Abstract
The reaction of protein bound iron-sulfur (Fe-S) clusters with nitric oxide (NO) plays key roles in NO-mediated toxicity and signaling. Elucidation of the mechanism of the reaction of NO with DNA regulatory proteins that contain Fe-S clusters has been hampered by a lack of information about the nature of the iron-nitrosyl products formed. Here, we report nuclear resonance vibrational spectroscopy (NRVS) and density functional theory (DFT) calculations that identify NO reaction products in WhiD and NsrR, regulatory proteins that use a [4Fe-4S] cluster to sense NO. This work reveals that nitrosylation yields multiple products structurally related to Roussin's Red Ester (RRE, [Fe2(NO)4(Cys)2]) and Roussin's Black Salt (RBS, [Fe4(NO)7S3]. In the latter case, the absence of 32S/34S shifts in the Fe-S region of the NRVS spectra suggest that a novel species, Roussin's Black Ester (RBE), may be formed, in which one or more of the sulfide ligands is replaced by Cys thiolates.
Keywords: iron-sulfur cluster, nitric oxide, gene regulation, nuclear vibrational resonance spectroscopy, DFT, synchrotron radiation
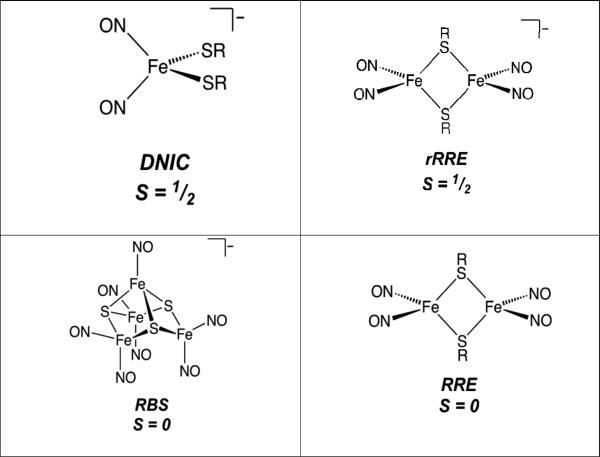
Iron-sulfur (Fe-S) proteins fulfil a wide range of biological functions, including electron transport, catalysis of chemical reactions, and chemical sensing.[1] Fe-S proteins vary in their sensitivity to NO and O2, but many are susceptible to damage under conditions of oxidative and nitrosative stress , resulting, for example, in deficiencies in respiration and DNA replication.[2] The intrinsic sensitivity of the cluster is exploited through the evolution of Fe-S proteins that function as sensors for these molecules.[3] Reactions of NO with Fe-S clusters usually result in the transformation of the Fe-S core into nitrosyl iron complexes.[4] The simplest of these are mononitrosyl iron complexes (MNICs) and dinitrosyl iron complexes (DNICs).[5] More complex non-homoleptic iron-nitrosyl complexes include the multi-iron Roussin's red ester (RRE), and Roussin's black salt (RBS) (Fig. 1).
Figure 1.
The major iron-nitrosyl species.
Early studies of nitrosative stress using EPR spectroscopy led to the conclusion that DNIC is the principal product of Fe-S nitrosylation, mainly because DNICs are paramagnetic (S= ½) and give rise to a readily detectable signal at g = 2.03.[6] However, quantification of the EPR showed only a fraction of the total iron content.[7] Other forms of spectroscopy including FTIR, 57Fe Mössbauer, UV-visible, Raman, and X-ray absorption have been used to study iron nitrosyls.[4, 8] However, these techniques lack full diagnostic ability to discriminate between different iron nitrosyl species.
Nuclear resonance vibrational spectroscopy (NRVS)[9] has recently been applied to iron proteins.[10] Like IR and Raman spectroscopies, data from NRVS can be converted into 57Fe partial vibrational density of states (PVDOS), but the selection rules differ such that it provides a full set of the vibrational modes that involve significant displacement of the 57Fe nucleus along the direction of the incident x-ray.[10a, 11] Consequently, NRVS is also 57Fe specific, making it ideal for probing the intermediates and products of Fe-S cluster nitrosylation. It was recently applied to study the reactions with NO of a [4Fe-4S] ferredoxin[12] and a [2Fe-2S] Rieske-type protein.[13] The fragility of Fe-S cluster proteins that function as physiological sensors of NO have so far hampered efforts to apply techniques such as NRVS, that require millimolar concentrations and 57Fe enrichment.
