Abstract
Brain insulin resistance (IR), which depends on Insulin-Receptor-Substrate-1 (IRS-1) phosphorylation, is characteristic of Alzheimer's disease (AD). Previously, we demonstrated higher pSer312-IRS-1 (ineffective insulin signaling) and lower p-panTyr-IRS-1 (effective insulin signaling) in neural origin-enriched plasma exosomes of AD patients vs. controls. Here, we hypothesized that these exosomal biomarkers associate with brain atrophy in AD. We studied 24 subjects with biomarker-supported probable AD (low CSF Aβ42). Exosomes were isolated from plasma, enriched for neural origin using immunoprecipitation for L1CAM, and measured for pSer312- and p-panTyr-IRS-1 phosphotypes. MPRAGE images were segmented by brain tissue type and voxel-based morphometry (VBM) analysis for gray matter against pSer312- and p-panTyr-IRS-1 was conducted. Given the regionally variable brain expression of IRS-1, we used the Allen Brain Atlas to make spatial comparisons between VBM results and IRS-1 expression. Brain volume was positively associated with P-panTyr-IRS-1 and negatively associated with pSer312-IRS-1 in a strikingly similar regional pattern (bilateral parietal-occipital junction, R middle temporal gyrus). This volumetric association pattern was spatially correlated with Allen Human Brain atlas normal brain IRS-1 expression. Exosomal biomarkers of brain IR are thus associated with atrophy in AD as could be expected by their pathophysiological roles and do so in a pattern that reflects regional IRS-1 expression. Furthermore, neural-origin plasma exosomes may recover molecular signals from specific brain regions.
Keywords: Alzheimer's Disease, Insulin Resistance, Magnetic Resonance Imaging
1. Introduction
Decades of epidemiological studies have established a compelling link between insulin resistance (IR) and Alzheimer's disease (AD) (Crane et al., 2013; Ott et al., 1999; Schrijvers et al., 2010). Once believed to be insulin insensitive, the brain is now known to use insulin delivered through the blood-brain barrier (BBB) or produced locally via insulin (INS) gene transcription (Gray, Meijer, & Barrett, 2014). Normal insulin signaling inhibits beta amyloid (Aβ) oligomerization and tau phosphorylation (Talbot & Wang, 2014), whereas mounting evidence suggests that several pathological processes active in AD converge to impair insulin signaling early in disease pathogenesis (Chua et al., 2012; Keeney, Ibrahimi, & Zhao, 2015; Pearson-Leary & McNay, 2012; Talbot et al., 2012). This results in a feed-forward loop exacerbating both IR and AD pathogenic processes.
A limitation for studying the clinical and epidemiological effects of brain IR in AD has been the absence of brain-specific IR biomarkers. Consequently, earlier studies have relied on measures of systemic IR based on blood glucose and insulin values, such as the Homeostatic Model Assessment of Insulin Resistance (HOMA-IR). Peripheral and brain IR do overlap to some degree (de la Monte, 2009), and this may explain why associations have been observed between HOMA-IR and brain glucose hypometabolism in cognitively normal elderly subjects (Baker et al., 2011; Willette, Bendlin, et al., 2015), patients with AD (Willette, Modanlo, Kapogiannis, & Alzheimer's Disease Neuroimaging Initiative, 2015), Aβ deposition (Willette, Johnson, et al., 2015), and atrophy in AD (Willette et al., 2013). The biological plausibility of these associations is supported by their general overlap with regions preferentially vulnerable to AD pathology, most notably the substantial grey matter atrophy that consistently occurs in the parieto-temporal and posterior cingulate regions of the AD brain (Pini et al., 2016; Wang et al., 2015). Nevertheless, post-mortem brain tissue from patients with AD and animal models manifests molecular changes characteristic of IR even in the absence of systemic IR (Chua et al., 2012; Craft, 2012; de la Monte, 2012; Talbot & Wang, 2014; Talbot et al., 2012), underlining the need for a brain-IR-specific biomarker.
