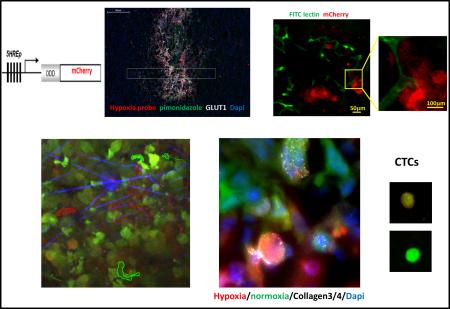
Abstract
Tumor hypoxia is linked to tumor progression, metastasis, and therapy resistance. However, the underlying mechanisms behind this linkage are not fully understood. Here we present a novel fluorescent mCherry hypoxia-responsive marker that can be used in real time imaging to specifically and sensitively identify hypoxic cells in vivo at single cell resolution. Tumors derived from triple negative tumor cells expressing the hypoxia marker reveal that the hypoxic tumor cells congregate near flowing blood vessels. Using multiphoton microscopy, hypoxic MDA-MB-231 cells were directly visualized and showed a more persistent slow migration phenotype as compared to normoxic cells in the same field in vivo. Hypoxic tumor cells are enriched in the cell population that migrates toward human epithelial growth factor gradients in vivo, and has increased collagen degradation and intravasation activity, characteristics of dissemination and metastasis competent tumor cells. The hypoxia probe introduced in this study provides a specific reporter of hypoxic cell phenotypes in vivo which reveals new insights into the mechanisms by which hypoxia is linked to metastasis.
Keywords: metastasis, tumor cell dissemination, invadopodia, intravital imaging, directional migration
Graphical abstract
Introduction
Metastasis, which accounts for more than 90% of all cancer related deaths, is a multistep process that includes tumor cell invasion of basement membranes and the surrounding tissue, intravasation into blood vessels, survival in blood circulation, extravasation, and growth at different organ sites 1. All of these steps are influenced by the tumor microenvironment, involving cell-cell and cell-matrix interactions in vivo. Under microenvironmental control, only a small subpopulation of tumor cells inside the primary tumor initiate invasion and disseminate, thereby seeding metastases in distant organs 2, 3. An improved understanding of how the microenvironment dominates the metastatic process is critical for the rational development of prognostics and treatments for patients with the potential of disseminating systemic disease.
Hypoxia is a common feature of solid tumors resulting from an imbalance between oxygen supply and consumption. The transcriptional activator hypoxia-inducible factor 1 (HIF-1) is the major regulator of hypoxia. HIF-1 is a heterodimeric basic helix–loop–helix (bHLH) protein consisting of two subunits, an O2-regulated HIF1α and a constitutively expressed HIF-1β. In normoxic conditions, O2-dependent hydroxylation of proline in HIF1α by prolyl hydroxylase 2 and 3 (PHD2, 3) leads to the recognition of HIF-1α by the von Hippel – Lindau (VHL) protein, which then leads to degradation through the ubiquitin-proteosome pathway. Therefore, under normoxic conditions, HIF-1α is rapidly degraded and thus undetectable. Under hypoxic conditions however, proline hydroxylation decreases and VHL cannot bind to HIF-1α, resulting in a decreased rate of HIF1α degradation. Thus, HIF1α is highly expressed under hypoxic conditions. The stabilized HIF1α subunits translocate from the cytoplasm to the nucleus where they are dimerized with HIF1β to form HIF1. Then HIF1 binds to the hypoxia-responsive element (HRE) in the enhancer region of its target genes, turning on the transcription of genes involved in angiogenesis, glucose transport, tyrosine hydroxylase and erythropoietin (Epo), cancer cell invasion and metastasis4-7. Although low O2 stress is the principal regulator of HIF1α activity, genetic alterations such as mutations of VHL 8, PTEN 9 or p53 10 are associated with increased levels of HIF1α transcriptional activity, even in aerobic conditions. This is consistent with the finding that HIF1α activity levels vary significantly in different cancer cell lines in response to the same degree of hypoxia11.
Significant work has been done to characterize the role of HIF1α in vitro and in vivo. Inhibition of HIF1α expression impairs tumor growth and lung metastasis in the MDAMB-231 breast xenograft tumor mouse model12. In the PyMT transgenic breast cancer mouse model, conditional deletion of HIF1α in the mammary epithelium resulted in delayed tumor onset and retarded tumor growth. Deletion of HIF-1α in the mammary epithelium resulted in decreased pulmonary metastases13. Recent studies have found that enhanced HIF1α expression promotes extracellular matrix (ECM) remodeling to facilitate tumor cell invasion and intravasation by upregulating intracellular collagen-modifying enzymes (i.e. P4HA1, P4HA2, PLOD1 and PLOD2)14-17. Since hypoxia can promote both tumor progression and resistance to radiation and chemotherapy, tumor hypoxia is of major clinical significance. However, the kinetic relationship of the hypoxic cell phenotype to oxygen tension in vivo remains unclear 18.
Live imaging of hypoxia at single cell resolution is essential to understand the tumor cell phenotypes resulting from hypoxia and whether the malignant progression correlated with hypoxia is the result of changes in the behavior of tumor cells that is specifically due to hypoxia. Several hypoxia responsive reporters with multiple HRE repeats as promoter have been generated using luciferase, eYFP or eGFP as a reporter fluorophore in order to perform live imaging. However, the in vivo imaging techniques were limited to in vivo imaging system (IVIS) with a CCD camera, positron emission tomography (PET), and wide field epi fluorescence microscopy 11, 19-21. The highest resolution imaging reported so far was multiphoton microscopy of monolayer cells or spheroids grown in culture to evaluate hypoxia activity and its spatial distribution in vitro22. To our knowledge, there is no report in the literature of in vivo imaging at single cell resolution in live tissue in real time of hypoxic tumor cells.
Intravital imaging using multiphoton microscopy has provided real time single cell resolution with visualization at great depths inside living tissues and has played a pivotal role in defining the specific microenvironments associated with invasion and intravasation in mammary tumors3,23. In order to study the spontaneous cell behaviors associated with hypoxia in mammary tumors, we have designed a hypoxia-responsive marker which labels breast cancer cells in low oxygen stress in real time, and validated this probe in vitro and in vivo. Using multiphoton microscopy, we were able to visualize and assess the phenotypes of hypoxic tumor cells relative to normoxic tumor cells at single cell resolution in live breast xenograft tumors derived from MDA–MB-231 which express our hypoxia probe (GFP MDA-MB-231-5HRE-ODD-mCherry).
Results
Generation of hypoxia reporter cell lines using a new hypoxia probe HRE-ODD-mCherry
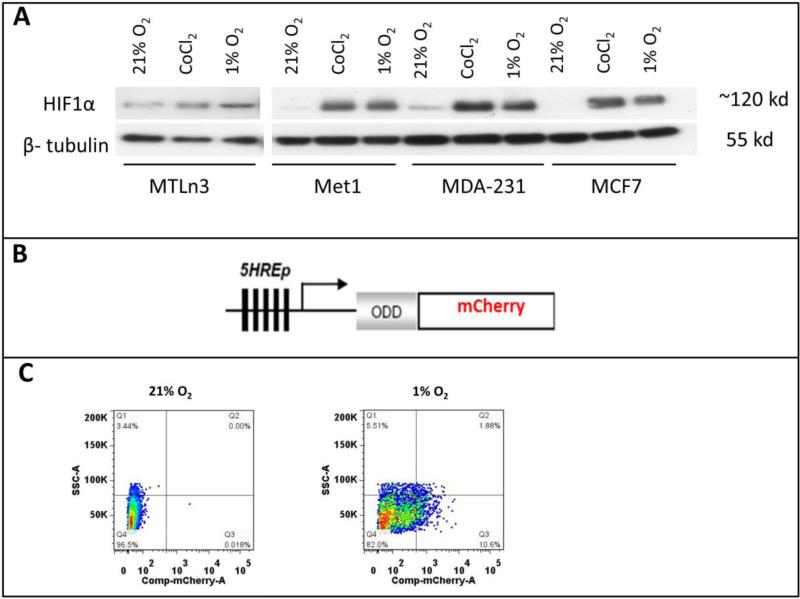
To generate a hypoxia responsive reporter, we investigated the HIF1α expression level in normoxic and hypoxic conditions for a number of metastatic cancer cell lines in order to select cell lines that exhibit a low basal level of HIF1α expression in normoxia and a significant increase in expression under hypoxia. This ensures that the selected cell lines are hypoxia-responsive and have low background and high contrast signals for a hypoxia reporter. MCF7, a luminal breast ductal carcinoma cell line, is known for low normoxic and high hypoxic HIF1α expression24, but is poorly invasive and rarely metastasizes25. Thus, we used MCF7 as a control in Western blots against the HIF1α antibody for selection of triple negative breast cancer (TNBC) cells that would be suitable for use with the hypoxia reporter. The cell lysates were collected from the cells being exposed to 21% O2, 1% O2 and 0.2mM CoCl2 for 20 hours. The Western blots revealed that MET1 and MDA-MB-231 cells have low HIF1α expression in normoxia, with dramatically increasing levels after hypoxic induction with either 1% O2 or CoCl2, while MTLn3 cells have a high HIF1α basal expression level in normoxia and very little to no increase of this level under 1% O2 or CoCl2 (Fig. 1A). Based upon this data, we chose to pursue MET1 and MDA-MB-231 as cell lines to use with the reporter to study metastatic cancer.
