Abstract
Biliary atresia (BA) is a neonatal obstructive cholangiopathy which progresses to end stage liver disease, often requiring transplantation. The murine model of BA, employing rhesus rotavirus (RRV), parallels human disease and has been used to elucidate mechanistic aspects of a virus induced biliary cholangiopathy. We previously reported that RRV VP4 gene plays an integral role in activating the immune system and induction of BA. Utilizing rotavirus binding and blocking assays, this study elucidated how RRV VP4 protein governs cholangiocyte susceptibility to infection both in vitro and in vivo in the murine model of BA. We identified the amino acid sequence on VP4 and its cholangiocyte binding protein, finding that the sequence is specific to those rotavirus strains which cause an obstructive cholangiopathy. Pretreatment of murine and human cholangiocytes with this VP4 derived peptide (TRTRVSRLY), significantly reduced RRV’s ability to bind and infect the cells. However, the peptide did not block cholangiocyte binding of TUCH and Ro1845, strains which do not induce murine BA. The SRL sequence within TRTRVSRLY is required for cholangiocyte binding and viral replication. The cholangiocyte membrane protein bound by SRL was found to be Hsc70. Inhibition of Hsc70 by siRNAs reduced RRV’s ability to infect cholangiocytes. This virus-cholangiocyte interaction is also seen in vivo in the murine model of BA, where inoculation of mice with TRTRVSRLY peptide significantly reduced symptoms and mortality in RRV-injected mice.
Conclusion
The tri-peptide SRL on RRV VP4 binds to the cholangiocyte membrane protein Hsc70 defining a novel binding site governing VP4 attachment. Investigations are underway to determine the cellular response following this interaction to understand how it contributes to the pathogenesis of BA.
Keywords: RRV, Biliary atresia, cholangiocyte, Hsc70
Introduction
Biliary atresia (BA) is an inflammatory cholangiopathy unique to infancy and is the most common cause of pathologic jaundice in the pediatric population, leading to extra-hepatic biliary obstruction. Surgical reconstruction of the extrahepatic biliary tree may restore drainage in some; however, many infants progress to cirrhosis and end stage liver disease. Due to the progressive nature of the disease, BA is the most common indication for pediatric liver transplantation in the United States (1).
The etiology of BA remains unclear. In 1974, Landing proposed that a viral insult leads to a progressive inflammatory process, resulting in cholangiopathic disease (2). Subsequent studies found multiple viruses including rotavirus group C (3), reovirus type 3 (4), Epstein-Barr virus (5), cytomegalovirus (6) and human papillomavirus (7) in explanted livers of infants with BA. In the murine model of BA, rhesus rotavirus (RRV) infection of newborn BALB/c mice results in an obstructive cholangiopathy (8, 9). Symptoms include bilirubinuria, jaundice, acholic stools and growth retardation that mirrors the human disease (9). In this model, the morphologic and histologic changes in the biliary tree (10) parallel those found in human BA (11).
Rotaviruses, including RRV, are 65–75nm icosahedral, non-enveloped viruses containing 11 dsRNA gene segments encoding six structural proteins (VP1-4, VP6 and VP7) and 6 non-structural (NSP1-6) proteins. The outer layer is comprised of a protein capsid consisting of VP4 and VP7 proteins, which are involved in cellular binding (12). In previous studies we found that one of the cellular targets of RRV is the biliary epithelial cell (cholangiocyte) (13). The mechanism by which RRV attaches to a host cell is a multi-step process involving sialic acid labeled glycoproteins (14, 15) and the integrins α2β1, α4β1, αVβ3 and αXβ2 (15–21). We have previously shown that expression of the α2β1 integrin plays a role in cholangiocyte susceptibility to RRV infection (22). Although blocking of the α2β1 integrin reduced cholangiocyte vulnerability by 50%, RRV was still able to infect the cell, suggesting the existence of other, undefined cholangiocyte cell surface receptors which may govern susceptibility.
We have previously used reassortant viruses to study the specific RRV genes involved in cholangiocyte susceptibility and viral pathogenicity. Using single-gene reassortants created from RRV and TUCH (a simian rotavirus strain capable of infecting cholangiocytes without causing murine BA), we found that the gene segment 4 of RRV encoding for the VP4 protein confers the ability of the virus to attach to and infect cholangiocytes, causing murine BA (19, 23). In a subsequent study, we found that virus strains with high homology to the RRV VP4 protein are capable of inducing murine BA (24).
The primary goal of this study was to determine the mechanism by which RRV VP4 governs the murine model of BA. Computer-aided analysis was utilized to identify potential protein-protein interaction sites on the RRV VP4 protein. A specific sequence (SRL) was identified on this protein that binds to cholangiocytes. It was then determined that the cholangiocyte binding site for this peptide is the Hsc70 protein.
Methods
Ethics Statement
All animal research was performed in accordance with regulations and protocols approved by the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital Medical Center (protocol number IACUC2013-0039) which adheres to the NIH OLAW regulation (Animal Assurance number A3108) and the Animal Welfare Act (certification number 31-8-001).
Viruses and Cells
The mouse cholangiocyte cell line generously provided by Dr. James Boyer (Yale Liver Care Center, Hartford, CT), MA104 cells (BioWhittaker, Walkersville, MD), and human cholangiocytes (H69 cells) kindly provided by Douglas Jefferson (New England Medical Center, Tufts University, Boston, MA) were cultured as described previously (22, 25). We used several rotavirus strains: RRV, a simian strain (kindly provided by H. Greenberg, Stanford University, Palo Alto, CA), TUCH (named after the locations where the strain was isolated: Tulane National Primate Research Center and Cincinnati Children’s Hospital), a simian strain [24], Ro1845 a human strain (generously provided by Dr. Yasutaka Hoshino, NIAID) and GRV a goat strain (graciously provided by Dr. Osamu Nakagomi, Nagasaki University, Japan). Virus stocks were grown in MA104 cells as previously described and kept frozen at −80°C until thawed for use (13). Gene sequences of gene segment 4 from each strain were obtained from GenBank: RRV [AY033150], TUCH [AY596189], Ro1845 [EU708893], and GRV [AB055967]
Structural modeling and interaction sites mapping
In order to map putative functional sites that may play a role in RRV VP4 interactions with cholangiocytes, a number of structural bioinformatics tools were employed to analyze and extrapolate from available structural data on entry apparatus. This included several solved conformations of the spike protein VP4 (PDB entries 1SLQ, 2B4H, 2B4I, 4V7Q). Additional protein-protein interaction sites were predicted using SPPIDER (http://sppider.cchmc.org) that combines sequence-based solvent accessibility prediction and data on unbound as well as known bound structures to identify sites likely to get buried upon complex formation (26). These predictions were further analyzed in the context of existing structural and sequence data using POLYVIEW-3D (http://polyview.cchmc.org), ConSurf (http://consurf.tau.ac.il/) and Evolutionary Trace Annotation Server (http://mammoth.bcm.tmc.edu/eta/index.html).