The Wbl family of regulators are found only in the actinobacteria[14], a phylum of Gram-positive bacteria that includes Streptomyces and important pathogens such as Mycobacterium tuberculosis. Wbl proteins are required for sporulation in Streptomyces and for the ability of the M. tuberculosis pathogen to persist within its host for long periods, as well as its remarkable tolerance to a wide range of antibiotics .[15] M. tuberculosis WhiB3, a member of the Wbl family, is believed to function as a sensor of O2 and NO to control expression of genes involved in intermediary metabolis m[16] and M. tuberculosis WhiB1 is known to bind DNA specifically following cluster nitrosylation,[17] suggesting that the reaction of Wbl proteins with NO may be a common signaling process. NsrR is a widespread NO-sensing protein that regulates the response of many bacteria to stress due to NO.[18]
NsrR and the Wbl protein WhiD (both from S. coelicolor) each contain a [4Fe-4S] cluster. The former is bound by three cysteine ligands plus one oxygen ligand,[19] and the latter by four cysteines,[20] thus giving a comparison between the NO reactivity of two clusters with different ligation, as well as biological function. Previous studies showed that both clusters react only slowly with O2, but rapidly with 8-10 NO in a multi-phase reaction that results in protein bound iron-nitrosyl species.[19-21] EPR studies showed that DNICs are not major products. Absorbance characteristics of the iron-nitrosyl products suggested the presence of RRE or RBS type species but their precise nature was not determined. A more definitive and iron specific method of analyzing Fe-S protein nitrosylated products is required.
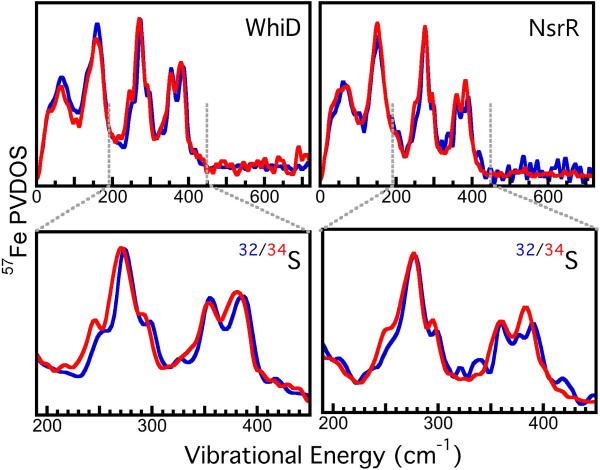
Mössbauer spectra and NRVS of WhiD and NsrR, as-isolated, were collected before and after NO treatment. The isomer shift (δ) and quadrupole splitting (ΔEQ) parameters from the Mössbauer spectra, shown in Fig. S1 and summarized in Table S1, are consistent with the presence of two pairs of high spin Fe2+/Fe3+ ions in each [4Fe-4S]2+ cluster.[22] Following addition of excess NO, the much lower isomer shift (δ 1 of ~0.16 compared to ~0.4 mm/s) and quadrupole splitting (ΔEQ of ~0.8 compared to >1) (Fig. S1 and Table S1) indicate a significant change in the iron environment, consistent with the formation of iron-nitrosyl species.[12, 23] NRVS spectra are shown in Fig. S2, where they are compared to the previously published NRVS spectrum of D14C variant of Pyrococcus furiosus [4Fe-4S] ferrodoxin.[12] All the spectra are very similar, demonstrating that the [4Fe-4S] core is intact in both WhiD and NsrR proteins. Although bands are due to collective modes involving different atomic groups, in some cases these collective motions are dominated by vibrations that are similar to local motions. Hence, by comparison with the previous work,[24] we can assign modes and regions of the spectrum that predominantly correspond to general cluster torsion, Fe-S bending, and Fe-S stretching, along with the region in which N-Fe-N modes are observed in iron nitrosyl