Abstract
Bacterial biofilms underlie many persistent infections, posing major hurdles in antibiotic treatment. Here we design and demonstrate ‘tug-of-war’ optical tweezers that can facilitate the assessment of cell–cell adhesion—a key contributing factor to biofilm development, thanks to the combined actions of optical scattering and gradient forces. With a customized optical landscape distinct from that of conventional tweezers, not only can such ‘tug-of-war’ tweezers stably trap and stretch a rod-shaped bacterium in the observing plane, but, more importantly, they can also impose a tunable lateral force that pulls apart cellular clusters without any tethering or mechanical movement. As a proof of principle, we examined a Sinorhizobium meliloti strain that forms robust biofilms and found that the strength of intercellular adhesion depends on the growth medium. This technique may herald new photonic tools for optical manipulation and biofilm study, as well as other biological applications.
Keywords: bacterial adhesion, biofilm, optical forces, optical manipulation, tug-of-war optical tweezers
INTRODUCTION
Antimicrobial resistance is a troubling and growing public health threat1. In addition to resistance mechanisms that can develop at the cellular level, the propensity of bacteria to form biofilms further protects them from environmental assaults, including antibiotics and the host immune system2–4. In fact, bacterial surface-attachment and subsequent biofilm formation are considered important hallmarks of the capacity of microbial communities to cause persistent infections. Understanding factors that contribute to bacterial aggregation during biofilm maturation is critical to the study of microbial physiology and ecology, as well as to the advancement of new treatments for chronic infections and novel strategies to prevent biofilm-associated problems5,6. Thus far, the primary optical tool for studying biofilms has been laser scanning microscopy, from single-photon, two-photon to multi-photon excitation microscopy7. Although these imaging techniques have reified our current view of complex and heterogeneous biofilm structures, new optical tools are still desired for characterization and manipulation of biofilms, especially since their structures vary over time and under different environmental conditions8.
Optical tweezers with fine-shaped light beams provide excellent tools for trapping and manipulating bacteria as well as micro- and nano-scale particles9–20. Over the past decades, optical tweezers have been routinely used for single-molecule force spectroscopy to understand the mechanics of biological processes. Recently, they have also been employed to study bacterial aggregation, as well as to better characterize bacterial motility and flagellar rotation21–25. In conventional single-beam gradient optical tweezers9 (Figure 1a), however, a rod-shaped object or bacterium (such as a Bacillus thuringiensis (B. thuringiensis) cell) tends to align preferentially towards the propagation direction of a trapping beam, preventing direct in-plane observation and manipulation of the trapped object. In single-molecule force measurements, a molecule of interest is often tethered to a surface at one end and attached to a trapped dielectric bead at the other end, or both ends are attached to simultaneously trapped beads in dual-beam (dumbbell) optical tweezers13,26,27. At the single-cell level, a dual-beam optical trap (Figure 1b) is also necessary for orientation and manipulation of individual cells in space23. The need for active control of single or numerous trapped objects has motivated the development of multi-trap optical tweezers, including dynamic holographic tweezers and those created with complex beam-shaping techniques28–32. Nevertheless, available methods for optical trapping and manipulation of bacteria still have substantial limitations in quantitative characterization of bacterial motility and intercellular interaction. For example, dual-beam optical tweezers rely on a pair of perpendicularly polarized beams and significant user control of each trapping beam: a rod-shaped bacterium has to be trapped and flipped by one of the beams first, and then the user needs to manually control the other trap to orient the cell into the desired observing plane. In addition, the use of optical tweezers to stretch a bacterial cell directly or to break up cellular clusters still remains a challenge.
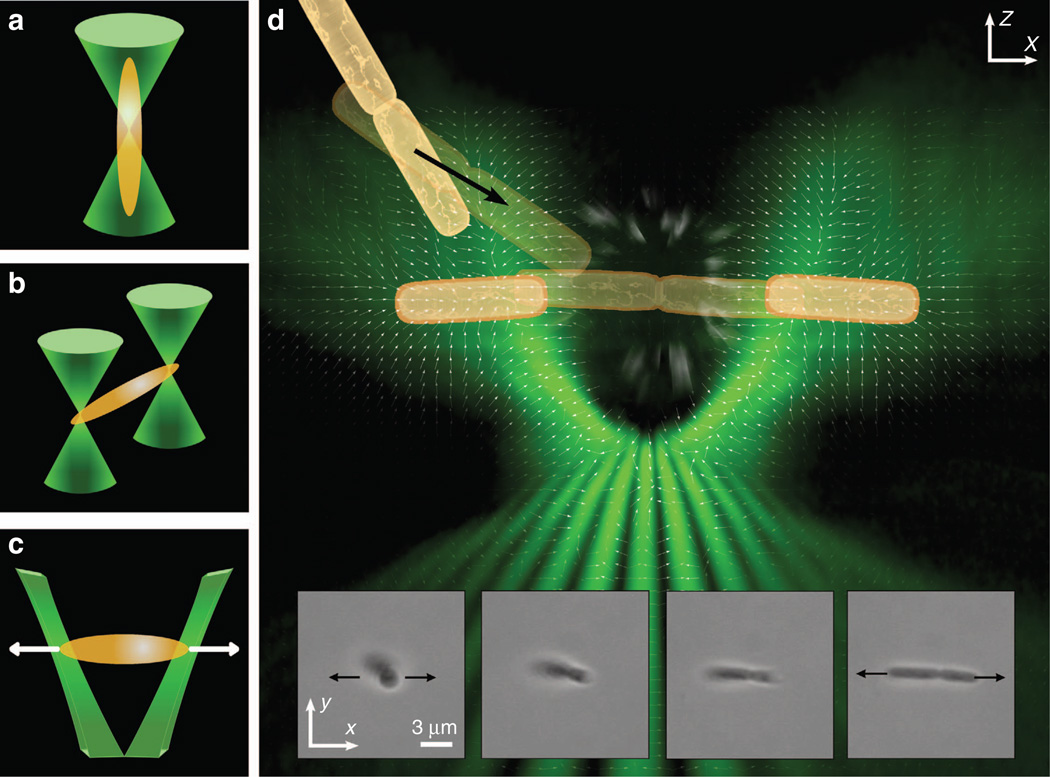
Figure 1.