Figure 1. MDA-MB-231 and MET1 cell lines are suitable to use with the hypoxia reporter.
A) Western blot with anti-HIF1α and anti-β-tubulin for MTLn3, MDA-MB-231, MCF7 and MET1 breast tumor cells in normoxia, 0.2mM CoCl2 and 1% O2 culturing conditions. Cells were cultured at test conditions for 20 hours before being lysed. B) Schematic representation of 5HRE-ODD-mCherry hypoxia reporter plasmid construct. C) Representative FACS sorting plots for mCherry positive hypoxic MET1-5HREODD mCherry cells at the 3rd cycle of FACS sorting. Left panel shows cells under normoxic culturing conditions as a gating control. Right panel shows cells cultured under 1% O2, 5% CO2, and 94% N2 for approximately 20 hours. The top 10% mCherry positive cells from the 1% O2 cultured cells were collected.
To ensure the best dynamic range of signal for our hypoxia reporter, we adopted the technique published by Harada et al. 26 in which a hypoxia-inducible luciferase (5HRE-ODD-luc) was constructed. In their work, which was a modification of the luciferace reporter (5HRE-luc) developed by Shibata, et al. (19), the addition of an oxygen-dependent degradation domain (ODD) was utilized to minimize the background level of luminescence under normoxia, This dramatically increased the hypoxia-responsiveness (the contrast between luciferase activity at normoxia and hypoxia) of the reporter compared to the original Shibata construct without the ODD domain (5HRE-luc) by providing a direct oxygen dependent degradation of reporter in cases where HIF1α might have escaped its own oxygen dependent degradation. Thus, we added the ODD domain upstream from the reporter mCherry coding sequence in our construct (Fig. 1B). As can be seen in Fig. 1C, excellent contrast in mCherry signals between normoxic and hypoxic states can be observed. Therefore we were able to enrich the hypoxia responsive cell population. While several fluorescent protein-based hypoxia markers have been developed in the past11, 27-34, we chose for the first time to use the red fluorescent protein mCherry as a hypoxia marker. The use of mCherry is advantageous over the use of other fluorescent proteins such as GFP and YFP which require more oxygen for proper folding 35, and hence show reduced stability and brightness under hypoxic conditions. As described in the Methods section, transfected cells were FACS sorted for low background mCherry expression in normoxia and high mCherry expression in hypoxia using 3 cycles of normoxic and hypoxic cell culture. In order to visualize the entire cell population under either normoxic or hypoxic conditions, we also transduced the reporter cells with a constitute GFP expression lenti viral vector as described in the methods section. Thus, we obtained the two hypoxia reporter cell lines, GFP MET1-5HRE ODD-mCherry and GFP MDA–MB-231-5HRE ODD-mCherry.
Validation of the hypoxia inducible reporters in vitro
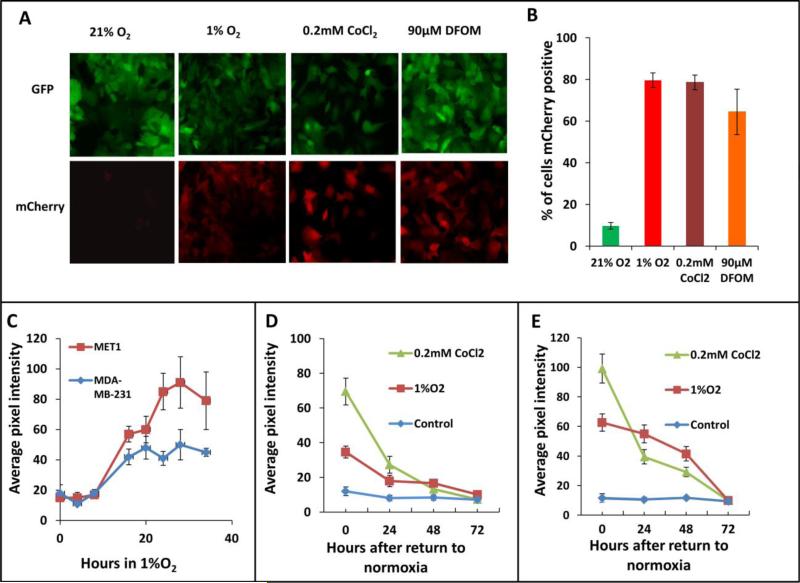
We performed 3 cycles of FACS sorting for the reporter cell lines because the cells expressing the hypoxia construct were not homogenous in their responses to hypoxia. The percentage of the cells responding to 1% O2 was enriched in each FACS sorting cycle. About 50% of the cells responded to 1% O2 at the third cycle of FACS (Fig. 1C). To determine the efficiency of the final cell lines in responding to hypoxic microenvironments, we cultured the cells for 20 hours either in a 1% O2 hypoxia chamber or under mimetic hypoxia conditions by adding CoCl2 or DFOM to the culture medium before taking images (Fig. 2A). The mCherry hypoxia indicator was induced to detectable levels under all conditions, with 80% of cells responding to 1% O2, 65% responding to 90μM DFOM, and 79% responding to 0.2mM CoCl2. At normoxia, approximately 10% of cells still expressed a very low but detectable level of mCherry (Fig. 2B). The hypoxia responsive cells were enriched dramatically after 3 rounds of FACS sorting. Thus, the heterogeneity of cells responding to hypoxia was less of a concern in later studies.
Figure 2. Hypoxia reporter characterization in vitro.
A) Representative images of GFP MET1-5HRE-ODD-mCherry cells in vitro in normoxia and hypoxia induced by 1% O2, 0.2 mM CoCl2 and 90μM DFOM for 20 hours respectively. B) Quantification of percent of cells expressing mCherry in normoxia, or after 20 hours of hypoxia induction with 1% O2, 5% CO2 and 94% N2 in a gas chamber or with 0.2mM CoCl2 or with 90μM DFOM. Error bars: mean ± s.e.m. C) mCherry expression kinetics of MET1 and MDAMB-231 hypoxia reporter cells upon 1% O2 induction. Error bars: mean ± s.e.m. D) mCherry expression in MET1 and E) MDA-MB-231 hypoxia reporter cells. Images analyzed were taken at the times shown after the cells were returned to normoxia after hypoxia treatment of 1%O2 or 0.2mM CoCl2. error bars: mean ± s.e.m.
The mCherry signal was induced at a significant high level at 16 hours of 1% O2 incubation and stayed at high levels to 34 hours tested (Fig. 2C). But the normoxic oxygen level in vivo is at 5-10% O2 36. So we tested the cells in 5% O2, and found that the mCherry signal didn't change when the cells were cultured in 5% O2 for up to 28 hours making this a good hypoxia probe for use in tissue (Fig. S1). To determine if the hypoxia induced mCherry protein level was capable of reverting back to its normoxic background state after reoxygenation, we imaged the cells on a fluorescence microscope at 0, 24, 48 and 72 hours after return to a normoxic environment. The mCherry pixel intensity was then quantified for both MET1 and MDA-MB-231 hypoxia cells (Fig. 2D and 2E). For both the MET1 and the MDA-MB-231 hypoxia reporter cell lines, reoxygenation of the cells exposed to hypoxic conditions for 20 hours resulted in a progressive decrease of mCherry expression that reached the normoxia control level in 72 hours. Together, these data confirm that the mCherry expression in both GFP MET1-5HRE ODD-mCherry and GFP MDA-MB-231-5HRE ODD-mCherry cell lines is responsive to hypoxia and is predictably reversible upon return to normoxia.
Validation of the hypoxia inducible reporter in vivo
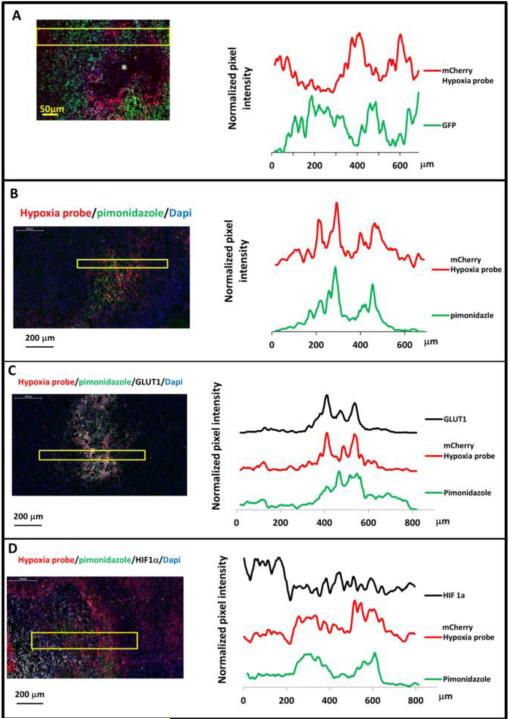
Prolonged hypoxia of tumor tissue often leads to necrosis, and necrotic areas are typically surrounded with hypoxic tumor cells37, 38. An initial approach to evaluate our hypoxia-responsive reporter in vivo was to assess the spatial distributions of mCherry fluorescence relative to the necrotic regions of tumor tissues. In mammary tumors derived from the MDA-MB-231 hypoxia probe cells, frozen sections showed mCherry positive hypoxic tumor cells around the necrotic core (Fig. 3A left panel). Interestingly, throughout the tumor sections, there was a diminution of GFP signal in high mCherry cells (Fig. 3A) consistent with the known decrease of GFP brightness in low oxygen levels39, 40. To demonstrate the different intensities of GFP and mCherry, line scans for both the green and red channel fluorescence intensities were extracted from the image, averaged (Fig. 3A, left panel, yellow box indicates lines averaged), normalized, and plotted (Fig. 3A right panel). The GFP and mCherry signals were oppositely related in normoxic and hypoxic areas. This phenomenon illustrates that mCherry indeed is ideal as a hypoxia probe and supports the ideas that it is both the “best general-purpose red momomer” 41 and, compared to GFP, folds well in hypoxic conditions 42.
