabsorption, GLP-1 secretion and altering the microbiome33. In leptin-deficient (ob/ob) mice, metformin has been shown to increase translocation of Glut2 to the apical surface resulting in improved glucose homeostasis33. In light of the interaction we report between rs8192675 and BMI on metformin response, obese humans are reported to have altered GLUT2 localisation in the fasting state compared to nonobese humans33, suggestive of dysregulation of glucose sensing and transport in obese individuals. If reduced SLC2A2 expression owing to rs819265 were to result in reduced apical GLUT2, metformin could potentially overcome this by restoring GLUT2 transport in the enterocytes and improving glucose homeostasis.
Finally, given that metformin is transported into different tissues by several organic cation transporters, including OCTs, MATEs and THTR234, we examined whether GLUT2 can transport metformin in Xenopus laevis oocytes. Our results suggested that metformin was not a substrate or an inhibitor of GLUT2 (Supplementary Figure 5). Detailed human physiological studies, as well as functional exploration in animal and cellular model systems, are required to fully elucidate the role of GLUT2 in metformin response, and whether this is mediated via a hepatic, intestinal or other mechanism.
We examined the potential clinical impact of rs8192675. An unbiased (from the nondiscovery cohorts) estimate of its allelic effect was a 0.15% absolute reduction in percentage of HbA1c. This is equivalent to the pharmacological impact of taking 250 mg extra metformin per day, which is 26% of the average daily dose. We saw more clinical potential in obese patients as the honozygote carriers of the C allele had a 0.33% (s.e.m. = 0.09%, P = 6.6×10−4) greater reduction in percentage of HbA1c than those carrying the T allele; this equates to 24% of the average glycaemic reduction seen with metformin treatment in the MetGen cohorts and is equivalent to the impact of 550mg extra metformin. Given that newer agents such as DPP-4 inhibitors only reduce HbA1c by 0.6-0.8% on average36, this genetic effect is large and has potential to be of clinical utility. C-allele homozygotes could be treated with lower doses, and be exposed to fewer side effects; conversely T-allele carriers could be treated with doses higher than normally recommended to achieve a response. This may be of particular importance in African Americans where 49% of the population are C-allele homozygotes, in contrast to only 9% in European Americans. Stratified clinical trials, in different ethnic groups, are required to evaluate the potential for this pharmacogenetic variant to influence clinical care.
In conclusion, we have established a robust association between rs8192675 and metformin-induced HbA1c reduction with a large multiethnic cohort. rs8192675 was the top cis-eQTL for SLC2A2 in the liver and potentially islets, kidney and intestine. Reduced SLC2A2 expression resulted in a defect in glucose homeostasis in type 2 diabetes before initiation of therapy, which could be ameliorated by metformin treatment. The clinically appreciable impact in obese patients suggests rs8192675 has the potential to be a biomarker for stratified medicine.
METHODS
Data access
The three liver eQTL data sets published previously are available with Gene Expression Ombibus (GEO) accession numbers: GSE39036, GSE25935 and GSE9588.
Studies and Samples
Both GWAS screening and the first-stage replication analyzed participants with type 2 diabetes of European ancestry from the GoDARTS cohort. The current GWAS screening used 1,373 participants, which included data from 345 samples released after our initial GWAS report on 1,028 participants12. The first-stage replication included up to 1,473 samples from the remaining GoDARTS participants depending on the call rate and genotyping assay. The second-stage replication consisted of 1,223 participants of European ancestry from the UKPDS study. The final replication and meta-analysis was conducted within the MetGen Consortium which included an extra 6,488 participants of European ancestry and 2,566 participants of non-European ancestry. Detailed information on the MetGen participants is provided in Supplementary Table 2. Of note, about 50% of the MetGen cohort is from PMT, which represents ethnically diverse U.S. populations. These cohorts were used extensively in our multi-ethnic analysis for replication purposes. Participants from the largest PMT cohort, PMT2, were selected from the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort, a subsample of the Kaiser Permanente Research Program on Genes, Environment, and Health (RPEGH) 36. Three MetGen cohorts, GoDARTS, UKPDS and DCS also provided data on response to sulfonylureas. All human research was approved by the relevant institutional review boards, and all participants provided written informed consent.
