Abstract
Hantavirus structural proteins are believed to localize to intracellular membranes often identified as Golgi membranes, in virus-infected cells. After virus budding into the Golgi luminal space, virus-containing vesicles are transported to the plasma membrane via trafficking pathways that are not well defined. Using the New World hantavirus, Andes virus, we have investigated the role of various Rab proteins in the release of hantavirus particles from infected cells. Rabs 8 and 11 were found to colocalize with Andes virus proteins in virus infected cells and when expressed from cDNA, implicating the recycling endosome as an organelle important for hantavirus infection. Small interfering RNA-mediated downregulation of Rab11a alone or Rab11a and Rab11b together resulted in a decrease in infectious virus particle secretion from infected cells. Downregulation of Rab8a did not alter infectious virus release but reduction of both isoforms did. These data implicate the recycling endosome and the Rab proteins associated with vesicular transport to or from this intracellular organelle as an important pathway for hantavirus trafficking to the plasma membrane.
Keywords: hantavirus, Andes virus, recycling endosome, Golgi, nucleocapsid
Introduction
Viruses must target structural proteins and genetic material to a specific location in infected cells for efficient assembly of new particles. After assembly, the virus particles must exit the cell in order to gain access to other susceptible cells or to spread from the infected host. Viruses that assemble at the plasma membrane exit the cell by budding directly into the extracellular milieu. Alternatively, some viruses assemble by budding through intracellular membranes and must then be transported to the plasma membrane – a process called virus egress (Ochsenbauer-Jambor et al., 2001). The assembly of several important human pathogens - hepatitis C virus (Lindenbach and Rice, 2002), rotavirus (LeBouder et al., 2008), herpes simplex virus types 1 and 2 (Mettenleiter, 2004), SARS coronavirus (Ksiazek et al., 2003) and various poxviruses (Moss, 2001; Smith and Law, 2004) - occurs at intracellular membranes, but little is known about the trafficking pathways used to deliver the newly formed viral particles to the plasma membrane (Ksiazek et al., 2003; Lai, 2001; Lindenbach and Rice, 2002; Moss, 2001).
Hantaviruses are members of the Bunyaviridae family that are further subdivided into Old World and New World groupings (Schmaljohn and Hjelle, 1997). The Old World hantaviruses are primarily found in Asia and Europe and include Hantaan virus, the causative agent of hemorrhagic fever with renal syndrome (HFRS) (Lee et al., 1978; Schmaljohn and Hjelle, 1997). New world hantaviruses, including Sin Nombre and Andes viruses, are found in North and South America, respectively, and are the causative agents of hantavirus pulmonary syndrome (HPS) (Elliott et al., 1994; Nichol et al., 1993; Toro et al., 1998).
A hantavirus genome consists of three single-stranded, negative-sense RNA segments (Hooper et al., 2001) and the entire replication cycle takes place in the cytoplasm (Hooper et al., 2001) with little cytopathic effects on the infected cell (Hardestam et al., 2005; Meyer and Schmaljohn, 2000; Rowe and Pekosz, 2006). Only four proteins are known to be expressed from the genome - the RNA-dependent RNA polymerase (L), two glycoproteins (GN and GC) produced by proteolytic cleavage of a precursor protein, and the nucleocapsid protein (N) (Hooper et al., 2001). The viral proteins and RNA localize to intracellular membranes, believed to be part of the Golgi apparatus, particles then bud into the luminal space and are transported to the plasma membrane via a vesicular trafficking pathway that has not been characterized to date (Goldsmith et al., 1995; Hooper et al., 2001; Ravkov and Compans, 2001; Salanueva et al., 2003; Shi and Elliott, 2002).
RabGTPases (Rabs) are a large family of small molecular weight proteins (Stenmark and Olkkonen, 2001) that are critical mediators of vesicle formation, trafficking, and fusion (Slimane et al., 2003; Zerial and McBride, 2001). Rabs gain their specificity for various membranes through interactions with effector molecules (Grosshans et al., 2006; Pfeffer and Aivazian, 2004) and play key roles in many important vesicular transport pathways, including Golgi to plasma membrane transport (Slimane et al., 2003; Zerial and McBride, 2001).
Rab8 and Rab11 have been shown to be critical in trafficking proteins from the Golgi to plasma membrane (Ang et al., 2003; Chen et al., 1998; Li et al., 1993; Zhang et al., 2005). Rab8 and Rab11 are found at the trans-Golgi and the recycling endosome, a post-Golgi compartment that serves as a transport intermediate for some cargo en route from the TGN to the plasma membrane (Ang et al., 2004; Chen et al., 1998; Li et al., 1993). Disruption of either Rab8- or Rab11-specific pathways leads to inhibition of recycling endosome-dependent TGN to plasma membrane transport as well as plasma membrane recycling (Ang et al., 2003; Ang et al., 2004; Brock et al., 2003; Chen et al., 1998; Li et al., 1993; Neznanov et al., 2003; Schlierf et al., 2000; Zhang et al., 2005). However, the recycling endosome, Rab8, and Rab11 are not required for trafficking of all proteins from the TGN to plasma membrane as plasma membrane delivery of the Influenza A virus hemagglutinin protein is unaffected by inhibition of Rab8 or Rab11 trafficking pathways (Chen et al., 1998; Li et al., 1993; Yoshimori et al., 1996). This indicates that specific sorting events occur to direct cargo into multiple transport pathways (i.e. recycling endosome-dependent and independent) to facilitate delivery from the TGN to the plasma membrane.
Viruses often hijack existing cellular pathways in order to complete various stages of their life cycle. We investigated the vesicular trafficking pathways important for ANDV release from infected cells and describe a role for the recycling endosome, and specifically Rab8 and Rab11, during ANDV exit from non-polarized epithelial cells.
Results
ANDV N forms cytoplasmic inclusions and localizes to intracellular vesicular membranes
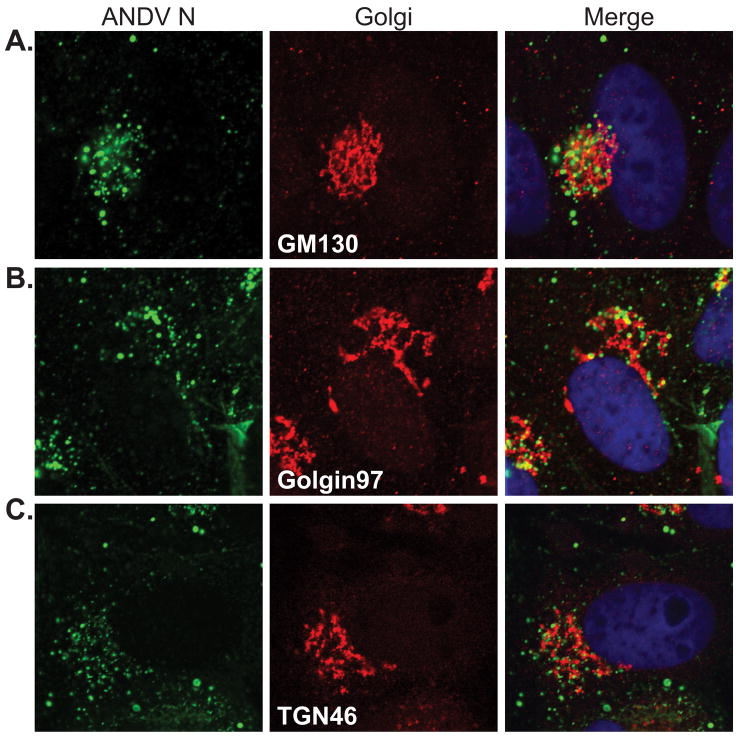
Viruses in the family Bunyaviridae are thought to primarily assemble at membranes of the Golgi and trans-Golgi network (TGN) and these cellular compartments likely play a critical role in ANDV replication and assembly. We focused our studies on the nucleocapsid protein because it is the most abundant protein in the virion (Kaukinen et al., 2005) and localizes to the site of viral RNA replication and particle assembly (Goldsmith et al., 1995; Hooper et al., 2001; Ravkov and Compans, 2001). While the ANDV glycoproteins have been shown to localize to the Golgi and TGN during infection and from cDNA expression (Deyde et al., 2005), little is known about the localization of ANDV N with this compartment. To determine the intracellular localization of ANDV N during infection, immunofluorescence confocal microscopy was performed on ANDV-infected Vero cells immunostained for ANDV N and various markers of the Golgi and TGN (Fig. 1). ANDV N showed partial overlap with GM130, Golgin-97 and TGN46, however the extent of colocalization differed slightly. The greatest degree of colocalization was observed with GM130 (Fig. 1A), a protein associated with the cis-Golgi (Pfeffer, 2001). Less colocalization was observed with Golgin97 (Fig. 1B), a membrane-associated protein of the TGN (Gu et al., 2001; Yoshino et al., 2003). ANDV N consistently showed more colocalization with membranes positive for TGN46 (Fig. 1C) than membranes positive for Golgin97. TGN46 is an integral membrane protein that recycles between the plasma membrane and TGN, but the recycling kinetics result in TGN enrichment (Greaves and Chamberlain, 2007; Roquemore and Banting, 1998). While there was only partial overlap of ANDV N with the Golgi markers, the remaining ANDV N was consistently in close proximity to all of the Golgi markers, suggesting a close spatial relationship between nucleocapsid and the Golgi. This data indicates that while a portion of ANDV N does localize with Golgi and TGN membranes, the majority of the protein appears to be proximal but not associated with these membrane compartments.
Figure 1. ANDV N partially localizes to the Golgi and trans-Golgi during infection.
Vero cells were infected with ANDV and fixed for immunofluorescence at 2 dpi. Cells were immunostained for ANDV N (green, Alexa Fluor 488) and the following Golgi and trans-Golgi markers (red, Alexa Fluor 555) (A) GM130, (B) Golgin97, and (C) TGN46. Cells were visualized using confocal microscopy. Nuclei (blue) were counterstained with TO-PRO-3 and are shown in the merged panels only. All images are confocal single planes acquired at a 63x optical magnification and 3x digital zoom.