Peptides
Synthetic peptides spanning different regions on the VP4 protein of RRV, TUCH and Ro1845 strains were synthesized and purified by high-performance liquid chromatography (HPLC) and kept lyophilized (GenScript, Piscataway, NJ). Unrelated peptides (GHRP) of the same size and random sequence peptides with identical amino acid (aa) compositions were synthesized for use as controls. Peptides were resuspended in Dulbecco’s modified Eagle’s medium before use in replication assays. Biotinylated TRTRVSRLY peptides were synthesized using Fmoc-Lys(Fmoc)-OH strategy where two biotin moieties are attached to each motif separated by hexanoic acid (GenScript).
Viral binding assay
The ability of the virus to attach to the cholangiocytes was assessed by binding assays as described previously (22). Briefly, cells were grown to confluence in 24-well plates. The cells, medium and inoculating virus were cooled to 4°C. Cells were then inoculated with varying amounts of virus and incubated at 4°C for one hour. The inoculum was removed and cells were washed twice to remove unbound virus. The cells then underwent two freeze/thaw cycles and virus within the final cell fraction represented attached virus. The washes and residual inoculum were combined. This combined solution represented all unbound virus. The amount of bound and unbound virus was determined by Focus Forming Assay (FFA) analysis. The amount of attached virus was expressed as percent of total amount of virus used to inoculate cells.
Quantification of infectious rotavirus
FFA was used to determine the presence of infectious rotavirus as previously described (27). Briefly, 96-well plates were treated with serially-diluted virus and incubated at 37°C for 14–16 hours. After acetone fixation, Guinea pig anti-rotavirus immunoglobulin G (IgG) primary antibody (1:1000) was added and incubated for 30 minutes. Wells were aspirated and washed with phosphate-buffered saline and fluorescein isothiocyanate (FITC)-tagged goat anti-guinea pig IgG secondary antibody (1:500) was added for 30 minutes at 37°C. Wells were then washed twice and plates scored using a UV microscopy (10× objective). Quantities of infectious virus were reported as FFU per milligram of tissue.
In vivo use of TRTRVSRLY peptide in the murine model of BA
All animal studies were performed in accordance with institutional animal welfare guidelines using an approved animal protocol. The experimental model of biliary atresia was induced in BALB/c mice (Envigo Labs, Indianapolis, IN). Newborn pups were injected intraperitoneally (ip.) with 500 ug of TRTRVSRLY or SSASAW peptide diluted in saline along with RRV at 7.5×105 ffu/pup. A subsequent injection of peptide was given on days 1 and 2 of life. Pups were monitored for 21 days for clinical signs of hepatobiliary injury (i.e. acholic stools, bilirubinuria and jaundice in non-fur covered skin) and survival. Subsets of these mice were sacrificed at 7 days post-infection and their extra-hepatic biliary tract harvested. The tissues were analyzed for the presence of replication competent virus by FFA.
Protein precipitation with biotinylated TRTRVSRLY peptides
Biotinylated TRTRVSRLY peptide was synthesized by GenScript. Peptides were designed with an N-terminal biotin linker. For affinity pull-downs, 2 mg of immobilized peptide was added to confluent monolayer of cholangiocytes and incubated at 4°C for 1 hour. After incubation, cells were lysed and the membrane fraction was isolated using 0.38M NaCl buffer. This membrane fraction was later treated with streptavidin agarose (Peirce, Rockford, IL) to pull down any bound protein (28). Control samples for each lysis buffer used included a sample in which no peptide was added. After extensive washes, bound proteins were eluted from the immobilized peptides by boiling in SDS sample buffer and run in 4–20% polyacrylamide gel.
Mass Spectrometry Analysis of cholangiocyte membrane proteins as they interact with the TRTRVSRLY peptide
Silver-stained bands were excised from the gels and tryptic-digested as described in Eismann et al. (29). The mass lists were generated by MALDI-TOF mass spectrometry on a 4800 MALDI-TOF (Applied Biosystems, Forest City, CA). The identity search was performed using the MASCOT search algorithm (Matrix Science, Boston, MA) by scanning the current version of NCBI sequence database. The significance of the identity was judged from the search engine’s scoring system.
Transfection of siRNA against Hsc70
Transfection of siRNA to reduce Hsc70 expression was carried out as previously described (22). The siRNA sequences were as follows: sequence 1, GCUUGAGUAAGGAAGAUAU; sequence 2, GAUUAAGUCAGUCCAAGAA; sequence 3, UCACGGACACAGAGAGAUU; sequence 4, CCACAGGGACCCAAAACAA (Dharmacon, Lafayette, CO). Non-targeting siRNA was purchased from Dharmacon. Immunohistochemistry and Western blot analysis was used to detect reduced membrane expression of Hsc70. Immunocytochemistry was performed by harvesting cholangiocytes before and after siRNA transfection and cytospin centrifugation of 10,000 cells onto slides. Slides were fixed in 4% paraformaldehyde and were blocked with 5% normal donkey serum. Slides were incubated at 4°C overnight with rat anti-mouse Hsc70 antibody diluted 1:200 in DAKO antibody diluent. After incubation, the slides were washed with PBS and further incubated with anti-rat Alexafluor 594 conjugated secondary antibody and incubated for 1.5 hours at room temperature. The slides were washed and embedded with DAPI mounting media for visualization under fluorescent microscope (Nikon Eclipse 90).
Statistical analysis
Analysis of non-continuous variables was performed by chi-square and Fisher Exact tests. Results of continuous variables were expressed as means ± standard error and were analyzed by ANOVA with post hoc testing, where appropriate. A p-value < 0.05 was considered significant. All statistical analysis was performed using Prism 7 (GraphPad Software, Inc., La Jolla, CA).
Results
Analysis of RRV VP4 protein-protein interactions through SPPIDER program
We utilized the SPPIDER program to identify possible protein-protein interaction sites on VP4, which predicted a number of putative protein interaction sites (Supplemental Fig. 1). Of the 9 possible sites of interest, 6 were determined to have analogous aa sequences to those strains, which do not cause murine BA (TUCH and Ro1845) and were excluded from further analysis. Synthetic peptides were generated of the 3 remaining peptide sequences (PTAAGV, SSASAW, and TRTRVSRLY) for use in subsequent studies (Fig. 1A).
Fig. 1. Viral binding and replication in cholangiocytes treated with synthetic peptides.

(A) Table illustrating the 9 potential protein-protein interaction sites on RRV VP4 as determined by the SPPIDER program. The bolded sequences (PTAAGV, SSASAW and TRTRVSRLY) represent the region where RRV differ from TUCH and Ro1845. (B, C) Mouse cholangiocytes were treated with peptides PTAAGV, SSASAW, or TRTRVSRLY at varying concentrations followed by RRV infection at 4°C. DGEA and GHRP were used as a positive and negative control respectively. (B) Cholangiocytes treated with TRTRSVRLY or DEGA showed a significant dose-dependent decrease in RRV binding. (C) Cholangiocytes also revealed a significant dose-dependent decrease in viral yield when treated with TRTRVSRLY while PTAAGV and SSASAW had no effect on viral replication. * = p<0.05 compared to serum free media.