species, see Fig. S2. In Fig. 2, the NRVS spectra for unlabeled and 34S labeled WhiD and NsrR proteins are compared. Since only the bridging S atoms are 34S labeled, the isotopic shifts further assist in normal mode identification, especially for the vibrational modes(s) related to the bridging S. The spectral region of importance, arising predominantly from Fe-S stretching modes, is 200 – 450 cm−1 (illustrated in Fig. S2). Being mostly collective vibrations, the magnitude of the shifts observed depends on the degree of sulfur motion in the particular vibration, meaning that isotopic substitution can result in some previously overlapping features becoming resolvable, or appearing to change in linewidth. For WhiD there is a clear shift to lower energy for two major peaks of the Fe-S stretching region (ca. 240 and 390 cm−1) and a smaller shift at ca. 355 nm. In NsrR the lower energy shifts are not as evident. This may be due in part to the symmetric “breathing modes” of the protein, not present in the WhiD protein,[10b] which occur at a lower range of energies ca. 314 – 370 cm−1.[25] However, a shift of the higher energy peak of the Fe-S stretching mode (ca. 390 cm−1) is observed for NsrR. These modes occur outside of the “breathing” region and therefore reflect the asymmetric nature of the Fe-S stretching mode, which are found to be 395 cm−1 and 385 cm−1 for 32S (unlabeled) and 34S labeled NsrR, respectively.
Figure 2. NRVS spectra of [4Fe-4S] WhiD and NsrR.
TOP: 57Fe PVDOS for: (Left) (—) Unlabeled (32S) WhiD (—), 34S labeled WhiD; (Right) (—) Unlabeled (32S) NsrR (—) 34S labeled NsrR. BOTTOM: Magnified view of the 200-450 cm−1 regions of the 32/34S WhiD (left) and NsrR (right) spectra.
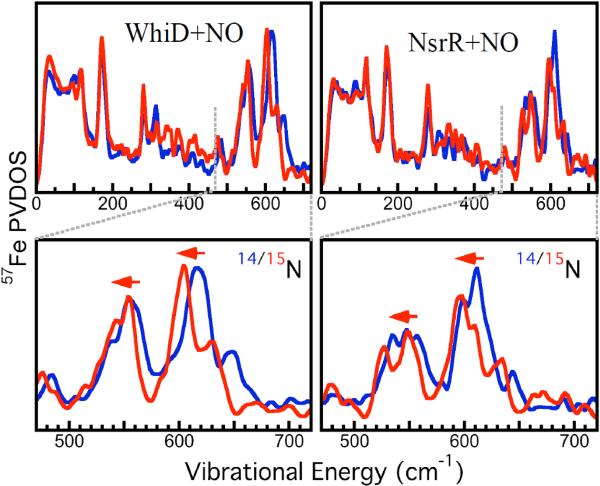
Extensive changes were observed in the thiolate and N-Fe-N regions of the NRVS PVDOS for [4Fe-4S] WhiD and NsrR following reaction with excess NO, see Fig. 3. For WhiD, the absorbance spectrum (Fig. S3) confirmed that the nitrosylation product is the same as that previously observed at ~11 NO per cluster.[21a] From previous studies of the reaction of NO with other [4Fe-4S] containing proteins it has been established that the Fe-N and N-Fe-N regions do not overlap with Fe-S regions.[12] For both WhiD and NsrR, the characteristic Fe-S peaks described above are much less intense or replaced, while there is significant new intensity in the Fe -N-Fe region, particularly at 530 – 580 cm−1 and 600 – 640 cm−1 (Fig. 3). The peak at ca. 170 cm−1 for all of the spectra corresponds to Fe-S bending modes. However, the peak is significantly sharper following reaction with NO, indicating the formation of nitrosylated products with a more limited range of Fe-S bending motions than the initial Fe-S clusters, resulting in a narrower feature observed at ca. 170 cm−1. The N-Fe-N region for 14NO/15NO-treated WhiD and NsrR proteins (Fig. 3) illustrate the shift in the Fe-NO related peaks to lower energy upon 15NO labeling. For WhiD, NRVS following addition of lower levels of NO (Fig. S4) revealed a transition between the spectra of Fig. 2 and 3.