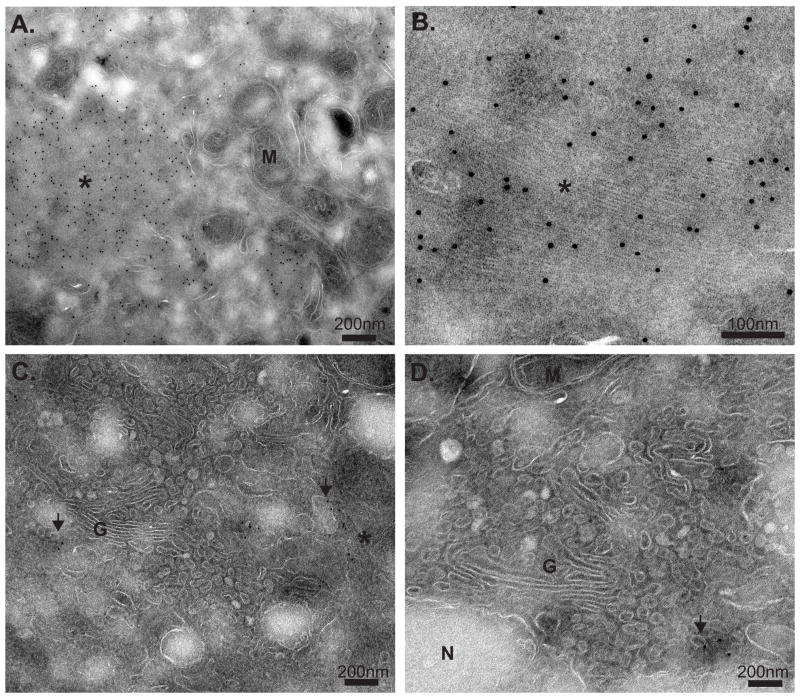
To gain additional insights on the location of ANDV N during infection, immuno-electron microscopy was performed on ANDV-infected cells (Fig. 2). ANDV N was localized throughout the cytoplasm but concentrated in the perinuclear region (Fig. 2). ANDV N was primarily localized to large, granulofilamentous structures surrounded by large numbers of mitochondria (Fig. 2 A and B). These structures were not membrane-bound, but were proximal to intracellular membranes resembling Golgi cisternae (Fig. 2 C). Smaller accumulations of ANDV N, not in filamentous structures, also localized to the membranes of small vesicles in close proximity to the Golgi (Fig. 2 C and D). Additionally, the structure of the Golgi was altered, with extensive small vesicles surrounding the cisternae (Fig. 2 C and D). There was no ANDV N found present at the plasma membrane or enclosed within membrane structures (~75 cells observed, data not shown), and even with large amounts of ANDV N present in the infected cells, no obvious structures resembling viral particles were detected. This data suggests that there are two cellular pools of ANDV N, one that is present in large, granulofilamentous structures not significantly associated with membranes and one that is membrane-associated.
Figure 2. Ultrastructural analysis of ANDV N during infection by immunoelectron microscopy.
At 2d (A and B) and 4d (C and D) post infection, ANDV-infected Vero cells were and immunolabeled using ANDV N-specific rat immunesera followed by secondary antibody conjugated to 12 nm colloidal gold particles. Granulofilamentous structures are indicated with an asterisk (*), while vesicle-associated ANDV N is noted by arrowheads. Cellular structures are denoted as follows: mitochondria (M), Golgi (G), and nucleus (N). Scale bars represent 100 or 200 nm as indicated. The image in panel (B) is a cropped and digital zoom of a region in panel (A), and was performed using Adobe Photoshop.
ANDV N colocalizes with Rabs of the recycling endosome
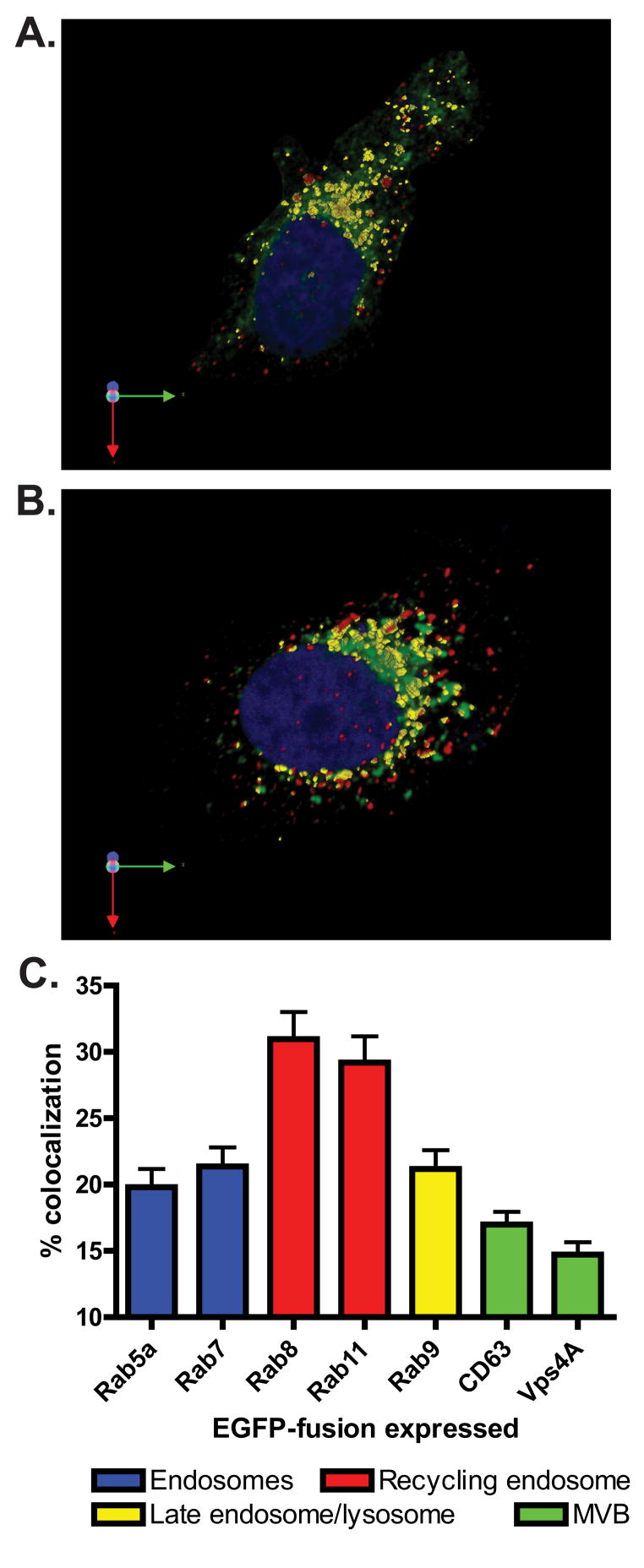
In order to determine the role of various cellular trafficking pathways in ANDV replication we focused our remaining studies on the portion of ANDV N localized to intracellular vesicular membranes. ANDV N is predicted to be a peripheral membrane protein that exists in a dynamic equilibrium between membrane bound and cytosolic pools (Hooper et al., 2001; Kaukinen et al., 2005; Ravkov and Compans, 2001). To identify specific vesicular populations during infection, ANDV-infected cells were transfected with cDNAs encoding eGFP-fusion proteins associated with various host cell trafficking pathways and analyzed by confocal microscopy for colocalization of eGFP with ANDV N protein Using 3-D reconstructions of serial Z-stack images acquired by confocal microscopy (data not shown) we were able to focus on the portion of membrane-associated ANDV N in infected cells expressing cDNAs of proteins associated with various vesicular transport pathways. ANDV-infected cells expressing eGFP-Rab 8 (Fig. 3A) or eGFP- Rab11 (Fig. 3B) show a high degree of colocalization between ANDV N and either Rab protein. We measured the degree of colocalization using parameters within the Volocity software that allowed us to focus on the viral antigen that was associated with the vesicular trafficking proteins (described in the Materials and Methods). The percent of eGFP colocalizing with ANDV N was determined by dividing the total colocalization volume by the volume of eGFP, and expressed as percent colocalization (Fig. 3C). Using this calculation, Rab8 and Rab11 had the highest level of colocalization with ANDV N, with 30.1% and 29.2% of the eGFP-Rab proteins colocalizing with ANDV N, respectively. Rabs associated with early endosomes (eGFP-Rab5a), late endosomes (eGFP-Rab7), and late endosomes/lysosomes (Rab9) showed colocalization that was clearly less than that observed with eGFP-Rab8 and eGFP-Rab11. Proteins associated with the multivesicular bodies (CD63-eGFP and eGFP-Vps4A) showed limited colocalization (Fig. 3C). Taken together, these results suggest that the recycling endosome-associated Rabs 8 and 11 colocalize with ANDV N and therefore may play a role in ANDV replication.
Figure 3. ANDV N localizes to the recycling endosome and has little colocalization with proteins of other vesicular trafficking pathways.
(A and B) Vero cells were infected with ANDV, then transfected at 3 dpi with cDNAs expressing (A) eGFP-Rab8 or (B) eGFP-Rab11 fusion proteins (green). Cells were fixed 18 hpt, immunostained for ANDV N (red, AlexaFluor 594), counterstained for the nucleus with TO-PRO-3 (blue), and visualized using confocal microscopy. 3-D reconstructions were created from confocal z-stacks using the Improvision Volocity imaging software. The regions of colocalization between eGFP and ANDV N can be seen in yellow. (C) 3-D regions of colocalization between ANDV N and eGFP-fusion proteins were measured as described in the Materials and Methods. The volume of colocalization was normalized to the volume of eGFP and expressed as percent colocalization. The trafficking molecules were grouped into the following trafficking pathways: endosome (blue bars), recycling endosome (red bars), late endosome/lysosome (yellow bars), and multvesicular body (MVB, green bars). Data presented is the mean of four independent experiments and at least 5 cells per cDNA transfection in each experiment.
Many viral proteins show similar subcellular localization during infection and when expressed independently from cDNA. In order to determine if viral proteins colocalized with Rab8 and Rab11 when expressed from cDNA, Vero cells were transfected with cDNAs expressing either ANDV N (Supplemental Fig. 1A and D) or the ANDV glycoprotein ORF (M), which expresses both GN and GC (Supplemental Fig. 1B and E), with either eGFP-Rab8 (Supplemental Fig. 1A and B) or eGFP-Rab11 (Supplemental Fig. 1D and E). Cells were then immunostained for either ANDV N or ANDV glycoproteins and analyzed by confocal microscopy. ANDV N and the ANDV glycoproteins colocalized with eGFP-Rab8 and eGFP-Rab11, suggesting that both proteins can localize to cellular membranes enriched in Rab8 and Rab11 independently of virus infection. The vesicular stomatitis virus G (VSV G) protein has been shown to traffic from the Golgi to the plasma membrane via Rab8- (Ang et al., 2003; Ang et al., 2004; Li et al., 1993) and Rab11-dependent pathways (Chen et al., 1998). As expected, VSV G colocalized extensively with both eGFP-Rab8 and eGFP-Rab11 (Supplemental Fig. 1C and F).
ANDV N colocalizes with endogenous Rab8 and Rab11 during infection
The data in Fig. 3 indicated that ANDV N could localize to cellular membranes containing cDNA expressed eGFP-Rab8 or eGFP-Rab11. To determine if the ANDV N colocalized with the endogenous forms of Rab8 and Rab11, mock- or ANDV-infected Vero cells were immunostained at 3 dpi for ANDV N and either endogenous Rab8 (Fig. 4A) or Rab11 (Fig. 4B). The signal intensity for immunostaining of endogenous Rabs was lower when compared to the cDNA-expressed eGFP proteins – most likely a combination of higher expression levels of the plasmid expressed fusion proteins and the intense fluorescence signal of eGFP. While endogenous Rab8 was readily detectable, endogenous Rab11 consistently showed low fluorescent intensity (compare green fluorescence images in Fig. 4). Despite the low level of signal intensity there were notable regions of colocalization between ANDV N and both Rab8 (Fig. 4a) and Rab11 (Fig. 4b), suggesting that a portion of ANDV N expressed during infection was present at membranes containing Rab8 and Rab11 and providing further support for a role for Rab8 and Rab11 during ANDV infection. ANDV N protein also colocalized with fluorescently labeled transferrin (data not shown), further supporting a role for the recycling endosome in ANDV infection.
