Abstract
NK cells play a role in many cancer immunotherapies. NK cell activity is tightly regulated by Killer Immunoglobulin-like Receptors (KIR) and KIR-ligand interactions. Inhibitory KIR-Ligands have been identified as HLA molecules, while activating KIR-ligands are largely unknown. Individuals that have not inherited the corresponding KIR-ligand for at least one inhibitory KIR gene are termed the “KIR-ligand missing” genotype, and they are thought to have a subset of NK cells that express inhibitory KIRs for which the corresponding KIR-ligand is missing on autologous tissue, and thus will not be inhibited through KIR-ligand recognition. In some settings where an anti-cancer immunotherapeutic effect is likely mediated by NK cells, individuals with a KIR-ligand missing genotype have shown improved clinical outcome compared to individuals with an “all KIR-ligand present” genotype. In addition, patients receiving hematopoietic stem cell transplants for leukemia may do better if their donor has more activating KIR genes (i.e. KIR haplotype-B).
In a recent multi-institution clinical trial of patients with metastatic renal cell carcinoma (mRCC) receiving high dose-IL2 (HD-IL2), 25% of patients showed a complete or partial tumor response to this therapy. We genotyped KIR and KIR-ligand genes for these patients (n=107) and tested whether KIR/KIR-Ligand genotypes correlated with patient clinical outcomes. In these analyses we did not find any significant association of KIR/KIR-ligand genotype (either KIR-Ligand missing or the presence of KIR haplotype-B) with patient outcome in response to the HD-IL2 therapy.
Keywords: Renal cell carcinoma, high-dose IL2, killer immunoglobulin-like receptors, HLA, NK cells
INTRODUCTION
Metastatic renal cell carcinoma (mRCC) is a disease with a poor prognosis. Cytokine based immunotherapy with IL-2 was approved by the FDA to treat patients with mRCC in 1992 [1]. The “SELECT” clinical trial was designed to validate predictive markers for patient response to high-dose IL2 (HD-IL2), and the study achieved a higher response rate than historical trials (25% vs. 14%) [2]. NK cell activation is regulated, in part, by Killer Immunoglobulin-like Receptors (KIRs) [3], and KIR and KIR-ligand genotypes have been shown to play a role in patient response to immunotherapies involving NK cells [4–7]. In this “SELECT” trial where patient NK cells have the potential to be activated by IL2 treatment, we performed correlative analyses to evaluate the potential role of KIR and KIR-Ligand genotypes in patient response to HD-IL2.
KIR genes are a family of receptors expressed on most NK cells and on some subsets of T cells. There are 15 different KIR genes, which can be inhibitory or activating depending on the KIR cytoplasmic signaling motifs [8]. The KIR genes are located on chromosome 19 and based on the KIR repertoire an individual inherits, he is designated to be either KIR haplotype-A or KIR haplotype-B. Individuals that have inherited KIR haplotype-A (genotype KIR haplotype-A/A), are designated as haplotype-A individuals, and they have fewer activating KIR genes than individuals that have inherited at least one KIR haplotype-B (genotype KIR Haplotype-B/x), and thus designated as haplotype-B individuals [8]. The Activating KIR genes are believed to have played a role in acute myeloid leukemia patients receiving unrelated hematopoietic cell transplantation where patients showed better outcome if their donor was a haplotype-B [9–11].
Most inhibitory KIRs recognize certain HLA epitopes as their ligand. HLA-C alleles with a lysine at amino acid position 80 (HLA-C2) are ligands for inhibitory KIR2DL1 [12]. This HLA-C2 is also a ligand for the activating KIR2DS1 with less strong binding [13]. HLA-C alleles with an asparagine at amino acid position 80 (HLA-C1) are ligands for inhibitory KIR2DL2 and 2DL3 [14]. Both HLA-A and HLA-B alleles with a Bw4 motif (HLA-A-Bw4 and HLA-B-Bw4) are ligands for inhibitory KIR3DL1 [15]. Except for the recognition of HLA-C2 by activating KIR2DS1, the ligands for most of the activating KIRs have not yet been identified.
Inhibitory KIR/KIR-ligand interactions play an important role in NK cell education and function. KIR-ligand mismatch between donor and recipient (recipient missing KIR-ligand for donor inhibitory KIR genes) in stem cell transplantation was associated with improved clinical outcome for leukemia patients [16,17]. Since KIR and HLA are inherited independently on chromosome 19 and chromosome 6, respectively [8], an individual may lack one or more HLA epitopes for their inhibitory KIR genes; such individuals are referred to as having a “KIR-ligand missing” genotype. This KIR/KIR-ligand genotype is present in more than 60% of the population [18,6]. Alternatively, those individuals that have HLA epitopes for all of their inherited inhibitory KIR genes are referred to as having “all KIR-ligands present”. The KIR-ligand missing genotype has also been shown to correlate with better clinical outcomes for patients receiving immunotherapies that involve NK cell function, such as anti-GD2 mAb for neuroblastoma patients and rituximab therapy for patients with B-cell lymphoma [4,6,7,5].