Figure 3. Hypoxia reporter signal co-localizes with hypoxia response markers in mammary tumors.
A) Representative image of GFP MDA-MB-231-5HREODD-mCherry derived tumor frozen tumor sections within normoxic and hypoxic regions relative to the necrotic area (*) (left panel) and the quantification of normalized GFP or mCherry signal in the yellow box (right panel). Green=GFP, normoxia, red=mCherry, hypoxia. B) Representative image showing mCherry overexpressing cells colocalizing with pimonidazole adduct staining in MDA-MB-231-5HREODD-mCherry derived tumor frozen sections (left panel), and the quantification of average pimonidazole or mCherry signal in the yellow box. Green=pimonidazole, red=mCherry, blue=nuclei. C) Representative image showing mCherry overexpressing cells correlated with GLUT1 antibody or pimonidazole adduct antibody staining in MDA-MB-231-5HREODD-mCherry derived tumor frozen sections (left panel), and the quantification of average mCherry or GLUT1 or pimonidazole signal in the yellow box (right panel). Green=pimonidazole, red=mCherry, grey=GLUT1, blue=nuclei. D) Representative image showing mCherry overexpressing hypoxic cells or pimonidazole adduct positive cells adjacent to high HIF1α expressing cells in MDA-MB-231-5HREODD-mCherry derived tumor frozen sections (left panel), and the quantification of average mCherry or pimonidazole or HIF1α signal in the yellow box (right panel). Green=pimonidazole, red=mCherry, grey= HIF1α, blue=nuclei.
To further validate our hypoxia cell reporter in vivo and to see whether the mCherry positive cells correlate with pimonidazole adduct (an exogenous marker of hypoxia used clinically), the tumors were removed one hour after pimonidazole injection in mice bearing a hypoxia reporter MDA-MB-231-5HREODD-mCherry derived tumor. Pimonidazole staining on the tumor frozen sections coincides with cells that were elevated in mCherry expression (Fig. 3B and S2). Again, averaged line scans of mCherry and pimonidazole staining (Fig. 3B left, yellow box indicates lines averaged) were normalized and plotted (Fig. 3B, right), showing the consistency of co-distribution of pimonodazole and mCherry overexpression.
In addition, to validate the mCherry hypoxia signal as an accurate marker for hypoxia in vivo we correlated it with genes upregulated by hypoxia. A major effect of activation of HIF1α is the stimulation of glycolytic energy production by upregulating the genes encoding the extracellular glucose transporters (GLUTs) in order to equilibrate O2 consumption with O2 supply43, 44. These transmembrane glycoproteins are omnipresent in normal tissue, facilitating glucose transport across the cell membrane. Malignant tumors generally have a higher rate of metabolism and have a higher glucose need. Under hypoxic conditions the cell's demand for glucose increases as anaerobic glycolysis becomes more important, leading to the recruitment and overexpression of the glucose transporters in malignant hypoxic tumor cells. The two glucose transporters most associated with invasive cancer are GLUT1 and GLUT3. It has been found that the pimonidazole uptake pattern is consistent with a GLUT1 staining pattern in MDA- MB-231 xenograft tumors, whereas the pattern of HIF1α rarely coincides as expected45. As such, we further stained frozen sections from the pimonidazole injected tumors with both GLUT1 and HIF1α antibodies and compared their staining to both the mCherry overexpression and pimonidazole adduct staining (Fig. 3C and 3D). As before, the averaged line scans from the imaged tissue (Fig. 3C left, yellow box indicates lines averaged) were normalized and plotted to show the relationship between the signals. As can be seen in Fig. 3C and S3, the overexpression of the mCherry hypoxia reporter co-localized with high GLUT1 membranous staining as well as to pimonidazole adduct. The positive GLUT1 staining in mCherry positive cells indicates that these cells indeed represent the hypoxia pathway activated tumor cell population. Compared with the GLUT1 staining, HIF1α staining was adjacent to the mCherry positive cell areas and was less co-localized with mCherry expression or pimonidazole adduct stained areas (Fig. 3D and S4) as expected45. Again, this pattern of HIF1α and GLUT1 expression relative to hypoxic cells defined by the hypoxic probe is consistent with previous findings with pimonidazole 45 and further validates the value of the mCherry hypoxia probe. The finding that HIF1α has a very short half-life (less than 5 min) 46, whereas GLUT1 has a half-life of about 72 hours 47 is also consistent with the different patterns of staining of HIF1α and GLUT1 relative to the hypoxia probe. Finally, the increased transcription of HIF-1α without stabilization in some hypoxic cells and stabilization of HIF-1α in severe hypoxic cells would also contribute to this difference in patterns48.
In MET1-reporter tumor sections, both GLUT1 and HIF1α expression were high at background throughout the sections. mCherry and GLUT1 were differentially expressed and overlapping at both peak expressions. As with the GFP MDA-MB-231-5HREODD-mCherry cell containing tumors there was correlation between mCherry and GLUT1 but less so with HIF1α expression pattern (Fig. S5). However, the tumors formed by MET1 cells became very necrotic at sizes as small as 0.2 mm in diameter in syngeneic FVB mice making them less useful for interrogating earlier stages of tumor progression. As such, we focused most of our in vivo studies on tumors containing the MDA-MB-231 hypoxia reporter cells.
Hypoxic cells can be found both near and away from blood vessels
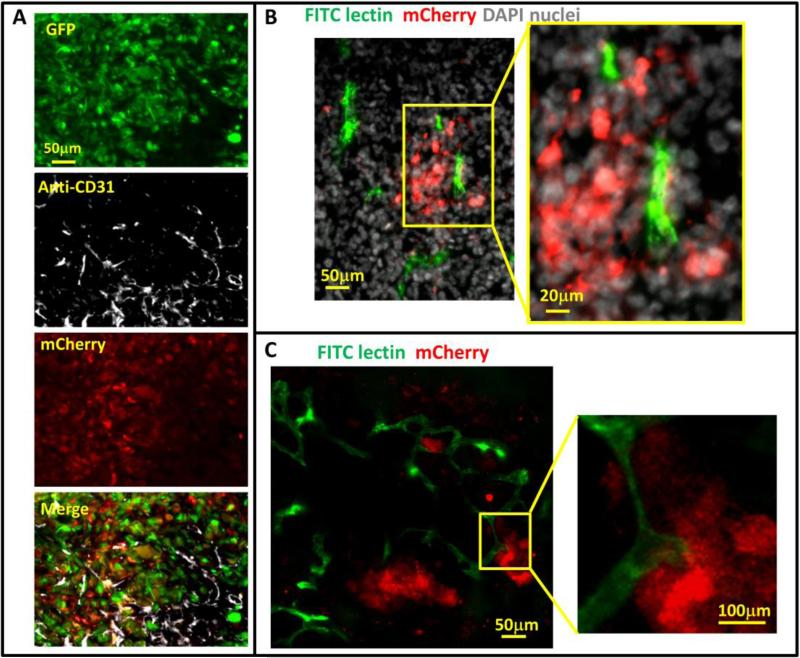
Tumor hypoxia is believed to be caused in part by the relative slow growth of blood vessels that cannot keep up with the demand of fast growing tumor cells resulting in the collection of hypoxic tumor cells in under vascularized regions of the tumor. To assess the spatial distribution of hypoxic cells in relation to vasculature within the tumors, we stained cryo-sections of GFP-MDA-MB-231-5HREODD-mCherry tumor with anti-CD31. As expected, some hypoxic tumor cells were found distant from blood vessels (Fig 4A). However, the frozen sections revealed that hypoxic tumor cells were also near and even in contact with a subset of blood vessels (Fig 4B and 4C), which is consistent with the literature 49. Since CD31 antibody staining is a marker for endothelial cells and does not distinguish between flowing and non-flowing vessels at the time of fixation, we marked flowing blood vessels by tail-vein injection of mice bearing MDA-MB-231 hypoxia reporter tumors (absent the GFP volume marker) with FITC labeled ricinus communis agglutinin I (RCA I). This was done 15 min before the mice were sacrificed and their tumors removed for ex vivo imaging using a confocal microscope. Only fully perfused vessels at the time of injection were labeled with FITC. GFP was not included in these tumors to avoid spectral overlap with the FITC RCA I. Surprisingly we observed that fully perfused vessels were associated with hypoxic tumor cells (Fig. 4B and 4C).
Figure 4. Hypoxic tumor cell distribution relative to blood vessels.