Figure 4. ANDV N colocalizes with endogenous Rab8 and Rab11 during infection.
Mock- or ANDV-infected Vero cells were fixed at 3 dpi and immunostained for ANDV N (red, AlexaFluor 594) and either (A) Rab8 (green, AlexaFluor 488) or (B) Rab11 (green, AlexaFluor 488). Nuclei were counterstained with TO-PRO-3 (blue) and are shown in the merged images only. All images were acquired at 63x magnification. Digital zoom (2x) images of the boxed area in the merged panels for (a) Rab8 and (b) Rab11 are also shown.
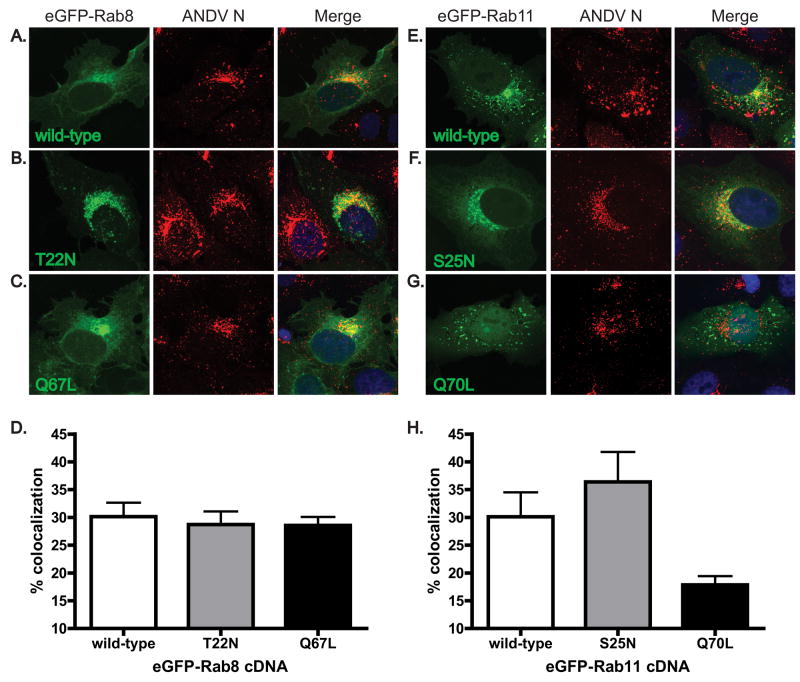
Dominant negative and constitutively active forms of Rab11 have altered levels of colocalization with ANDV N during infection
Amino acid substitutions at highly conserved residues within RabGTPases create proteins that are no longer capable of GDP-GTP cycling. A serine to asparagine mutation at position 25 (S25N) in Rab11 results in constitutive binding of GDP and locks the protein in a dominant negative form. In contrast, a glutamine to lysine mutation at position 70 (Q70L) prevents GTP hydrolysis, resulting in a constitutively active protein. The analogous mutations in Rab8 are T22N and Q70L. These mutant proteins often display altered subcellular localization due to augmented protein-protein interactions (Chen et al., 1998; Hattula et al., 2006; Ren et al., 1998; Scheiffele et al., 1995). To determine if expression of mutated eGFP-Rab8 and -Rab11 proteins during ANDV infection altered the colocalization of these proteins with ANDV N, infected Vero cells were transfected with cDNAs expressing eGFP-fusions of either wild-type, dominant negative (GDP-bound), or constitutively active (GTP-bound) mutants of eGFP-Rab8 and 11 (Fig. 5). There were notable differences in the subcellular distribution of the eGFP-Rab8 proteins. The GDP-bound form (Rab8T22N) had a tighter perinuclear localization and fewer effects on cell morphology (Fig. 5B), while the GTP-bound protein (Rab8Q67L, Fig. 5C) had an expression pattern similar to wild-type eGFP-Rab8 (Fig. 5A) (Hattula et al., 2006; Scheiffele et al., 1995). Interestingly, both forms of eGFP-Rab8 displayed similar levels of colocalization with ANDV N compared to wild-type Rab8 (Fig. 5 A–C, quantitated data in Fig. 5D). In contrast, eGFP-Rab11S25N showed increased levels of colocalization with ANDV N while the constitutively active eGFP-Rab11Q70L showed an approximately 50% decrease in colocalization when compared to eGFP-Rab11 (Fig. 5 E–G, quantitated data in Fig. 5H).
Figure 5. Dominant negative and constitutively active forms of Rab11 have altered levels of colocalization with ANDV N during infection.
ANDV-infected Vero cells were transfected with cDNAs expressing (A) wild-type, (B) T22N mutant, GDP-bound dominant negative, and (C) Q67L mutant, GTP-bound constitutively active, forms of eGFP-Rab8 (green); and (E) wild-type, (F) S25N mutant, GDP-bound dominant negative, and (G) Q70L mutant, GTP-bound constitutively active, forms of eGFP-Rab11 (green). At 18 hpt, cells were fixed and immunostained for ANDV N (red, AlexaFluor 555). Nuclei were counterstained with TO-PRO-3 (blue) and are shown in the merged images only. All images are flattened reconstructions of confocal z-stacks acquired at 63x optical magnification and 2x digital zoom. Levels of colocalization between ANDV N and (D) eGFP-Rab8 and (H) eGFP-Rab11 proteins were quantitated using the Volocity imaging software. Data presented is the mean of three independent experiments and at least 5 cells per cDNA transfection in each experiment.
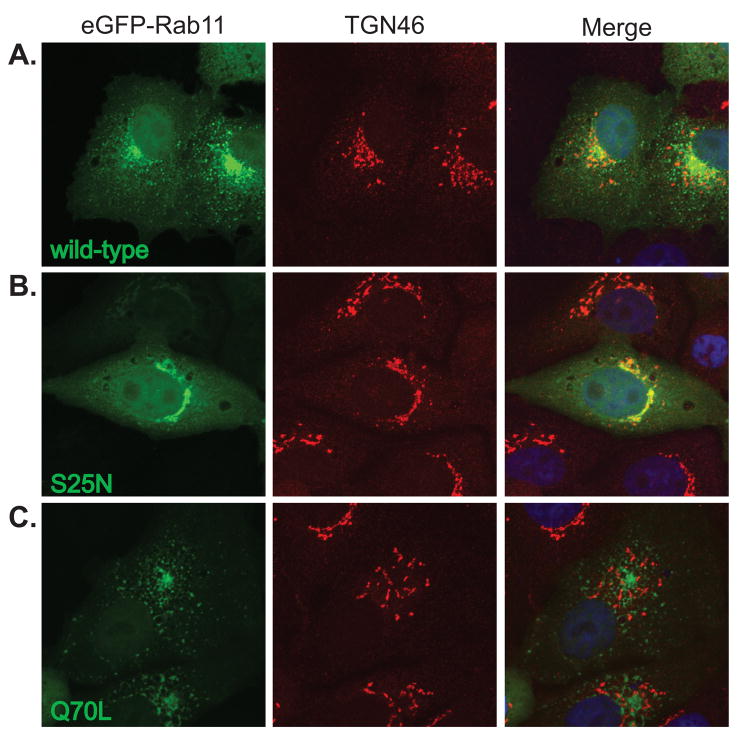
Rab11 has been reported to function in trafficking between the trans-Golgi and recycling endosome, therefore, we determined the localization of the eGFP-Rab11 proteins with respect to the trans-Golgi marker, TGN46 (Fig. 6). Wild-type eGFP-Rab11 showed partial localization with TGN46 (Fig. 6A), while the Q70L mutant showed limited overlap (Fig. 6C). In contrast, the membrane-associated portion of Rab11S25N showed almost complete localization with TGN46 (Fig. 6B). These data are consistent with previous reports that the dominant negative Rab11 localizes to the TGN, while the constitutive active form localizes at the recycling endosome (Chen et al., 1998; Neznanov et al., 2003).
Figure 6. Dominant negative and constitutively active forms of Rab11 localize to distinct membrane compartments.
Vero cells were transfected with cDNAs expressing eGFP-Rab11 (A) wild-type, (B) S25N, or (C) Q70L proteins (green). At 18 hpt, cells were fixed and immunostained for the trans-Golgi marker, TGN46 (red, AlexaFluor 594). Nuclei were counterstained with TO-PRO-3 (blue) and are shown in the merged images only. All images are flattened reconstructions of confocal z-stacks acquired at 63x optical magnification and 2x digital zoom.