Different designs of optical tweezers (left panels). (a) Single-beam optical tweezers tend to align a rod-shaped bacterium along the beam axis. (b) Dual-beam optical tweezers trap a bacterium at each end to orient the cell into the observing plane. (c) TOW optical tweezers trap a bacterium at each end but also exert lateral forces in opposite directions. Composite image illustrating the process of how a bacterial cluster is trapped, stretched and separated by the TOW tweezers (right panel). The diagram in (d) comprises the vector field of the intensity gradient of the trapping beam (white arrows), a volumetric rendering of the beam from experimental data near the focus of an objective lens (green shading), and a schematic representation of a pair of attached bacterial cells being trapped and pulled apart. The inserts in d show snapshots of a dividing B. thuringiensis cell that was aligned gradually onto the observing plane and stretched by the TOW tweezers (Supplementary Movie 1).
In this work, we describe an optical tweezer-based assay for the study of bacterial adhesion, relying on a ‘tug-of-war’ (TOW) design from novel shaping of light (Figure 1c). Instead of using two separate traps under independent control, we ‘split’ a single beam into a pair of elongated trapping beams propagating with a diverging angle. This TOW design has the following advantages over the conventional single- or dual-beam optical tweezers. First, it allows for stable in-plane trapping of a rod-shaped object with a single control implemented at will, without any mechanical movement or phase-sensitive interference. Second, and more importantly, such TOW tweezers can apply a tunable lateral pulling force on the trapped object, and the strength of the pulling force can be varied by changing the trapping beam intensity from femto- to piconewton levels. As an example, we employed the TOW tweezers to trap, stretch and even break apart Sinorhizobium meliloti (S. meliloti) cellular clusters aggregated under different conditions. We estimated the force needed for disassembly of adhesive S. meliloti cells, and found that the strength of bacterial adhesion is dependent on the growth medium. We believe this technique can bring about new avenues of exploration for optical manipulation and biophotonics.
MATERIALS AND METHODS
First, we discuss the design and demonstration of the TOW optical tweezers. Conceptually, the design of TOW optical tweezers relies on nontrivial shaping of a Gaussian beam into two elongated (stripe-like) beams with opposite transverse momenta. When applied in the optical tweezers setting, pulling forces arise on both sides of the trapped object, as in a TOW duel. In practice, the size, separation, and propagation direction of the two beams can be varied at will, as implemented by encoding the holographic information onto a LabView-assisted spatial light modulator (SLM), thus allowing interactive control of the directions and magnitudes of the pulling forces (Supplementary Information).
To better visualize the intensity distribution and structure of the resulting TOW beam, a technique for volumetric representations of holographic optical traps was used33. By acquiring a stack of two-dimensional images of the trapping beam near the focal plane, experimentally recorded data are replotted in Figure 1d, in which a composite picture shows the side-view of the beam structure, along with the calculated vector field distribution of the intensity gradient. This design of the TOW tweezers, although still based on the holographic principle, leads to an effective optical tool for trapping rod-shaped objects. Distinct from conventional dual-beam or holographic tweezers, the intensity landscaping in the TOW tweezers manifests a strong intensity gradient in the central ‘pulling’ region, with maximal momenta oriented in opposing directions as a result of the synergistic action of optical scattering and gradient forces. In addition, the two elongated beams in the TOW tweezers provide a better match to the bacterial morphology, enabling stable trapping of a rod-shaped bacterium even at low power levels, thus reducing the effects of photodamage on the trapped cell9. The vector field of the intensity gradient of the trapping beam in Figure 1 represents a useful description of the contribution from the gradient force that normally dominates in optical tweezers34. By reshaping the trapping beam (thus the force distribution), TOW tweezers can be optimized to trap rod-shaped objects of different sizes and compositions, including silica rods (used as a proof of principle), Escherichia coli (Gram-negative bacterium about 2 µm in length) and B. thuringiensis (Gram-positive bacterium with cell length ranging from 5 to 15 µm). In fact, TOW tweezers can be reconfigured to trap asymmetrically shaped particles with lengths varying from micrometre to sub-millimetre. As an example, we show in the bottom panels of Figure 1d experimental snapshots obtained from the TOW trapping and self-aligning of a dividing, rod-shaped B. thuringiensis cell. After entering the trap, the bacterium is reoriented onto the observing plane and stretched from two ends, unable to escape the trap.
RESULTS AND DISCUSSION
To illustrate the feasibility and potential application of the TOW design for biofilm study, we used TOW optical tweezers to disassemble clusters of S. meliloti cells and to show that the strength of cell–cell adhesion depends on the growth medium.