A) Representative images of CD31 antibody staining for blood vessel endothelium in GFPMDA-MB-231-5HREODD-mCherry tumor frozen sections showing the hypoxic cells are found both away from and adjacent to blood vessels. Green=GFP hypoxia reporter in normoxia; Red=mCherry hypoxia reporter in hypoxia, Gray=CD31. B and C) Representative images of tumor frozen sections (B) and ex vivo imaging (C) to show perfused blood vessels (green) relative to hypoxic cell distribution (red) in MDA-MB-231-5HREODD-mCherry derived tumors that were IV injected with lectin (green) before sacrificing. In areas of flowing blood vessels the hypoxic tumor cells are closely associated with a subset of blood vessels. Yellow boxes indicate regions shown at high magnification. In this tumor, tumor cell does not express the GFP volume marker so as to prevent overlap with FITC lectin signal. For B and C, Green =FITC lectin stained blood vessels, red = mCherry hypoxia, Gray=DAPI.
Hypoxic tumor cells exhibit an invasive phenotype with increased collagen degradation ability compared to normoxic tumor cells
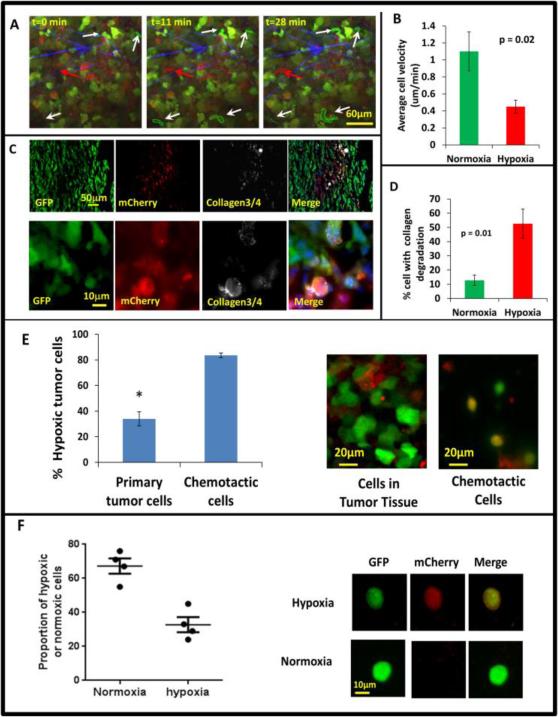
It has been often reported in the literature that increased tumor hypoxia is correlated with increased invasion and metastasis 50. In order to determine the invasive and motility phenotype of hypoxic cells in vivo as suggested by the above results (Fig 4) we used intravital multiphoton imaging to visualize hypoxic tumor cells in living tumors. Imaging was accomplished using a custom-built, two-laser multiphoton microscope51, and 4D data sets were each acquired over periods of 0.5 to 1 hour (Fig 5A, movies 1 and 2). To quantify hypoxic and normoxic cell behaviors, we analyzed 28 movies and measured tumor cell velocities. We found that the hypoxic tumor cells showed the slow migration phenotype observed previously in GFP MDA-MB-231 tumors3. These cells are characterized as cells moving less than 0.4 μm/min, with abundant invadopodia, most often seen in perivascular regions moving toward blood vessels 3, while normoxic cells fall into the fast tumor cell phenotype (Fig. 5B) characterized by cells without invadopodia moving faster than 1 μm/min where migration is directed along collagen fibers and not only toward blood vessels 3. In Fig. 5A, still images from movie 1, show a representative example where several normoxic cells are actively moving, while hypoxic cells move at reduced velocity.
Figure 5. Hypoxic tumor cells are enriched in the invasive and chemotactic tumor cell population in vivo.
A) Multiphoton microscopy images of GFP-MDA-MB-231-5HREODD-mCherry xenograft tumor cells at 0, 11 and 28 minutes. White arrows point to GFP-only normoxic cells. Red arrow points to a motile mCherry positive hypoxic cell. Each moving cell outline was traced. The green outlined cells are normoxic cells and the red outlined cell is a hypoxic cell. Green=GFP normoxic, Red=mCherry hypoxic, Blue=Second Harmonic Generation (SHG) from collagen. B) Quantification of average cell velocity in vivo migration for hypoxic and normoxic cells. Fields of view containing hypoxic cells were analyzed for motion of either green normoxic and/or red hypoxic cells, n=28. P=0.02 (Student's t-test), error bars: mean±s.e.m. C) Representative images of collagen ¾antibody staining on GFP MDA-MB-231 hypoxia reporter derived tumor tissue frozen sections showing overlap of collagen degradation with hypoxic tumor cells. The lower panel images are at higher magnification to have a better view of the collagen¾ antibody staining around single cells. Red=mCherry hypoxic, green=GFP normoxic, grey=collagen¾, blue=nuclei. D) Quantification of the percent of cells with collagen degradation within the normoxic or hypoxic tumor cell populations. Each bar is the percentage of the respective cell type. The green bar is normalized to the number of total green cells while the red bar is normalized to the total number of hypoxic cells. Fields of view containing collagen degradation were analyzed. P value is by Student's t-test, error bars: mean±s.e.m., n=5. E) Percentage of hypoxic cells in the population that migrated to HuEGF gradient (chemotactic cells) vs the percentage of hypoxic cells in primary tumor tissue where the needle was placed. Representative images are tumor cells in primary tumor tissue and chemotactic tumor cells collected using the in vivo invasion assay with 25nM huEGF. Red=mCherry hypoxic, green=GFP normoxic. Error bar: SEM. * p<0.05 (Student's t-test), n=3 mice. F) Analysis of circulating tumor cells. CTCs were imaged for mCherry and GFP signals. mCherry and GFP pixel intensity was normalized to the highest signal in each channel respectively among the CTCs. The ratio of the normalized mCherry to GFP signals in individual cells was calculated. Ratio of normalized mCherry to GFP below 0.5 = normoxia, above 0.5 = hypoxia. The percentage of hypoxic and normoxic cells were plotted. 4 mice were used. The representative images for hypoxic and normoxic cells are shown on the right.
The presence of abundant invadopodia is a defining characteristic of tumor cells with the slow and blood vessel-directed migration phenotype in vivo 3. Invadopodia are important for tumor cells to penetrate extracellular matrix and the basement membrane of blood vessels, and provide a way for tumor cells to precisely couple focal matrix degradation with directional movement52, 53. To study the relative abundance of invadopodia in hypoxic and normoxic tumor cells, we employed a thin gelatin matrix degradation assay in vitro to assess the ability of normoxic and hypoxic cells to form invadopodia and to degrade matrix. MDA-MB-231 cells were plated on 405 nm labeled gelatin thin matrix and subjected to a 21 % O2 or 1% O2 culturing condition for 24 hours to form invadopodia which were then scored as F-actin-rich structures co-localized with matrix degradation (arrow heads in Fig. S6C). There was higher percentage of hypoxic cells degrading gelatin matrix compared to normoxic cells (Fig. S6C). To compare levels of proteolytic degradation of normoxic and hypoxic tumor cells in vivo, we used an antibody against degraded collagen, Collagen 3/4 (i.e. Col2 3/4 short), which recognizes collagen fragments created by the proteolytic activity of MMP-1, MMP-2 or MMP-13 54, 55 in tumor cryosections (Fig 5C). Significantly, more hypoxic cells degraded collagen than normoxic cells in the same field in vivo (Fig. 5D), which is consistent with the presence of abundant invadopodia associated with the slow blood vessel directed tumor cells in vivo 3, 54 and the invadopdium assay results in vitro (Fig. S6C). Our finding that hypoxic cells fall into the slow migration invadopodium rich phenotype in vivo seems to contradict the in vitro scratch assays used in previous studies56-58. To investigate this, we time-lapse imaged both hypoxic and normoxic GFP MDA-MB-231-5HRE-ODD-mCherry cells in vitro over a period of 4 hours on glass bottom Matek dishes and measured the velocities of cells undergoing random motility. To maintain their oxygen status during the experiment, imaging was performed under the same hypoxic/normoxic conditions the cells had been cultured in for 20 hrs prior. As in the in vivo images, we observed a significant reduction in random cell motility for the hypoxic cells (Fig. S6A, movie 3, and movie 4), measuring an average cell random velocity of 0.23 μm/min for hypoxic cells and 0.49 μm/min for normoxic cells. However, the hypoxic cells show higher directionality of movement indicating that the hypoxic cells move more efficiently (Fig. S6B). By utilizing an uncoated transwell assay, we measured tumor cell chemotaxis toward a nutrient source of 10% FBS placed in the bottom of transwell in a 24-hour period in hypoxic or normoxic conditions. Consistent with published literature7, 59 and the higher directionality of the hypoxic tumor cells we observed, hypoxic cells were more chemotactic than normoxic cells (Fig. S6D).