siRNA-mediated downregulation of Rab8 and Rab11 block ANDV secretion
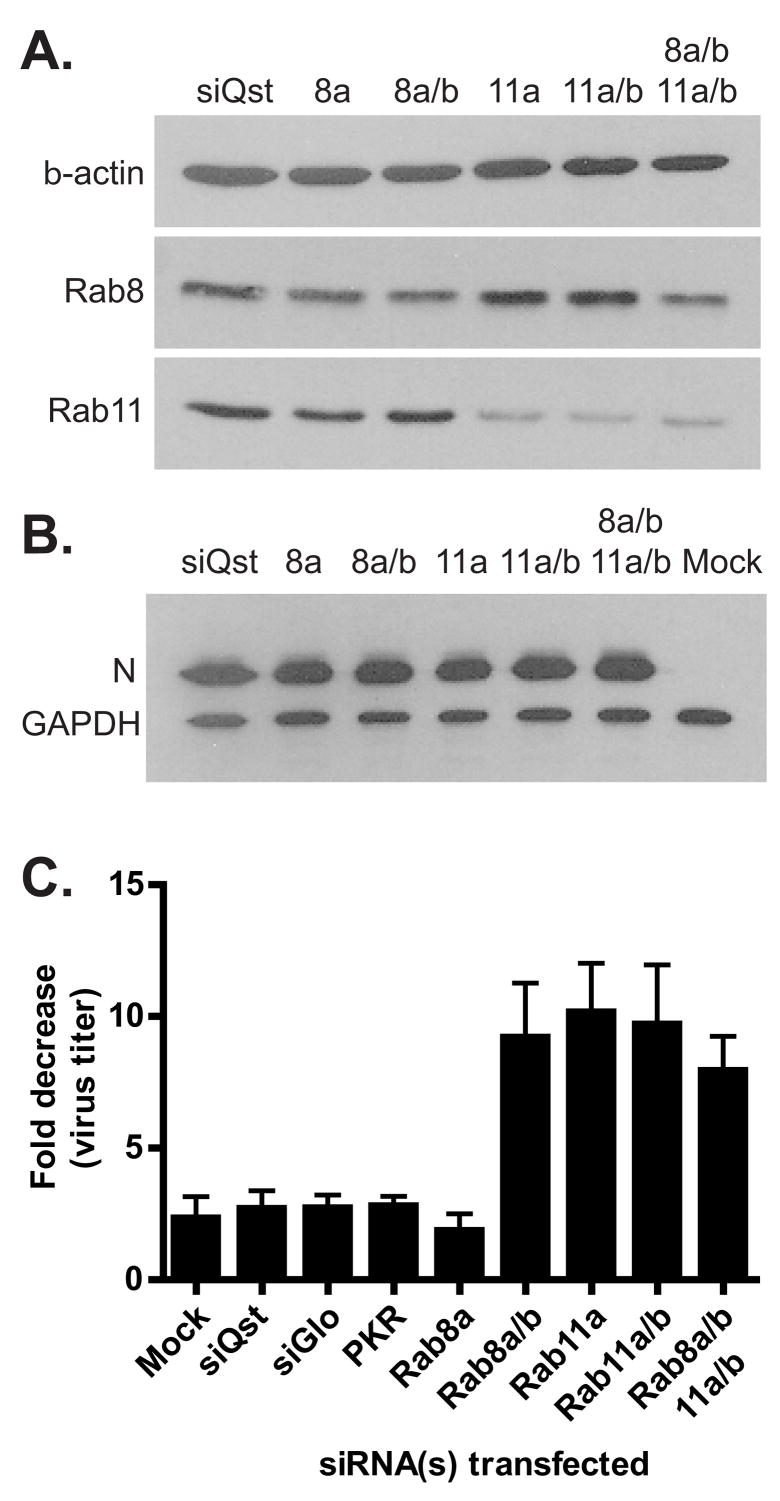
To determine if Rabs 8 and 11 played a functional role in ANDV release from infected cells, RNA interference (RNAi) (Elbashir et al., 2001) was utilized to downregulate Rab8 and Rab11 protein expression during infection. Many cell surface receptors, including the hantavirus receptor β3 integrin, are recycled through the recycling endosome (Jones et al., 2006), therefore downregulation of molecules within this pathway could alter surface expression of the viral receptor, thus affecting virus entry. To avoid altering early events in the virus life cycle, we took advantage of the fact that ANDV infection of Vero cells leads to prolonged, low level virus secretion with little obvious cytopathic effects (Hardestam et al., 2005; Meyer and Schmaljohn, 2000; Rowe and Pekosz, 2006). Seventy-two hours following ANDV infection, Vero cells were transfected with siRNAs specific for the a or b isoforms of Rabs 8 and 11, along with siRNA controls. At this time, over 90% of the cells are expressing viral antigen (data not shown). Since most cells express two isoforms (a and b) of Rab8 and Rab11 (Lai et al., 1994; Lau and Mruk, 2003), we utilized pooled siRNAs specific for either the a isoform or both the a and b isoforms. Four siRNAs per target transcript were used. At 48 hpt, there was a significant decrease in the Rab8 protein expression levels (Fig. 7A) after transfection with siRNAs targeting Rab8a (52.7% decrease) or both Rab8a and 8b isoforms (51.6% decrease). Transfection of siRNAs specific for Rab11a or Rab11a and 11b isoforms, as well as transfection reagent alone (siQst) resulted in no changes in Rab8 expression levels (Fig. 7A). There was also a significant decrease in the Rab11 protein expression levels (Fig. 7A) after transfection with siRNAs targeting Rab11a (78.1% decrease) or both Rab11a and 11b isoforms (61.9% decrease), again with no off-target effects from siRNAs targeting Rab8 isoforms. Unfortunately, the antibodies used cannot distinguish between the two isoforms, so we could not determine the specific downregulation of the b isoforms. Downregulation of all four targets (Rab8a and 8b, Rab11a and 11b) resulted in significant decreases in both Rab8 (48.8% decrease) and Rab11 (61.3% decrease) proteins (Fig. 7A). The level of β-actin expression was not altered after any siRNA transfection (Fig. 7A). The treatment of cells with structured, double stranded RNAs can lead to host cell antiviral responses that can inhibit viral replication (Stetson and Medzhitov, 2006). Intracellular ANDV N levels remained constant after siRNA transfection (Fig. 7B), indicating there were no adverse effects of siRNA transfection on viral protein production.
Figure 7. siRNA downregulation of Rabs of the recycling endosome block ANDV egress.
Vero cells were infected with ANDV and at 3 dpi were transfected with the following siRNAs: siGlo (100nM, RISC-binding deficient siRNA), murine PKR (100nM, unrelated siRNA), Rab8A (100nM), Rab8a and b (50nM each), Rab11a (100nM), Rab11a and b (50nM each), and quadruple knockdown Rab8a and b, and Rab11a and b (50nM each). (A) The cells were lysed at 48 hpt and the amount of Rab8, Rab11, and β-actin were determined by western blot analysis. (B) Intracellular ANDV N levels at 48 hpt were determined by western blot analysis. Protein levels were normalized to GAPDH. (C) Supernatants were harvested at the time of transfection (0h) and 48 hpt. Virus titer was determined by immune TCID50 analysis. The fold decrease in virus titer between 0h and 48h was determined. Data presented is the mean of four independent experiments with duplicate wells analyzed for each condition.
In order to assess if siRNA treatments altered the secretion of infectious virus particles, infected-cell supernatants were harvested immediately before and 48 hours after siRNA transfection. Infectious virus production was quantified by an immune TCID50 assay and expressed as a fold decrease in virus production (Fig. 7C). Downregulation of Rab11a or Rab11a and 11b resulted in a nearly 10 fold decrease in virus secretion, indicating Rab11 isoforms were functionally important for ANDV release from infected cells. Transfection with control siRNAs (siQst, transfection reagent alone; siGlo, RISC-binding deficient siRNA; and murine PKR siRNA, a protein not associated with vesicular transport) did not alter the amount of infectious virus production when compared to mock-transfected cells, indicating there were no non-specific effects of siRNA transfection on ANDV virus production. Interestingly, siRNAs specific for Rab8a did not alter ANDV virus secretion (Fig. 7C) even though Rab8 levels were decreased significantly (Fig. 7A). However, downregulation of both Rab8a and 8b led to a 10-fold decrease in virus secretion. Decreasing all four Rab isoforms did not enhance the decrease virus secretion compared to downregulating Rab8a/b, Rab11a, or Rab11a/b. The data in Fig. 7 demonstrates that Rabs associated with the recycling endosome play a functional role in ANDV secretion from epithelial cells, and specifically indicates Rab11a as a major host cell factor involved in this process.
Discussion
Many viruses in the family Bunyaviridae assemble at Golgi membranes and new particles are transported to the plasma membrane via vesicular transport (Nichol, 2001; Salanueva et al., 2003). The vesicular trafficking mechanism utilized during hantavirus egress is poorly understood but the data presented in this study suggests trafficking through the recycling endosome may be a key step in this process for ANDV.
The nucleocapsid proteins of viruses within Bunyaviridae have been localized to multiple intracellular compartments. The nucleocapsids of Uukuniemi virus (genus Phlebovirus) and Black Creek Canal virus (BCCV, genus Hantavirus) have both been shown to localize to Golgi membranes (Jantti et al., 1997; Kuismanen et al., 1982; Ravkov and Compans, 2001). In contrast, Hantaan virus (genus Hantavirus) and LaCrosse virus (genus Orthobunyavirus) nucleocapsids have been shown to localize to membranes of the endoplasmic reticulum (ER) and ER-Golgi intermediate compartment (ERGIC) (Ramanathan et al., 2007; Stertz et al., 2006). Even though ANDV N had partial colocalization with Golgi and trans-Golgi markers by immunofluorescence confocal microscopy, a significant portion did not overlap with these markers. Electron microscopy studies further indicated two populations of nucleocapsid. The majority of ANDV N was localized to large cytoplasmic structures not obviously associated with membranes, which have been previously described as inclusion bodies in studies of other hantaviruses (Goldsmith et al., 1995; Hung et al., 1985; Ravkov et al., 1997). These structures may be caused by accumulation of large amounts of N protein in infected cells or may be the site of RNA replication as described for the orthobunyavirus, Bunyamwera (Yan et al., 2002). Further studies will be required to determine the composition of these structures. A smaller portion of ANDV N was membrane-associated at vesicles proximal to the Golgi, which has been described for Uukuniemi virus N (Jantti et al., 1997). These vesicles could be part of the Golgi, trans-Golgi network, or post-Golgi secretory pathway. In order to identify these vesicles, we focused on proteins involved in host cell vesicular trafficking pathways. From these studies, the data suggests an involvement of the recycling endosome pathway and RabGTPases, Rab8 and Rab11, in ANDV release from infected cells.
Plasma membrane assembly has been noted for Sin Nombre virus (SNV) and BCCV, two New World hantaviruses (Goldsmith et al., 1995; Ravkov et al., 1997), however the lack of plasma membrane localization of ANDV N in the electron microscopy studies strongly suggests limited plasma membrane assembly. However, we could not confirm the site of ANDV assembly because no obvious structures resembling viral particles were observed by electron microscopy at high frequency. This is not surprising due to the low numbers of ANDV particles released per cell. It may also be possible that the anti-nucleocapsid sera may not recognize ribonucleocapsids (RNPs) present in budding particles and fully assembled virions. However, based on the localization of ANDV proteins, it is likely that ANDV assembles at membranes of the Golgi or trans-Golgi, similar to that observed for Hantaan virus and other viruses in Bunyaviridae (Chen et al., 1991; Hung et al., 1985; Kuismanen et al., 1982; Salanueva et al., 2003; Tao et al., 1987).
Our proposed model of ANDV assembly and egress starts with ANDV protein concentrated at the site of assembly to form new particles by budding into the host cell membrane, which is enriched in Rab8 and Rab11 thereby, facilitating the access of newly formed particles to the correct egress trafficking pathway. Egress could then occur via a Rab8- and/or Rab11-mediated mechanism which traffics virus-containing vesicles to the recycling endosome. The virus particles could then be sorted into vesicles destined for the apical or basolateral plasma membrane. It is important to note that our studies utilized non-polarized Vero cells. Intracellular trafficking in polarized versus non-polarized cells may utilize slightly different pathways so an analysis of ANDV trafficking in other relevant cell types (e.g. polarized epithelial cells, macrophages, endothelial cells) is warranted.
The recycling endosome regulates plasma membrane recycling of many cell surface receptors, such as transferrin (Ang et al., 2004; Ren et al., 1998; Schlierf et al., 2000; Sheff et al., 2002), low-density lipoprotein (LDLR) (Ang et al., 2004), and polyimmunoglobulin (pIg) receptors (Apodaca et al., 1994; Rojas and Apodaca, 2002; Sheff et al., 2002). In addition to receptor recycling, the transport of select cargo from the Golgi to the plasma membrane is also regulated by this compartment (Ang et al., 2003; Ang et al., 2004; Chen et al., 1998; Hattula et al., 2006; Ren et al., 1998). Our model proposes a similar role for the recycling endosome in the transport of ANDV particles to the plasma membrane. Both the viral nucleocapsid and glycoproteins colocalized with eGFP-Rab8 and eGFP-Rab11 when expressed from cDNA. However, the localization of the nucleocapsid protein varied from cell to cell with some cells showing high levels of colocalization and others showing minimal colocalization. This variation seemed to be related to high nucleocapsid expression levels in some cDNA transfected cells, therefore we focused our intracellular localization studies on cells that had levels of viral protein similar to that observed in ANDV-infected cells.