Blocking of RRV binding and replication by synthetic VP4 derived peptide
Through in vitro peptide binding assays, the synthetic peptides of interest derived from the VP4 protein (PTAAGV, SSASAW, and TRTRVSRLY) were used to determine which specific sequence was involved in cholangiocyte binding. Investigation of cholangiocyte binding determined that the TRTRVSRLY peptide was able to block RRV binding in a dose-dependent manner. Increased concentrations of the peptide (0.5mM, 1mM, 2mM) resulted in blocking of the virus, as a percent of control (media without peptides), to be significantly decreased compared to treatment with media alone (76.33%, 64.17% and 51.60%, respectively, P<0.05). Conversely, the PTAAGV and SSASAW peptides did not interfere with RRV- cholangiocyte binding. DGEA is the collagen-binding domain for the α2β1 cholangiocyte receptor and has previously shown to be involved in RRV infection (30). Thus, it was used as a positive control, resulting in decreased viral binding (61.40%). GHRP is a nonspecific peptide that was used as a negative control, which has been previously shown to have no ability to inhibit RRV binding (22) (Fig. 1B).
In order to determine if cholangiocyte surface blocking with synthetic peptides had any effect on viral replication, blocking assays were carried out followed by incubation at 37°C for 18 hours. Viral yield in cholangiocytes treated with increasing dosages of synthetic TRTRVSRLY peptide was significantly lower than media or GHRP peptide controls (69.97%, 66.23%, 59.90%, p<0.05), while the PTAAGV and SSASAW peptides had no effect on viral yield (Fig. 1C). TRTRVSRLY peptide demonstrated similar blocking in MA104 cells (Supplemental Fig. 2A, 2B).
Inhibition of viral binding and replication by the TRTRVSRLY peptide is strain specific
We have previously shown that the induction of BA in the murine model was rotavirus strain-specific (13). In these investigations, RRV and GRV (goat strain) were able to induce the murine model of BA (24), while TUCH (a simian strain) and Ro1845 (a human strain) could not. Blocking and replication assays were performed on cholangiocytes using the synthetic TRTRVSRLY peptide and these other rotavirus strains. The TRTRVSRLY peptide inhibited binding of RRV and GRV (BA-inducing strains) by 45.13% and 41.27%, respectively, when compared to control (p<0.05). Conversely, the peptide had no effect on viral binding of the TUCH and Ro1845 strains (Fig. 2A). Similarly, viral yield was reduced in RRV and GRV by 51.29% and 35.51% of control (p<0.05) respectively, while cholangiocytes infected with Ro1845 and TUCH showed no reduction in viral yield after treatment with TRTRVSRLY peptide (Fig. 2B).
Fig. 2. Viral binding and replication in cholangiocytes treated with TRTRVSRLY as a function of rotavirus strain.

Mouse cholangiocytes were pre-treated with 1 mM synthetic peptide TRTRVSRLY followed by infection with RRV, TUCH, Ro1845 or GRV. (A) Viral binding revealed significant inhibition of binding only in strains which induce murine biliary atresia (RRV and GRV). DGEA and GHRP are positive and negative controls, respectively. (B) A significant reduction in viral yield in RRV and GRV was measured, with no decrease in replication for TUCH or Ro1845. * = p<0.05 compared to serum free media.
TUCH and Ro1845 VP4 amino acid sequences (440–448) have no effect on virus binding and replication
TRTRVSRLY constitutes the 440–448 aa sequence on RRV VP4. The 440–448 aa sequences in other strains, including TUCH and Ro1845 differ from that of RRV (Fig. 3A). This information was used to synthesize peptides of those regions found in the respective viruses, in order to determine if the inhibition of binding and replication was sequence specific. Cholangiocytes were treated with MRTRVSGLY (Ro1845) and VRTRISGLY (TUCH) peptides. Binding and replication assays were carried out as previously described. TRTRVSRLY peptide was able to inhibit binding and replication of RRV, with no effect on Ro1845 and TUCH. MRTRVSGLY and VRTRISGLY had no effect on any of the strains assessed (Fig. 3B, 3C). This suggested that BA-inducing strains (RRV and GRV) utilize the TRTRVSRLY sequence as a strain-specific cholangiocyte binding site.
Fig. 3. Corresponding peptides (amino acids 440–448) on TUCH and Ro1845 have no effect on rotavirus binding and replication.
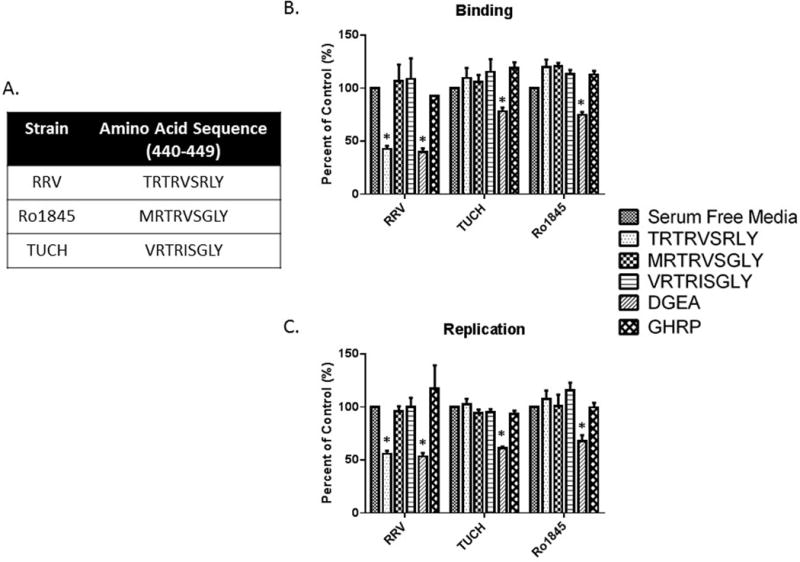
(A) Amino acids 440–448 of RRV, TUCH and Ro1845. (B, C) Treatment of mouse cholangiocytes with 1 mM MRTRVSGLY and VRTRISGLY had no effect on binding or replication in RRV, TUCH or Ro1845. In all strains, DGEA (positive control) significantly inhibited both viral binding and replication compared to media only control. Treatment with TRTRVSRLY significantly reduced RRV binding and viral yield with no effect on other rotavirus strains. * = p<0.05 as compared to serum free media pretreatment.
The SRL portion of the TRTRVSRLY peptide is integral to viral binding and replication
Once it was determined that the TRTRVSRLY peptide plays a role in RRV binding and cholangiocyte infection, the next objective was to identify the specific amino acids within this 9 aa peptide responsible for these functions. RRV and Ro1845 440–448 peptides differ by two aa. We synthesized peptides with each of those single aa changes (MRTRVSRLY, TRTRVSGLY) to evaluate the region of the peptide critical in binding and replication. Altering aa 440 (T→M) had no impact on inhibition of binding and replication, with both TRTRVSRLY and MRTRVSRLY significantly inhibiting RRV function. However, alteration at peptide 446 (R→G) inhibited the peptide’s ability to block binding and replication (Fig. 4A, 4B). This suggested that the critical portion of the 9 aa sequence lies within the last five residues. We then investigated the latter half of the TRTRVSRLY sequence to determine which aa govern the peptide’s function. Five-amino-acid peptides were synthesized with single aa changes at each position of the VP4 VSRLY sequence. Assaying rotavirus binding and replication revealed that peptides in which the SRL aa sequence was preserved (VSRLY, ISRLY and VSRLA), maintained the ability to inhibit virus binding and replication. Those peptides in which the S, R, or L peptide was modified (VVRLY, VSGLY, VSRAY) were unable to exhibit such inhibition (Fig. 4C, 4D). These results suggest that the presence of the “SRL” aa sequence is integral to RRV-cholangiocyte binding and viral replication.