Figure 3. NRVS spectra of nitrosylated [4Fe-4S] WhiD and NsrR.
57Fe PVDOS: (TOP) Left (—) unlabeled WhiD + 14NO; (—) unlabeled WhiD + 15NO. Right (—) unlabeled NsrR +14NO; (—) unlabeled NsrR + 15NO. (BOTTOM) Magnified view of N-Fe-N region (440 cm−1 – 740 cm−1).
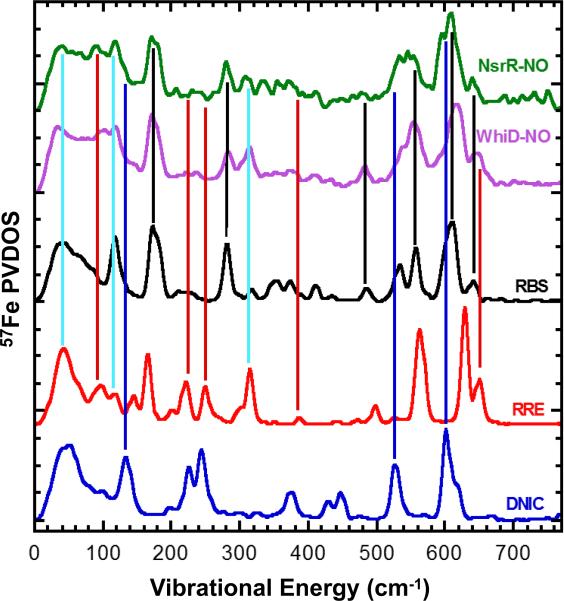
The NRVS spectra of the two proteins contain features that are characteristic of both RRE- and RBS-like species, see Fig. 4. For example, signals at 278, 481 and 537 cm−1 correspond to RBS, while features at 97, 318 and 643 cm−1 correspond to RRE. Furthermore, the bands in the N-Fe-N region of the WhiD and NsrR spectra are quite broad, consistent with the superposition of features due to RRE and RBS, see Fig. S5. In the case of NsrR, the RRE-like component appears to be smaller than in WhiD. Previous EPR studies showed that there is also a minor component of DNIC species present in these samples. This is more substantial for NsrR (~16% total iron), consistent with the presence of features in the nitrosylated NsrR NRVS spectrum at 137, 527 and 600 cm−1 that correspond to DNIC signals, see Fig. 4 and S5B.[21b]
Figure 4. Comparison of NRVS spectra of DNIC, RRE and RBS with those of WhiD-NO and NsrR-NO.
Black, red and blue lines indicate the signals that correspond to the RBS, RRE and DNIC model compounds, respectively. Light blue lines highlight the signals that appear in both WhiD-NO and NsrR-NO samples and both RBS and RRE model spectra. RRE and DNIC models utilised methyl thiolate ligands.
The observation of multiple iron-nitrosyl species accords with their chemical properties in solution that show an equilibrium exists between DNIC and RRE species, controlled by the availability of thiolate ligands ,[26] that Roussin's red salt (RRS, the sulfide bridged form of RRE) can undergo reversible conversion to RBS,[27] and that RRS complexes can readily undergo ligand exchange with thiolate ligands .[28] Conversion between different iron nitrosyl species depends on the concentration of NO, the redox states of iron and sulfur, and the availability of sulfide and thiol ligands. Our data show clearly that the Fe-S binding pockets of both WhiD (4 Cys ligands) and NsrR (3 Cys ligands) can accommodate a variety of iron-nitrosyl products related to those that can be generated as small molecule complexes. Previously only the DNIC species had been identified in these two proteins.[2c, 7a] These results raise the question of whether the different nitrosyl forms have distinct biological functions. We showed recently that, in vitro, the effect of NO on the binding of [4Fe-4S] NsrR to DNA is dependent on the promoter sequence.[21b] For the hmpA2 promoter, only ~2.5 NO per cluster were needed to abolish DNA binding, while for the hmpA1 and nsrR promoters, ~4 and ~8 NO per cluster, respectively, were needed to entirely abolish binding. This implies that different iron nitrosyl species are indeed involved in activating/deactivating DNA binding.