The expression of constitutively active and dominant negative Rab11 proteins resulted in altered colocalization of the eGFP-fusion proteins with ANDV N during infection. The dominant negative Rab11S25N protein showed an enhanced colocalization with ANDV N, localizing closely at a perinuclear location, and is similar to observations in which Rab11S25N retained VSV G intracellularly at a perinuclear site, resulting in decreased G surface expression (Chen et al., 1998). In stark contrast to Rab11S25N, the constitutively active Rab11Q70L showed a dramatic decrease in colocalization with ANDV N. It was further determined that the two mutant Rab11 proteins localize to distinct membrane compartments. Rab11S25N localized to the trans-Golgi, while Rab11Q70L showed limited localization with the trans-Golgi marker. Consistent with Rab11 function and our model of ANDV assembly and egress, the GDP-bound mutant localizes at the donor membrane (the trans-Golgi) (Chen et al., 1998; Neznanov et al., 2003) and may retain or enhance ANDV N localization to that membrane compartment. In contrast the GTP-bound mutant localizes to the acceptor membrane (the recycling endosome) and by sequestering transport machinery at the recycling endosome may reduce trafficking of important host cell proteins involved in ANDV replication or trafficking of the viral proteins or particles themselves.
By utilizing RNAi, we determined that both Rab8 and Rab11 play a functional role in ANDV release from infected cells. Downregulation of Rab8a showed minimal effects on virus secretion and was similar to control transfections, however in cells transfected with siRNAs targeting Rab11a, a 10–15 fold decrease in virus secretion was observed. Downregulation both Rab11 isoforms did not enhance the effect on ANDV release seen with Rab11a downregulation alone, suggesting that Rab11a plays the predominant role in ANDV infection. In contrast, downregulating Rab8a had little effect on ANDV secretion but reducing the expression of both isoforms led to a 10-fold decrease in virus release. This result indicates that there is either redundancy in the Rab8 pathway, or Rab8b is playing a more important role in ANDV replication. It is unlikely that the effects of siRNA downregulation of Rab8 and Rab11 on virus secretion are caused by a nonspecific block in all post-Golgi secretion, since numerous studies indicate that Rab8- and Rab11- independent pathways of Golgi to plasma membrane transport exist (Chen et al., 1998; Crespo et al., 2004; Li et al., 1993; Yoshimori et al., 1996). Consistent with our model, the siRNA studies indicate an important role for Rab8 and Rab11 in ANDV replication. Given the low level of ANDV particle formation (Fig. 2), we were unable to identify the precise step at which ANDV particle egress was blocked after treatment of infected cells with Rab8 and Rab11 specific siRNAs. The siRNA sequences used were designed to recognize human forms of Rab 8 and 11 and may not be optimal for recognizing the corresponding proteins from African green monkeys. However, we utilized four siRNAs per construct and did in fact see downregulation of some Rab isoforms, indicating that siRNA downregulation of Rab transcripts was in fact occurring. A more extensive downregulation of Rab 8 and 11 protein levels may be needed to find the relatively low numbers of virus particles that are in the intracellular trafficking pathway during any particular time after ANDV infection.
Rab11a has been shown to play a role in the replication of respiratory syncytial virus (RSV) (Brock et al., 2003), HIV-1 (Murray et al., 2005; Varthakavi et al., 2006), and another retrovirus, Mason-Pfizer monkey virus (M-PMV) (Sfakianos and Hunter, 2003). In these examples, trafficking of viral proteins through the recycling endosome is required for the proteins to reach the site of particle formation at the plasma membrane, thus the recycling endosome is functioning prior to virus assembly. Although the experiments we performed have not formally ruled out a role for Rab8 and Rab11 in ANDV assembly or budding, Rab8- and Rab11-coated vesicles bud away from the membrane compartment into the cytoplasm and therefore have a different topology from that required to promote virus assembly into the Golgi lumen. Alternatively, disruption of Rab8 and Rab11 pathways could disrupt the localization of a host protein critical for virus assembly, and further studies will need to be performed to determine this possibility. Since ANDV assembly is hypothesized to occur at the Golgi, Rab8 and Rab11 are presumably altering trafficking steps that occur after virus assembly. This leads us to propose a model in which Rab8 and Rab11 are playing roles in steps of egress, downstream of virus assembly. To our knowledge, a role for the recycling endosome in post-assembly trafficking of viruses to the plasma membrane has not been described to date.
Hantaviruses may be utilizing the recycling endosome, and its function in polarized trafficking, to direct particles from the host cell in a directional fashion. Infection of polarized epithelial cells with viruses in the family Bunyaviridae, including ANDV, leads to the polarized release of virus particles. This release can be strictly apical, strictly basolateral, or a combination of both (Chen et al., 1991; Connolly-Andersen et al., 2007; Gerrard et al., 2002; Ravkov et al., 1997; Rowe and Pekosz, 2006) Since Rab8 and Rab11 play roles in polarized trafficking (Ang et al., 2003; Ang et al., 2004 ; Brock et al., 2003), it is possible that ANDV utilizes these pathways to egress in a polarized fashion from cells. By utilizing the recycling endosome during egress, hantaviruses could be sorted in both the apical and basolateral direction and ensure appropriate viral spread and tissue tropism within the host.
Materials and Methods
Reagents and antibodies
Anti-ANDV human convalescent sera (1:150 immunofluorescence) was kindly provided by Stephen St. Jeor (University of Nevada, Reno, NV). Rabbit anti-ANDV N sera (1:500 immunofluorescence) was kindly provided by Colleen Jonsson (Southern Research Institute, Birmingham, AL). A rat anti-ANDV N sera was raised by immunizing rats with purified, bacterially expressed glutathione-S-transferase (GST)-ANDV N fusion protein (1:1500 immunofluorescence, 1:5000 western blotting and immune TCID50, cryoelectron microscopy 1:2000). Other primary antibodies were used as follows: mouse anti-GM130 (1:100 immunofluorescence, BDBiosciences, San Jose, CA); mouse anti-Golgin97 (1:50 immunofluorescence, Molecular Probes/Invitrogen); sheep anti-TGN46 (1:250 immunofluorescence, Serotec, Oxford, UK); mouse anti-Rab8 and mouse anti-Rab11 (1:100 immunofluorescence; 1:1000 western blotting; BDBiosciences, San Jose, CA); rabbit anti-vesicular stomatitis virus glycoprotein (VSV G) (1:100 immunofluorescence, Bethyl Laboratories, Montgomery, TX); rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1000 western blotting; Cell Signaling, Danvers, MA), and mouse anti-β-actin (1:15,000 western blotting, AbCam, Cambridge, MA). Secondary antibodies were used as follows: goat anti-rat Alexa Fluor 594, goat anti-rat Alexa Fluor 555, goat anti-mouse Alexa Fluor 488, goat anti-mouse Alexa Fluor 594, goat anti-human Alexa Fluor 633, and goat anti-rabbit Alexa Fluor 594, donkey anti-sheep Alexa Fluor 594 (1:500 immunofluorescence, Molecular Probes/Invitrogen, Carlsbad, CA); goat anti-rat horseradish peroxidase (HRP) (1:7500 western blot, 1:5000 immune TCID50), goat anti-mouse HRP and goat anti-rabbit HRP (1:7500 western blot, Jackson Immunoresearch, Westgrove, PA).
ANDV infection
Vero E6 cells (African green monkey kidney epithelial cells, American Type Culture Collection) were cultured as previously described and used under conditions were the cells were subconfluent and therefore not polarized (Rowe and Pekosz, 2006). Cells were infected with Andes virus, strain 9717869 (ANDV, courtesy of Stuart Nichol, Centers for Disease Control, Atlanta, GA) as previously described (Rowe and Pekosz, 2006). Infected cell supernatants were collected at the indicated times post infection or transfection and stored at −70°C until further analysis. All infections were performed using institution-approved biosafety level 3 (BSL3) containment procedures.
Immuno-electron microscopy
For ultrastructural analysis, ANDV-infected Vero cells were fixed at the indicated days post infection in 4% paraformaldehyde, 0.1% glutaraldehyde in (Polysciences, Warrington, PA) in 100mM PIPES, pH 7.2 with 0.5mM MgCl2 for 1h at 4°C. Samples were embedded in 10% gelatin and infiltrated with 2.3M sucrose/20% polyvinyl pyrrolidone in PIPES/MgCl2 overnight at 4°C. Samples were frozen in liquid nitrogen and 70 nm sections were cut using a Leica Ultracut UCT cryo-ultramicrotome (Leica Microsystems, Bannockburn, IL). Sections were blocked with 5% fetal bovine serum and 5% goat serum (blocking buffer) for 30 minutes and then incubated with primary antibody for 1h at room temperature. Sections were washed for 30 minutes in blocking buffer and probed with 12 nm colloidal gold-conjugated goat anti-rat secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1h at room temperature. Sections were washed in PIPES buffer, followed by an extensive water rinse and stained with 1% uranyl acetate and 1.6% methyl cellulose (Ted Pella, Redding, CA). Samples were visualized with a JEOL 1200EX transmission electron microscope (JEOL USA, Peabody, MA) (Beatty, 2006).
Immune TCID50 assay
Serial 5-fold dilutions of infected cell supernatants were made in Dulbecco’s modified Eagles medium (DMEM) containing 2% fetal bovine sera (FBS, Atlanta Biologicals, Atlanta, GA). Confluent monolayers of Vero cells in 96-well dishes were infected with 100μl/well in sextuplicate. The infected cells were incubated at 37°C, 5% CO2 for 7 days at which time cells were washed 1x with PBS and fixed with cold 95% ethanol/5% acetic acid solution. For immunostaining, cells were rehydrated with phosphate buffered saline (PBS) and incubated in 3% bovine serum albumin (BSA, Calbiochem, San Diego, CA) in PBS (blocking buffer). Cells were incubated with rat anti-ANDV N antibody diluted in blocking buffer for 2h at room temperature. Cells were washed with PBS and incubated with secondary antibody goat anti-rat HRP for 2h at room temperature. Cells were washed in PBS and nucleocapsid positive wells were visualized by addition of soluble HRP substrate (DakoCytomation, Carpenteria, CA). Titers were calculated using the Reed-Muench formula (Reed and Muench, 1938).