Fig. 4. The SRL portion of TRTRVSRLY has an effect on viral binding and replication.

(A, B) Mouse cholangiocytes pre-treated with TRTRSVRLY or synthetic peptides with altered aa at positions 440 (T→M) or 446 (R→G). Altering aa 440 had a similar effect as TRTRVSRLY while a peptide containing an altered aa 446 could not inhibit RRV binding or replication. (C, D) Cholangiocytes pre-treated with 1 mM of synthetic peptides with single aa changes from VSRLY. Peptides conserving the “SRL” portion maintained the peptide’s ability to block RRV binding and replication. Peptides with an altered single aa between 445–447 lost the ability to block RRV binding and replication. * = p<0.05 compared to serum free media.
TRTRVSRLY peptide inhibits RRV replication in vivo and attenuates BA phenotype
To prove that the TRTRVSRLY peptide played a similar role in vivo, pups were inoculated with the peptide and RRV at birth followed by peptide again on day of life 1 and 2. Symptoms (acholic stool, bilirubinuria, jaundice, and weight loss) were recorded for 21 days. Mice injected with TRTRVSRLY peptide along with RRV developed significantly less symptoms compared to those injected with RRV and SSASAW peptide or saline (21.1% vs 100.0% vs 100.0% respectively, p<0.05) (Fig. 5A). Survival was similarly improved when pups were given the TRTRVSRLY peptide, with only 26.3% mortality when compared to 92.9% with SSASAW peptide and 88.5% with saline (Fig. 5B).
Fig. 5. Effects of in vivo use of TRTRSVRLY peptide.
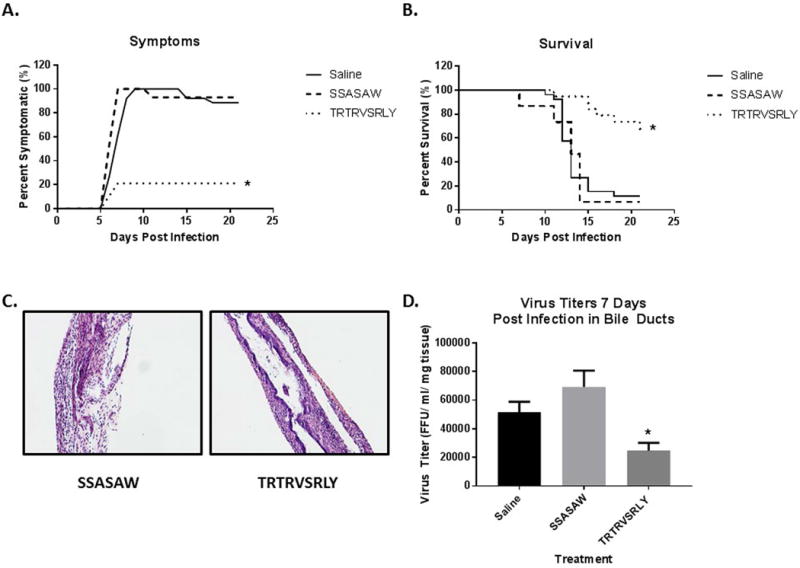
Mice were treated with the TRTRVSRLY peptide along with RRV. Saline (no peptide) or SSASAW peptides with RRV injection were used as control. (A) Symptoms were tracked in both sets of mice for 21 days. Symptoms in the TRTRVSRLY-treated group were 21.1% as compared to 100.0% in the saline control group. (B) Survival was noted to be significantly improved (26.3%) in mice treated with the synthetic peptide as compared to saline control (88.5%). (C) Histologic evaluation of the extrahepatic biliary tract 12 days after infection with RRV and peptide treatment. Hematoxylin and Eosin staining of extrahepatic bile ducts after infection with RRV and SSASAW peptide treatment resulted in complete obstruction of extrahepatic biliary duct. In contrast, those mice treated with TRTRVSRLY peptide had some mild inflammation with a patent bile duct. Magnified 10X. (D) Viral titers from extrahepatic bile ducts harvested seven days post infection were significantly lower in mice treated with the TRTRVSRLY peptide as compared to saline or those treated with SSASAW peptide. * = p<0.05
Bile ducts were harvested 12 days post infection from SSASAW and TRTRVSRLY peptide treated mice and stained with Hematoxylin and Eosin. Mice treated with SSASAW peptide exhibited a complete obstruction of their bile ducts while those treated with TRTRVSRLY had a mild inflammation with a patent lumen (Fig. 5C). Viral titers were obtained from extrahepatic bile ducts seven days post-injection with RRV. As expected, bile ducts of mice treated with the TRTRVSRLY peptide had significantly lower amounts of virus than those treated with SSASAW and the no peptide controls (Fig. 5D), thus proving that the synthetic peptide (TRTRVSRLY) competitively inhibited viral replication and subsequently attenuated the disease phenotype.
Inhibition of viral binding and replication by the TRTRVSRLY peptide on human cholangiocytes
To test the relevance of the TRTRVSRLY peptide on human rotavirus infection we next performed binding and replication assays on human cholangiocytes (H69 cells). Similar to the experiments carried out in mouse cholangiocytes, treatment with 1 mM TRTRVSRLY peptide significantly reduced RRV’s ability to bind to and replicate within human cholangiocytes to 65.9% and 57.9% of control, respectively (Fig. 6A, 6B).
Fig. 6. Viral binding and replication in Human cholangiocytes treated with TRTRVSRLY as a function of rotavirus strain.
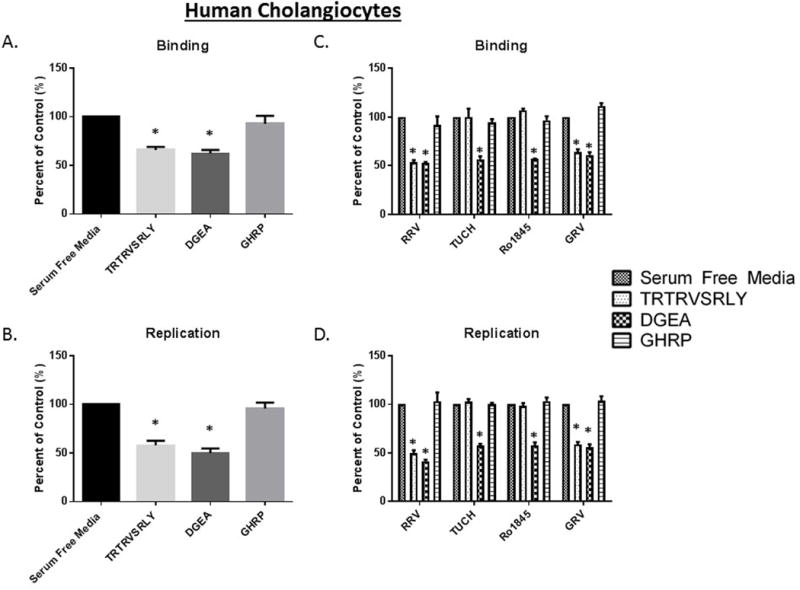
(A, B) Human cholangiocytes (H69) pre-treated with 1 mM TRTRVSRLY peptide demonstrated significant decrease in RRV binding and replication. DGEA and GHRP are positive and negative controls, respectively. (C) The TRTRVSRLY peptide significantly inhibit the binding of only those strains which induce murine biliary atresia (RRV and GRV) with no effect on TUCH or Ro1845. (D) Similarly, RRV and GRV viral yields were significant reduced, with no decrease in yield for TUCH or Ro1845. * = p<0.05 compared to serum free media control.