Insulin Receptor Substrate-1 (IRS-1) serves as the effector molecule of the insulin receptor. Specific changes in the phosphorylation pattern of IRS-1 resulting in impaired insulin signaling have long been established as pathological markers of IR in peripheral tissues (Copps & White, 2012; Hoehn et al., 2008; Taniguchi, Emanuelli, & Kahn, 2006). Talbot et al. recently introduced various differentially phosphorylated forms of IRS-1 as pathological markers of brain IR in AD, confirming previous findings (Ma et al., 2009), and demonstrated profoundly impaired insulin and IRS-1 signaling in postmortem AD brains (Talbot & Wang, 2014; Talbot et al., 2012). Nevertheless, this intracellular (and intraneuronal) molecule had not been accessible as a biomarker until we successfully recovered it from plasma exosomes enriched for neural origin.
In a recent study, we demonstrated that pSer312-IRS-1 and p-panTyr-IRS-1 are biomarkers for AD (Kapogiannis et al., 2015), perhaps because they reflect the IR that characterizes the AD brain. The pSer312-IRS-1 phosphotype stimulates uncoupling of IRS-1 and leads to its degradation (Pederson, Kramer, & Rondinone, 2001; Sun, Goldberg, Qiao, & Mitchell, 1999), while the p-panTyr-IRS-1 phosphotype promotes insulin-stimulated responses (Gual, Le Marchand-Brustel, & Tanti, 2005). Our team has pioneered a methodology for exosome isolation from plasma followed by immunoprecipitation against the cell surface adhesion protein L1-CAM to enrich for neural origin (Schmid & Maness, 2008). In a series of case-control studies of patients with AD and cognitively normal controls, we demonstrated how pathogenic and signaling peptides in L1-CAM/NCAM-expressing plasma exosomes effectively distinguish patients and controls and may predict disease diagnosis (Fiandaca et al., 2015; Goetzl, Boxer, Schwartz, Abner, Petersen, Miller, Carlson, et al., 2015; Goetzl, Boxer, Schwartz, Abner, Petersen, Miller, & Kapogiannis, 2015; Kapogiannis et al., 2015). In addition, by demonstrating differences in pSer312-IRS-1 and p-panTyr-IRS-1 in L1-enriched plasma exosomes between AD patients and cognitively normal control subjects with type 2 diabetes, we confirmed in vivo that peripheral IR is dissociable from the brain IR occurring in AD (Kapogiannis et al., 2015). These findings suggest the presence of brain IR in patients with AD and to a lesser extent in individuals with diabetes (Kapogiannis et al., 2015).
In the present study, we assess the external validity of exosomal brain IR biomarkers by examining their association with structural MRI-based brain atrophy, a surrogate of AD-related neurodegeneration (Pini et al., 2016), in the same group of subjects with early AD as in Kapogiannis et al. (Kapogiannis et al., 2015). In addition, we study the regional distribution of the ensuing associations to demonstrate their biological plausibility in terms of regional brain differences in gene expression.
2. Materials and Methods
2.1. Subjects (Table 1)
Table 1.
Subject Characteristics.
| Subject Characteristics | ||
|---|---|---|
| N | 24 | # |
| Age | 73 ± 8 | years |
| Sex | 14M:10F | |
| MMSE | 25.3 ± 4.3 | |
| BMI | 26.7 ± 4.6 | kg/m |
| CSF Aβ42 | 151.5± 31.7 | pg/dl |
| CSF p181 tau | 44 ± 23 | pg/dl |
| CSF total tau | 74 ± 28 | pg/dl |
| Fasting Glucose | 90.9 ± 9.1 | mg/dl |
| Fasting Insulin | 8.2 ± 4.5 | uIU/ml |
| HOMA2 %B | 96.3 ± 38.0 | |
| HOMA2 %S | 122.8 ± 60.3 | |
| HOMA2 IR | 1.1 ± 0.6 | |
Demographic and clinical data for all subjects. Values presented as mean ± standard deviation where applicable. MMSE: mini-mental state examination, BMI: body mass index, CSF Aβ42: cerebrospinal fluid B-amyloid42; p181 tau: tau phosphorylated on threonine 181, HOMA2 %B: Homeostasis Model Assessment 2 beta cell function, HOMA2 %S: Homeostasis Model Assessment 2 insulin sensitivity, HOMA2 IR: Homeostasis Model Assessment 2 insulin resistance.