Plasmids
The ANDV N open reading frame was amplified from ANDV vRNA and cloned into the pCAGGS (Niwa et al., 1991) and pCDNA3.1(+) (Invitrogen, Carlsbad, CA) mammalian expression vectors. Briefly, total cellular RNA was isolated from ANDV-infected Vero cells using the Qiagen cellular RNA mini prep kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). Using ANDV N ORF specific primers (Genebank accession: AF291702; forward primer: nucleotides (nts) 37–54 and a SacI restriction enzyme site; reverse primer: nts 1312–1330 and a SphI restriction enzyme site), the N ORF was amplified by reverse transcriptase polymerase chain reaction (RT-PCR) using Superscript One-step RT-PCR reagent (Invitrogen), digested using SacI and SphI restriction endonucleases (New England Biolabs, Ipswich, MA), and cloned into the identically digested pCAGGS vector. The ANDV N fragment was subcloned from the resulting plasmid into the pCDNA3.1(+) vector using EcoRI and XhoI restriction enzymes. The ANDV M segment ORF was amplified using M segment specific primers (Genebank accession: AF291703; forward primer: nts 46–63; reverse primer nts 3449- 3469 and a KpnI restriction enzyme site), cloned into the pCR4-Blunt-TOPO vector (Invitrogen) according to the manufacturer’s instructions. The M ORF fragment was excised using NotI and KpnI restriction endonucleases and this fragment was ligated to an EcoRI and KpnI digested pCAGGS vector after the EcoRI and NotI sites were blunt ended by a Klenow fill-in reaction. The sequences of both N and M ORF cDNAs were verified by DNA sequencing. The CD63-eGFP cDNA was kindly provided by David Sibley (Washington University, St. Louis, MO). The eGFP-Vps4A cDNA was kindly provided by Wes Sundquist (University of Utah, Salt Lake City, UT). The eGFP-Rab5a, eGFP-Rab7, eGFP-Rab11a, eGFP-Rab11S25N, and eGFP-Rab11Q70L, and Rab9 cDNAs were kindly provided by Phil Stahl (Washington University, St. Louis, MO). The VSV G cDNA was kindly provided by Garry Nolan (Stanford University, Palo Alto, CA). To construct the eGFP-Rab9 expression plasmid, Rab9 cDNA was amplified by PCR using canine Rab9 sequence specific primers (Genebank accession: X56386; forward primer: nts 1–28 and a XhoI restriction enzyme site; reverse primer: nts 585–606 and a KpnI restriction enzyme site) and cloned in frame with the eGFP ORF into the eGFP-C1 vector (Clontech, Mountain View, CA). The Rab8a cDNA was kindly provided by Ira Mellman (Yale University, New Haven CT). To construct N-terminal eGFP-fusions of Rab8a, canine Rab8 specific primers (Genebank accession: X56385; forward primer: nts 13–32 and containing a EcoRI restriction enzyme site; reverse primer: nts 613–633 and containing a XhoI restriction enzyme site) were used to amplify the Rab8 open reading frame by PCR before cloning into an EcoRI and SalI digested eGFP-C1 vector in frame with the eGFP ORF. The Rab8 dominant negative and constitutively active mutations were created using a four-primer PCR method. The Rab8T22N (GDP-bound) mutation was made using the following mutagenesis primers in addition to the outer primers above described to amplify the appropriate product from eGFP-Rab8wt: 5′ primer, 5′– GTGGGGAAGAATTGTGTCCTGTTC-3′; 3′ primer, 5′-GAACAGGACACAATTCTTCCCCAC-3′. The Rab8Q67L (GTP-bound mutation) was made using the following mutagenesis primers: 5′ primer, 5′-CACAGCTGGTCTAGAACGGTTT-3′; 3′ primer, 5′-AAACCGTTCTAGACCAGCTGTG-3′. The amplified fragments were cloned into the eGFP-C1 vector. All PCR amplified products and mutations were confirmed by sequencing.
DNA transfections
Vero cells were seeded at 7.5×104 cells/well on coverslips in 12-well dishes one day prior to transfection. Growth media was replaced with 0.5ml OptiMEM media (Invitrogen) prior to transfection. LT1 transfection reagent (Mirus, Madison, WI) was diluted into 100μL of OptiMEM, mixed and incubated for 15 minutes at room temperature. DNA was added to the LT1 solution, mixed and incubated for 15 minutes at room temperature. The cells were transfected with 1μg of DNA at a ratio of 3 μL LT1 to 1μg of DNA. For cotransfections, 0.5μg of each plasmid was used. The transfection mixture was added to each well, followed by 0.5ml of growth media at 4 hours post transfection (hpt). At 18–24 hpt, cells were washed 1x in PBS and fixed for 10 minutes in 2% paraformaldehyde in PBS. For ANDV infected cell transfections, cells were infected at MOI=0.5 and detached from the tissue culture dish at 72 hours post infection (hpi) with 1x trypsin-EDTA. Cells were pelleted at 300×g for 5 minutes, resuspended in growth media, and seeded on glass coverslips in a 12-well dish at a density of 1×105 cells/well. At 24 hours post seeding, cells were transfected as above described.
Immunofluorescence confocal microscopy
At the indicated time post infection or transfection cells were washed with PBS and fixed in 2% paraformaldehyde in PBS for 10 minutes at room temperature. After fixation, cells were washed extensively with PBS and permeabilized with PBS containing 0.2% TX-100 and 0.1% sodium citrate for 10 minutes at room temperature. The cells were incubated with PBS containing 3% normal goat or donkey serum and 0.5% BSA (blocking buffer) for 30 minutes at room temperature. Cells were incubated with primary antibodies followed by secondary antibodies. All antibodies were diluted in blocking buffer and incubation times were 1h at room temperature. Nuclei were stained with TO-PRO-3 (Molecular Probes/Invitrogen) for 15 minutes at room temperature. All washes were performed with PBS. Coverslips were mounted with Molecular Probes Prolong Gold antifade (Molecular Probes/Invitrogen). Cells were visualized using a Zeiss LSM 510 Meta confocal microscope. Z-stacks were acquired at a 63x magnification, 1024×1024 pixel resolution, using 2x digital zoom (unless otherwise noted), 0.7μm optical slice, and 0.3μm slice overlap.
Volocity quantitation of confocal microscopy
The amount of colocalization in the confocal z-stacks was quantitated using the Volocity (v3.0) imaging software (Improvision, Lexington, MA). Images were cropped to obtain the single cell to be quantitated. Classifiers were designed to measure three dimensional (3-D) regions according to signal intensity and region size. 3-D regions of intensity were measured using the following classifier parameters: intensity, >3 standard deviations above the mean (for eGFP signal), and >2 standard deviations above the mean for ANDV N; volume, >0.01μm3; separation of touching objects; and noise reduction. Classifiers were applied to measure each channel to be colocalized. The measurement calculations for each channel were colocalized using the “colocalize sessions” option to overlay the two data sets and measure the regions of colocalization. All regions less than 10 voxels were removed and the total volume of colocalization was then normalized to the total volume measured for eGFP and expressed as percent colocalization.
Small interfering RNA transfection
Small interfering RNA (siRNA) duplexes were purchased from Dharmacon (Lafayette, CO) and reconstituted at 20μM stock concentrations according to the manufacturer’s instructions. The siRNAs are designed against human Rab 8 and 11 isoforms and four siRNAs per transcript were used. Vero cells were infected in 3.5cm2 dishes with ANDV at MOI=0.5 and at 72 hpi, cells were detached from the tissue culture dish with 1x trypsin-EDTA and pelleted at 300 × g for 5 minutes. Cells were resuspended in growth media and seeded into 24-well dishes at a density of 5×104 cells/well. At 24h post seeding (considered the 0h timepoint), the cells were transfected with the siRNA duplexes using siQuest reagent (Mirus). For each well, 1.5μL of siQuest reagent was diluted in 50μL of OptiMEM media, mixed and incubated for 10 minutes at room temperature. The siRNAs were added to the siQuest solution at a final concentration of 50–100nM, mixed and incubated for 30 minutes at room temperature. The infected-cell supernatant was harvested (0h timepoint) and 250μL of growth media was added to each well. The transfection mixture was added to the cells and transfections were incubated at 37°C, 5% CO2. Supernatants and whole cell lysates were harvested at 48 hpt. Supernatants were titered by immune TCID50 and the fold reduction in titer was determined by dividing the titer at 0 hpt by the titer at 48 hpt (0h TCID50 units/ml)/(48h TCID50 units/ml). Virus titers were determined for duplicate siRNA transfections in four independent experiments.
SDS-PAGE and western blotting
Cells were washed once in PBS and lysed in 1% sodium dodecyl sulfate (SDS). Lysates were run through a 22-guage needle, sonicated for 15 minutes, and diluted in 2x SDS loading buffer (Paterson and Lamb, 1993). Samples were boiled for 15 minutes and run on a 15% polyacrylamide mini gel (BioRad, Hercules, CA) at 150 volts (V) through the stacking gel followed by an additional 2 hours at 100V. Proteins were transferred to PVDF membrane (Immobilon-FL, Millipore, Billerica, MA) overnight at 22V (Mini Transblot, BioRad). Membranes were blocked in 5% dry milk and incubated sequentially in primary and secondary antibodies for 1 hour at room temperature. Washes were performed in PBS supplemented with 0.3% Tween20. HRP-conjugated secondary antibodies were detected using the ECL-Plus substrate (Amersham, Buckinghamshire, UK). Protein band intensities were quantitated by phosphorimager analysis (Fuji Film FLA3000, Tokyo, Japan).
Supplementary Material
ANDV proteins colocalize with Rab8 and Rab11 when expressed from cDNA. Vero cells were transfected with cDNAs expressing either (A and B) eGFP-Rab8 or (D and E) eGFP-Rab11 in combination with (A and D) ANDV N (red, AlexaFluor 555) or (B and E) ANDV glycoproteins (M) (red, AlexaFluor 633). As a positive control for localization with these two Rabs, Vero cells were transfected with cDNAs expressing (C and F) VSV G (red, AlexaFluor 594) and (C) eGFP-Rab8 or (F) eGFP-Rab11. The cells were fixed at 18 hpt, immunostained for the viral proteins, and visualized by confocal microscopy. Images are flattened reconstructions of confocal z-stacks that were acquired at 63x optical magnification and 2x digital zoom.