[4Fe-4S] WhiD and NsrR labeled with 34S sulfides were also reacted with NO and NRVS spectra recorded, see Fig. S6 and S7. Unlike the 32S/34S [4Fe-4S] spectra, no clear shifts were observed in the spectra of the nitrosylated samples compared to NO-treated natural abundance (32S) samples. This is a key observation because it shows that there is no significant amount of 34S (i.e. bridging S) bound directly to Fe in the nitrosylated forms, ruling out RBS itself as a product of the nitrosylation.
In order to assist with spectral assignments, and in particular to determine the effects of isotopic substitution (34S and 15N), and to investigate the possibility of persulfide formation and coordination of iron-nitrosyls, we performed a normal mode analysis using density functional theory (DFT) calculations. Similar calculations were previously successfully carried out for a [4Fe -4S] cluster.[24] Calculations for RRE and RBS iron nitrosyl species are shown in Fig. S8 and S9. In general, the lower energy region of the spectra consists of peaks formed from a combination of different vibrations. The mid energy region is mostly S-Fe-S vibrations and the upper energy region mostly strong N-Fe-N motions. The RRE calculations (Fig. S8) show good agreement with the previously reported 57Fe PVDOS of [Fe2(μ-SPh)2(NO)4].[12] Calculations with 15N in place of 14N showed that bands in the 500-650 cm−1 region are sensitive to 14/15N substitution, as found experimentally for WhiD and NsrR. The RBS calculations (Fig. S9) also show good agreement with the previously reported 57Fe PVDOS of (Et4N)[Fe4(μ3-S)3(NO)7],[12] particularly in terms of the pattern of bands and their relative intensities right across the spectrum. Calculations with 15N in place of 14N and 34S in place of sulfide 32S (with Cys thiolate sulfur remaining as 32S) revealed that bands in the 500 – 650 cm−1 region are sensitive to 14/15N substitution, and bands in the 240 – 380 cm−1 region are sensitive to 32/34S (sulfide) substitution. While 14/15N shifts were observed experimentally for the products of the NO reactions of WhiD and NsrR, 32/34S shifts were not, providing further support for the conclusion that RBS is not a nitrosylation product. Indeed, because all of the coordination positions at the irons are satisfied by non-protein ligands (NO and bridging sulfides) the RBS ion can only be bound to a protein by ionic forces and/or H-bonding. Therefore, we considered whether Roussin's black ester (RBE) species, as shown in Fig. S10-S12, having one, two or three thiolate groups (from protein Cys residues) in place of bridging sulfides, might be present. DFT calculations of the 57Fe PVDOS for each of these putative complexes revealed only relatively minor changes due to these substitutions (Fig. S10-S12) and a diminishing 32/34S shift. Therefore, on the basis of the DFT calculations, RBS and RBE-type species, particularly those with only one or two thiolate bridges, are not readily distinguishable by NRVS, and so the data are not inconsistent with the presence of RBE-type species.
Nitrosylation of [4Fe-4S] WhiD was previously shown to result in oxidation of sulfide to S0.[21a] This has also been reported in studies of model complexes.[23a] For WhiD, at least some of the S0 is retained by the protein in the form of Cys persulfide, raising the possibility that iron nitrosyl species formed might have coordination by Cys persulfides in addition to Cys thiolates.[29] DFT calculation of the 57Fe PVDOS for a persulfide form of the mono-thiol RBE was performed, see Fig S13. There are relatively few differences between this and the calculated spectrum for the mono-thiol RBE (Fig. S10) and indeed that of RBS itself (Fig. S7). Similar calculations were performed for single and double persulfide forms of RRE, see Fig. S14 and S15. While the calculated spectrum of the single persulfide form is very similar to that of RRE, that of the double persulfide form has some significant differences, particularly in the 100 – 200 and >600 cm−1 regions, suggesting that this species is distinguishable from RRE and a single persulfide adduct.