Genotyping and quality control
Genotyping for the GWAS screening and the first-stage CardioMetabochip replication in GoDARTS cohort has been described before by WTCCC2 and DIAGRAM12,37. Standard quality control procedures were applied to both data sets to filter SNPs with minor allele frequency (MAF)<1% or call rate <98% or Hardy-Weinberg Equilibrium (HWE) deviation (p<10−4). Samples with call rate <98% or extra heterozygosity (more than 3 standard deviation away from the mean) or correlated with another sample (identity by descent [IBD]>0.125) were filtered out. In-house genotyping of the GoDARTS samples in the first-stage replication were performed with Sequenom MassArray for 66 SNPs and TaqMan based Allelic Discrimination assays for 9 SNPs. Details of the SNP selection procedure is described in Supplementary Data. All 75 SNPs had call rate >90% and no deviation from HWE (p>0.005). The second-stage genotyping of the UKPDS sample was carried out in duplicate runs using standard TaqMan assays. All the SNPs were in HWE (p>0.05) and only samples with concordant genotypes from both runs were analysed. The third-stage replication used high quality genotypes from either TaqMan assay or GWAS imputed data on rs8192675 (Supplementary Table 2).
Assessment of glycaemic response to metformin and sulfonylureas
As with our previous GWAS12, two correlated measures of glycaemic response to metformin were used in the current GWAS screening and the first-stage replication. A quantitative measure of HbA1c reduction (baseline minus on-treatment HbA1c) and a categorical measure of whether achieving a target of treatment HbA1c≤7% were used for genetic association tests. Therefore only participants with type 2 diabetes and a baseline HbA1c>7% were included. Baseline HbA1c was measured within 6 months prior to metformin start whilst on-treatment HbA1c was taken as the minimum achieved within 18 months after metformin start.
In the second-stage replication and the meta-analysis in the third-stage replication, we opted to maximize the sample size by synchronizing the measurement of metformin efficacy in a wider spectrum of participants with type 2 diabetes (including those with baseline HbA1c<7%) across the MetGen. Therefore only the quantitative outcome of HbA1c reduction was used to assess the glycaemic response to metformin. To maintain relative clinical homogeneity, only participants with type 2 diabetes on metformin monotherapy or using metformin as an add-on therapy to another oral agent were included.
Data from two MetGen cohorts, which used alternative measures of glycaemic response, were not included in the current meta-analyses, but the results are shown in Supplementary Table 4. In the DPP cohort of pre-diabetes participants, Cox proportional hazards regression was used to evaluate the genetic impact on the time to diabetes incidence8. In the HOME cohort, a multiple linear regression was used to test the genetic association with the difference in daily dose of insulin because metformin was used in conjunction with insulin in these participants38.
Assessment of glycaemic response to sulfonylureas adopted a similar approach as the quantitative outcome of metformin response in the MetGen. Baseline HbA1c and on-treatment HbA1c were captured in a similar manner as those in defining metformin response. Only participants with type 2 diabetes who were on sulfonylureas monotherapy or using sulfonylureas as an add-on therapy to metformin were included. All participants had a baseline HbA1c>7%.
Statistical Analysis
In the GWAS screening and first-stage replication, each SNP was tested for association with the continuous measure and categorical measure of glycaemic response to metformin separately with PLINK software using linear and logistic regression respectively39. Baseline HbA1c, adherence, metformin dose, creatinine clearance and treatment scheme (whether on metformin monotherapy or dual therapy of metformin add-on to sulfonylureas) and the first 10 principle component from EIGENSTRAT were used as covariates40. Statistical evidence of the two associations at each SNP was averaged by taking the geometric mean of the two p-values in cases in which the direction of effect was consistent (for example more HbA1c reduction and more likely to achieve the treatment target both indicate better response).
In the second and third stage replications, association with HbA1c reduction was tested with multiple linear regression. Within each cohort, two linear models were fitted either with or without adjustment for baseline HbA1c. Baseline HbA1c has been shown as the strongest predictor of metformin induced HbA1c reduction in pharmaco-epidemiological studies41. Adjusting for baseline HbA1c could reduce the confounding of measurement error in baseline HbA1c and increase the statistical power for pharmacogenetic studies42. However, if a variant is associated with baseline HbA1c, adjusting for baseline HbA1c would lead to a reduced estimate of its pharmacogenetic effect compared to a model that did not adjust for the baseline HbA1c. Therefore we presented both models in the current study. Other clinical factors such as creatinine clearance (or other measurement of kidney function) and treatment scheme were included as covariates where available (Supplementary Table 2). Combining the association results from individual cohort was conducted by a fixed-effect inverse-variance–weighted meta-analysis as applied in GWAMA43. Cochran's heterogeneity statistic's p-value was reported as Phet.