Acknowledgments
This work was supported by Public Health Service grant R21 AI058353, The Eliasberg Foundation and The Marjorie Gilbert Foundation. We acknowledge the Molecular Microbiology Imaging Facility for technical support and all the members of the Pekosz laboratory for insightful comments and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed R, Oldstone MBA, Palese P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat Immunol. 2007;8(11):1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang AL, Folsch H, Koivisto U-M, Pypaert M, Mellman I. The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin-Darby canine kidney cells. J Cell Biol. 2003;163(2):339–350. doi: 10.1083/jcb.200307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang AL, Taguchi T, Francis S, Folsch H, Murrells LJ, Pypaert M, Warren G, Mellman I. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol. 2004;167(3):531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca G, Katz LA, Mostov KE. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J Cell Biol. 1994;125(1):67–86. doi: 10.1083/jcb.125.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbero P, Bittova L, Pfeffer SR. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J Cell Biol. 2002;156(3):511–518. doi: 10.1083/jcb.200109030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty WL. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J Cell Sci. 2006;119(Pt 2):350–9. doi: 10.1242/jcs.02733. [DOI] [PubMed] [Google Scholar]
- Brock SC, Goldenring JR, Crowe JE., Jr Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. PNAS. 2003 doi: 10.1073/pnas.2434327100. 2434327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE, Wang X, Kumar R, Bhartur SG, Navarre J, Woodrum JE, Altschuler Y, Ray GS, Goldenring JR. Association of Rab25 and Rab11a with the Apical Recycling System of Polarized Madin-Darby Canine Kidney Cells. Mol Biol Cell. 1999;10(1):47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabrillat ML, Wilhelm C, Wasmeier C, Sviderskaya EV, Louvard D, Coudrier E. Rab8 Regulates the Actin-based Movement of Melanosomes. Mol Biol Cell. 2005;16(4):1640–1650. doi: 10.1091/mbc.E04-09-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Matsuoka Y, Compans RW. Assembly and polarized release of Punta Toro virus and effects of brefeldin A. J Virol. 1991;65(3):1427–39. doi: 10.1128/jvi.65.3.1427-1439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Feng Y, Chen D, Wandinger-Ness A. Rab11 Is Required for Trans-Golgi Network-to-Plasma Membrane Transport and a Preferential Target for GDP Dissociation Inhibitor. Mol Biol Cell. 1998;9(11):3241–3257. doi: 10.1091/mbc.9.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly-Andersen AM, Magnusson KE, Mirazimi A. Basolateral Entry and Release of Crimean-Congo Hemorrhagic Fever Virus in Polarized MDCK-1 Cells. J Virol. 2007;81(5):2158–64. doi: 10.1128/JVI.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo PM, Iglesias-Bartolome R, Daniotti JL. Ganglioside GD3 traffics from the trans-Golgi network to plasma membrane by a Rab11-independent and brefeldin A-insensitive exocytic pathway. J Biol Chem. 2004;279(46):47610–8. doi: 10.1074/jbc.M407181200. [DOI] [PubMed] [Google Scholar]
- Deyde VM, Rizvanov AA, Chase J, Otteson EW, St Jeor SC. Interactions and trafficking of Andes and Sin Nombre Hantavirus glycoproteins G1 and G2. Virology. 2005;331(2):307–315. doi: 10.1016/j.virol.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Ding W, Zhang LN, Yeaman C, Engelhardt JF. rAAV2 traffics through both the late and the recycling endosomes in a dose-dependent fashion. Mol Ther. 2006;13(4):671–82. doi: 10.1016/j.ymthe.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elliott LH, Ksiazek TG, Rollin PE, Spiropoulou C, Morzunov S, Monroe M, Goldsmith C, Humphrey C, Zaki SR, Krebs JW, Maupin G, Gage K, Childs JE, Nichol ST, Peters CJ. Isolation of the causative agent of hantavirus pulmonary syndrome. Am J Trop Med Hyg. 1994;51(1):102–108. doi: 10.4269/ajtmh.1994.51.102. [DOI] [PubMed] [Google Scholar]
- Galeno H, Mora J, Villagra E, Fernandez J, Hernandez J, Mertz GJ, Ramirez E. First human isolate of Hantavirus (Andes virus) in the Americas. Emerg Infect Dis. 2002;8(7):657–61. doi: 10.3201/eid0807.010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppert M, Bolshakov VY, Siegelbaum SA, Takei K, De Camilli P, Hammer RE, Sudhof TC. The role of Rab3A in neurotransmitter release. Nature. 1994;369(6480):493–7. doi: 10.1038/369493a0. [DOI] [PubMed] [Google Scholar]
- Geppert M, Goda Y, Stevens CF, Sudhof TC. The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature. 1997;387(6635):810–4. doi: 10.1038/42954. [DOI] [PubMed] [Google Scholar]
- Gerrard SR, Rollin PE, Nichol ST. Bidirectional Infection and Release of Rift Valley Fever Virus in Polarized Epithelial Cells. Virology. 2002;301(2):226–235. doi: 10.1006/viro.2002.1588. [DOI] [PubMed] [Google Scholar]
- Goldsmith CS, Elliott LH, Peters CJ, Zaki SR. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch Virol. 1995;140(12):2107–22. doi: 10.1007/BF01323234. [DOI] [PubMed] [Google Scholar]
- Gomez-Puertas P, Mena I, Castillo M, Vivo A, Perez-Pastrana E, Portela A. Efficient formation of influenza virus-like particles: dependence on the expression levels of viral proteins. J Gen Virol. 1999;80(Pt 7):1635–45. doi: 10.1099/0022-1317-80-7-1635. [DOI] [PubMed] [Google Scholar]
- Greaves J, Chamberlain LH. Palmitoylation-dependent protein sorting. J Cell Biol. 2007;176(3):249–254. doi: 10.1083/jcb.200610151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. PNAS. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Crump CM, Thomas G. Trans-Golgi network sorting. Cell Mol Life Sci. 2001;58(8):1067–84. doi: 10.1007/PL00000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardestam J, Klingstrom J, Mattsson K, Lundkvist A. HFRS causing hantaviruses do not induce apoptosis in confluent Vero E6 and A-549 cells. J Med Virol. 2005;76(2):234–40. doi: 10.1002/jmv.20347. [DOI] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpaa K, Laakkonen P, Peranen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J Cell Sci. 2006;119(23):4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- Hooper JW, Larsen T, Custer DM, Schmaljohn CS. A lethal disease model for hantavirus pulmonary syndrome. Virology. 2001;289(1):6–14. doi: 10.1006/viro.2001.1133. [DOI] [PubMed] [Google Scholar]
- Hung T, Chou ZY, Zhao TX, Xia SM, Hang CS. Morphology and morphogenesis of viruses of hemorrhagic fever with renal syndrome (HFRS). I. Some peculiar aspects of the morphogenesis of various strains of HFRS virus. Intervirology. 1985;23(2):97–108. doi: 10.1159/000149591. [DOI] [PubMed] [Google Scholar]
- Izumi T, Gomi H, Kasai K, Mizutani S, Torii S. The roles of Rab27 and its effectors in the regulated secretory pathways. Cell Struct Funct. 2003;28(5):465–74. doi: 10.1247/csf.28.465. [DOI] [PubMed] [Google Scholar]
- Jantti J, Hilden P, Ronka H, Makiranta V, Keranen S, Kuismanen E. Immunocytochemical analysis of Uukuniemi virus budding compartments: role of the intermediate compartment and the Golgi stack in virus maturation. J Virol. 1997;71(2):1162–72. doi: 10.1128/jvi.71.2.1162-1172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Park J, Lee S, Park B, Shin J, Song KJ, Ahn TI, Hwang SY, Ahn BY, Ahn K. Hantaan virus enters cells by clathrin-dependent receptor-mediated endocytosis. Virology. 2002;294(1):60–9. doi: 10.1006/viro.2001.1303. [DOI] [PubMed] [Google Scholar]
- Jones JC, Turpin EA, Bultmann H, Brandt CR, Schultz-Cherry S. Inhibition of Influenza Virus Infection by a Novel Antiviral Peptide That Targets Viral Attachment to Cells. J Virol. 2006;80(24):11960–11967. doi: 10.1128/JVI.01678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukinen P, Vaheri A, Plyusnin A. Hantavirus nucleocapsid protein: a multifunctional molecule with both housekeeping and ambassadorial duties. Arch Virol. 2005;150(9):1693–713. doi: 10.1007/s00705-005-0555-4. [DOI] [PubMed] [Google Scholar]
- Khan AS, Ksiazek TG, Peters CJ. Hantavirus pulmonary syndrome. The Lancet. 1996;347(9003):739–741. doi: 10.1016/s0140-6736(96)90082-3. [DOI] [PubMed] [Google Scholar]
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, the SARS Working Group A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Kuismanen E, Hedman K, Saraste J, Pettersson RF. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol Cell Biol. 1982;2(11):1444–58. doi: 10.1128/mcb.2.11.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Stubbs L, Artzt K. Molecular analysis of mouse Rab11b: a new type of mammalian YPT/Rab protein. Genomics. 1994;22(3):610–6. doi: 10.1006/geno.1994.1434. [DOI] [PubMed] [Google Scholar]
- Lai MMC. In: Coronaviridae: The Viruses and Their Replication. Fields Virology. 4. Knipe DM, Howley PM, editors. Lippincott Williams and Wilkins; Philadelphia Pa: 2001. pp. 1163–1186. [Google Scholar]
- Lau AS, Mruk DD. Rab8B GTPase and junction dynamics in the testis. Endocrinology. 2003;144(4):1549–63. doi: 10.1210/en.2002-220893. [DOI] [PubMed] [Google Scholar]
- LeBouder F, Morello E, Rimmelzwaan GF, Bosse F, Pechoux C, Delmas B, Riteau B. Annexin II Incorporated into Influenza Virus Particles Supports Virus Replication by Converting Plasminogen into Plasmin. J Virol. 2008;82(14):6820–6828. doi: 10.1128/JVI.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Lee PW, Johnson KM. Isolation of the etiologic agent of Korean Hemorrhagic fever. J Infect Dis. 1978;137(3):298–308. doi: 10.1093/infdis/137.3.298. [DOI] [PubMed] [Google Scholar]
- Li Y, Luo L, Schubert M, Wagner RR, Kang CY. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J Virol. 1993;67(7):4415–20. doi: 10.1128/jvi.67.7.4415-4420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. RNAi targeting an animal virus: news from the front. Mol Cell. 2002;9(5):925–7. doi: 10.1016/s1097-2765(02)00539-7. [DOI] [PubMed] [Google Scholar]
- Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. Embo J. 1993;12(2):677–82. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Miki T, Shibasaki T, Kawaguchi M, Shinozaki H, Nio J, Saraya A, Koseki H, Miyazaki M, Iwanaga T, Seino S. Noc2 is essential in normal regulation of exocytosis in endocrine and exocrine cells. PNAS. 2004;101(22):8313–8318. doi: 10.1073/pnas.0306709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC. Budding events in herpesvirus morphogenesis. Virus Res. 2004;106(2):167–80. doi: 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Meyer BJ, Schmaljohn CS. Persistent hantavirus infections: characteristics and mechanisms. Trends Microbiol. 2000;8(2):61–7. doi: 10.1016/s0966-842x(99)01658-3. [DOI] [PubMed] [Google Scholar]
- Mills JN, Corneli A, Young JC, Garrison LE, Khan AS, Ksiazek TG. Hantavirus pulmonary syndrome--United States: updated recommendations for risk reduction. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2002;51(RR9):1–12. [PubMed] [Google Scholar]
- Moss B. Poxviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4. Lippincott Williams and Wilkins; Philidelphia Pa: 2001. pp. 2849–2883. [Google Scholar]