In summary, the combination of 57Fe Mossbauer, NRVS and DFT, along with 14NO/15NO and 32S/34S (sulfide) substitutions have revealed that nitrosylation of the NO-sensing [4Fe-4S] cluster containing regulatory proteins WhiD and NsrR gives rise to a range of products, including variable but small (≤16%) amounts of DNIC, but principally to species related to RBS and RRE. The former are not RBS itself because no 32/34S sulfide shifts were observed in NRVS spectra. To explain these findings we suggest the formation of a novel Black Salt in which one or more of the sulfides are substituted by a Cys thiolate, resulting in a so-called Roussins’ Black Ester (RBE). We have also uncovered evidence of Cys persulfides that may ligate cores of RRE and RBE in these proteins. We await the preparation of inorganic model compounds that will help further in characterising these novel protein-bound forms of iron-nitrosyls.
Supplementary Material
Footnotes
This work was supported by Biotechnology and Biological Sciences Research Council grants BB/J003247/1 (to NLB, MIH and AJT), BB/L007673/1 (to NLB, JCC and AJT) and BB/K02115X/1 (a US Partnering award to facilitate exchange between the NLB and SPC laboratories), and National Institutes of Health Grant GM65440 to SPC. We thank UEA for funding the purchase of the mass spectrometer instrument and for access to the High Performance Computer Cluster (Grace).
Supporting information for this article is available on the WWW under http://.
Contributor Information
Pauline N. Serrano, Department of Chemistry, University of California, Davis, CA 95616
Dr Hongxin Wang, Department of Chemistry, University of California, Davis, CA 95616; Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720.
Dr Jason C. Crack, Centre for Molecular and Structural Biochemistry, School of Chemistry, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK.
Christopher Prior, Centre for Molecular and Structural Biochemistry, School of Chemistry, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK.
Prof. Matthew I Hutchings, School of Biological Sciences, University of East Anglia, Norwich, NR4 7TJ, UK
Prof. Andrew J. Thomson, Centre for Molecular and Structural Biochemistry, School of Chemistry, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK
Dr Saeed Kamali, University of Tennessee Space Institute, Tullahome, TN 37388-9700.
Dr Yoshitaka Yoda, Research and Utilization Division, SPring-8/JASRI, 1-1-1 Kouto, Sayo, Hyogo 679-5198, JAPAN.
Dr Jiyong Zhao, Advanced Photon Source, Argonne National Laboratory, Argonne, IL 60439.
Dr Michael Y. Hu, Advanced Photon Source, Argonne National Laboratory, Argonne, IL 60439.
Dr Ercan E. Alp, Advanced Photon Source, Argonne National Laboratory, Argonne, IL 60439.
Dr. Vasily S. Oganesyan, Centre for Molecular and Structural Biochemistry, School of Chemistry, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK
Prof Nick E. Le Brun, Centre for Molecular and Structural Biochemistry, School of Chemistry, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK.
Prof Stephen P. Cramer, Department of Chemistry, University of California, Davis, CA 95616; Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720.
References
- 1.Johnson DC, Dean DR, Smith AD, Johnson MK. Ann. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 2.a Ding HG, Demple B. Proc. Nat. Acad. Sci. U. S. A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Drapier JC. Methods. 1997;11:319–329. doi: 10.1006/meth.1996.0426. [DOI] [PubMed] [Google Scholar]; c Crack JC, Green J, Thomson AJ, Le Brun NE. Acc. Chem. Res. 2014;47:3196–3205. doi: 10.1021/ar5002507. [DOI] [PubMed] [Google Scholar]