In addition to tumor-reactive mAb, IL2 is also a potent activator of NK cells, in vitro and in vivo [19,20]. Here we examined whether KIR/KIR-ligand genotypes were associated with the clinical anti-tumor activity of HD-IL2, potentially indicating a role for IL2-activated NK cells in vivo. The KIR/KIR-ligand genotypes we evaluated here mainly focused on those with past observations in the literature that did show some prior correlation with cancer immunotherapy outcomes, or those with some prior biological observation that led to the hypothesis examined: overall KIR-ligand status (KIR-ligand missing vs. KIR-ligands present) [4,16,6], individual KIR-ligand missing vs. present [5,21], and KIR haplotypes [9,10]. More specifically, three distinct levels of analyses were performed to assess: 1) association between clinical outcome and KIR/KIR-ligand genotypes; 2) association between KIR/KIR-ligand genotypes and tumor PDL1 expression and 3) association between clinical outcome and KIR/KIR-ligand genotypes/PDL1 interactions.
MATERIALS AND METHODS
Patients and clinical treatment
All patients evaluated in this study participated in the SELECT trial of HD-IL2 for patients with mRCC. The clinical details of this study and its clinical conclusions were reported by McDermott DF et al. [2].
DNA isolation and whole-genome amplification
Genomic DNA was isolated from patient PBMCs using the DNeasy Blood and Tissue Kit (Qiagen). DNA was then whole genome-amplified using the REPLI-g Mini Kit per the manufacturer’s instructions (Qiagen). Whole-genome amplified DNA samples were used for KIR and KIR-ligand genotyping.
KIR/KIR-ligand genotyping
KIR genes were genotyped using real-time SYBR green PCR melt curve analyses as developed by Vilches et al. [22]. KIR-ligand genotypes were determined using a sequence specific primer PCR kit, SSP KIR HLA ligand, per the manufacturers protocol (Olerup), as previously reported [4] .
Histologic analyses
Tumor samples from patients were analyzed by immunohistochemistry for expression of various histological markers, as reported elsewhere [2].
Data management
Genotyping data from our lab were collected and managed using the REDCap system hosted at the University of Wisconsin-Madison. REDCap (Research Electronic Data Capture) is a secure, HIPAA-compatible, web-based application designed to support data capture for research studies, providing: 1) an intuitive interface for validated data entry; 2) audit trails for tracking data manipulation, algorithm input for data analysis, and export procedures; and 3) procedures for importing data from external sources [23]. The clinical outcome data from the mRCC study (which is HIPAA protected) were merged with the genotyping data in REDCap to create a SAS dataset for analysis.
Algorithms for genotype categorization
KIR-ligand missing vs. KIR-ligands present
If an individual’s genotype was found to be negative for any one of the KIR-ligand genes for the inhibitory KIR genes (KIR2DL1, 2DL2, 2DL3 or 3DL1) that were found to be present in their genome, their genotype was defined as “KIR-ligand missing”. If an individual’s genotype is positive for KIR-ligand genes for all the inhibitory KIR genes studied here (KIR2DL1, 2DL2, 2DL3 or 3DL1) that are present in that individual’s genome, this genotype is defined as “all KIR-ligands present”, and will be referred to in this report as “KIR-ligands present” [4].
KIR haplotype-A vs. KIR haplotype-B
Individuals with a KIR haplotype-A/A genotype are categorized as KIR haplotype-A. Individuals with at least one KIR haplotype-B (KIR haplotype-B/A or KIR haplotype B/B genotype) are categorized as KIR haplotype-B. KIR haplotype-A or -B were determined as published by Cooley et al. [9].
Statistical analyses
Genotyping results were analyzed for association with the following clinical outcomes: 1) maximum % tumor shrinkage; 2) progression-free survival (PFS); and 3) overall survival (OS). The maximum % tumor shrinkage represents the peak shrinkage in tumor burden when combining all target lesions (measured as the product of the longest tumor diameter and longest perpendicular diameter) for each patient according to WHO criteria.
Statistical analyses were performed using SAS v9.4 (SAS Institute, Cary, NC). Linear regression models for maximum % tumor shrinkage were used to evaluate association with genotyping predictors. The Kaplan-Meier method was used for estimation of the survival distribution for both PFS and OS. Log-rank tests and Cox proportional hazards regression models for PFS and OS were used to evaluate associations with genotyping predictors. Fisher’s exact tests or chi-square tests were used to assess the association between KIR/KIR-ligand genotype and histological markers. For interaction analyses between KIR and KIR-ligand genes, or between KIR/KIR-ligand genotype and histological markers, further intergroup comparisons were performed if the p-value was ≤ 0.10. As all analyses in this report are exploratory, p-values are reported as calculated by the methods described above, without corrections for multiple comparisons.
RESULTS
Clinical characteristics of patient cohort analyzed for KIR/KIR-ligand genotypes
The SELECT clinical trial enrolled a total of 120 patients for treatment with HD-IL2 [2]. Of these, 107 patients had PBMCs available for DNA isolation and were genotyped for KIR and KIR-ligand genes. There were no major differences observed in the clinical characteristics from the total SELECT patient cohort (n=120) as compared to the subset of SELECT patients (n=107) genotyped for KIR/KIR-ligand genes (Table 1).
Table 1.