- Murray JL, Mavrakis M, McDonald NJ, Yilla M, Sheng J, Bellini WJ, Zhao L, Le Doux JM, Shaw MW, Luo C-C, Lippincott-Schwartz J, Sanchez A, Rubin DH, Hodge TW. Rab9 GTPase Is Required for Replication of Human Immunodeficiency Virus Type 1, Filoviruses, and Measles Virus. J Virol. 2005;79(18):11742–11751. doi: 10.1128/JVI.79.18.11742-11751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neznanov N, Neznanova L, Kondratov RV, Burdelya L, Kandel ES, O’Rourke DM, Ullrich A, Gudkov AV. Dominant Negative Form of Signal-regulatory Protein-alpha (SIRPalpha/SHPS-1) Inhibits Tumor Necrosis Factor-mediated Apoptosis by Activation of NF-kappa B. J Biol Chem. 2003;278(6):3809–3815. doi: 10.1074/jbc.M210698200. [DOI] [PubMed] [Google Scholar]
- Nichol ST. Bunyaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 4. Lippincott Williams and Wilkins; Philidelphia Pa: 2001. pp. 1603–1634. [Google Scholar]
- Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262(5135):914–7. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Dudziak D, Dirmeier U, Hobom G, Riedel A, Schlee M, Staudt LM, Rosenwald A, Behrends U, Bornkamm GW, Mautner J. Active NF-{kappa}B signalling is a prerequisite for influenza virus infection. J Gen Virol. 2004;85(8):2347–2356. doi: 10.1099/vir.0.79958-0. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–9. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Ochsenbauer-Jambor C, Miller DC, Roberts CR, Rhee SS, Hunter E. Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J Virol. 2001;75(23):11544–54. doi: 10.1128/JVI.75.23.11544-11554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padula PJ, Edelstein A, Miguel SDL, Lopez NM, Rossi CM, Rabinovich RD. Hantavirus Pulmonary Syndrome Outbreak in Argentina: Molecular Evidence for Person-to-Person Transmission of Andes Virus. Virology. 1998;241(2):323–330. doi: 10.1006/viro.1997.8976. [DOI] [PubMed] [Google Scholar]
- Paterson RG, Lamb RA. Molecular Virology: A Practical Approach. Oxford University Press; Oxford, U.K: 1993. The molecular biology of influenza viruses and paramyxoviruses; pp. 35–73. [Google Scholar]
- Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5(11):886–96. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Constructing a Golgi complex. J Cell Biol. 2001;155(6):873–876. doi: 10.1083/jcb.200109095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan HN, Chung DH, Plane SJ, Sztul E, Chu YK, Guttieri MC, McDowell M, Ali G, Jonsson CB. Dynein-dependent transport of the hantaan virus nucleocapsid protein to the endoplasmic reticulum-Golgi intermediate compartment. J Virol. 2007;81(16):8634–47. doi: 10.1128/JVI.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauma T, Tuukkanen J, Bergelson JM, Denning G, Hautala T. rab5 GTPase Regulates Adenovirus Endocytosis. J Virol. 1999;73(11):9664–9668. doi: 10.1128/jvi.73.11.9664-9668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravkov E, Nichol S, Compans R. Polarized entry and release in epithelial cells of Black Creek Canal virus, a New World hantavirus. J Virol. 1997;71(2):1147–1154. doi: 10.1128/jvi.71.2.1147-1154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravkov EV, Compans RW. Hantavirus Nucleocapsid Protein Is Expressed as a Membrane-Associated Protein in the Perinuclear Region. J Virol. 2001;75(4):1808–1815. doi: 10.1128/JVI.75.4.1808-1815.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating 50 percent endpoints. Am J Hyg. 1938;27:493–499. [Google Scholar]
- Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini DD. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc Natl Acad Sci U S A. 1998;95(11):6187–92. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, Apodaca G. Immunoglobulin transport across polarized epithelial cells. Nat Rev Mol Cell Biol. 2002;3(12):944–55. doi: 10.1038/nrm972. [DOI] [PubMed] [Google Scholar]
- Roquemore EP, Banting G. Efficient trafficking of TGN38 from the endosome to the trans-Golgi network requires a free hydroxyl group at position 331 in the cytosolic domain. Mol Biol Cell. 1998;9(8):2125–44. doi: 10.1091/mbc.9.8.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RK, Pekosz A. Bidirectional Virus Secretion and Nonciliated Cell Tropism following Andes Virus Infection of Primary Airway Epithelial Cell Cultures. J Virol. 2006;80(3):1087–1097. doi: 10.1128/JVI.80.3.1087-1097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanueva IJ, Novoa RR, Cabezas P, Lopez-Iglesias C, Carrascosa JL, Elliott RM, Risco C. Polymorphism and Structural Maturation of Bunyamwera Virus in Golgi and Post-Golgi Compartments. J Virol. 2003;77(2):1368–1381. doi: 10.1128/JVI.77.2.1368-1381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Peranen J, Simons K. N-glycans as apical sorting signals in epithelial cells. Nature. 1995;378(6552):96–8. doi: 10.1038/378096a0. [DOI] [PubMed] [Google Scholar]
- Schlierf B, Fey GH, Hauber J, Hocke GM, Rosorius O. Rab11b Is Essential for Recycling of Transferrin to the Plasma Membrane. Experimental Cell Research. 2000;259(1):257–265. doi: 10.1006/excr.2000.4947. [DOI] [PubMed] [Google Scholar]
- Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3(2):95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Resistance of cell membranes to different detergents. PNAS. 2003;100(10):5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos JN, Hunter E. M-PMV Capsid Transport Is Mediated by Env/Gag Interactions at the Pericentriolar Recycling Endosome. Traffic. 2003;4(10):671–680. doi: 10.1034/j.1600-0854.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- Sheff DR, Kroschewski R, Mellman I. Actin dependence of polarized receptor recycling in madin-darby canine kidney cell endosomes. Mol Biol Cell. 2002;13(1):262–75. doi: 10.1091/mbc.01-07-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Elliott RM. Golgi localization of Hantaan virus glycoproteins requires coexpression of G1 and G2. Virology. 2002;300(1):31–8. doi: 10.1006/viro.2002.1414. [DOI] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR. Differential Requirements of Rab5 and Rab7 for Endocytosis of Influenza and Other Enveloped Viruses. Traffic. 2003;4(5):333–343. doi: 10.1034/j.1600-0854.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- Slimane TA, Trugnan G, van IJzendoorn SCD, Hoekstra D. Raft-mediated Trafficking of Apical Resident Proteins Occurs in Both Direct and Transcytotic Pathways in Polarized Hepatic Cells: Role of Distinct Lipid Microdomains. Mol Biol Cell. 2003;14(2):611–624. doi: 10.1091/mbc.E02-08-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GL, Law M. The exit of vaccinia virus from infected cells. Virus Res. 2004;106(2):189–97. doi: 10.1016/j.virusres.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149(4):901–14. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2(5):reviews3007.1–3007.7. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S, Reichelt M, Krijnse-Locker J, Mackenzie J, Simpson JC, Haller O, Kochs G. Interferon-Induced, Antiviral Human MxA Protein Localizes to a Distinct Subcompartment of the Smooth Endoplasmic Reticulum. Journal of Interferon & Cytokine Research. 2006;26(9):650–660. doi: 10.1089/jir.2006.26.650. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I Interferons in Host Defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Su X, Lodhi IJ, Saltiel AR, Stahl PD. Insulin-stimulated Interaction between insulin receptor substrate 1 and p85alpha and activation of protein kinase B/Akt require Rab5. J Biol Chem. 2006;281(38):27982–90. doi: 10.1074/jbc.M602873200. [DOI] [PubMed] [Google Scholar]
- Tao H, Xia SM, Chan ZY, Song G, Yanagihara R. Morphology and morphogenesis of viruses of hemorrhagic fever with renal syndrome. II. Inclusion bodies--ultrastructural markers of hantavirus-infected cells. Intervirology. 1987;27(1):45–52. doi: 10.1159/000149714. [DOI] [PubMed] [Google Scholar]
- Toro J, Vega JD, Khan AS, Mills JN, Padula P, Terry W, Yadón Z, Valderrama R, Ellis BA, Carlos Pavletic Cerda R, Zaki S, Wun-Ju S, Meyer R, Tapia M, Mansilla C, Baro M, Vergara JA, Concha M, Calderon G, Enria D, Peters CJ, Ksiazek TG. An Outbreak of Hantavirus Pulmonary Syndrome, Chile, 1997. Emerging Infectious Diseases. 1998;4(4):687–94. doi: 10.3201/eid0404.980425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trischler M, Stoorvogel W, Ullrich O. Biochemical analysis of distinct Rab5- and Rab11-positive endosomes along the transferrin pathway. J Cell Sci. 1999;112(Pt 24):4773–83. doi: 10.1242/jcs.112.24.4773. [DOI] [PubMed] [Google Scholar]
- van Weeren L, de Graaff AM, Jamieson JD, Batenburg JJ, Valentijn JA. Rab3D and Actin Reveal Distinct Lamellar Body Subpopulations in Alveolar Epithelial Type II Cells. Am J Respir Cell Mol Biol. 2004;30(3):288–295. doi: 10.1165/rcmb.2003-0264OC. [DOI] [PubMed] [Google Scholar]
- Varthakavi V, Smith RM, Martin KL, Derdowski A, Lapierre LA, Goldenring JR, Spearman P. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic. 2006;7(3):298–307. doi: 10.1111/j.1600-0854.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- Vidricaire G, Tremblay MJ. Rab5 and Rab7, but Not ARF6, Govern the Early Events of HIV-1 Infection in Polarized Human Placental Cells. J Immunol. 2005;175(10):6517–6530. doi: 10.4049/jimmunol.175.10.6517. [DOI] [PubMed] [Google Scholar]
- Vonderheit A, Helenius A. Rab7 Associates with Early Endosomes to Mediate Sorting and Transport of Semliki Forest Virus to Late Endosomes. PLoS Biology. 2005;3(7):e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis BC, Kim KJ, Li X, Liebler J, Crandall ED, Borok Z. Modulation of ion conductance and active transport by TGF-{beta}1 in alveolar epithelial cell monolayers. Am J Physiol Lung Cell Mol Physiol. 2003;285(6):L1192–1200. doi: 10.1152/ajplung.00379.2002. [DOI] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. PNAS. 2002;99(25):15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Keller P, Roth M, Simons K. Different biosynthetic transport routes to the plasma membrane in BHK and CHO cells. J Cell Biol. 1996;133(2):247–256. doi: 10.1083/jcb.133.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino A, Bieler BM, Harper DC, Cowan DA, Sutterwala S, Gay DM, Cole NB, McCaffery JM, Marks MS. A role for GRIP domain proteins and/or their ligands in structure and function of the trans Golgi network. J Cell Sci. 2003;116(21):4441–4454. doi: 10.1242/jcs.00746. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–17. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhang W, Yang H, Kong X, Mohapatra S, Juan-Vergara HS, Hellermann G, Behera S, Singam R, Lockey RF, Mohapatra SS. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. 2005;11(1):56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ANDV proteins colocalize with Rab8 and Rab11 when expressed from cDNA. Vero cells were transfected with cDNAs expressing either (A and B) eGFP-Rab8 or (D and E) eGFP-Rab11 in combination with (A and D) ANDV N (red, AlexaFluor 555) or (B and E) ANDV glycoproteins (M) (red, AlexaFluor 633). As a positive control for localization with these two Rabs, Vero cells were transfected with cDNAs expressing (C and F) VSV G (red, AlexaFluor 594) and (C) eGFP-Rab8 or (F) eGFP-Rab11. The cells were fixed at 18 hpt, immunostained for the viral proteins, and visualized by confocal microscopy. Images are flattened reconstructions of confocal z-stacks that were acquired at 63x optical magnification and 2x digital zoom.