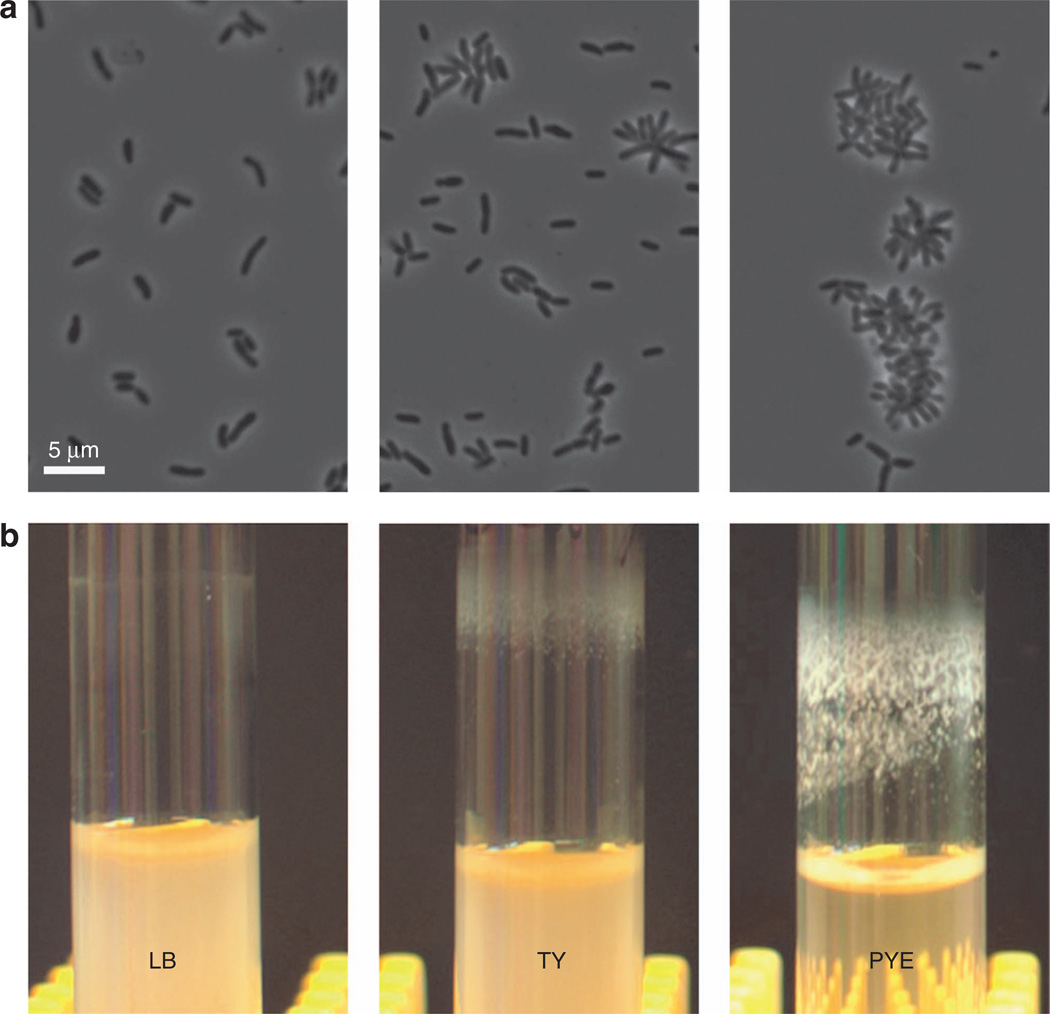
S. meliloti is a Gram-negative soil bacterium capable of establishing endosymbiosis with compatible host plants35. It serves as an advantageous model for investigating microbe–host interactions and shares with related bacteria, including those that act as pathogens, many critical factors that regulate cellular differentiation and organelle development36,37. Recent analysis revealed that a common laboratory strain of S. meliloti, Rm1021, possesses a nonsense mutation in the podJ gene, which encodes a conserved polarity factor that influences various cell envelope-associated functions38. Correcting the mutation and restoring the gene to that seen in environmental isolates of S. meliloti (Supplementary Information) resulted in a strain that develops robust biofilms in select liquid medium. This podJ+ strain forms cellular clusters that resemble those of related alphaproteobacteria, suggesting that the strain synthesizes an adhesive organelle at one pole of the cell, similar to the holdfast of Caulobacter crescentus and unipolar polysaccharide of Agrobacterium tumefaciens39–41. The amount of biofilms formed by the podJ+ strain depends on the growth medium and correlates with the extent of cellular aggregation observed (Figure 2). Specifically, the strain forms large cellular clusters and heavy biofilm in peptone yeast extract (PYE) medium, while the clusters tend to be smaller and the biofilm lighter in tryptone yeast extract (TY) medium. There is no or very weak biofilm formation and aggregation in lysogeny broth (LB) medium. Thus, the degree of biofilm formation appears to reflect the strength of intercellular attachment.
Figure 2.
S. meliloti podJ+ cells display distinct assemblages, as well as different levels of flocculation and biofilm formation, in three growth media (LB, TY and PYE). Top panels (a) show phase contrast images of the S. meliloti cells and aggregates, while bottom panels (b) show photographs of corresponding bacterial biofilms formed in 16-mm-diameter glass tubes.