Patient clinical characteristics from original SELECT Trial and the subset of patients analyzed in this study
| Total Patients Enrolled | KIR/KIR-ligand Genotyped Patients | |
|---|---|---|
|
| ||
| Characteristics | n = 120 | n = 107 |
|
| ||
| Median age, y (range) | 56 (28–70) | 56 (28–70) |
|
| ||
| ECOG performance status (0/1), % | 72/24 | 71/24 |
|
| ||
| Prior nephrectomy, % | 99 | 99 |
|
| ||
| MSKCC risk factor, n (%) | ||
| 0 (favorable) | 23 (19) | 22 (21) |
| 1–2 (intermediate) | 84 (70) | 73 (68) |
| ≥3 (poor) | 13 (11) | 12 (11) |
|
| ||
| UCLA SANI Score, n (%) | ||
| Low | 10 (8) | 10 (9) |
| Intermediate | 102 (85) | 89 (83) |
| High | 8 (7) | 8 (7) |
KIR and KIR-ligand genotype frequency
Among the 107 patients genotyped for KIR and KIR-ligand, one DNA sample did not result in any discernible KIR or KIR-ligand genotypes. Another DNA sample had repeatable KIR genotype results but no KIR-ligand genotype results. Therefore, the former was excluded for any statistical analyses, and the latter was excluded for any analyses involving KIR-ligand genotypes.
Genotype frequencies of KIR2DL1, 2DL2, 2DL3, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1 and 3DS1 as well as for KIR-ligands HLA-C1, HLA-C2 and HLA-Bw4 in the cohort of patients with mRCC that were genotyped for this study are shown in Table 2. Eighty-four percent of the patients had HLA-C1, the ligand for KIR2DL2/3, and 65% of the patients had HLA-C2, the ligand for KIR2DL1. Seventy percent of the patients had HLA-Bw4, the ligand for KIR3DL1 (Table 2). The KIR and KIR-ligand genotype frequencies are similar to what is observed in the healthy population [24].
Table 2.
KIR and KIR-ligand genotype frequency
| Number of patientsa | % | ||
|---|---|---|---|
|
| |||
| 2DL1 | + | 103 | 97 |
| − | 3 | 3 | |
|
| |||
| 2DL2 | + | 52 | 49 |
| − | 54 | 51 | |
|
| |||
| 2DL3 | + | 92 | 87 |
| − | 14 | 13 | |
|
| |||
| 2DL5 | + | 51 | 48 |
| − | 55 | 52 | |
|
| |||
| 2DS1 | + | 35 | 33 |
| − | 71 | 67 | |
|
| |||
| 2DS2 | + | 50 | 48 |
| − | 56 | 52 | |
|
| |||
| 2DS3 | + | 28 | 26 |
| − | 78 | 74 | |
|
| |||
| 2DS4 | + | 102 | 96 |
| − | 4 | 4 | |
|
| |||
| 2DS5 | + | 31 | 29 |
| − | 75 | 71 | |
|
| |||
| 3DL1 | + | 103 | 97 |
| − | 3 | 3 | |
|
| |||
| 3DS1 | + | 32 | 30 |
| − | 74 | 70 | |
|
| |||
| HLA-C | C1/C1 | 37 | 35 |
| C1/C2 | 51 | 49 | |
| C2/C2 | 17 | 16 | |
|
| |||
| HLA-Bw4 | + | 74 | 70 |
| − | 31 | 30 | |
One of the 107 patients was excluded from KIR analyses; two of the 107 patients were excluded from KIR-ligand analyses.
Association of KIR-ligand missing and KIR haplotypes with patient outcome
Inhibitory KIR-ligand missing status has correlated with improved outcomes for patients receiving certain immunotherapies [4,6,7]. In this study of mRCC patients treated with HD-IL2, we did not find any significant associations between KIR/KIR-ligand status (any KIR-ligand missing vs. all KIR-ligands present, see method section “Algorithms for genotype categorization”) and patient clinical outcomes (Table 3). In addition, when separate analyses were performed looking at outcome (tumor shrinkage, PFS and OS) we found no differences (or trends) between individuals with 0, 1 or 2 KIR-ligands missing (data not shown). Analyses of the individual KIR-ligand genotypes also revealed no association with patient clinical outcomes (Supplemental Table 1).
Table 3.
No association between KIR-ligand genotype and KIR haplotype with clinical outcome for RCC patients receiving HD-IL2 was found.
| KIR-ligands present† | KIR-ligand missing† | p-value* | Haplotype-A | Haplotype-B | p-value* | ||
|---|---|---|---|---|---|---|---|
| Maximum tumor shrinkage [% shrinkage] | n | 41 | 62 | 0.45 | 36 | 68 | 0.65 |
| Mean | 19.0 | 9.2 | 9.0 | 15.0 | |||
| SD | 63.4 | 64.0 | 61.7 | 64.5 | |||
|
| |||||||
| Progression-free survival [in months] | n | 42 | 63 | 0.47 | 37 | 69 | 0.38 |
| # of Events | 37 | 60 | 36 | 62 | |||
| Median PFS (95% CI) | 4.2 (2.3–5.9) | 4.0 (2.3–4.8) | 4.2 (2.3–5.3) | 4.0 (2.3–5.9) | |||
|
| |||||||
| Overall survival [in months] | n | 42 | 63 | 0.63 | 37 | 69 | 0.74 |
| # of Events | 27 | 43 | 26 | 45 | |||
| Median OS (95% CI) | 38.1 (26.7–61.1) | 48.8 (32.5–56.0) | 42.6 (22.5–61.1) | 47.7 (32.5–56.0) | |||
All KIR-ligands present vs. Any KIR-ligand missing (see method section “Algorithms for genotype categorization”);
Linear regression models were used for maximum tumor shrinkage analyses; Log-rank tests and Cox proportional hazards regression models were used for PFS and OS analyses.