- 3.a McCleverty JA. Chem. Rev. 2004;104:403–418. doi: 10.1021/cr020623q. [DOI] [PubMed] [Google Scholar]; b Ignarro LJ. Nitric Oxide: Biology and Pathobiology. Academic Press; San Diego: 2004. [Google Scholar]
- 4.Foster MW, Cowan JA. J. Am. Chem. Soc. 1999;121:4093–4100. [Google Scholar]
- 5.a Butler AR, Megson IL. Chem. Rev. 2002;102:1155–1166. doi: 10.1021/cr000076d. [DOI] [PubMed] [Google Scholar]; b Enemark JH, Feltham RD. Coord. Chem. Rev. 1974;13:339–406. [Google Scholar]
- 6.a Reddy D, Lancaster JR, Cornforth DP. Science. 1983;221:769–770. doi: 10.1126/science.6308761. [DOI] [PubMed] [Google Scholar]; b Vanin AF, Aliev DI. Studia Biophys. 1983;97:223–229. [Google Scholar]
- 7.a Cruz-Ramos H, Crack J, Guanghui W, Hughes MM, Scott C, Thomson AJ, Green J, Poole RK. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kennedy MC, Anthonline WE, Beinert H. J. Biol. Chem. 1997;272:20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- 8.a Lu S, Libby E, Saleh L, Xing G, Bollinger JM, Moenne-Loccoz P. J. Biol. Inorg. Chem. 2004;9:818–827. doi: 10.1007/s00775-004-0582-8. [DOI] [PubMed] [Google Scholar]; b Sedney D, Reiff WM. Inorg. Chim. Acta. 1979;34:231–236. [Google Scholar]; c Lobysheva II, Serezhenkov VA, Vanin AF. Biochemistry (Moscow) 1997;62:801. [Google Scholar]; d Dai RJ, Ke SC. J. Phys. Chem. B. 2007;111:2335–2346. doi: 10.1021/jp066964f. [DOI] [PubMed] [Google Scholar]; e Tsai MC, Tsai FT, Lu TT, Tsai ML, Wei YC, Hsu IJ, Lee JF, Liaw WF. Inorg. Chem. 2009;48:9579–9591. doi: 10.1021/ic901675p. [DOI] [PubMed] [Google Scholar]
- 9.Seto M, Yoda Y, Kikuta S, Zhang XW, Ando M. Phys. Rev. Lett. 1995;74:3828–3831. doi: 10.1103/PhysRevLett.74.3828. [DOI] [PubMed] [Google Scholar]
- 10.a Wang H, Alp E, Yoda Y, Cramer S. In: Metalloproteins. Fontecilla-Camps JC, Nicolet Y, editors. Vol. 1122. Humana Press; 2014. pp. 125–137. [Google Scholar]; b Xiao Y, Wang H, George SJ, Smith MC, Adams MWW, Francis J, Jenney E, Sturhahn W, Alp EE, Zhao J, Yoda Y, Dey A, Solomon EI, Cramer SP. J. Am. Chem. Soc. 2005;127:14596–14606. doi: 10.1021/ja042960h. [DOI] [PubMed] [Google Scholar]; c Ogata H, Kramer T, Wang H, Schilter D, Pelmenschikov V, van Gastel M, Neese F, Rauchfuss TB, Gee LB, Scott AD, Yoda Y, Tanaka Y, Lubitz W, Cramer SP. Nat. Commun. 2015;6:7890. doi: 10.1038/ncomms8890. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Lauterbach L, Wang H, Horch M, Gee LB, Yoda Y, Tanaka Y, Zebger I, Lenz O, Cramer SP. Chem. Sci. 2015;6:1055–1060. doi: 10.1039/c4sc02982h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Kamali S, Wang H, Mitra D, Ogata H, Lubitz W, Manor BC, Rauchfuss TB, Byrne D, Bonnefoy V, Jenney FE, Jr., Adams MWW, Yoda Y, Alp E, Zhao J, Cramer SP. Angew. Chemie Int. Ed. 2013;52:724–728. doi: 10.1002/anie.201204616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Petrenko T, Sturhahn W, Neese F. Hyperfine Int. 2007;175:165–174. [Google Scholar]; b Do LH, Wang H, Tinberg CE, Dowty E, Yoda Y, Cramer SP, Lippard SJ. Chem. Commun. 2011;47:10945–10947. doi: 10.1039/c1cc13836g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tonzetich ZJ, Wang H, Mitra D, Tinberg CE, Do LH, Jenney FE, Jr., Adams MWW, Cramer SP, Lippard SJ. J. Am. Chem. Soc. 2010;132:6914–6916. doi: 10.1021/ja101002f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tinberg CE, Tonzetich ZJ, Wang H, Do LH, Yoda Y, Cramer SP, Lippard SJ. J. Am. Chem. Soc. 2010;132:18168–18176. doi: 10.1021/ja106290p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soliveri JA, Gomez J, Bishai WR, Chater KF. Microbiol. 