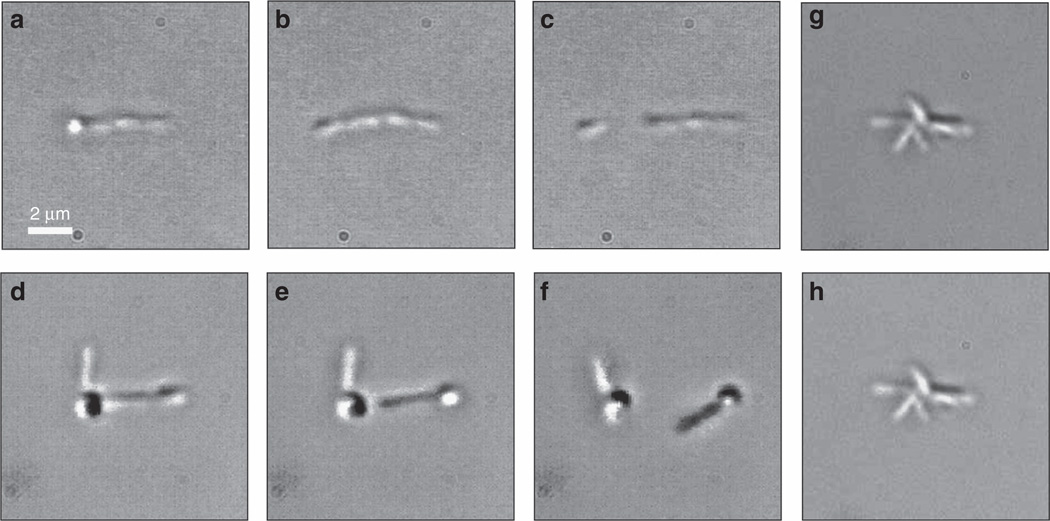
With the TOW optical tweezers, we can administer an adhesive strength assay by directly trapping and stretching a cellular cluster to estimate the underlying force. Typical experimental results are presented in Figure 3. As shown in the left panels of Figure 3a–3f, cells attached to one another in the TY medium could be trapped by the TOW tweezers, stretched gradually from two ends, and, most importantly, broken apart eventually. Note that the whole disassembly process does not need any tethering or mechanical movement, nor does it require recalibration of trapping power with beam positions. The lateral pulling force can be ‘tuned’ merely by varying the trapping beam power at the focal point and/or SLM-controlled angle and the spacing of the TOW beams. (For the results shown in Figure 3, the length of the two stripe-like beams was about 1.5 µm each, while the spacing between the two beams was about 5 µm). The beam power used to break apart the S. meliloti clusters in TY medium was only about 20 mW. This simply cannot be achieved with conventional dual-trap tweezers42 or an optical stretcher created with two counter-propagating beams43. By reconfiguring the TOW beams, even an asymmetrically shaped cellular cluster in TY medium was disassembled (Figure 3d–3f). In contrast, S. meliloti clusters formed in PYE medium remained intact even when the power of the TOW beam was increased to more than 5 times higher (Figure 3g and 3h), indicating stronger adhesion among the cells. From our estimate, the pulling force required to break up a S. meliloti cluster in TY medium should be at least 5 pN from each side of the TOW tweezers. A much stronger force would be needed in PYE medium for similar disassembly. These results illustrate that the TOW tweezers allow us to characterize quantitatively the effects of growth media on cell–cell adhesion, thus facilitating the elucidation of environmental factors that affect bacterial aggregation and biofilm formation.
Figure 3.
(a–c) Snapshots of a trapped S. meliloti aggregate consisting of four attached, self-aligned cells in TY medium, being gradually stretched from two ends and eventually broken up into two pieces by the TOW optical tweezers. (d–f) Similarly dynamic process for the disassembly of an asymmetrically shaped S. meliloti cluster grown in TY medium (Supplementary Movie 2). (g, h) Strong binding of several S. meliloti cells in PYE medium into a cluster g, which remains intact under the action of the TOW tweezers h.
We now discuss the forces involved in TOW tweezers. Direct measurement of the forces from the TOW tweezers acting on the bacteria is challenging since the forces involved in separating and trapping bacterial cells are of vastly varying magnitudes. In order to have a better understanding of the lateral forces in the TOW optical trap, we utilized a few different measurement techniques. Theoretically, in contrast to the familiar case of a spherical particle in the single-beam trap, precise calculation of stiffness and strength of compound traps such as the TOW tweezers is very complicated. In fact, only recently have theoretical models been put forth for the study of trapping forces with asymmetrically shaped particles, and for shape-induced force fields in general44. The non-conservative nature of the optical force fields certainly manifests itself directly in the stiffness of trapped aspherical objects, such as a rod-shaped bacterium.