KIR haplotypes were previously shown to influence the clinical outcome of leukemia patients following allogeneic HSCT [9–11]. Our assessment of KIR haplotypes (haplotype-A vs. haplotype-B) for these patients with mRCC revealed no association with clinical outcomes (Table 3). Similarly, when the presence or absence of individual activating KIR were analyzed, there were no significant associations seen for any clinical outcome (data not shown).
Since activating KIRs that are primarily present in haplotype-B may potentially alter the impact from inhibitory KIR/KIR-ligand interactions, we also assessed whether KIR haplotype-A or B status might influence whether KIR-ligand missing or present status was associated with outcome for these patients with mRCC treated with HD-IL2, but no significant association was found (data not shown).
Association between KIR/KIR-ligand genotype and PDL1 expression on tumors
In evaluating IHC analyses of several clinically relevant markers on tumors from patients participating in this clinical trial, McDermott et al. noted that the presence of Programmed Death-Ligand 1 (PDL1) expression on patient’s tumors was associated with improved response rate and PFS [2]. We thus evaluated whether an individual’s KIR/KIR-ligand genotype might be associated with PDL1 expression on a patient’s tumor. We found that individuals with KIR haplotype-B were more likely to express PDL1 on their tumors (Table 4). As KIR genes 2DS2 and 2DL2 demonstrate linkage disequilibrium [25], and about 72% of haplotype-B patients are also 2DS2/2DL2+ in this patient cohort, these are not independent observations, but reflect overlapping sets of patients.
Table 4.
Association of PDL1 expression on tumors by IHC with certain KIR/KIR-ligand genotypes
| KIR genotype | PDL1− n |
PDL1+ n |
p-value* |
|---|---|---|---|
| Haplotype | |||
|
| |||
| Haplotype A | 35 | 2 | 0.03 |
| Haplotype B | 50 | 15 | |
|
| |||
| KIR2DS2 | |||
|
| |||
| 2DS2+ | 35 | 12 | 0.03 |
| 2DS2− | 50 | 5 | |
|
| |||
| KIR2DL2 | |||
|
| |||
| 2DL2+ | 36 | 12 | 0.04 |
| 2DL2− | 48 | 5 | |
Fisher’s exact tests or chi-square tests were used to assess the association between KIR/KIR-ligand genotype and histological markers.
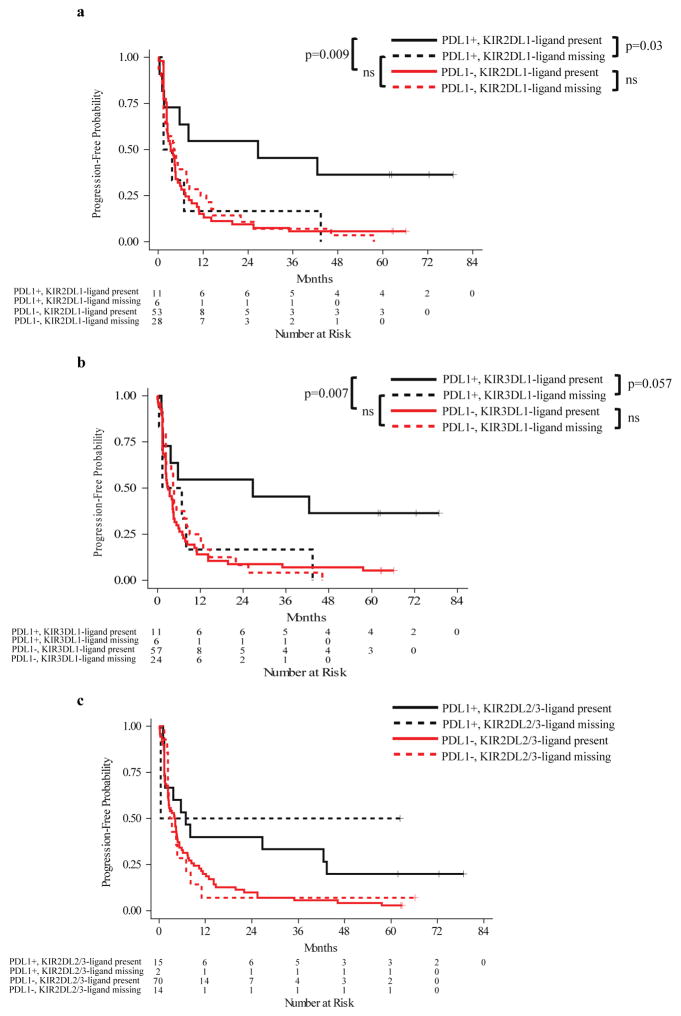
While our study did not show an association of KIR/KIR-ligand genotype with outcome overall, our additional analyses did indicate that the influence of KIR/KIR-ligand genotype on outcome might be PDL1 dependent. Individuals that were PDL1+ and KIR2DL1-ligand present (i.e. 2DL1+, HLA-C2+) had a significantly prolonged PFS as compared to patients that were PDL1+ and KIR2DL1-ligand missing (i.e. 2DL1+, HLA-C2-) (Figure 1a). The same association was also seen when evaluating KIR3DL1-ligand status (Figure 1b). There was no significant interaction between PDL1 status and KIR2DL2/3-ligand genotype in terms of their influence on patient outcome (Figure 1c), indicating that the beneficial impact of PDL1+ status on patient outcome was independent of KIR2DL2/3-ligand present or missing genotype. Similar results to those for PFS in Figures 1a and 1b were also observed for maximum % tumor shrinkage and for overall survival (data not shown). Even though 2DL1-ligand and 3DL1-ligand were assessed independently, we found substantial overlap of these populations. Nine out of 11 PDL1+ 2DL1-ligand present patients were also PDL1+ 3DL1-ligand present; 4 out of 6 PDL1+ 2DL1-ligand missing patients were also PDL1+ 3DL1-ligand missing. Therefore, the data shown in Figures 1a and 1b are not independent observations.