We studied 24 subjects (14 women and 10 men; age = 73 ± 8 years old (mean ± SD)) fulfilling criteria for Mild Cognitive Impairment or mild dementia due to AD with high probability. All subjects had CSF Aβ42 < = 192 pg/dl, as well as high CSF total-tau (74 ± 28 pg/dl) and/or p181-tau (44 ± 23 pg/dl) (Albert et al., 2011; McKhann et al., 2011). Thirteen subjects had Clinical Dementia Rating (CDR) global score of 0.5 (subjects with MCI) and 11 subjects had CDR global score of 1 (subjects with mild dementia). Twelve of the subjects were previously diagnosed with hypertension. Seventeen of the 24 subjects in this study overlap with the 26-subject AD cohort in the first report of exosomal IRS-1 biomarkers (Kapogiannis et al., 2015), Nine of the subjects included in the prior study did not have MRI acquisitions and were excluded from the present analysis and 7 additional subjects were subsequently recruited to complement the dataset. Control subjects included in (Kapogiannis et al., 2015) did not have MR scans and could not be included in the present study. All data were acquired as part of the baseline/screening visit for a clinical trial (NCT01255163) approved by the CNS Institutional Review Board of the NIH. All participants provided informed consent.
2.2. Exosome isolation protein assays
The methodology for exosome isolation and L1 CAM enrichment has been described in detail previously (Fiandaca et al., 2015; Goetzl, Boxer, Schwartz, Abner, Petersen, Miller, Carlson, et al., 2015; Goetzl, Boxer, Schwartz, Abner, Petersen, Miller, & Kapogiannis, 2015; Kapogiannis et al., 2015). Briefly, plasma was mixed with Thromboplastin-D followed by ExoQuick exosome precipitation solution (System Biosciences, Inc., Mountainview, CA). Exosome suspensions were incubated, centrifuged, re-suspended in medium with protease and phosphatase inhibitors inhibitor cocktails and immunoprecipitated using mouse anti-human CD171 (L1 CAM) biotinylated antibody (clone 5G3, eBioscience, San Diego, CA). Exosomes were lysed after pellet re-suspension and biotin elution. For p-panTyr-IRS-1 quantification, we used MesoScale Discovery plates for IRS-1 (catalog N450HLA-1) with Anti-Phospho-Tyrosine Antibody Sulfo-TAG Labeled detection antibody (catalog R32AP-5), and for Ser312-IRS-1 quantification we used Phospho-IRS-1 (Ser312) Kit (catalog K150HLD-2). For normalization, we used measurements of exosome marker human CD81 (Hölzel Diagnostika-Cusabio, Cologne, Germany), with verification of the CD81 antigen standard curve using human purified recombinant CD81 antigen (Origene Technologies, Inc., Rockville, MD). The mean value for all determinations of CD81 in each assay group was set at 1.00 and the relative values for individual samples were used to normalize their recovery.