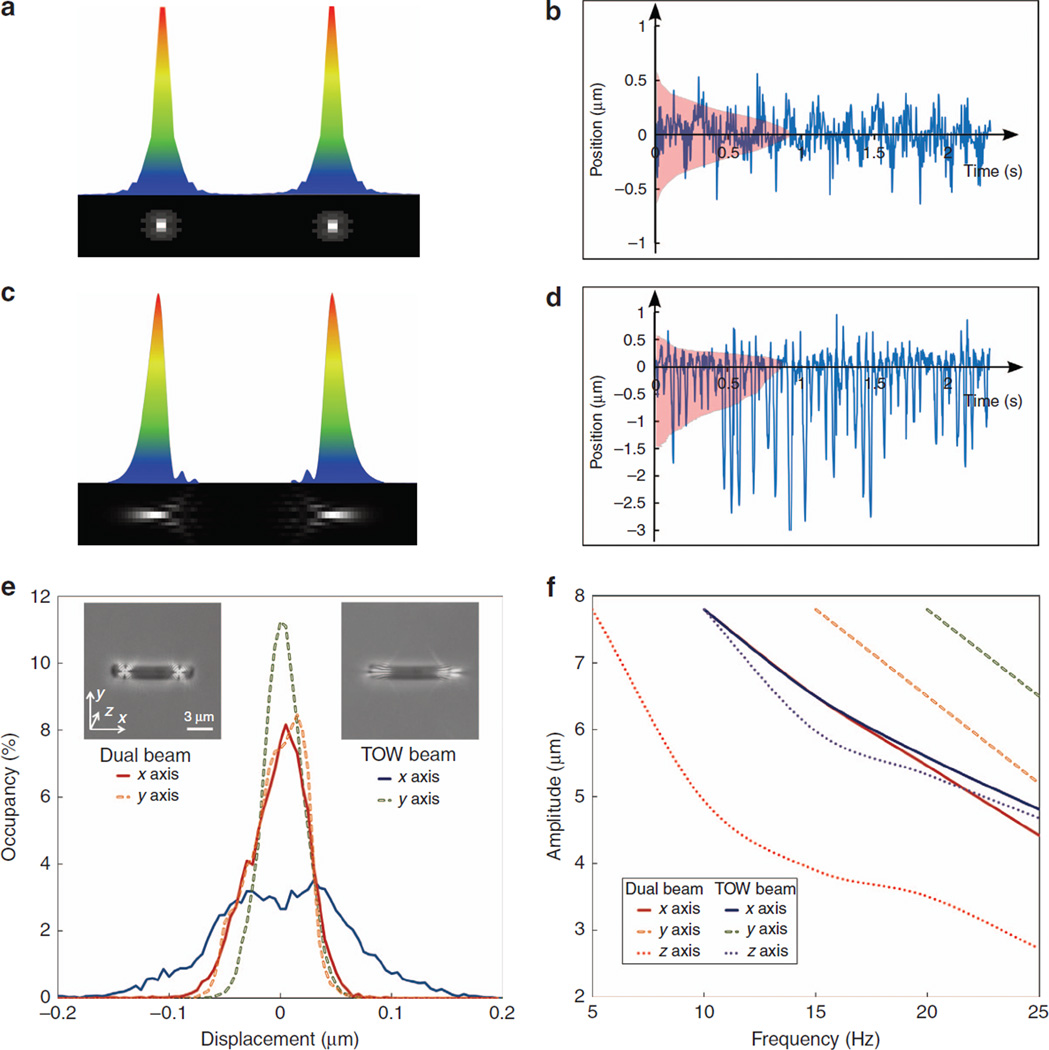
First, to substantiate that an outward pulling or splitting force indeed exists in the TOW trap, we used the method of particle image velocimetry to estimate the magnitude and direction of the flow of suspended polystyrene beads driven by the TOW tweezers. To illustrate the concept, the two beams (diverging in the x-direction) constituting the tweezers have a large separation of about 5 µm at the trapping plane, and the beads have an average size of about 500 nm. As seen in Figure 4a and 4b, the TOW tweezer behave as two micropumps for a thin sample of aqueous suspension of the beads: the beads flow away from the central region along two opposite directions due to the scattering force exerted by the diverging beams (Supplementary Movie 3). The time-averaged velocity of the particle flow is replotted in Figure 4b, where arrowed lines mark the particle flow velocity distribution. Clearly, this diagram of hydrodynamic particle flow illustrates a transverse momentum leading to pulling in opposite directions, giving rise to the TOW action mediated by our judiciously shaped optical beam shown in Figure 1d. Moreover, since the hydrodynamic driving of particles infers the direct relation to the force, it also provides information about the force distribution (magnitude and direction) from the TOW tweezers. As seen in Figure 4a and 4b, the pulling forces drive the particles towards the two sides rather than the central region between the two traps.
Figure 4.
(a) A snapshot of suspended polystyrene beads (500 nm in diameter) driven laterally to two opposite sides by the TOW tweezers. The locations of the two main intensity spots from the ‘diverging’ TOW beams are marked by two dashed circles, from where the beads are pushed away (Supplementary Movie 3). (b) Illustration of time-averaged direction and magnitude of the flowing beads when the two beams constituting the TOW tweezers are 5 µm apart at the focal plane. The colour represents the magnitude of the normalized average particle velocity, and arrows indicate the direction of particle flow. (c) Trapping force resulting from only one arm of the TOW tweezers while the other is absent (that is, that part of the beam is blocked). Measured results are plotted in solid curves with shaded area representing the error. The theoretically calculated force in x-direction is shown on the same graph (dashed curve) for comparison. (d) The force profile of a 1-µm ‘bacterium-like’ particle with a refractive index 1.38 calculated via the generalized Lorenz–Mie theory. Red and blue colours represent the magnitude of the force in positive and negative directions, respectively. The overlay shows the forces (in normalized units) along the dotted line. The hair-like wisps represent paths taken by simulated particles in the absence of Brownian motion. The dotted box indicates the region similar to that shown in c.
Second, to obtain an estimate of the magnitude of the trapping forces from the TOW tweezers, we analysed the time-dependent positions of a single S. meliloti cell trapped by only one side of the TOW tweezers using the established method of ‘optical potential analysis’45. This position distribution of the trapped cell is obtained by extracting data from video microscopy using a particle tracking software46. From the occupancy probability and by employing the Boltzmann statistics theorem, the potential energy, and thus, the force distribution in space can be deduced47. Figure 4c shows the position distribution of the cell when it is trapped by only one arm of the TOW tweezers. As seen in Figure 4c, force distribution in the transverse y-direction (that is, perpendicular to the pulling direction) is not notably different from that resulting from a standard Gaussian trap; in contrast, in the x-direction, the distribution becomes highly asymmetric. In other words, even when just one arm of the TOW tweezers is present, the bacterium experiences a net force in a preferred direction (that is, along the pulling direction). Although the bacterium is not necessarily stably trapped at the point where the peak force is applied, we can still obtain a value for the peak force by fitting experimental data to the theoretical model. The results from a theoretical estimate of the force are also plotted in Figure 4c for comparison, which gives a force of about 0.35 pN per 0.1 µm. With this method, the peak force from one arm of the tweezers is estimated to be at least 5 pN for a rod-shaped cell ~ 1.5 µm in length when displaced to the centre of the TOW trap. Of course, when both traps are present under the TOW action as shown in Figure 3a–3c for breaking up the S. meliloti cluster, the actual pulling forces from both sides could be much larger than this estimated value.