Previously we have reported strains specific infectability of human cholangiocytes mirroring that of mouse cholangiocytes (25). To prove that the TRTRVSRLY peptide strain specific blocking was consistent in human cholangiocytes we performed binding and replication assays on H69 cells infected with RRV, TUCH, Ro1845 and GRV. In agreement with our mouse data, the TRTRVSRLY peptide was able to block the binding of RRV and GRV to H69 cells while having no effects on TUCH and Ro1845 binding (Fig 6C). An identical effect was observed with replication as RRV and GRV exhibited reduced viral yields following treatment with the TRTRVSRLY peptide (Fig 6D).
The Hsc70 protein on the cell membrane is involved in RRV VP4 binding through SRL
In order to identify the SRL peptide’s corresponding cholangiocyte receptor, cell membrane fractions were treated with a synthetic biotinylated TRTRVSRLY peptide, as previously described. After isolation of the bound protein from a silver-stained polyacrylamide gel, MALDI-TOF analysis identified the protein of interest to be 70 KDa protein, Hsc70 (Fig. 7A). Interestingly, Hsc70 has previously been implicated in rotavirus cell entry through a secondary interaction, involving the VP4 protein (31).
Fig. 7. The role of Hsc70 in binding to TRTRVSRLY.
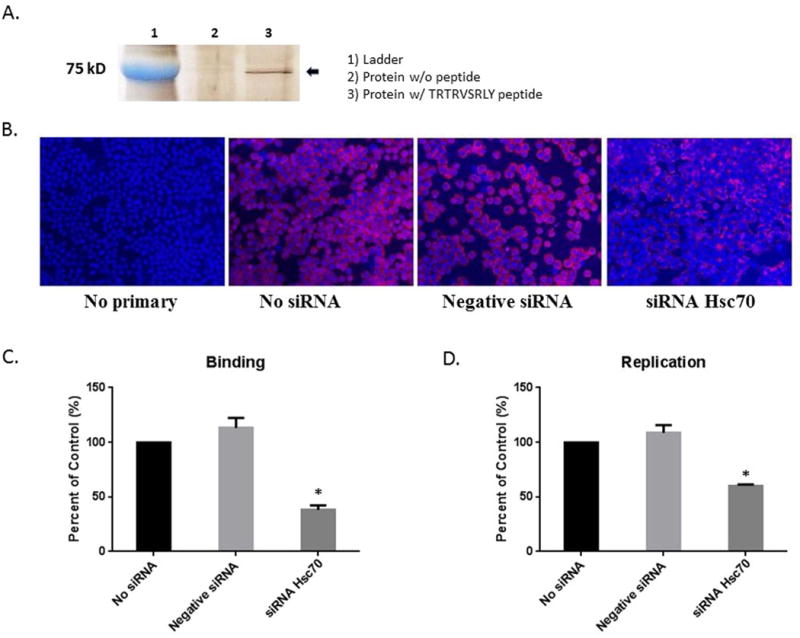
(A) Silver-stained band of protein immunoprecipitated from mouse cholangiocytes treated with no peptide (Lane 2) and biotinylated TRTRVSRLY peptide (Lane 3). (B) Cholangiocytes were treated with transfection agent in the absence of siRNA, negative (scrambled) siRNA or Hsc70 siRNA. “No siRNA” and “negative siRNA” were negative controls. siRNA against Hsc70 was able to block Hsc70 expression on the cholangiocyte cell surface. (C, D) When infected with RRV, cholangiocytes treated with Hsc70 siRNA showed a significant reduction in virus binding and replication as compared to negative controls. * = p<0.05 as compared to No siRNA.
Hsc70 knockdown in cholangiocyte blocks the action of TRTRVSRLY peptide
To determine if Hsc70 was a cholangiocyte receptor for the SRL sequence in RRV VP4, siRNA was utilized to knockdown Hsc70 expression. A siRNA combination of four sequences targeting Hsc70 gene was transfected into cholangiocytes, followed by immunostaining for Hsc70. “No siRNA” referred to cells treated with a transfection agent without siRNA, used as a negative control. A combination of scrambled sequences, or negative siRNA, which are incapable of blocking any cellular function, were utilized as a second negative control. Forty-eight hours after transfection of siRNA against Hsc70, a decrease of cholangiocyte surface expression of Hsc70 was detected (Fig. 7B). Binding assays were performed on cholangiocytes infected with RRV to validate the role of Hsc70 in viral binding. Only those treated with Hsc70 siRNA displayed a significant decrease in RRV binding and replication compared to cholangiocytes treated with negative controls (Fig. 7C, D).
Hsc70 siRNA-transfected cholangiocytes underwent TRTRVSRLY peptide binding assays, revealing that downregulation of Hsc70 expression in cholangiocytes lead to inability of TRTRVSRLY to block RRV binding, while those treated with negative control siRNA remained susceptible to inhibition of RRV attachment. All cells remained sensitive to DGEA blocking regardless of treatment (Fig. 8A). Percent of control is normalized to cells treated with serum free media in the absence of peptide (presented as 100%).
Fig. 8. The role of Hsc70 in RRV binding and replication.
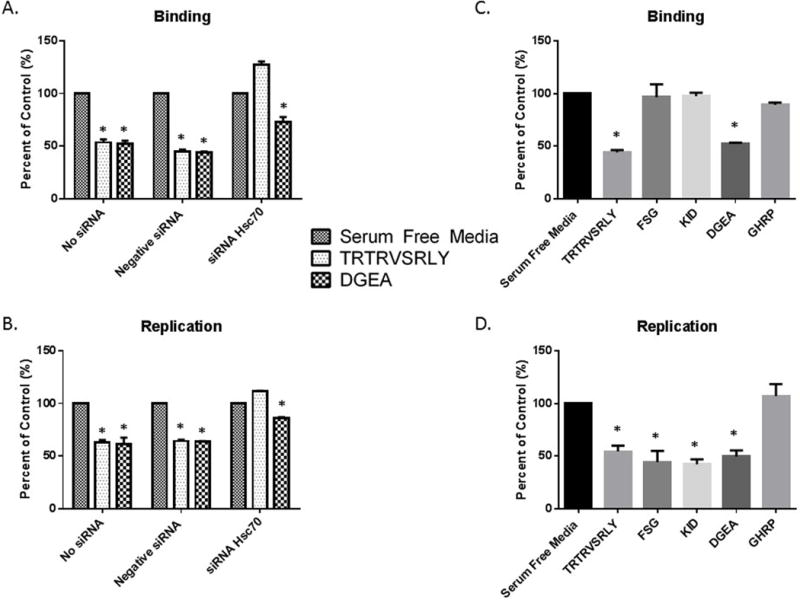
(A, B) Hsc70 siRNA-transfected mouse cholangiocytes treated with 1 mM TRTRVSRLY were unable to inhibit RRV binding or replication, while DGEA displayed significant inhibition as compared to serum free media in all treatments. (C) Cholangiocytes pre-treated with synthetic 1 mM TRTRVSRLY, FSGIKSTIDAAKSMATNVMKKFKK (“FSG”) and KTKIDRSTQISPNTLPD (“KID”) were infected with RRV. A significant decrease in viral binding with TRTRVSRLY was seen, with no change in binding with FSG or KID. (D) Viral replication assays with synthetic peptides revealed an inhibition of viral replication in cholangiocytes treated with TRTRVSRLY, FSG and KID. * = p<0.05 compared to serum free media.