Figure 1. Progression-free survival: tumor PDL-1 expression and KIR-ligand genotypes.
The interaction analysis between tumor PDL1 phenotype and KIR2DL1 (interaction p=0.041) or 3DL1-ligand genotype (interaction p=0.054) was evaluated by Cox proportional hazards analyses with further subgroup comparisons shown below. a) While PDL1+ patients had prolonged PFS than PDL1− patients if they were KIR2DL1-ligand present genotype (p=0.009), there was no difference between PDL1+ and PDL1− patients if they were KIR2DL1-ligand missing. In addition, individuals that were PDL1+ and KIR2DL1-ligand present showed prolonged PFS than patients that are PDL1+ and KIR2DL1-ligand missing (p=0.03); b) While PDL1+ patients had prolonged PFS than PDL1− patients if they were KIR3DL1-ligand present genotype (p=0.007), there was no difference between PDL1+ and PDL1− patients if they were KIR3DL1-ligand missing. In addition, individuals that were PDL1+ and KIR3DL1-ligand present showed a trend towards prolonged PFS than patients that are PDL1+ and KIR3DL1-ligand missing (p=0.057). c) Since the interaction p value between PDL1 and KIR2DL2/3-ligand missing is above 0.1 (interaction p=0.64), the impact from PDL1 expression and KIR2DL2/3-ligand genotypes on PFS are independent from each other. Therefore, no pairwise comparisons were made between the subgroups of patients.
DISCUSSION
In IL2-treated patients with advanced cancer, West WH et al. have shown that the mononuclear cells from responders had slightly increased cytotoxicity in vitro than those cells from non-responders, although the difference was not significant [26], which suggests that NK cells may play a role in patient response to IL2 treatment. However, such correlation was not seen in other early phase clinical trials with different regimens of IL2 treatment [27,28]. Since the role that NK cells play in IL2 treatment is still unclear, we assessed the correlation between patient KIR/KIR-ligand genotypes and their response to HD-IL2 treatment in the mRCC study [2].
An inhibitory KIR-ligand missing genotype has been shown to result in a subset of NK cells that express inhibitory KIRs that are not inhibited by self-HLA [29]. Due to reduced inhibition for these NK cells, when stimulatory signals are present, such as tumor-reactive mAb, these uninhibited NK cells exhibit higher cytotoxic capability as compared to NK cells that are inhibited by self-HLA [6]. Inhibitory KIR-ligand missing status was associated with better clinical outcomes in patients with neuroblastoma who received anti-GD2 mAb therapies [4,6,7]. In addition, in a study of patients with lymphoma receiving rituximab therapy, we found a haplotype-dependent influence of KIR-ligand missing (unpublished data). However, in this cohort of mRCC patients that received HD-IL2, we did not find any statistically significant associations between patient response and KIR-ligand missing genotype or KIR haplotypes, either alone (Table 2) or in combination (data not shown). Unlike patients with neuroblastoma or lymphoma who received tumor-reactive mAb as immunotherapy [4–6], there was no RCC-reactive mAb infused into these patients with mRCC as part of their immunotherapy treatment in this clinical trial. Therefore, even though inhibitory KIR-ligand missing may be predictive of patient response to mAb therapy where NK cells are activated via tumor-reactive mAb through Fc receptors, this KIR-ligand missing status appears to not be associated with anti-tumor activity in the setting of IL2 activation, which is mediated via IL2 receptors [30]. Based on these experiments, we can’t confirm a role for KIR or KIR-ligand genotypes related to NK cell function in the anti-tumor activity of HD IL-2 therapy for patients with mRCC.
In addition to overall KIR-ligand missing status, the presence or absence of certain individual inhibitory KIR-ligand missing, for a patient’s KIR/KIR-ligand genotype, may also be associated with response to certain cancer immunotherapies. Du et al. reported that HLA-C2 and Bw4 missing in patients with follicular lymphoma receiving rituximab mAb therapy significantly improved progression-free survival [5]. However, our results indicate that missing C2 for 2DL1+ patients, missing C1 for 2DL2+ or 2DL3+ patients, or missing Bw4 for 3DL1+ patients is not associated with improved response for these patients with mRCC treated with HD-IL2 (Supplemental Table 1).