Third, we provided a theoretical analysis of the forces mediated by the optical landscape of the TOW tweezers. Since the beam shaped by the SLM is strongly focused by an objective lens to achieve a high-field gradient, it is necessary to use a rigorous vectorial electromagnetic (EM) treatment to facilitate the modelling. As such, the Debye–Wolf integral was used to construct the field in the focal plane47,48. Following a similar method used previously for SLM-assisted beam shaping47,49, the radiation in the focal plane was calculated as the integral of spherical vectors waves emanating from the objective lens. To simplify the calculation, we treated the trapped bacterium approximately as a spherical particle with a size of 1 µm and a refractive index of 1.38. The forces acting on the ‘particle’ by the TOW beams were calculated via the T-matrix method50,51, derived from the generalized Lorenz–Mie theory: at each point of the trapping plane, the incident EM field around the particle was calculated based on the actual holograms used experimentally on the SLM and the optical parameters of the system, and the scattered field was then calculated via the T-matrix. By comparing the incident and scattered fields, the forces on the particle can then be found by integrating fields around the particle. A typical calculated force distribution around such a particle at the trapping plane is plotted in Figure 4d, showing a clear pulling effect on the trapped particle: the optical force is particularly strong in the middle of the TOW duel where particles will be strongly pushed away from the centre of the beam.
In order to categorize the forces at work in the TOW duel, the Q value (trapping efficiency) for the optical trap was calculated. This corresponds to the amount of power required to provide a particular trapping force for a given particle. Our analyses show that, under the experimental conditions, the peak Q-value of the TOW beam is 1.5 times larger than that of a similarly positioned Gaussian trap. In other words, in TOW tweezers, a trapped particle will experience a pulling force 1.5 times greater that in Gaussian beam-based tweezers. In addition, the TOW beam has a stronger force differential between the positive and negative sides of the trap when compared with a Gaussian equivalent, leading to enhanced lateral pulling forces. Nevertheless, due to the elongated shaping of the TOW beams, the intensity threshold for bacterial photodamage could be higher as compared with that for Gaussian traps. The range of forces that the trap can exert is bounded on the lower end by the thermal force (in the low femtonewton regime, depending on temperature), and on the upper end by undesirable photodamage of bacterial cells (in the high piconewton regime, which typically occurs at a relatively high power level, depending on the species and the trapping wavelength).
Finally, to highlight the difference between conventional dual-beam tweezers and our TOW tweezers, two approaches were used. One was to compare the position distribution of a S. meliloti cell trapped by only one side of the dual-traps, and the other was to compare the stability of a micro-rod trapped by both sides with two different tweezers systems. In both cases, a strongly oscillating environment52 was provided to test the stability of the traps and to simulate ambient perturbation for motile bacteria. Although both types of tweezers can trap and hold a rod-shaped object or a bacterial cluster in the observing plane, the TOW system has obvious advantages. First, unlike in a single trap based on a symmetric Gaussian beam, a trapped object in one side of the TOW beam has a preferred direction of displacement due to its asymmetrically shaped intensity profile (Figure 5a–5d). As shown in Figure 5b and 5d, under a periodic perturbation (for example, the sample is oscillated sinusoidally), a trapped S. meliloti cell moves around its central equilibrium position evenly in the Gaussian trap, but it shows up more in a preferred direction in the TOW trap, indicating the pulling effect from the latter configuration. Second, to stretch the trapped object, at least one of the Gaussian-beam-based dual traps has to be moved laterally, while in the TOW tweezers, one does not need to translate the objective lens, and the object still experiences a constant stretching force due to the asymmetric intensity gradient. Third, an object trapped by TOW tweezers exhibits increased stability and resistance to ambient perturbation when compared against conventional dual-beam tweezers.
Figure 5.
Comparison between conventional dual-beam tweezers and TOW tweezers. (a) Intensity profile of the Gaussian dual-beam trap. (b) Position distribution of a S. meliloti cell trapped by only one side of the dual-trap in an oscillating environment (that is, the sample is sinusoidally dragged at a frequency of 20 Hz and an amplitude of 2 µm), overlaid with the intensity profile of the trap (in pink). (c, d) Corresponding results for the TOW tweezers. (e) Occupation probability of a trapped silica rod around its equilibrium position in the two different traps. With the TOW tweezers, the micro-rod is more stably trapped in the y-direction yet still has the flexibility to move around its equilibrium position in the x-direction (desirable for stretching). Inserts show the location of each beam on the silica rod. (f) Plot of maximum oscillating amplitude vs. frequency at which the silica rod can stay in the three-dimensional trap, under the action of either dual-beam or TOW tweezers. Enhanced stability in the TOW tweezers is evident.