2.3. Plasma and CSF Measures
Blood samples were collected by venipuncture into a vacutainer EDTA tube. Plasma was immediately separated from cells by centrifugation at 3,000 rpms for 30 seconds at 25°C, then realiquoted and stored at −80°C immediately afterwards. Samples were thawed to room temperature before assaying. The Mercodia Insulin assay (Uppsala, Sweden) was used to provide a measure of fasting insulin (uIU/ml). Coefficient of variation within assay was 2.8-4.0 % and 2.6-3.6% between assays, with standards ranging from 0-200 mU/L. Fasting plasma glucose was measured from the same aliquot used for insulin via the YSI 2300 STAT PLUS™ Glucose Analyzer by Yellow Springs Instrument (YSI; YSI Inc, Yellow Springs, Ohio, USA). The YSI 2300 STAT PLUS™ uses a steady state measurement methodology, where membrane based glucose oxidase catalyzes the oxidation of glucose to gluconic acid and hydrogen peroxide. The difference between the sample generated plateau current and the initial baseline current is proportional to the glucose concentration. Homeostatic Model Assessment (HOMA2) values for were obtained by entering the fasting glucose and insulin measures into the HOMA2 Calculator, available at https://www.dtu.ox.ac.uk/homacalculator/.
Cerebrospinal fluid was obtained via lumbar puncture and stored at −80 °C prior to sample processing. The levels (pg/ml) of beta-amyloid1-42 (Aβ1-42), phosphorylated tau (tau181P) and total tau (t-tau) were processed at the University of Pennsylvania Biomarker Research Laboratory using Luminex xMAP technology with INNO-BIA Alz Bio3 kits provided by Fujirebio.
2.4. MRI acquisition and pre-processing
T1-weighted magnetization-prepared rapid gradient-echo (MPRAGE) images were acquired on a 3T Phillips Achieva scanner with an 8-channel SENSE head coil. A Turbo Field Echo acquisition sequence was used with the following parameters: TR = 6.803 ms, TE = 3.19 ms, number of excitations = 1, flip angle = 8°, acquisition matrix = 256 × 256 × 170, resolution = 1 × 1 × 1.2 mm. Images were preprocessed using the Voxel Based Morphometry 8 (VBM8) toolbox in SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/). VBM8 employs a unified segmentation methodology, registering and warping native-space images into Montreal Neurological Institute (MNI) template space using affine and nonlinear parameters (Ashburner & Friston, 2005). Simultaneously, prior probability maps are applied to each subject's image to segment the brain into GM, white matter, CSF, bone, and fat tissue classes. Derived GM maps were modulated and normalized to preserve individual differences in regional volumes. GM maps were inspected to ensure success of the segmentation algorithm. An 8-mm full-width half-maximum smoothing kernel was applied prior to analysis.
2.5. VBM Analysis
Using SPM8, we conducted a whole-brain voxel-wise VBM regression analysis (Ashburner & Friston, 2000) of modulated normalized GM images with age, sex, fasting insulin, pSer312-IRS-1 and p-panTyr-IRS-1 as covariates. These five variables comprised a single model in SPM8. The uncorrected p statistic threshold was set to < 0.001 in concordance with standard and recent recommendations for cluster-extent based thresholding in fMRI analyses (Woo, Krishnan, & Wager, 2014). A cluster size of > 300 voxels (corresponding to uncorrected cluster-level p < 0.05); family-wise error (FWE) correction at cluster-level p < 0.05 and false-discovery rate (FDR) correction at cluster-level p < 0.05 were also applied to the reported results. Alternative models included i) age, sex, HOMA-IR2, pSer312-IRS-1 and p-panTyr-IRS-1 and ii) age, sex, CSF Aβ1-42, pSer312-IRS-1 and p-panTyr-IRS-1 (results reported as supplemental data).