Hypoxic cells in vivo are more chemotactic to signals associated with blood vessels and enter blood vessels
We previously showed that we can collect the migratory cells from primary tumors in response to chemotactic signals associated with blood vessels such as human epidermal growth factor (EGF) 60-63. In brief, microneedles containing Matrigel and huEGF are inserted into the primary tumors to set up a chemotactic gradient that attracts actively migrating tumor cells. This assay mimics natural blood vessels inside the primary tumor in which EGF secreted by peri-vascular macrophages attracts the invasive tumor cells to blood vessels, a movement that eventually will lead to intravasation and hematogenous metastasis23. Using this assay in the MDA-MB-231 hypoxia probe xenograft tumors, we collected the chemotactic tumor cells that actively responded to huEGF in vivo, extruded them out of the needles onto slides, imaged and scored the normoxic cells and hypoxic cells on a confocal microscope. In order to compare the % hypoxic tumor cells in the chemotactic population of cells to the % of hypoxic tumor cells in the primary tumor tissue, we imaged the tumor for hypoxic and normoxic cells in situ. In the subpopulation of chemotactic tumor cells that were collected in response to huEGF, about 83% of the cells were hypoxic, while only about 33% of the tumor cells were hypoxic in the primary tumor (Fig. 5E). To see if the normoxic cells become hypoxic during the 4 hours of collection, we left normoxic cells in the needle for 4 hours, and extruded them on a slide to compare the cells directly prepared from normoxic culture. We found there was no change in mCherry expression between the cells collected in the needle and the cells prepared from normoxic cell culture (Fig. S7). These results suggest that hypoxic tumor cells are more chemotactic to blood vessel associated signals like EGF in vivo than normoxic cells. To see if the hypoxic cells are actually migrating into blood vessels to become circulating tumor cells, we collected the blood from the mouse right ventricle of heart, lysed the red blood cells with RBC lysis buffer, then fixed and imaged for GFP and/or mCherry positive cells. The percentage of hypoxic or normoxic circulating tumor cells (CTCs) (i.e. the ratio of normalized mCherry to GFP signals) was plotted in Fig. 5F. Here, the CTCs were classified into two categories, with a ratio of mCherry to GFP below 0.5 for normoxic CTCs, and above 0.5 for hypoxic CTCs. Representative images are shown next to each category (Fig. 5F, right panel). About 67% of the CTCs were normoxic and 33% of CTCs were hypoxic. Although we are not able to determine the exact location in the primary tumor acting as the source of the CTCs, this result confirms that the hypoxic cells intravasate into blood circulatory system.
Discussion
We have designed and validated a unique, hypoxia-response reporter to specifically identify hypoxic cells caused by physiological low oxygen stress. Use of mCherry as the fluorescent hypoxia probe has improved oxygen sensitivity since the mCherry protein requires less oxygen to fold properly than the previously reported hypoxia probes using GFP or YFP. To further improve the sensitivity and specificity of our hypoxia reporter, cells were FACS sorted for hypoxia probe+ cells with 3 cycles of enforced normoxia and hypoxia. The resulting MDA-MB-231 and MET1 hypoxia reporter cell lines demonstrated low background mCherry expression in normoxia and a high contrast in mCherry expression between normoxia and hypoxia. Importantly, the mCherry reporter cell lines showed that the mCherry signal is reversible upon reoxygenation. In vivo, the hypoxia reporter cells accurately identify the hypoxic tumor cells in the tumor. In addition, mcherry expression is coincident with GLUT1 expression in hypoxia reporter cell line derived tumors, which further validates that mCherry positive hypoxic cells are activated on the hypoxia pathway since GLUT1 is a direct target of HIF1α. Taken together, these results demonstrate that our hypoxia reporter is a faithful probe of hypoxic tumor cells and identifies individual hypoxic cells in live tissue in real time in vivo.
This probe is a sensor for the identification of the hypoxic phenotype of the cells themselves which is retained for 48-72 hours after exposure to a hypoxic microenvironment. Given the long decay time of our probe, there are two explanations for the localization of the mCherry hypoxic cells near and even in contact with a subset of blood vessels (Fig. 4B and 4C). One is that the observed environment was hypoxic 48-72 hours earlier and the cells remained in place. As shown by Helmlinger et al.49, the presence of blood vessels is not a guarantee of normoxia. Many hypoxic regions are directly adjacent to blood vessels. The second is that it is known that tumor cells migrate to a subset of blood vessels that contain tumor microenvironment of metastasis (TMEM) sites 23, 61 and this is consistent with the enhanced collection of hypoxic tumor cells in response to EGF, a signal associated with blood vessel-directed migration 60. The migration of tumor cells to a subset of vessels with TMEM sites is also consistent with the observation that only some vessels have hypoxic cells with them. It is established that hypoxic cells secrete VEGF to promote vessel growth in the tumor and blood vessels might grow into hypoxic areas64. However, migrating tumor cells move at 0.4 - 1 μm/min in vivo65, which is much faster than that of endothelial migration at about 0.035 μm/min66, 67. Considering that the mCherry hypoxia probe expression level reverses back to background level 48-72 hours after reoxygenation in vitro (Fig. 2D), the results shown in Fig. 4B suggest that hypoxic cells actively move to vessels from a hypoxic region in less than 48-72 hours. Furthermore, the long duration after the onset of hypoxia may explain the mixing of hypoxic and normoxic cells (red and green) observed in vivo (Fig. 5A, Supplemental Movies 1& 2). In addition, because of the slow dynamics of the hypoxia probe, the hypoxic tumor cells found in blood circulation (CTC) might have been hypoxic for hours before intravasation. An instantaneous reporter of hypoxia would likely highlight where the initiation of hypoxia occurred instead of the phenotype of the hypoxic cells as reported by our hypoxia sensor.
We utilized intravital multiphoton microscopy to characterize the in vivo migration properties of hypoxic cells within breast tumors derived from the GFP MDA-MB-231 reporter cell line. Interestingly, we found that hypoxic cells display the slow migratory phenotype previously associated with tumor cells moving less than 0.4 μm/min that are enriched for invadopodia which are most often seen in perivascular regions moving toward blood vessels3. This result is consistent with the in vivo invasion assay results where the migratory cell population collected in response to the blood vessel associated signal EGF is enriched for hypoxic tumor cells compared to the average primary tumor cell density of the whole tumor, and with the finding of hypoxic CTCs. Of particular interest is the previous finding that tumor cells in the slow migration category that are enriched for invadopodia are the tumor cells that disseminate systemically via the blood vessels3. This suggests that the hypoxic subpopulation of tumor cells in the primary tumor are disseminating tumor cells.
To gain insights into the mechanisms by which hypoxia is linked to these tumor cell phenotypes, we investigated MDA-MB-231 cell behavior in vitro and found that hypoxic MDA-MB-231 cells move slower than normoxic cells in vitro but move more efficiently with higher directionality and chemotaxis. These data are consistent with in vitro scratch assays reported by others showing that hypoxia promotes gap closure in a period of 24 hrs 7, 57, 58. We propose that the increased ability of hypoxic cells to promote faster gap closure in vitro is the net result of their higher directionality of migration.
Invadopodia are defined as invasive actin polymerization-dependent protrusions found specifically on cancer cells that degrade matrix. Invadopodium formation is potentiated by hypoxia in the fibrosarcoma HT-1080, pancreas BxPC3, head and neck SCC61, and lung H1792 cell lines 68, 69. Recently, Gligorijevic et al. found that the slow, but not the fast, migratory tumor cells in MDA-MB-231 tumors have abundant invadopodia3. The correlation between the tumor cell velocity and the collagen degradation ability described in that study 3 resembles what we have found here in hypoxic tumor cells in vivo and in vitro, where hypoxic cells have reduced velocity and increased degradation ability compared to nomoxic cells. In particular, we found that approximately 30% more hypoxic tumor cells form matrix-degrading invadopodia (Fig. S6C). This finding is also similar to what was observed in DMOG treated MDA-MB-231 cells by others 70
In general, invadopodia are mechanistically linked to directionality and chemotaxis. In 3D, MDA-MB-231 cell matrix-degrading invadopodia are localized to the leading edge of the invading cell where the secretory machinery and metalloproteases are located 71. In addition, invadopodium assembly is required for chemotaxis of tumor cells to growth factor signals such as EGF53. Thus, although hypoxic tumor cells move slowly in terms of velocity, they are more directional and efficient at moving directionally toward attractants and invasive allowing them to deal with blocking ECM barriers. Indeed, the in vivo invasion assay reveals that hypoxic cells are enriched in the migratory cell population in vivo, a population known to undergo intravasation and hematogenous metastasis 2, 3, 72. In addition, CTCs collected from tumors expressing the hypoxia reporter contain a large percentage of hypoxic probe positive cells consistent with the above findings that hypoxic tumor cells in the slow migration category that are enriched for invadopodia are tumor cells that disseminate systemically via blood vessels. Therefore, the hypoxia marker described in this study will be valuable in further studies of the mechanisms by which the hypoxic microenvironment dominates tumor cell phenotype in vivo and tumor progression.
Materials and Methods
Cell culture
The mouse mammary tumor Met-1 cell line, and the human breast tumor MCF7 and MDA-MB-231 cell lines were maintained in DMEM (Life Technologies, Inc.) supplemented with 10% fetal bovine serum and antibiotics (penicillin and streptomycin). Live-cell imaging experiments were conducted at 37°C using L15 media and 10% FBS.
Western blot
Cells were cultured in either normoxia or hypoxia conditions for 20 hours, then washed in cold phosphate-buffered saline (PBS) and lysed in cell lysis (loading) buffer (160 mM Tris pH 6.8, 20% glycerol, 10% beta-mercaptoethanol, 4% SDS). The cell lysates were resolved by SDS–PAGE, transferred to nitrocellulose, blocked in 5% milk in TBS-T for 1 hour, incubated in primary anti- HIF1α (Novus Biologicals, NB100-449) and anti-beta-tubulin (Santa Cruz Biotechnology, Sc-33749) overnight at 4 °C, followed by the secondary antibodies for 1 h at room temperature, and finally analyzed using the Odyssey (LI-COR Biosciences, Lincoln, NE). Visualization and processing of images was performed with ImageJ (NIH).