2000;146:333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 15.Saini V, Farhana A, Glasgow JN, Steyn AJ. Curr. Opin. Chem. Biol. 2012;16:45–53. doi: 10.1016/j.cbpa.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Jr., Steyn AJ. Proc. Nat. Acad. Sci. U. S. A. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, Le Brun NE, Hunt DM, Harvey E, Adinolfi S, Buxton RS, Green J. Biochem. J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker NP, Le Brun NE, Dixon R, Hutchings MI. Trends Microbiol. 2010;18:149–156. doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Crack JC, Dodd EL, Knowles F, Al Bassam MM, Kamali S, Holland AA, Cramer SP, Hamilton CJ, Johnson MK, Thomson AJ, Hutchings MI, Le Brun NE. J. Biol. Chem. 2015;290:12689–12704. doi: 10.1074/jbc.M115.643072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crack JC, den Hengst CD, Jakimowicz P, Subramanian S, Johnson MK, Buttner MJ, Thomson AJ, Le Brun NE. Biochemistry. 2009;48:12252–12264. doi: 10.1021/bi901498v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a Crack JC, Smith LJ, Stapleton MR, Peck J, Watmough NJ, Buttner MJ, Buxton RS, Green J, Oganesyan VS, Thomson AJ, Le Brun NE. J. Am. Chem. Soc. 2011;133:1112–1121. doi: 10.1021/ja109581t. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Crack JC, Svistunenko DA, Munnoch J, Thomson AJ, Hutchings MI, Le Brun NE. J. Biol. Chem. 2016;291:8663–8672. doi: 10.1074/jbc.M115.693192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandelia ME, Bykov D, Izsak R, Infossi P, Giudici-Orticoni MT, Bill E, Neese F, Lubitz W. Proc. Nat. Acad. Sci. 2012;110:483–488. doi: 10.1073/pnas.1202575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a Harrop TC, Tonzetich ZJ, Reisner E, Lippard SJ. J. Am. Chem. Soc. 2008;130:15602–15610. doi: 10.1021/ja8054996. [DOI] [PubMed] [Google Scholar]; b Hess JL, Hsieh CH, Brothers SM, Hall MB, Darensbourg MY. J. Am. Chem. Soc. 2011;133:20426–20434. doi: 10.1021/ja208384d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra D, Pelmenschikov V, Guo Y, Case DA, Wang H, Dong W, Tan M-L, Ichiye T, Francis J, Jenney E, Adams MWW, Yoda Y, Zhao J, Cramer SP. Biochemistry. 2011;50:5220–5235. doi: 10.1021/bi200046p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith MC, Xiao Y, Wang H, George SJ, Coucovanis D, Koutmos M, Sturhahn W, Alp EE, Zhao J, Cramer SP. Inorg. Chem. 2005;44:5562–5570. doi: 10.1021/ic0482584. [DOI] [PubMed] [Google Scholar]
- 26.Fitzpatrick J, Kalyvas H, Shearer J, Kim E. Chem. Commun. 2013;49:5550–5552. doi: 10.1039/c3cc40352a. [DOI] [PubMed] [Google Scholar]
- 27.a Butler AR, Glidewell C, Li MH. Adv. Inorg. Chem. Rad. 1988;32:335–393. [Google Scholar]; b Yeh SW, Tsou CC, Liaw WF. Dalton Trans. 2014;43:9022–9025. doi: 10.1039/c4dt00450g. [DOI] [PubMed] [Google Scholar]
- 28.Chen TN, Lo FC, Tsai ML, Shih KN, Chiang MH, Lee GH, Liaw WF. Inorg. Chim. Acta. 2006;359:2525–2533. [Google Scholar]
- 29.a Crack JC, Stapleton MR, Green J, Thomson AJ, Le Brun NE. J. Biol. Chem. 2013;288:11492–11502. doi: 10.1074/jbc.M112.439901. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhang B, Crack JC, Subramanian S, Green J, Thomson AJ, Le Brun NE, Johnson MK. Proc. Nat. Acad. Sci. U. S. A. 2012;109:15734–15739. doi: 10.1073/pnas.1208787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.