Other factors beyond KIR-ligand missing may also contribute to the degree of response that may be attributed to NK cells in patients with mRCC, such as the copy number of both inhibitory KIR and KIR-ligand, as well as other receptors expressed on NK cells. For example, Programmed Cell Death Protein 1 (PD-1), another inhibitory receptor expressed on NK cells, recognizes PDL1 expressed on tumor tissue [31]. PD-1 expression on peripheral NK cells has been reported for patients with RCC and is correlated with disease stage [32]. In addition, McDermott et al. found that, in the same cohort of patients enrolled in the SELECT study, the expression of PDL1 on patient tumors was associated with a higher response rate and prolonged PFS following HD-IL2 treatment [2]. In this study, our data suggest that certain KIR/KIR-ligand interactions are associated with better outcome for those patients with PDL1+ tumors. Specifically, a subgroup of patients with 2DL1-ligand or 3DL1-ligand present and PDL1+ tumors had significantly prolonged PFS compared to the rest of the patients. The presence of PDL1 on a variety of tumors has previously been associated with a greater degree of immune cell infiltrate into tumors [33,34]. This might reflect a stronger pre-existing immune response and could account, in part, for our data indicating that PDL1 expression (in the context of having the KIR gene and its ligand present) may be associated with better outcome.
In certain other settings (particularly allogeneic bone marrow transplantation, autologous bone marrow transplantation for solid tumors, and anti-GD2 mAb treatment for neuroblastoma), there has been benefit for patients that have KIR-ligand missing genotype [4,35,16]. This in part results from NK mediated killing of tumor cells under conditions where the inhibitory KIRs on the NK cells are not seeing their corresponding inhibitory ligand on the tumor cells (and thus not inhibited) [6]. However, this is not always observed [36], and we do not see a benefit in outcome for KIR-ligand missing genotype here in our analyses of these mRCC patients. In addition, while the KIR-ligands can inhibit NK cells through their inhibitory KIRs, those same KIR-ligands (HLA class-I molecules) are also important targets for T cell recognition. Moreover, the interactions of KIR on NK cells with their KIR-ligands during NK development are also associated with greater subsequent NK cell function, through NK cell licensing [37]. This licensing may be playing a role in potentially accounting for the better outcome we observe for patients with PDL1+ tumors that have KIR-ligands present for KIRs 2DL1 and 3DL1 (Figure 1). However, due to a limited number of patients that have PDL1+ tumors in this study, this observation requires further validation in future studies.
Multiple other factors may also contribute to the absence of evidence supporting an association of KIR/KIR-ligand genotype with clinical outcome for patients with mRCC receiving HD-IL2 from this study. Analyses of KIR/KIR-ligand genotypes and clinical outcome are revealing different patterns of associations for different diseases and for different forms of immunotherapy [4–6,38,16]. Different types of tumor cells may have differential susceptibility to NK recognition or NK-mediated cytotoxicity. If so, the level of resistance to NK lysis may influence how much KIR/KIR-ligand inhibition may play a role in immunotherapeutic effect. Different forms of immunotherapy may also have widely different roles for NK cells. While NK cells (and KIR/KIR-ligand interactions) appear to be involved in the clinical response to the use of anti-GD2 or anti-CD20 based immunotherapies for neuroblastoma and follicular lymphoma [4–6], respectively, our data suggest that the role of NK cells, and of KIR, seems less evident in the clinical responses of patients with mRCC to HD-IL2.
Overall, our data show that inhibitory KIR/KIR-ligand genotypes or KIR haplotype (A/A vs. B/x), either alone or in combination, do not correlate with mRCC patient response to HD-IL2.
Supplementary Material
Acknowledgments
The authors thank the medical and nursing staff that participated in the care of patients in this study, and especially all patients participating in this study. Thanks also to Bartosz Grzywacz and Patrick Reville for their helpful discussions and input. This research was supported by Hyundai Hope on Wheels Grant; Midwest Athletes Against Childhood; Stand Up 2 Cancer; The St. Baldrick’s Foundation; American Association of Cancer Research; University of Wisconsin-Madison Carbone Cancer Center; by NCI-Cytokine Working Group and supported in part by Public Health Service Grants CA014520, CA021115, CA023318, CA066636, CA180820, CA180794, CA021076, CA180799, CA14958, CA180816, CA166105, and CA197078, from the National Cancer Institute; the National Institutes of Health and the Department of Health and Human Services. Its content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Abbreviations
- HD-IL2
High-dose IL2
- KIR
Killer immunoglobulin-like receptor
- mRCC
Metastatic renal cell carcinoma
- OS
Overall survival
- PD-1
Programmed cell death protein 1
- PDL-1
Programmed death-ligand 1
- PFS
Progression-free survival
Footnotes
CONFLICT OF INTEREST
The authors have no financial conflicts of interest.