To better illustrate the advantage in trap stability, a rigid silica micro-rod was used as a test object instead of a bacterial cell to prevent any possible damage- or deformation-induced effects. Experimental results are presented in Figure 5e and 5f. In Figure 5e, the position distributions of the trapped rod in both the dual-beam and the TOW tweezers are plotted, where the occupancy probability is obtained by taking 10 000 snapshots from a recorded video of the micro-rod in the trap. Clearly, in the TOW tweezers, the micro-rod is better confined in the y-direction (that is, the direction perpendicular to the stretching direction) than in the x-direction, whereas in the dual traps based on the symmetric Gaussian beams there is no such difference. Thus, when applied to a bacterial cell or a cellular cluster, the TOW tweezers give rise to a stable trapping in the y-direction along with the flexibility to move around its equilibrium position in the x-direction, which offers an advantage for stretching. In Figure 5f, the sample experiences sinusoidal oscillation in three dimensions, as driven by a piezoelectric transducer (PZT)-actuated vibration control, and the cutoff amplitude and frequency of oscillation at which the silica rod can no longer stay in the trap are plotted for comparison. From these results, we can see that the silica rod, while it is being ‘stretched’ in the transverse x-direction, is much more stably trapped in the y- and z-directions in the TOW tweezers than it is in the dual Gaussian beam tweezers. Clearly, both approaches coherently show that a rod-shaped object exhibits better stability when being trapped and stretched by the TOW tweezers, as compared with the conventional dual-beam tweezers.
CONCLUSIONS
In summary, we have demonstrated that judiciously shaped light beams can stretch and even break apart bacterial clusters, leading to a simple assay for cellular adhesion. In particular, we have shown that our specially designed TOW optical tweezers can be used as an effective tool for evaluating S. meliloti cell adhesion under different growth conditions. We have estimated the optical forces needed to disassemble S. meliloti flocs and determined that the trapping stability of TOW tweezers exceeds that of conventional dual-beam tweezers. This work represents another successful example of using static optical forces and novel beam shaping to perform diagnostic mechanical tests at the cellular level, and the technique can be readily adopted for studying the mechanical properties and dynamics of various living cells53–55. Since cellular adhesion has a crucial role in biofilm development, our technique suggests exciting possibilities of developing new optical tools for investigating biofilm formation and related biomedical applications. Finally, this technique might be employed in single-molecule force microscopy, for example, to stretch DNA molecules without the need for positional calibration of paired traps.
Supplementary Material
Acknowledgments
This work was supported by NIH and NSF. We thank R Gautam and D Gallardo for assistance. All authors contributed significantly to this work.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization; 2014. [Google Scholar]
- 2.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Penesyan A, Gillings M, Paulsen IT. Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules. 2015;20:5286–5298. doi: 10.3390/molecules20045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock REW. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol. 2013;16:580–589. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 6.O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 7.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 8.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 9.Ashkin A, Dziedzic JM. Optical trapping and manipulation of viruses and bacteria. Science. 1987;235:1517–1520. doi: 10.1126/science.3547653. [DOI] [PubMed] [Google Scholar]
- 10.Block SM, Blair DF, Berg HC. Compliance of bacterial flagella measured with optical tweezers. Nature. 1989;338:514–518. doi: 10.1038/338514a0. [DOI] [PubMed] [Google Scholar]
- 11.Dholakia K, Reece P, Gu M. Optical micromanipulation. Chem Soc Rev. 2008;37:42–55. doi: 10.1039/b512471a. [DOI] [PubMed] [Google Scholar]
- 12.Padgett M, Bowman R. Tweezers with a twist. Nat Photonics. 2011;5:343–348. [Google Scholar]
- 13.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friese MEJ, Nieminen TA, Heckenberg NR, Rubinsztein-Dunlop H. Optical alignment and spinning of laser-trapped microscopic particles. Nature. 1998;394:348–350. [Google Scholar]
- 15.Fällman E, Schedin S, Jass J, Andersson M, Uhlin BE, et al. Optical tweezers based force measurement system for quantitating binding interactions: system design and application for the study of bacterial adhesion. Biosens Bioelectron. 2004;19:1429–1437. doi: 10.1016/j.bios.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 16.Čižmár T, Garcés-Chávez V, Dholakia K, Zemánek P. Optical conveyor belt for delivery of submicron objects. Appl Phys Lett. 2005;86:174101. [Google Scholar]
- 17.Pauzauskie PJ, Radenovic A, Trepagnier E, Shroff H, Yang PD, et al. Optical trapping and integration of semiconductor nanowire assemblies in water. Nat Mater. 2006;5:97–101. doi: 10.1038/nmat1563. [DOI] [PubMed] [Google Scholar]
- 18.Di Leonardo R, Ianni F, Ruocco G. Computer generation of optimal holograms for optical trap arrays. Opt Express. 2007;15:1913–1922. doi: 10.1364/oe.15.001913. [DOI] [PubMed] [Google Scholar]
- 19.Dholakia K, Zemánek P. Gripped by light: optical binding. Rev Mod Phys. 2010;82:1767–1791. [Google Scholar]
- 20.Bendix PM, Jauffred L, Norregaard K, Oddershede LB. Optical trapping of nanoparticles and quantum dots. IEEE J Select Top Quant Electron. 2014;20:15–26. [Google Scholar]