For the genetic association tests with response to sulfonylureas, multiple linear regression was used to assess the association between rs8192675 and baseline HbA1c, on-treatment HbA1c, HbA1c reduction and baseline adjusted HbA1c reduction. Treatment scheme (whether on sulfonylureas monotherapy or using sulfonylureas as add-on treatment to metformin) was included as a covariate when modelling sulfonylureas induced HbA1c reduction. Association test results from the three cohorts were combined with fixed-effect inverse-variance–weighted meta-analysis in GWAMA.
Locus-wise association was performed with GWAS imputed data of 7,223 participants available in the GoDARTS and PMT2-EU. Software IMPUTE2 was used to impute the post quality control GWAS data at 1Mb flank of rs8192675 against the 1000 Genomes reference panel44. Only SNPs with high imputation quality (info>0.9 and MAF>0.02) in both cohorts were tested for association with SNPTEST 45. Summary statistics from GoDARTS and PMT2-EU were combined with fixed-effect inverse-variance–weighted meta-analysis in GWAMA.
To evaluate the translational potential of rs8192675, we derived an unbiased estimate of its allelic effect by excluding the discovery cohort in the meta-analysis. This effect size was aligned to the clinical impact observed in the PMT2-EU which was the biggest replication cohort and used the median average daily dose in the MetGen. The average daily dose and dosing impact in PMT2-EU were 962mg/day and an extra 0.6% HbA1c reduction per gram metformin respectively. The evaluation of rs8192675 genotype by BMI group interaction was performed with linear regression by adjusting for treatment group, sex and study cohort.
Expression quantitative trait locus (eQTL) analyses
We used four liver eQTL datasets comprising a total number of 1,226 livers samples from individuals of European ancestry (Supplementary Table 8). Tissue procurement, gene expression analysis, genotyping and eQTL analyses have been described previously for three of the datasets46-48. The fourth data set was contributed by E. Schadt (E. Schadt, C. Molony, E. Chudin, K. Hao, X. Yang et al., personal communication). Genotypes were imputed to the 1000 Genome reference panel with IMPUTE2. Expression probe sequences were mapped to ENSEMBL genes and only the common genes across all datasets were included for subsequent analyses. Within each dataset, the genome-wide eQTL analysis was run with an additive genetic model including dataset specific covariates to examine cis-associations within a 100kb flanking window. Results from the four datasets were then combined with a modified meta test statistic which was calculated using the following approach: tmeta=(Σwiti)/√(Σwi2), w=√(n−(#covariates)−1) where i=data sets 1-4 and n=sample size49. This method Generation of p-values was accomplished by assuming the meta test statistics were normally distributed; a Benjamini-Hochberg multiple testing correction was applied to the p-values. For the current study, we extended the cis-association tests to all SNPs within 1Mb window of SLC2A2 and report the locus-wise p-values of the meta test statistic.
We investigated whether rs8192675 is a cis-QTL in other tissues in the GTEx data release V6. Due to the sample size limitation, rs8192675 is not a genomewide significant cis-eQTL for SLC2A2 in any of tissues examined. However, given the strong evidence of the variant being a cis-eQTL in the large liver samples reported in this study, we considered a directionally consistent association with p<0.05 as supportive evidence. The eQTL data for islet and intestine were acquired through contacting the authors of the original publications21,22. The eQTL data for kidney were obtained by quantitative real-time PCR of 44 kidney samples genotyped with the Affymetrix Axiom array. Sample acquirement and tissue preparation was described previously50. The transcript levels of SLC2A2 were determined using TaqMan probe (ID Hs01096908_m1). The relative expression level of SLC2A2 transcript was calculated by the comparative method (ΔΔCt) normalized to the housekeeping gene GAPDH, as described previously51.
Supplementary Material
Acknowledgement
We acknowledge G.I. Bell (University of Chicago) for providing the expression vector for SLC2A2 (pSP64T-SLC2A2), and D.L. Minor and F. Findeisen for their guidance in performing oocyte injection and preparing cRNA. For full acknowledgments, see the Supplementary Note.