References
- 1.Bhatia S, Tykodi SS, Thompson JA. Treatment of metastatic melanoma: an overview. Oncology (Williston Park) 2009;23(6):488–496. [PMC free article] [PubMed] [Google Scholar]
- 2.McDermott DF, Cheng SC, Signoretti S, Margolin KA, Clark JI, Sosman JA, Dutcher JP, Logan TF, Curti BD, Ernstoff MS, Appleman L, Wong MK, Khushalani NI, Oleksowicz L, Vaishampayan UN, Mier JW, Panka DJ, Bhatt RS, Bailey AS, Leibovich BC, Kwon ED, Kabbinavar FF, Belldegrun AS, Figlin RA, Pantuck AJ, Regan MM, Atkins MB. The high-dose aldesleukin “select” trial: a trial to prospectively validate predictive models of response to treatment in patients with metastatic renal cell carcinoma. Clin Cancer Res. 2015;21(3):561–568. doi: 10.1158/1078-0432.CCR-14-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasser S, Raulet DH. Activation and self-tolerance of natural killer cells. Immunol Rev. 2006;214:130–142. doi: 10.1111/j.1600-065X.2006.00460.x. [DOI] [PubMed] [Google Scholar]
- 4.Delgado DC, Hank JA, Kolesar J, Lorentzen D, Gan J, Seo S, Kim K, Shusterman S, Gillies SD, Reisfeld RA, Yang R, Gadbaw B, DeSantes KB, London WB, Seeger RC, Maris JM, Sondel PM. Genotypes of NK Cell KIR Receptors, Their Ligands, and Fc Receptors in the Response of Neuroblastoma Patients to Hu14.18-IL2 Immunotherapy. Cancer Res. 2010;70(23):9554–9561. doi: 10.1158/0008-5472.can-10-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Lopez-Verges S, Pitcher BN, Johnson J, Jung SH, Zhou L, Hsu K, Czuczman MS, Cheson B, Kaplan L, Lanier LL, Venstrom JM. CALGB 150905 (Alliance): Rituximab Broadens the Antilymphoma Response by Activating Unlicensed NK Cells. Cancer Immunol Res. 2014;2(9):878–889. doi: 10.1158/2326-6066.cir-13-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarek N, Luduec JL, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, Modak S, Heller G, Dupont B, Cheung NV, Hsu KC. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest. 2012;122(9):3260–3270. doi: 10.1172/jci62749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venstrom JM, Zheng J, Noor N, Danis KE, Yeh AW, Cheung IY, Dupont B, O’Reilly RJ, Cheung NKV, Hsu KC. KIR and HLA Genotypes Are Associated with Disease Progression and Survival following Autologous Hematopoietic Stem Cell Transplantation for High-Risk Neuroblastoma. Clin Cancer Res. 2009;15(23):7330–7334. doi: 10.1158/1078-0432.ccr-09-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashirova AA, Martin MP, McVicar DW, Carrington M. The Killer Immunoglobulin-Like Receptor Gene Cluster: Tuning the Genome for Defense. Annu Rev Genomics Hum Genet. 2006;7(1):277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 9.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, Marsh SGE, Guethlein LA, Parham P, Miller JS, Weisdorf DJ. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2008;113(3):726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, Marsh SGE, Geraghty D, Spellman S, Haagenson MD, Ladner M, Trachtenberg E, Parham P, Miller JS. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116(14):2411–2419. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SGE, Spellman S, Haagenson MD, Saeturn K, Ladner M, Trachtenberg E, Parham P, Miller JS. Donor Killer Cell Ig-like Receptor B Haplotypes, Recipient HLA-C1, and HLA-C Mismatch Enhance the Clinical Benefit of Unrelated Transplantation for Acute Myelogenous Leukemia. J Immunol. 2014;192(10):4592–4600. doi: 10.4049/jimmunol.1302517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almeida CR, Ashkenazi A, Shahaf G, Kaplan D, Davis DM, Mehr R. Human NK cells differ more in their KIR2DL1-dependent thresholds for HLA-Cw6-mediated inhibition than in their maximal killing capacity. PLoS ONE. 2011;6(9):e24927. doi: 10.1371/journal.pone.0024927.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J Immunol. 2007;179(2):854–868. doi: 10.4049/jimmunol.179.2.854. [DOI] [PubMed] [Google Scholar]
- 14.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180(6):3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 15.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180(2):537–543. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of Donor Natural Killer Cell Alloreactivity in Mismatched Hematopoietic Transplants. Science. 2002;295(5562):2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Martínez A, Leung W, Muñoz E, Iyengar R, Ramírez M, Vicario JL, Lassaletta Á, Sevilla J, González-Vicent M, Madero L, Díaz-Pérez MÁ. KIR-HLA receptor-ligand mismatch associated with a graft-versus-tumor effect in haploidentical stem cell transplantation for pediatric metastatic solid tumors. Pediatr Blood Cancer. 2009;53(1):120–124. doi: 10.1002/pbc.21955. [DOI] [PubMed] [Google Scholar]
- 18.Linn YC, Phang CY, Lim TJ, Chong SF, Heng KK, Lee JJ, Loh Y, Hwang W, Goh YT, Koh M. Effect of missing killer-immunoglobulin-like receptor ligand in recipients undergoing HLA full matched, non-T-depleted sibling donor transplantation: a single institution experience of 151 Asian patients. Bone Marrow Transplant. 2010;45(6):1031–1037. doi: 10.1038/bmt.2009.303. [DOI] [PubMed] [Google Scholar]