DNA constructs
Hypoxia-response elements (HREs) are responsible for activating gene transcription in response to hypoxia. To construct a hypoxia-responsive red fluorescence protein reporter, we started with the pcDNA3.1 (+) plasmid as backbone. The pCMV promoter was removed using Mlu + Nhe I and replaced with 5HRE-CMVmp taken from the pHRP-Luc+ plasmid19. The ODD-mCherry fusion gene was generated by overlapping PCR and inserted at Kpn I site of plasmid above. Primers used for overlapping PCR were: HIF530F (5′-3′): CCCAAGCTTGGATCCGAATTCGCCACCATGGAATTCAAGTTGGAATTGGTAG; HIF653B: AGTAGTTTCTTTATGTATGTGGG; ODD-Mcherry5: CCCACATACATAAAGAAACTACTGTGAGCAAGGGCGAGGAGG; Mcherry3: CCCTCTAGACTACTTGTACAGCTCGTCCATGCC. Overlapping PCR was done following the standard protocol. Briefly, 1) use primers HIF530F and HIF653B to amplify PCR product 1 (ODD domain) from HIF-1α plasmid; 2) use primers ODD-Mcherry5 and Mcherry3 to amplify PCR product 2 from mCherry plasmid. Underlined sequences above indicate the overlapping part between product 1 and 2; 3) Mix purified products 1 and 2 together (about 10-100 ng DNA) and use primers HIF530F and Mcherry3 to generate fusion gene ODD-mCherry. The resulting construct named 5HRE-ODD-mCherry was sequence confirmed (Fig. S8) and tested by transient transfection into cells, followed by 150-200 uM Cobalt (II) chloride (CoCl2) treatment to mimic hypoxia condition. 16 hours after CoCl2 treatment, the cells were imaged under fluorescent microscope for hypoxia-induced mCherry expression.
Establishment of double-fluorescent reporter cells
The 5HRE-ODD-mCherry DNA plasmid was transfected into cancer cell lines using Lipofectamin transfection reagent (Invitrogen), following the protocol in the product user manual. Three days after transfection, clonal cell populations that had stably integrated the DNA vectors were selected by G418 for the 5HRE-ODD-MCherry reporter. Colonies were selected and further enriched by repeated FACS gated for hypoxia-induced mCherry expression. In brief, the drug resistant cells were pooled and subjected to three cycles of FACS selection as follows. The normoxic cells were sorted for negative mCherry cells. After recovering from the sorting, the cells were cultured in 1% O2 hypoxia chamber for 20 hours and then were sorted for mCherry positive cells (Fig. 1C). For the GFP expressing hypoxia reporter cell lines, the cells selected from the third cycle sorting experiment were transduced with a constitute GFP expression lenti viral vector into the hypoxia reporter cells to serve as a tumor cell volume marker. After viral transduction, the cells were cultured under selection of 3μg/ml puromycin and then sorted for the brightest 10% of the GFP expressing hypoxia reporter cells. The established cells with or without GFP expression were frozen as stocks. All hypoxia reporter cell lines were thrown out after 10 passages from thaw.
Time Lapse imaging
Cells were cultured in MatTek dishes under either normoxic or hypoxic conditions by incubation within a hyperbaric chamber filled with a 1% O2, 5% CO2, 94% N2 gas mixture. At the time of imaging, cells were placed on the stage of an inverted microscope (DeltaVision, Applied Instruments) fitted with a temperature control chamber and media was replaced with L15 with 10%FBS. A custom designed gas flow chamber maintained the oxygen status by providing a continual flow of positive pressure gas of the appropriate composition. Time lapse images were taken every 2 min for 4 hours. The hypoxic status of the cells was confirmed before and after the time lapse imaging.
Invadopodial matrix degradation assay
The invadopodial matrix degradation assay was conducted as described previously73, 74. Briefly, gelatin was conjugated with an Alexa 405 dye (Molecular Probes, Eugene, OR). MatTek dishes (MatTek Corporation, Ashland, MA) were treated with 1 N HCl and coated with 50 μg/ml poly-l-lysine, which was cross-linked with 0.01% glutaraldehyde. A 0.2% gelatin solution was prepared in phosphate-buffered saline (PBS), and a 1:40 mixture of Alexa 405–labeled gelatin : unlabeled gelatin was warmed to 37°C before addition to the poly-l-lysine. The dish was then quenched with 5 mg/ml sodium borohydride. 2 × 105 MDA-MB-231 were plated on Alexa 405–labeled gelatin and incubated in 21% O2 or 1% O2 respectively for 24 h. Cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.1% Triton X-100, and blocked with 1% FBS and 1% BSA in PBS. Then the cells were stained with phalloidin. Mature invadopodia were scored as F-actin rich structures that colocalize with a degradation hole in the Alexa 405–labeled gelatin.
Animal models
All procedures were conducted in accordance with the National Institutes of Health regulations and approved by the Albert Einstein College of Medicine Animal Use Committee. The reporter cells were thawed from the frozen vial made after the three cycles of FACS sorting for the no-GFP expressing cells or from GFP FACS sorting for the GFP expressing cells. Within 10 passages in cell culture, a total of 2×106 MDA-MB-231 hypoxia reporter cells per mouse were suspended in sterile PBS with 20% collagen I (BD Biosciences, cat # 354249) and injected into the lower left mammary fat pad of severe combined immunodeficiency mice (SCID) (NCI, Frederick, MD, USA). For MET-1 hypoxia reporter cells, injected 1×106 cells suspended in sterile PBS into each FVB mouse mammary fat pad (NCI, Frederick, MD, USA). All experiments, unless otherwise stated, were performed on tumors that were 1–1.2 cm in diameter.
Frozen section preparation and imaging of hypoxia induced MCherry expression
When tumors grew to about 1-1.2cm cm in diameter, they were dissected and fixed with 5% formalin, 20% sucrose at 4°C overnight, frozen in OCT on dry ice, and then stored at −80°C. For pimonidazole injection, mice bearing a no-GFP hypoxia reporter cell derived tumor were injected intraperitoneally with pimonidazole at 60mg/kg (Hypoxyprobe, Cat# HP6-100Kit). The mice were euthanized after one hour of pimonidazole injection. The tumor was removed, fixed and frozen as above. Sections (7 μm) were cut on a cryostat and dried onto positively charged glass slides. The sections were mounted with DAPI fluorescent mounting medium (Vector Laboratories, Burlingame, CA) or stained with GLUT1, pimonidazole, CD31 or HIF1α antibodies. Sections were stored in the dark at 4°C until imaging.
Immunofluorescence (IF)
Frozen sections were permeabilized with 0.1% Triton X-100 or cold aceton for 10 min followed by 3 times washes with PBS. Nonspecific binding was blocked by 1% BSA and 1% FBS for 1 hour, followed by primary antibody incubation at 4°C overnight (rabbit anti-GLUT1 at 1:200, Abcam, cat# ab652; rabbit anti-HIF1α at 1:200, Novus Biologicals, NB100-449; rat anti-CD31, BD Bioseciences, cat# 550274; FITC-conjugated anti-pimonidazole antibody at 1:50, Hypoxyprobe, Cat# HP6-100Kit). For degraded collagen antibody (Ibex Pharmaceuticals, C1,2C (Col 2 3/4Cshort) polyclonal rabbit antibody, 1:100; we refer to this antibody as collagen3/4 antibody), sections were blocked by 1% BSA and 1% FBS at 4°C for overnight, followed by 1 hour collagen3/4 antibody incubation at room temperature 54. After the primary antibody incubations, the sections were washed with PBS (0.05% Tween 20) and exposed to Alexa fluor 647-conjugated secondary antibodies (anti-rabbit IgG, 1:200, Life Technologies, cat# A31573) for 30 min, followed by 5 washes. The sections were mounted with DAPI fluorescent mounting medium (Vector Laboratories, Burlingame, CA) beforeimaged. The GLUT1, HIF1α or pimonidazole antibody stained slides were scanned at 20X on a PerkinElmer P250 High Capacity Slide Scanner. In order to quantify the relative pixel intensity of GFP, mCherry and antibody staining of GLUT1 or HIF1α or pimonidazole the averaged pixel intensity in the selected yellow boxes was measured in ImageJ using the “Plot Profile” macro under “Analyze” menu. Then each averaged pixel intensity was normalized to the highest pixel intensity in each respective channel. The histogram of normalized pixel intensity in each channel was plotted and stacked on top of each other for better comparison.
Intravital imaging
Intravital multiphoton imaging of the MDA-MB-231 hypoxia reporter tumor-bearing mice was performed with methods similar to previous studies, using a custom-built two-laser multiphoton microscope fitted with a 25X, 1.05NA water immersion objective lens with a correction collar51. The laser-light source consisted of a both a standard femtosecond-pulsed laser system (Mai Tai HP, Newport/Spectra-Physics) used for excitation of GFP fluorophore and an optical parametric oscillator (Opal, Newport/Spectra-Physics). The fluorescence and second-harmonic signals generated were collected via a dichroic mirror and sent to three photomultiplier tube (PMT) detectors. 880nm light was used to excite GFP and SHG and 1170nm was used to excite mCherry. To generally characterize in vivo migration, we imaged random fields of 512×512 μm containing mCherry expressing cells for a depth of 100 μm (21 slices at steps of 5 μm) beginning at the surface of the tumor. Tumor areas with any signs of necrosis or apoptosis were excluded from the analysis. Images were collected at 2 min intervals for 30 min to 1 hour except where indicated otherwise.