Footnotes
Author Contributions
Conception and design of the study: E.R.P. and K.M.G.; Data analysis: K.Z., S.W.Y, E.L.S., N.V.L., A.V.H., J.W.B., C.E.K., L.Z., D.M.R., M.O., K.A.J., L.C., M.J., A.M.L., L.K.W., T.D., A.A.M.; data collection and genotyping: S.W.Y., C.S., R.T.,A.J.B.,C.J.G., R.L.C., L.L., L.K.W., T.D., S.S., M.K., M.M.H., H.C.C., F.I., S.M., J.S.W., L.W., J.Z., I.T., A.K., R.H.S., C.D.S., J.K., V.P., A.H., B.H.S., M.J.W., L.M.H., J.C.F., R.R.H., M.I.M. and C.N.A.P.; Manuscript writing: E.R.P., K.Z., S.W.Y., K.M.G. with contributions from all authors on the final version.
Competing financial interests
The authors have declared that no competing interests exist.
Accession codes.
Part of the unpublished eQTL data set 4 (Supplementary Table 8), which covers the SLC2A2 locus, has been deposited in the Figshare: http://dx.doi.org/10.6084/m9.figshare.3438362. Phenotype and genotype data used in the first-stage GWAS screening have been deposited at the European Genome-phenome Archive: EGAS00001001875 and EGAD00010000282, respectively.
References
- 1.Madiraju AK, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeFronzo RA, Goodman AM. Efficacy of metformin in patients with non-insulin-dependent diabetes mellitus. N. Engl. J. Med. 1995;333:541–549. doi: 10.1056/NEJM199508313330902. [DOI] [PubMed] [Google Scholar]
- 3.UK Prospective Diabetes Study (UKPDS) Group Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (UKPDS 34). Lancet. 1998;352:854–865. [PubMed] [Google Scholar]
- 4.Zhou K, et al. Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol. 2014;2:481–487. doi: 10.1016/S2213-8587(14)70050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawlyk AC, Giacomini KM, McKeon C, Shuldiner AR, Florez JC. Metformin pharmacogenomics: current status and future directions. Diabetes. 2014;63:2590–2599. doi: 10.2337/db13-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tkáč I, et al. Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes. Metab. 2013;15:189–191. doi: 10.1111/j.1463-1326.2012.01691.x. [DOI] [PubMed] [Google Scholar]
- 7.Stocker SL, et al. The effect of novel promoter variants in MATE1 and MATE2 on the pharmacokinetics and pharmacodynamics of metformin. Clin. Pharmacol. Ther. 2013;93:186–194. doi: 10.1038/clpt.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jablonski KA, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–2681. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu Y, et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 2008;83:273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou K, et al. Reduced-function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes. 2009;58:1434–1439. doi: 10.2337/db08-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Leeuwen N, et al. A gene variant near ATM is significantly associated with metformin treatment response in type 2 diabetes: a replication and meta-analysis of five cohorts. Diabetologia. 2012;55:1971–1977. doi: 10.1007/s00125-012-2537-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group Wellcome Trust Case Control Consortium 2 & MAGIC investigators. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet. 2011;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou K, et al. Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes: a Go-DARTS study. Clin. Pharmacol. Ther. 2010;87:52–56. doi: 10.1038/clpt.2009.176. [DOI] [PubMed] [Google Scholar]
- 14.UK Prospective Diabetes Study (UKPDS) Group Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). Lancet. 1998;352:837–853. [PubMed] [Google Scholar]
- 15.Diabetes Prevention Program Research Group Long-term safety, tolerability, and weight loss associated with metformin in the Diabetes Prevention Program Outcomes Study. Diabetes Care. 2012;35:731–737. doi: 10.2337/dc11-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diabetes Prevention Program Research Group 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet. 2009;374:1677–1686. doi: 10.1016/S0140-6736(09)61457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn SE, et al. Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N. Engl. J. Med. 2006;355:2427–2443. doi: 10.1056/NEJMoa066224. [DOI] [PubMed] [Google Scholar]
- 18.Scott RA, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranzo N, et al. Common variants at 10 genomic loci influence hemoglobin AC levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.GTEx Consortium The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van de Bunt M, et al. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet. 2015;11:e1005694. doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabakchiev B, Silverberg MS. Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology. 2013;144:1488–496. doi: 10.1053/j.gastro.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manz F, et al. Fanconi–Bickel syndrome. Pediatr. Nephrol. 1987;1:509–518. doi: 10.1007/BF00849262. [DOI] [PubMed] [Google Scholar]