RRV replication was similarly assessed using negative, no-siRNA and Hsc70 siRNA. Once again, negative and no siRNA remained susceptible to both DGEA and synthetic TRTRVSRLY peptide viral blockade. Viral replication in Hsc70 siRNA treated cells resulted in no change in viral yield for TRTRVSRLY peptide when normalized to serum free control. Without the presence of Hsc70, viral binding and yield are lower in all treatment groups (Fig. 8B).
The SRL peptide plays a role as a primary binding site in rotavirus entry
Previous studies have shown a secondary, post-attachment interaction to exist between some rotavirus strains and Hsc70. In these studies, the post attachment secondary interaction with Hsc70 was mediated by the aa sequence within VP4 spanning aa 531–554 termed “FSG” (FSGIKSTIDAAKSMATNVMKKFKK) (32) and 642–658, termed “KID” (KTKIDRSTQISPNTLPD) (33). In these previous experiments, MA104 cells treated with these peptides display reduced viral replication due to decreased viral entry (32, 33). We used the FSG and KID peptides to determine if they were involved in primary Hsc70 binding and replication in cholangiocytes similarly to the SRL peptide. Blocking assays using the three different peptides demonstrated that only TRTRVSRLY was capable of significantly inhibiting primary viral binding (44.35%) whereas FSG and KID had no effect (Fig. 8C). In addition, all three peptides inhibited RRV replication (44.53%, 42.43% and 54.50%, respectively). These results confirm that TRTRSVRLY blocks RRV primary attachment on cholangiocytes, resulting in reduced replication rates, while FSG and KID peptides had no effect on primary binding, but reduced post attachment interaction, reflected by reduced viral yield (Fig. 8D).
Discussion
BA is a devastating disease, affecting newborns in the first months of life. It is characterized by bile duct obstruction, liver injury, progressing to fibrosis and without treatment, death. Although the etiology of BA remains unclear, perinatal viral infection has been implicated in its pathogenesis (34). Several viruses, including rotavirus, may play a role in the activation of both inflammatory and autoimmune pathways (35). In the murine model of BA, RRV contributes to a release of pro-inflammatory cytokines and immune cell activation (36, 37). Although the exact mechanism through which rotavirus plays a role has not yet been elucidated, current studies indicate that the virus targets biliary epithelium, leading to inflammation and extrahepatic biliary obstruction (13, 36).
We have previously shown that the VP4 gene of RRV plays an integral role in the murine BA model induction through cholangiocyte infection (23) and immune cell activation (24). In 1997, Coulson et al. (16) discovered the collagen peptide-binding sequence DGE on rotavirus. This was then determined to bind the α2β1 integrin on cholangiocytes. This receptor, utilized in primary rotavirus binding, was not found on hepatocytes or Cho cells, less susceptible lines, suggesting that other receptors may govern RRV infectivity (20).
Using sequence and structure based modeling approaches, we determined three peptide sequences within the RRV VP4 protein that map into putative protein-protein binding sites. Through selective treatment with these peptides (PTAAGV, TRTRVSRLY, SSASAW), we determined that only TRTRVSRLY (aa 440–448) was able to decrease RRV binding and subsequent replication in murine cholangiocytes along with MA104 cells, the cell type rotavirus replicates efficiently. This peptide was only capable of inhibiting RRV and GRV, two strains with homologous TRTRVSRLY sequences, both of which can induce murine BA. However, it was not capable of inhibiting binding of TUCH and Ro1845, viruses with dissimilar peptide sequences that cannot induce murine BA. This suggested that TRTRVSRLY was an essential peptide that plays a role in RRV-specific cellular binding. Within the TRTRVSRLY sequence, it was determined that the 3 aa sequence “SRL” plays a critical role. This established a RRV-specific peptide, governing binding and entry, specific to rotavirus strains capable of inducing murine BA (16).
In order to determine the role of the RRV TRTRVSRLY peptide in human BA, we performed similar experiments in human cholangiocyte cells (H69 cells). Through this analysis, we determined that the TRTRVSRLY peptide is also capable of diminishing both RRV and GRV binding and viral replication, both strains capable of inducing the murine model of BA. It is important to note that there has been human rotavirus strains isolated which contain the TRTRVSRLY sequence in their VP4 protein (38, 39). Unfortunately, these strains were not maintained and as a result, are not available for study.
Once we established the importance of the TRTRVSRLY peptide in the in vitro infection of cholangiocytes, we evaluated its function in the RRV-induced in vivo model of BA. Through our investigations, we determined that the TRTRVSRLY peptide plays a role in the murine model of BA, lessening symptoms and mortality as well as resulting in lower viral titers in the bile ducts of mice.
Following determination of SRL specificity, we sought the corresponding cholangiocyte membrane receptor for this peptide. We found that in cholangiocytes, SRL attachment is through the Hsc70 protein. Hsc70 is a constitutively expressed chaperone within the Hsp70 family. This protein is involved in various cellular functions, including uncoating during clathrin-mediated endocytosis (40), protein folding (41), protein degradation through ubiquitin-proteasome degradation (42) and the import of proteins into organelles (43). This protein is also upregulated during inflammation, infection or malignant processes (40). Through interaction with antigen-presenting cells through LRP1/CD91 (low density lipoprotein receptor-related protein 1), it may result in translocation of NF-kB1 into the nucleus and dendritic cell maturation (44).
Rotavirus cell entry has previously been determined to be a complex, multistep process with various cellular receptors including integrins and Hsc70 (45, 46). Hsc70 is present on the cell surface of multiple rotavirus-susceptible and non-susceptible cell types including monkey MA104 and human Caco-2 cells and is believed to be essential to infection in susceptible cells. When blocked with antibodies, the ability of rotavirus to infect the cell is diminished (31). The Hsc70 protein has been shown to be involved in secondary rotavirus binding through a post-attachment step (18, 31, 47) via the peptide sequences, KID (642–658) and FSG peptides (531–554) (32, 33). The rotavirus capsid protein, composed of VP4 (later cleaved into VP5 and VP8) and VP7, is responsible for initial cell interaction. VP4 has previously been shown to play an essential role in cellular binding and attachment (46). Previous investigations of aa sequences within the VP4 protein by Zarate et al. provided knowledge that specific Hsc70 binding occurs through the KID (aa 642–658) domain on VP5. Blocking this domain impedes viral replication, though not binding, implying that this is a secondary binding site (33). Similarly, Gualtero et al. showed the FSG peptide (aa 531–554) within VP4 is implicated in Hsc70 binding and also plays a role in rotavirus replication, though not primary binding (32). Through our investigation of the TRTRVSRLY peptide, we determined that this unique peptide, unlike previously described peptides, interacts with Hsc70 during the primary binding phase of the VP4 domain of RRV, while the previously established peptides are involved in a post-attachment step. Thus, blocking Hsc70 through siRNA had significant effect in blocking RRV binding. Replication of RRV however, was similar to previous reports and inhibited not only by TRTRSVRLY, but also KID and FSG peptides (31, 33). Studies are under way to elucidate these differences.