Transwell Assay
2.5 × 104 cells were plated inside 8.0 um pore size un-coated transwells (#354483, BD Biosciences) in 0.5% FBS/DMEM. Then the transwells were put into the wells of a 24-well plate filled with 10% FBS/DMEM. Cells were allowed to cross the transwell membrane for 24 hours in 21% O2 or 1% O2 culturing condition, fixed with 4% paraformaldehyde, stained with DAPI for nuclei, and imaged at 10X. For each of the 3 independent experiments, two transwells per condition were divided into 6 fields each and were analyzed.
In vivo invasion assay
Cell collection into needles placed into live anesthetized animals was carried out as described previously2, 75. Migratory cells enter the needles containing Matrigel and huEGF only in response to a chemoattractant and only by active migration toward the chemotactic gradient. Six needles were inserted into a tumor per mouse at random locations, and 3 mice were used for in vivo invasion assay. After 4 hours of collection, the needles were removed and cells were expelled with PBS onto cover glass for imaging. In the meantime, the tumor was collected, sliced at needle insertion sites, and put into a MaTek dish with about 50μl of PBS. The needle collected cells and the sliced tumor tissue were imaged on a confocal microscope for GFP and mCherry signal. The chemoattractant used in this study was human recombinant EGF (Invitrogen) at final concentration of 25 nM.
Intravasation assay
The number of circulating tumor cells was collected in mice bearing a tumor at about 1-1.2 cm as previously described75. Briefly, blood was drawn for the right heart ventricle of anesthetized mice, the red blood cells were lysed with 10ml of RBC lysis buffer in room temperature for 10 minutes, spun down at 1000rpm for 3 minutes. Then the pellet was resuspended with 100 μl of 4% paraformaldehyde with DAPI, transferred to a MaTek dish and left in 4°C overnight for the cells to settle onto the bottom of the dish for imaging. Images were taken on a fluorescent microscope (DeltaVision, Applied Instruments).
Supplementary Material
Acknowledgements
The authors thank Drs Julio Aguirre-Ghiso, and Patricia Keely for helpful discussions. For the technical help, the authors thank Dr. Bojana Gligorijevic for collagen ¾ staining, the Einstein FACS facility for the cell sorting, and the Einstein Analytical Imaging Facility (AIF) for imaging. The pHRP-Luc+ construct was a generous gift from Dr. Ziqiang Yuan. This work was supported by NIH CA1664468 (to JC and DE), NIH CA100324 (to YW and AX), the Integrated Imaging Program at Einstein College of Medicine (to DE, YW), and NIH SIG#1S100D019961-01 for use of PE-250 Slide Scanner.
Footnotes
Disclosure of Potential Conflicts of Interest
J.S. Condeelis has ownership interest (including patents) in MetaStat; and is a consultant/advisory board member for Deciphera and MetaStat. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–69. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patsialou A, Wang Y, Lin J, Whitney K, Goswami S, Kenny PA, Condeelis JS. Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012;14:R139. doi: 10.1186/bcr3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gligorijevic B, Bergman A, Condeelis J. Multiparametric classification links tumor microenvironments with tumor cell phenotype. PLoS Biol. 2014;12:e1001995. doi: 10.1371/journal.pbio.1001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8:967–75. doi: 10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan DA, Giaccia AJ. Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 2007;26:333–9. doi: 10.1007/s10555-007-9063-1. [DOI] [PubMed] [Google Scholar]
- 6.Gort EH, Groot AJ, van der Wall E, van Diest PJ, Vooijs MA. Hypoxic regulation of metastasis via hypoxia-inducible factors. Curr Mol Med. 2008;8:60–7. doi: 10.2174/156652408783565568. [DOI] [PubMed] [Google Scholar]
- 7.Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 2013;15:R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425:307–11. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 9.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk AR, Ryan HE, Johnson RS, Jefferson AB, et al. Loss of PTEN facilitates HIF-1- mediated gene expression. Genes Dev. 2000;14:391–6. [PMC free article] [PubMed] [Google Scholar]
- 10.Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273:11995–8. doi: 10.1074/jbc.273.20.11995. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W, Dosey TL, Biechele T, Moon RT, Horwitz MS, Ruohola-Baker H. Assessment of hypoxia inducible factor levels in cancer cell lines upon hypoxic induction using a novel reporter construct. PLoS One. 2011;6:e27460. doi: 10.1371/journal.pone.0027460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Wong CC, Wei H, Gilkes DM, Korangath P, Chaturvedi P, Schito L, Chen J, Krishnamachary B, Winnard PT, Jr., et al. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31:1757–70. doi: 10.1038/onc.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1alpha is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67:563–72. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- 14.Eisinger-Mathason TS, Zhang M, Qiu Q, Skuli N, Nakazawa MS, Karakasheva T, Mucaj V, Shay JE, Stangenberg L, Sadri N, et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013;3:1190–205. doi: 10.1158/2159-8290.CD-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilkes DM, Bajpai S, Wong CC, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol Cancer Res. 2013;11:456–66. doi: 10.1158/1541-7786.MCR-12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Aro E, Khatri R, Gerard-O'Riley R, Mangiavini L, Myllyharju J, Schipani E. Hypoxia-inducible factor-1 (HIF-1) but not HIF-2 is essential for hypoxic induction of collagen prolyl 4-hydroxylases in primary newborn mouse epiphyseal growth plate chondrocytes. J Biol Chem. 2012;287:37134–44. doi: 10.1074/jbc.M112.352872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentovim L, Amarilio R, Zelzer E. HIF1alpha is a central regulator of collagen hydroxylation and secretion under hypoxia during bone development. Development. 2012;139:4473–83. doi: 10.1242/dev.083881. [DOI] [PubMed] [Google Scholar]
- 18.Koh MY, Powis G. HAF : the new player in oxygen-independent HIF-1alpha degradation. Cell Cycle. 2009;8:1359–66. doi: 10.4161/cc.8.9.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata T, Giaccia AJ, Brown JM. Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther. 2000;7:493–8. doi: 10.1038/sj.gt.3301124. [DOI] [PubMed] [Google Scholar]
- 20.He F, Deng X, Wen B, Liu Y, Sun X, Xing L, Minami A, Huang Y, Chen Q, Zanzonico PB, et al. Noninvasive molecular imaging of hypoxia in human xenografts: comparing hypoxia-induced gene expression with endogenous and exogenous hypoxia markers. Cancer Res. 2008;68:8597–606. doi: 10.1158/0008-5472.CAN-08-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Qu R, Ogura M, Shibata T, Harada H, Hiraoka M. Real-time imaging of hypoxia-inducible factor-1 activity in tumor xenografts. J Radiat Res. 2005;46:93–102. doi: 10.1269/jrr.46.93. [DOI] [PubMed] [Google Scholar]
- 22.Indovina P, Collini M, Chirico G, Santini MT. Three-dimensional cell organization leads to almost immediate HRE activity as demonstrated by molecular imaging of MG-63 spheroids using two-photon excitation microscopy. FEBS Lett. 2007;581:719–26. doi: 10.1016/j.febslet.2007.01.040. [DOI] [PubMed] [Google Scholar]
- 23.Harney AS, Arwert EN, Entenberg D, Wang Y, Guo P, Qian BZ, Oktay MH, Pollard JW, Jones JG, Condeelis JS. Real-Time Imaging Reveals Local, Transient Vascular Permeability, and Tumor Cell Intravasation Stimulated by TIE2hi Macrophage- Derived VEGFA. Cancer Discov. 2015;5:932–43. doi: 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doublier S, Belisario DC, Polimeni M, Annaratone L, Riganti C, Allia E, Ghigo D, Bosia A, Sapino A. HIF-1 activation induces doxorubicin resistance in MCF7 3-D spheroids via P-glycoprotein expression: a potential model of the chemo-resistance of invasive micropapillary carcinoma of the breast. BMC Cancer. 2012;12:4. doi: 10.1186/1471-2407-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249–89. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 26.Harada H, Kizaka-Kondoh S, Itasaka S, Shibuya K, Morinibu A, Shinomiya K, Hiraoka M. The combination of hypoxia-response enhancers and an oxygen-dependent proteolytic motif enables real-time imaging of absolute HIF-1 activity in tumor xenografts. Biochem Biophys Res Commun. 2007;360:791–6. doi: 10.1016/j.bbrc.2007.06.149. [DOI] [PubMed] [Google Scholar]
- 27.Krishnamachary B, Penet MF, Nimmagadda S, Mironchik Y, Raman V, Solaiyappan M, Semenza GL, Pomper MG, Bhujwalla ZM. Hypoxia regulates CD44 and its variant isoforms through HIF-1alpha in triple negative breast cancer. PLoS One. 2012;7:e44078. doi: 10.1371/journal.pone.0044078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potzkei J, Kunze M, Drepper T, Gensch T, Jaeger KE, Buchs J. Real-time determination of intracellular oxygen in bacteria using a genetically encoded FRET- based biosensor. BMC Biol. 2012;10:28. doi: 10.1186/1741-7007-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang L, Greenwood TR, Artemov D, Raman V, Winnard PT, Jr., Heeren RM, Bhujwalla ZM, Glunde K. Localized hypoxia results in spatially heterogeneous metabolic signatures in breast tumor models. Neoplasia. 2012;14:732–41. doi: 10.1593/neo.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman V, Artemov D, Pathak AP, Winnard PT, Jr., McNutt S, Yudina A, Bogdanov A, Jr., Bhujwalla ZM. Characterizing vascular parameters in hypoxic regions: a combined magnetic resonance and optical imaging study of a human prostate cancer model. Cancer Res. 2006;66:9929–36. doi: 10.1158/0008-5472.CAN-06-0886. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi E, Sato M. Imaging of oxygen gradients in monolayer cultured cells using green fluorescent protein. Am J Physiol Cell Physiol. 2010;299:C1318–23. doi: 10.1152/ajpcell.00254.2010. [DOI] [PubMed] [Google Scholar]