- 24.Fanconi G, Bickel H. Chronic aminoaciduria (amino acid diabetes or nephrotic glucosuric dwarfism) in glycogen storage and cystine disease. Helv. Paediatr. Acta. 1949;4:359–396. [PubMed] [Google Scholar]
- 25.Miller RA, et al. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosokawa M, Thorens B. Glucose release from GLUT2-null hepatocytes: characterization of a major and a minor pathway. Am. J. Physiol. Endocrinol. Metab. 2002;282:E794–E801. doi: 10.1152/ajpendo.00374.2001. [DOI] [PubMed] [Google Scholar]
- 27.Burcelin R, Dolci W, Thorens B. Glucose sensing by the hepatoportal sensor is GLUT2-dependent: in vivo analysis in GLUT2-null mice. Diabetes. 2000;49:1643–1648. doi: 10.2337/diabetes.49.10.1643. [DOI] [PubMed] [Google Scholar]
- 28.Seyer P, et al. Hepatic glucose sensing is required to preserve β cell glucose competence. J. Clin. Invest. 2013;123:1662–1676. doi: 10.1172/JCI65538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hundal RS, et al. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes. 2000;49:2063–2069. doi: 10.2337/diabetes.49.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fullerton MD, et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 2013;19:1649–1654. doi: 10.1038/nm.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pau CT, Keefe C, Duran J, Welt CK. Metformin improves glucose effectiveness, not insulin sensitivity: predicting treatment response in women with polycystic ovary syndrome in an open-label, interventional study. J. Clin. Endocrinol. Metab. 2014;99:1870–1878. doi: 10.1210/jc.2013-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCreight LJ, Bailey CJ, Pearson ER. Metformin and the gastrointestinal tract. Diabetologia. 2016;59:426–435. doi: 10.1007/s00125-015-3844-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ait-Omar A, et al. GLUT2 accumulation in enterocyte apical and intracellular membranes: a study in morbidly obese human subjects and ob/ob and high fat–fed mice. Diabetes. 2011;60:2598–2607. doi: 10.2337/db10-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong L, Goswami S, Giacomini KM, Altman RB, Klein TE. Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet. Genomics. 2012;22:820–827. doi: 10.1097/FPC.0b013e3283559b22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey CJ. The current drug treatment landscape for diabetes and perspectives for the future. Clin. Pharmacol. Ther. 2015;98:170–184. doi: 10.1002/cpt.144. [DOI] [PubMed] [Google Scholar]
- 36.Banda Y, et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics. 2015;200:1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kooy A, et al. Long-term effects of metformin on metabolism and microvascular and macrovascular disease in patients with type 2 diabetes mellitus. Arch. Intern. Med. 2009;169:616–625. doi: 10.1001/archinternmed.2009.20. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Sherifali D, Nerenberg K, Pullenayegum E, Cheng JE, Gerstein HC. The effect of oral antidiabetic agents on A1C levels: a systematic review and meta-analysis. Diabetes Care. 2010;33:1859–1864. doi: 10.2337/dc09-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postmus I, et al. Pharmacogenetic meta-analysis of genome-wide association studies of LDL cholesterol response to statins. Nat. Commun. 2014;5:5068. doi: 10.1038/ncomms6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 46.Innocenti F, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schröder A, et al. Genomics of ADME gene expression: mapping expression quantitative trait loci relevant for absorption, distribution, metabolism and excretion of drugs in human liver. Pharmacogenomics J. 2013;13:12–20. doi: 10.1038/tpj.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia K, et al. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012;28:451–452. doi: 10.1093/bioinformatics/btr678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dahlin A, et al. Gene expression profiling of transporters in the solute carrier and ATP-binding cassette superfamilies in human eye substructures. Mol. Pharm. 2013;10:650–663. doi: 10.1021/mp300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang X, et al. Metformin is a substrate and inhibitor of the human thiamine transporter, THTR-2 (SLC19A3). Mol. Pharm. 2015;12:4301–4310. doi: 10.1021/acs.molpharmaceut.5b00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.