Interestingly, through our analysis of the NCBI blastp database (www.NCBI/BLAST/blastp), the presence of a VSRLY peptide was also found on other viruses including Reovirus, Cytomegalovirus, Human Papillomavirus, Epstein Barr Virus, Bluetongue virus, Polyomavirus, Coronavirus, Respiratory Syncytial Virus, Adenovirus, Rodent Paramyxovirus, Cyprinid, Canine picodicistrovirus, Herpes Simplex virus 1, Torovirus and Chicken gallivirus 1. Several of these (CMV, EBV, HPV and Reovirus) have previously been found in explanted livers of infants with BA (3–7). This amino acid sequence was found on viral attachment proteins in CMV and reovirus. In EBV and HPV, it is also located on the viral capsid. Thus, this sequence may be involved in cholangiocyte binding in a similar fashion to the RRV VSRLY peptide. The role of Hsc70 binding and human BA induction as a function of these proteins has not yet been established.
Our findings support the concept that the rotavirus binding and entry process is multifaceted and involves binding of several VP4 peptides in a primary and secondary fashion to Hsc70 and integrins. Further investigations are underway to identify the cellular response following interaction of the peptide/virus with Hsc70 to understand the pathogenesis of BA.
Supplementary Material
Acknowledgments
The sequence and structure analyses were performed with the help of Cincinnati Children’s Hospital Medical Center Protein Informatics Core, and University of Cincinnati Bioinformatics Core.
JM gratefully acknowledges partial support by NIH grants R21AI097936, R01CA122346, P30ES006096, and U54HL127624
Funding: Funded by National Institutes of Health (NIH) R01 DK-091566
This project was supported in part by National Institutes of Health P30 DK-078392
Abbreviations
- RRV
rhesus rotavirus
- BA
Biliary atresia
- aa
amino acid
- ip
intraperitoneally
- CMV
Cytomegalovirus
- EBV
Epstein Barr Virus
- HPV
Human Papillomavirus
Footnotes
Conflict of interest statement: No conflicts of interest exist for any of the authors
References
- 1.Balistreri WF, Grand R, Hoofnagle JH, Suchy FJ, Ryckman FC, Perlmutter DH, Sokol RJ. Biliary atresia: current concepts and research directions. Summary of a symposium. Hepatology. 1996;23:1682–1692. doi: 10.1002/hep.510230652. [DOI] [PubMed] [Google Scholar]
- 2.Landing BH. Considerations of the pathogenesis of neonatal hepatitis, biliary atresia and choledochal cyst–the concept of infantile obstructive cholangiopathy. Prog Pediatr Surg. 1974;6:113–139. [PubMed] [Google Scholar]
- 3.Riepenhoff-Talty M, Gouvea V, Evans MJ, Svensson L, Hoffenberg E, Sokol RJ, Uhnoo I, et al. Detection of group C rotavirus in infants with extrahepatic biliary atresia. J Infect Dis. 1996;174:8–15. doi: 10.1093/infdis/174.1.8. [DOI] [PubMed] [Google Scholar]
- 4.Glaser JH, Balistreri WF, Morecki R. Role of reovirus type 3 in persistent infantile cholestasis. J Pediatr. 1984;105:912–915. doi: 10.1016/s0022-3476(84)80076-1. [DOI] [PubMed] [Google Scholar]
- 5.Fjaer RB, Bruu AL, Nordbo SA. Extrahepatic bile duct atresia and viral involvement. Pediatr Transplant. 2005;9:68–73. doi: 10.1111/j.1399-3046.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 6.Domiati-Saad R, Dawson DB, Margraf LR, Finegold MJ, Weinberg AG, Rogers BB. Cytomegalovirus and human herpesvirus 6, but not human papillomavirus, are present in neonatal giant cell hepatitis and extrahepatic biliary atresia. Pediatr Dev Pathol. 2000;3:367–373. doi: 10.1007/s100240010045. [DOI] [PubMed] [Google Scholar]
- 7.Drut R, Drut RM, Gomez MA, Cueto Rua E, Lojo MM. Presence of human papillomavirus in extrahepatic biliary atresia. J Pediatr Gastroenterol Nutr. 1998;27:530–535. doi: 10.1097/00005176-199811000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Petersen C, Grasshoff S, Luciano L. Diverse morphology of biliary atresia in an animal model. J Hepatol. 1998;28:603–607. doi: 10.1016/s0168-8278(98)80283-3. [DOI] [PubMed] [Google Scholar]
- 9.Riepenhoff-Talty M, Schaekel K, Clark HF, Mueller W, Uhnoo I, Rossi T, Fisher J, et al. Group A rotaviruses produce extrahepatic biliary obstruction in orally inoculated newborn mice. Pediatr Res. 1993;33:394–399. doi: 10.1203/00006450-199304000-00016. [DOI] [PubMed] [Google Scholar]
- 10.Czech-Schmidt G, Verhagen W, Szavay P, Leonhardt J, Petersen C. Immunological gap in the infectious animal model for biliary atresia. J Surg Res. 2001;101:62–67. doi: 10.1006/jsre.2001.6234. [DOI] [PubMed] [Google Scholar]
- 11.Petersen C, Grasshoff S, Luciano L. Diverse morphology of biliary atresia in an animal model. Journal of Hepatology. 1998;28:603–607. doi: 10.1016/s0168-8278(98)80283-3. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. Rotavirus architecture at subnanometer resolution. J Virol. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen SR, Jafri M, Donnelly B, McNeal M, Witte D, Bezerra J, Ward R, et al. Effect of rotavirus strain on the murine model of biliary atresia. J Virol. 2007;81:1671–1679. doi: 10.1128/JVI.02094-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez S, Arias CF. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 2004;12:271–278. doi: 10.1016/j.tim.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Arias CF, Isa P, Guerrero CA, Mendez E, Zarate S, Lopez T, Espinosa R, et al. Molecular biology of rotavirus cell entry. Arch Med Res. 2002;33:356–361. doi: 10.1016/s0188-4409(02)00374-0. [DOI] [PubMed] [Google Scholar]
- 16.Coulson BS, Londrigan SL, Lee DJ. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc Natl Acad Sci U S A. 1997;94:5389–5394. doi: 10.1073/pnas.94.10.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham KL, Halasz P, Tan Y, Hewish MJ, Takada Y, Mackow ER, Robinson MK, et al. Integrin-using rotaviruses bind alpha2beta1 integrin alpha2 I domain via VP4 DGE sequence and recognize alphaXbeta2 and alphaVbeta3 by using VP7 during cell entry. J Virol. 2003;77:9969–9978. doi: 10.1128/JVI.77.18.9969-9978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerrero CA, Mendez E, Zarate S, Isa P, Lopez S, Arias CF. Integrin alpha(v)beta(3) mediates rotavirus cell entry. Proc Natl Acad Sci U S A. 2000;97:14644–14649. doi: 10.1073/pnas.250299897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewish MJ, Takada Y, Coulson BS. Integrins alpha2beta1 and alpha4beta1 can mediate SA11 rotavirus attachment and entry into cells. J Virol. 2000;74:228–236. doi: 10.1128/jvi.74.1.228-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Londrigan SL, Graham KL, Takada Y, Halasz P, Coulson BS. Monkey rotavirus binding to alpha2beta1 integrin requires the alpha2 I domain and is facilitated by the homologous beta1 subunit. J Virol. 2003;77:9486–9501. doi: 10.1128/JVI.77.17.9486-9501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarate S, Espinosa R, Romero P, Guerrero CA, Arias CF, Lopez S. Integrin alpha2beta1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology. 2000;278:50–54. doi: 10.1006/viro.2000.0660. [DOI] [PubMed] [Google Scholar]