- 21.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, et al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dienerowitz M, Cowan LV, Gibson GM, Hay R, Padgett MJ, et al. Optically trapped bacteria pairs reveal discrete motile response to control aggregation upon cell-cell approach. Curr Microbiol. 2014;69:669–674. doi: 10.1007/s00284-014-0641-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min TL, Mears PJ, Chubiz LM, Rao CV, Golding I, et al. High-resolution, long-term characterization of bacterial motility using optical tweezers. Nat Methods. 2009;6:831–835. doi: 10.1038/nmeth.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altindal T, Chattopadhyay S, Wu XL. Bacterial chemotaxis in an optical trap. PLoS One. 2011;6:e18231. doi: 10.1371/journal.pone.0018231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martínez IA, Campoy S, Tort M, Llagostera M, Petrov D. A simple technique based on a single optical trap for the determination of bacterial swimming pattern. PLoS One. 2013;8:e61630. doi: 10.1371/journal.pone.0061630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brzobohatý O, Arzola AV, Šiler M, Chvátal L, Jákl P, et al. Complex rotational dynamics of multiple spheroidal particles in a circularly polarized, dual beam trap. Opt Express. 2015;23:7273–7287. doi: 10.1364/OE.23.007273. [DOI] [PubMed] [Google Scholar]
- 28.Curtis JE, Koss BA, Grier DG. Dynamic holographic optical tweezers. Opt Commun. 2002;207:169–175. [Google Scholar]
- 29.Simpson SH, Hanna S. Holographic optical trapping of microrods and nanowires. J Opt Soc Am A. 2010;27:1255–1264. doi: 10.1364/JOSAA.27.001255. [DOI] [PubMed] [Google Scholar]
- 30.Hörner F, Woerdemann M, Müller S, Maier B, Denz C. Full 3D translational and rotational optical control of multiple rod-shaped bacteria. J Biophotonics. 2010;3:468–475. doi: 10.1002/jbio.201000033. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P, Hernandez D, Cannan D, Hu Y, Fardad S, et al. Trapping and rotating microparticles and bacteria with Moiré-based optical propelling beams. Biomed Opt Express. 2012;3:1891–1897. doi: 10.1364/BOE.3.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao J, Chremmos ID, Song D, Christodoulides DN, Efremidis NK, et al. Curved singular beams for three-dimensional particle manipulation. Sci Rep. 2015;5:12086. doi: 10.1038/srep12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roichman Y, Cholis I, Grier DG. Volumetric imaging of holographic optical traps. Opt Express. 2006;14:10907–10912. doi: 10.1364/oe.14.010907. [DOI] [PubMed] [Google Scholar]
- 34.Harada Y, Asakura T. Radiation forces on a dielectric sphere in the Rayleigh scattering regime. Opt Commun. 1996;124:529–541. [Google Scholar]
- 35.Galibert F, Finan TM, Long SR, Pühler A, Abola P, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 36.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, et al. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol. 2010;4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fields AT, Navarrete CS, Zare AZ, Huang ZZ, Mostafavi M, et al. The conserved polarity factor PodJ1 impacts multiple cell envelope-associated functions in Sinorhizobium meliloti. Mol Microbiol. 2012;84:892–920. doi: 10.1111/j.1365-2958.2012.08064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsang PH, Li GL, Brun YV, Freund LB, Tang JX. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci USA. 2006;103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, et al. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol. 2013;89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, et al. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet. 2014;10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fällman E, Axner O. Design for fully steerable dual-trap optical tweezers. Appl Opt. 1997;36:2107–2113. doi: 10.1364/ao.36.002107. [DOI] [PubMed] [Google Scholar]
- 43.Guck J, Ananthakrishnan R, Mahmood H, Moon TJ, Cunningham CC, et al. The optical stretcher: a novel laser tool to micromanipulate cells. Biophys J. 2001;81:767–784. doi: 10.1016/S0006-3495(01)75740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips DB, Padgett MJ, Hanna S, Ho YLD, Carberry DM, et al. Shape-induced force fields in optical trapping. Nat Photonics. 2014;8:400–405. [Google Scholar]
- 45.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preece D, Bowman R, Linnenberger A, Gibson G, Serati S, et al. Increasing trap stiffness with position clamping in holographic optical tweezers. Opt Express. 2009;17:22718–22725. doi: 10.1364/OE.17.022718. [DOI] [PubMed] [Google Scholar]
- 47.Török P, Varga P, Laczik Z, Booker GR. Electromagnetic diffraction of light focused through a planar interface between materials of mismatched refractive indices: an integral representation. J Opt Soc Am A. 1995;12:325–332. [Google Scholar]
- 48.Richards B, Wolf E. Electromagnetic diffraction in optical systems. II. structure of the image field in an aplanatic system. Proc Roy Soc London. A. Mathem Phys Eng Sci. 1959;253:358–379. [Google Scholar]
- 49.Sun B, Roichman Y, Grier DG. Theory of holographic optical trapping. Opt Express. 2008;16:15765–15776. doi: 10.1364/oe.16.015765. [DOI] [PubMed] [Google Scholar]
- 50.Nieminen TA, Loke VLY, Stilgoe AB, Kröner G, Brańczyk AM, et al. Optical tweezers computational toolbox. J Opt A Pure Appl Opt. 2007;9:S196–S203. [Google Scholar]
- 51.Mishchenko MI, Travis LD, Lacis AA. Absorption, and Emission of Light by Small Particles. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- 52.Carmon G, Feingold M. Controlled alignment of bacterial cells with oscillating optical tweezers. J Nanophotonics. 2011;5:051803. [Google Scholar]
- 53.Gu M, Kuriakose S, Gan XS. A single beam near-field laser trap for optical stretching, folding and rotation of erythrocytes. Opt Express. 2007;15:1369–1375. doi: 10.1364/oe.15.001369. [DOI] [PubMed] [Google Scholar]
- 54.Zhong MC, Wei XB, Zhou JH, Wang ZQ, Li YM. Trapping red blood cells in living animals using optical tweezers. Nat Commun. 2013;4:1768. doi: 10.1038/ncomms2786. [DOI] [PubMed] [Google Scholar]
- 55.Kisung L, Danilina AV, Kinnunen M, Priezzhev AV, Meglinski I. Probing the red blood cells aggregating force with optical tweezers. IEEE J Select Top Quant Electron. 2016;22:1–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.