- 19.Hank JA, Robinson RR, Surfus J, Mueller BM, Reisfeld RA, Cheung NK, Sondel PM. Augmentation of antibody dependent cell mediated cytotoxicity following in vivo therapy with recombinant interleukin 2. Cancer Res. 1990;50(17):5234–5239. [PubMed] [Google Scholar]
- 20.Hank JA, Surfus J, Gan J, Chew TL, Hong R, Tans K, Reisfeld R, Seeger RC, Reynolds CP, Bauer M, et al. Treatment of neuroblastoma patients with antiganglioside GD2 antibody plus interleukin-2 induces antibody-dependent cellular cytotoxicity against neuroblastoma detected in vitro. J Immunother Emphasis Tumor Immunol. 1994;15(1):29–37. doi: 10.1097/00002371-199401000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Forlenza CJ, Boudreau JE, Zheng J, Le Luduec JB, Chamberlain E, Heller G, Cheung NK, Hsu KC. KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma. J Clin Oncol. 2016;34(21):2443–2451. doi: 10.1200/JCO.2015.64.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilches C, Castaño J, Gómez-Lozano N, Estefanía E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70(5):415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 23.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2010;39(Database):D913–D919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gourraud PA, Meenagh A, Cambon-Thomsen A, Middleton D. Linkage disequilibrium organization of the human KIR superlocus: implications for KIR data analyses. Immunogenetics. 2010;62(11–12):729–740. doi: 10.1007/s00251-010-0478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West WH, Tauer KW, Yannelli JR, Marshall GD, Orr DW, Thurman GB, Oldham RK. Constant-infusion recombinant interleukin-2 in adoptive immunotherapy of advanced cancer. N Engl J Med. 1987;316(15):898–905. doi: 10.1056/NEJM198704093161502. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell W, McDevitt J, Reid I, Sharpe I, Feighery C, Tanner WA, Emmons R, Monson JR. Changes in immunological parameters during interleukin 2 and interferon 2 alpha treatment of recurrent renal cell carcinoma and malignant melanoma. Eur J Surg Oncol. 1993;19(3):265–272. [PubMed] [Google Scholar]
- 28.Thompson JA, Lee DJ, Cox WW, Lindgren CG, Collins C, Neraas KA, Dennin RA, Fefer A. Recombinant interleukin 2 toxicity, pharmacokinetics, and immunomodulatory effects in a phase I trial. Cancer Res. 1987;47(15):4202–4207. [PubMed] [Google Scholar]
- 29.Becker S, Tonn T, Füssel T, Uhrberg M, Bogdanow M, Seifried E, Seidl C. Assessment of killer cell immunoglobulinlike receptor expression and corresponding HLA class I phenotypes demonstrates heterogenous KIR expression independent of anticipated HLA class I ligands. Hum Immunol. 2003;64(2):183–193. doi: 10.1016/s0198-8859(02)00802-9. [DOI] [PubMed] [Google Scholar]
- 30.Kehrl JH, Dukovich M, Whalen G, Katz P, Fauci AS, Greene WC. Novel Interleukin-2 (Il-2) Receptor Appears to Mediate Il-2-Induced Activation of Natural-Killer Cells. J Clin Invest. 1988;81(1):200–205. doi: 10.1172/Jci113295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benson DM, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, Baiocchi RA, Zhang JY, Yu JH, Smith MK, Greenfield CN, Porcu P, Devine SM, Rotem-Yehudar R, Lozanski G, Byrd JC, Caligiuri MA. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood. 2010;116(13):2286–2294. doi: 10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacFarlane AW, Jillab M, Plimack ER, Hudes GR, Uzzo RG, Litwin S, Dulaimi E, Al-Saleem T, Campbell KS. PD-1 Expression on Peripheral Blood Cells Increases with Stage in Renal Cell Carcinoma Patients and Is Rapidly Reduced after Surgical Tumor Resection. Cancer Immunol Res. 2014;2(4):320–331. doi: 10.1158/2326-6066.CIR-13-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson ED, Zahurak M, Murphy A, Cornish T, Cuka N, Abdelfatah E, Yang S, Duncan M, Ahuja N, Taube JM, Anders RA, Kelly RJ. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut. 2016 doi: 10.1136/gutjnl-2015-310839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung W, Handgretinger R, Iyengar R, Turner V, Holladay MS, Hale GA. Inhibitory KIR–HLA receptor–ligand mismatch in autologous haematopoietic stem cell transplantation for solid tumour and lymphoma. Br J Cancer. 2007;97(4):539–542. doi: 10.1038/sj.bjc.6603913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scquizzato E, Zambello R, Teramo A, Baesso I, Varotto S, Albergoni MP, Boscaro E, Cesaro S, Pillon M, Calore E, Gazzola MV, Semenzato G, Messina C, Trentin L. KIR/HLA-I mismatching and risk of relapse in paediatric patients undergoing non-haploidentical allogeneic haematopoietic stem cell transplantation. Pediatr Transplant. 2011;15(2):198–204. doi: 10.1111/j.1399-3046.2010.01447.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M, Mohanakumar T, Hsu KC, Dupont B, Yokoyama WM. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A. 2008;105(8):3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M, Gallagher MM, Malkki M, Petersdorf E, Dupont B, Hsu KC. HLA-C–Dependent Prevention of Leukemia Relapse by Donor ActivatingKIR2DS1. N Engl J Med. 2012;367(9):805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.