- 22.Jafri M, Donnelly B, Allen S, Bondoc A, McNeal M, Rennert PD, Weinreb PH, et al. Cholangiocyte expression of alpha2beta1-integrin confers susceptibility to rotavirus-induced experimental biliary atresia. Am J Physiol Gastrointest Liver Physiol. 2008;295:G16–G26. doi: 10.1152/ajpgi.00442.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Donnelly B, Bondoc A, Mohanty SK, McNeal M, Ward R, Sestak K, et al. The rhesus rotavirus gene encoding VP4 is a major determinant in the pathogenesis of biliary atresia in newborn mice. J Virol. 2011;85:9069–9077. doi: 10.1128/JVI.02436-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walther A, Mohanty SK, Donnelly B, Coots A, Lages CS, Lobeck I, Dupree P, et al. Rhesus Rotavirus Vp4 Sequence-Specific Activation of Mononuclear Cells Is Associated with Cholangiopathy in Murine Biliary Atresia. Am J Physiol Gastrointest Liver Physiol. 2015 doi: 10.1152/ajpgi.00079.2015. ajpgi 00079 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coots A, Donnelly B, Mohanty SK, McNeal M, Sestak K, Tiao G. Rotavirus infection of human cholangiocytes parallels the murine model of biliary atresia. J Surg Res. 2012;177:275–281. doi: 10.1016/j.jss.2012.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porollo A, Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins. 2007;66:630–645. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
- 27.Mohanty SK, Donnelly B, Bondoc A, Jafri M, Walther A, Coots A, McNeal M, et al. Rotavirus replication in the cholangiocyte mediates the temporal dependence of murine biliary atresia. PLoS One. 2013;8:e69069. doi: 10.1371/journal.pone.0069069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto Y, Shindo Y, Takakusagi Y, Takakusagi K, Tsukuda S, Kusayanagi T, Sato H, et al. Screening of a library of T7 phage-displayed peptides identifies alphaC helix in 14-3-3 protein as a CBP501-binding site. Bioorg Med Chem. 2011;19:7049–7056. doi: 10.1016/j.bmc.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Eismann T, Huber N, Shin T, Kuboki S, Galloway E, Wyder M, Edwards MJ, et al. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staatz WD, Fok KF, Zutter MM, Adams SP, Rodriguez BA, Santoro SA. Identification of a tetrapeptide recognition sequence for the alpha 2 beta 1 integrin in collagen. J Biol Chem. 1991;266:7363–7367. [PubMed] [Google Scholar]
- 31.Guerrero CA, Bouyssounade D, Zarate S, Isa P, Lopez T, Espinosa R, Romero P, et al. Heat shock cognate protein 70 is involved in rotavirus cell entry. J Virol. 2002;76:4096–4102. doi: 10.1128/JVI.76.8.4096-4102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gualtero DF, Guzman F, Acosta O, Guerrero CA. Amino acid domains 280–297 of VP6 and 531–554 of VP4 are implicated in heat shock cognate protein hsc70-mediated rotavirus infection. Arch Virol. 2007;152:2183–2196. doi: 10.1007/s00705-007-1055-5. [DOI] [PubMed] [Google Scholar]
- 33.Zarate S, Cuadras MA, Espinosa R, Romero P, Juarez KO, Camacho-Nuez M, Arias CF, et al. Interaction of rotaviruses with Hsc70 during cell entry is mediated by VP5. J Virol. 2003;77:7254–7260. doi: 10.1128/JVI.77.13.7254-7260.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezerra JA. Potential etiologies of biliary atresia. Pediatr Transplant. 2005;9:646–651. doi: 10.1111/j.1399-3046.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 35.Mack CL. The pathogenesis of biliary atresia: evidence for a virus-induced autoimmune disease. Semin Liver Dis. 2007;27:233–242. doi: 10.1055/s-2007-985068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shivakumar P, Campbell KM, Sabla GE, Miethke A, Tiao G, McNeal MM, Ward RL, et al. Obstruction of extrahepatic bile ducts by lymphocytes is regulated by IFN-gamma in experimental biliary atresia. J Clin Invest. 2004;114:322–329. doi: 10.1172/JCI21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxena V, Shivakumar P, Sabla G, Mourya R, Chougnet C, Bezerra JA. Dendritic cells regulate natural killer cell activation and epithelial injury in experimental biliary atresia. Sci Transl Med. 2011;3:102ra194. doi: 10.1126/scitranslmed.3002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khamrin P, Maneekarn N, Peerakome S, Yagyu F, Okitsu S, Ushijima H. Molecular characterization of a rare G3P[3] human rotavirus reassortant strain reveals evidence for multiple human-animal interspecies transmissions. J Med Virol. 2006;78:986–994. doi: 10.1002/jmv.20651. [DOI] [PubMed] [Google Scholar]
- 39.Li K, Lin XD, Huang KY, Zhang B, Shi M, Guo WP, Wang MR, et al. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology. 2016;494:168–177. doi: 10.1016/j.virol.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stricher F, Macri C, Ruff M, Muller S. HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy. 2013;9:1937–1954. doi: 10.4161/auto.26448. [DOI] [PubMed] [Google Scholar]
- 41.Frydman J, Hartl FU. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science. 1996;272:1497–1502. doi: 10.1126/science.272.5267.1497. [DOI] [PubMed] [Google Scholar]
- 42.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuno Y, Imamoto N, Yoneda Y. 70-kDa heat-shock cognate protein colocalizes with karyophilic proteins into the nucleus during their transport in vitro. Exp Cell Res. 1993;206:134–142. doi: 10.1006/excr.1993.1129. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava P. Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol. 2002;2:185–194. doi: 10.1038/nri749. [DOI] [PubMed] [Google Scholar]
- 45.Lopez S, Arias CF. Early steps in rotavirus cell entry. Curr Top Microbiol Immunol. 2006;309:39–66. doi: 10.1007/3-540-30773-7_2. [DOI] [PubMed] [Google Scholar]
- 46.Arias CF, Guerrero CA, Mendez E, Zarate S, Isa P, Espinosa R, Romero P, et al. Early events of rotavirus infection: the search for the receptor(s) Novartis Found Symp. 2001;238:47–60. doi: 10.1002/0470846534.ch4. discussion 60–43. [DOI] [PubMed] [Google Scholar]
- 47.Arias CF, Guerrero CA, Mendez E, Zarate S, Isa P, Espinosa R, Romero P, et al. Early events of rotavirus infection: the search for the receptor(s) Novartis Found Symp. 2001;238:47–60. doi: 10.1002/0470846534.ch4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