- 32.Cao Y, Li CY, Moeller BJ, Yu D, Zhao Y, Dreher MR, Shan S, Dewhirst MW. Observation of incipient tumor angiogenesis that is independent of hypoxia and hypoxia inducible factor-1 activation. Cancer Res. 2005;65:5498–505. doi: 10.1158/0008-5472.CAN-04-4553. [DOI] [PubMed] [Google Scholar]
- 33.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–41. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 34.Vordermark D, Shibata T, Brown JM. Green fluorescent protein is a suitable reporter of tumor hypoxia despite an oxygen requirement for chromophore formation. Neoplasia. 2001;3:527–34. doi: 10.1038/sj.neo.7900192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–91. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 36.McKeown SR. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br J Radiol. 2014;87:20130676. doi: 10.1259/bjr.20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 38.Gorin F, Harley W, Schnier J, Lyeth B, Jue T. Perinecrotic glioma proliferation and metabolic profile within an intracerebral tumor xenograft. Acta Neuropathol. 2004;107:235–44. doi: 10.1007/s00401-003-0803-1. [DOI] [PubMed] [Google Scholar]
- 39.Drepper T, Huber R, Heck A, Circolone F, Hillmer AK, Buchs J, Jaeger KE. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl Environ Microbiol. 2010;76:5990–4. doi: 10.1128/AEM.00701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coralli C, Cemazar M, Kanthou C, Tozer GM, Dachs GU. Limitations of the reporter green fluorescent protein under simulated tumor conditions. Cancer Res. 2001;61:4784–90. [PubMed] [Google Scholar]
- 41.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 42.Carroll P, Schreuder LJ, Muwanguzi-Karugaba J, Wiles S, Robertson BD, Ripoll J, Ward TH, Bancroft GJ, Schaible UE, Parish T. Sensitive detection of gene expression in mycobacteria under replicating and non-replicating conditions using optimized far-red reporters. PLoS One. 2010;5:e9823. doi: 10.1371/journal.pone.0009823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia signalling controls metabolic demand. Curr Opin Cell Biol. 2007;19:223–9. doi: 10.1016/j.ceb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Behrooz A, Ismail-Beigi F. Stimulation of Glucose Transport by Hypoxia: Signals and Mechanisms. News Physiol Sci. 1999;14:105–10. doi: 10.1152/physiologyonline.1999.14.3.105. [DOI] [PubMed] [Google Scholar]
- 45.Ameri K, Luong R, Zhang H, Powell AA, Montgomery KD, Espinosa I, Bouley DM, Harris AL, Jeffrey SS. Circulating tumour cells demonstrate an altered response to hypoxia and an aggressive phenotype. Br J Cancer. 2010;102:561–9. doi: 10.1038/sj.bjc.6605491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rademakers SE, Lok J, van der Kogel AJ, Bussink J, Kaanders JH. Metabolic markers in relation to hypoxia; staining patterns and colocalization of pimonidazole, HIF-1alpha, CAIX, LDH-5, GLUT-1, MCT1 and MCT4. BMC Cancer. 2011;11:167. doi: 10.1186/1471-2407-11-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heilig C, Brosius F, Siu B, Concepcion L, Mortensen R, Heilig K, Zhu M, Weldon R, Wu G, Conner D. Implications of glucose transporter protein type 1 (GLUT1)- haplodeficiency in embryonic stem cells for their survival in response to hypoxic stress. Am J Pathol. 2003;163:1873–85. doi: 10.1016/S0002-9440(10)63546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaluz S, Kaluzova M, Chrastina A, Olive PL, Pastorekova S, Pastorek J, Lerman MI, Stanbridge EJ. Lowered oxygen tension induces expression of the hypoxia marker MN/carbonic anhydrase IX in the absence of hypoxia-inducible factor 1 alpha stabilization: a role for phosphatidylinositol 3′-kinase. Cancer Res. 2002;62:4469–77. [PubMed] [Google Scholar]
- 49.Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177–82. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 50.Rundqvist H, Johnson RS. Hypoxia and metastasis in breast cancer. Curr Top Microbiol Immunol. 2010;345:121–39. doi: 10.1007/82_2010_77. [DOI] [PubMed] [Google Scholar]
- 51.Entenberg D, Wyckoff J, Gligorijevic B, Roussos ET, Verkhusha VV, Pollard JW, Condeelis J. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat Protoc. 2011;6:1500–20. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paz H, Pathak N, Yang J. Invading one step at a time: the role of invadopodia in tumor metastasis. Oncogene. 2013 doi: 10.1038/onc.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desmarais V, Yamaguchi H, Oser M, Soon L, Mouneimne G, Sarmiento C, Eddy R, Condeelis J. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil Cytoskeleton. 2009;66:303–16. doi: 10.1002/cm.20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gligorijevic B, Wyckoff J, Yamaguchi H, Wang Y, Roussos ET, Condeelis J. N- WASP-mediated invadopodium formation is involved in intravasation and lung metastasis of mammary tumors. J Cell Sci. 2012;125:724–34. doi: 10.1242/jcs.092726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolde O, Folk P. Stress-induced expression of p53 target genes is insensitive to SNW1/SKIP downregulation. Cell Mol Biol Lett. 2011;16:373–84. doi: 10.2478/s11658-011-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagelkerke A, Bussink J, van der Kogel AJ, Sweep FC, Span PN. The PERK/ATF4/LAMP3-arm of the unfolded protein response affects radioresistance by interfering with the DNA damage response. Radiother Oncol. 2013;108:415–21. doi: 10.1016/j.radonc.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 57.Salnikov AV, Liu L, Platen M, Gladkich J, Salnikova O, Ryschich E, Mattern J, Moldenhauer G, Werner J, Schemmer P, et al. Hypoxia induces EMT in low and highly aggressive pancreatic tumor cells but only cells with cancer stem cell characteristics acquire pronounced migratory potential. PLoS One. 2012;7:e46391. doi: 10.1371/journal.pone.0046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raheja LF, Genetos DC, Wong A, Yellowley CE. Hypoxic regulation of mesenchymal stem cell migration: the role of RhoA and HIF-1alpha. Cell Biol Int. 2011;35:981–9. doi: 10.1042/CBI20100733. [DOI] [PubMed] [Google Scholar]
- 59.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085–99. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–9. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 61.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–56. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 62.Roussos ET, Balsamo M, Alford SK, Wyckoff JB, Gligorijevic B, Wang Y, Pozzuto M, Stobezki R, Goswami S, Segall JE, et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci. 2011;124:2120–31. doi: 10.1242/jcs.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69:9498–506. doi: 10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–84. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 65.Patsialou A, Bravo-Cordero JJ, Wang Y, Entenberg D, Liu H, Clarke M, Condeelis JS. Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. Intravital. 2013;2:e25294. doi: 10.4161/intv.25294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nehls V, Herrmann R, Huhnken M. Guided migration as a novel mechanism of capillary network remodeling is regulated by basic fibroblast growth factor. Histochem Cell Biol. 1998;109:319–29. doi: 10.1007/s004180050232. [DOI] [PubMed] [Google Scholar]
- 67.Secomb TW, Alberding JP, Hsu R, Dewhirst MW, Pries AR. Angiogenesis: an adaptive dynamic biological patterning problem. PLoS Comput Biol. 2013;9:e1002983. doi: 10.1371/journal.pcbi.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diaz B, Yuen A, Iizuka S, Higashiyama S, Courtneidge SA. Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J Cell Biol. 2013;201:279–92. doi: 10.1083/jcb.201209151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lucien F, Brochu-Gaudreau K, Arsenault D, Harper K, Dubois CM. Hypoxia-induced invadopodia formation involves activation of NHE-1 by the p90 ribosomal S6 kinase (p90RSK). PLoS One. 2011;6:e28851. doi: 10.1371/journal.pone.0028851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Md Hashim NF, Nicholas NS, Dart AE, Kiriakidis S, Paleolog E, Wells CM. Hypoxia-induced invadopodia formation: a role for beta-PIX. Open Biol. 2013;3:120159. doi: 10.1098/rsob.120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bravo-Cordero JJ, Marrero-Diaz R, Megias D, Genis L, Garcia-Grande A, Garcia MA, Arroyo AG, Montoya MC. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W, Di Modugno F, Nistico P, Gertler FB, Condeelis JS. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis. 2009;26:153–9. doi: 10.1007/s10585-008-9225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mader CC, Oser M, Magalhaes MA, Bravo-Cordero JJ, Condeelis J, Koleske AJ, Gil-Henn H. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–41. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma VP, Eddy R, Entenberg D, Kai M, Gertler F, Condeelis J. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr Biol. doi: 10.1016/j.cub.2013.08.044. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wyckoff JB, Segall JE, Condeelis JS. The collection of the motile population of cells from a living tumor. Cancer Res. 2000;60:5401–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.