2.6. Allen Atlas Expression-Volumetrics Correlation
Microarray expression values and their tissue sample coordinates were downloaded for the total current set of six human brain specimens included in the Allen Human Brain Map project (Hawrylycz et al., 2012). These specimens (see Table 3) are “normal” controls with no known history of neuropsychiatric or neurological conditions, long term illnesses, or hypoxic events lasting over an hour. Case qualification and donor profiles are available on the Allen human brain map website (http://human.brain-map.org). A non-linearly corrected set of MRI coordinates for the probe samples was obtained from github.com/chrisfilo/alleninf. These expression intensity values (log2) were then correlated with their matching coordinates on z-transformed SPM T-images. The MATLAB script for this expression-to-volumetric correlation was adapted from www.brain-map.org/api/examples/examples/spm/index.html, with substantial changes made to provide detailed results for specific probe to map correlations. The Allen atlas contains three different probe sequence results for IRS-1: A_24_P225679, A_32_P113646 & A_24_P802145. IRS-1 probe A_24_P225679 was selected for this analysis over A_32_P113646 & A_24_P802145 due to having the greatest number of samples above background signal (92% vs. 78% & 55%, respectively), strongest expression levels, and lowest standard deviation (5.81 ± 0.2 vs. 4.75 ± 0.34 & 4.52 ± 0.2, respectively). Only samples with expression significantly above background signal were included in the analysis. An approximate random effects analysis was used to analyze the group correlation results from the six subjects (Gorgolewski et al., 2014; Penny & Holmes, 2004), which consisted of performing a one-sample t-test on the slopes of the six individual Allen specimen correlations.
Table 3.
Allen Atlas Correlations.
| Allen Specimen | Age (yrs) | p-panTyr-IRS-1 to VBM Correlation |
|||
|---|---|---|---|---|---|
| Sex | Samples | r | p | ||
| H0351.2001 | 24 | M | 471 | 0.283 | 3.74E-10 |
| H0351.2002 | 39 | M | 418 | 0.111 | 2.40E-02 |
| H0351.1009 | 57 | M | 260 | 0.172 | 0.005 |
| H0351.1012 | 31 | M | 402 | 0.247 | 5.54E-07 |
| H0351.1015 | 49 | F | 390 | 0.155 | 0.0022 |
| H0351.1016 | 55 | M | 428 | 0.276 | 6.40E-09 |
Pearson correlations between matched coordinates on each of six Allen Specimens and the p-panTyrIRS-1 SPM T-image. R and p values show a strong positive correlation between brain IRS-1 levels and p-panTyrIRS-1 associated reductions in atrophy.
3. Results
3.1. VBM results (Table 2 and Figure 1)
Table 2.
Grey matter VBM results.
| Peak-voxel |
Cluster-level |
||||||
|---|---|---|---|---|---|---|---|
| BA | MNI x, y, z | t-Value | Size (k) | p Unc. | p FDR | p FWE | |
| p-panTyr-IRS-1 | |||||||
| positive association: | |||||||
| (R) middle / superior temporal gyri | 21, 22 | 48, −45, 4 | 6.17 | 1956 | <0.001 | <0.001 | <0.001 |
| (L) angular gyrus | 39 | -42, −60, 26 | 6.32 | 642 | 0.006 | 0.046 | 0.05 |
| (R) superior parietal lobule | 40 | 26, −42, 54 | 5.4 | 515 | 0.012 | 0.061 | 0.097 |
| (R) fusiform gyrus | 37 | 24, −56, −6 | 4.44 | 304 | 0.045 | 0.169 | 0.314 |
| pSer312-IRS-1 | |||||||
| negative association: | |||||||
| (R) middle /superior temporal gyri | 21, 22 | 48, −45, 4 | 6.68 | 1978 | <0.001 | <0.001 | <0.001 |
(R) = right; (L) = left; BA = Brodmann's area; MNI = Montreal Neurological Institute; Unc. = uncorrected; FWE = family wise error; FDR = false discovery rate
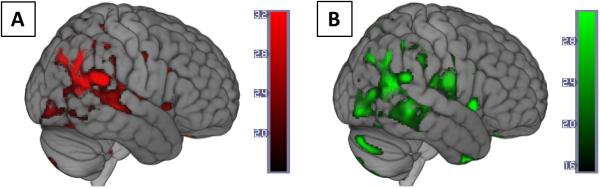
Figure 1. Voxel-wise regression of p-panTyr-IRS-1 and pSer312-IRS-1.
Color map overlays represent brain regions where higher gray matter volume was associated with A) higher exosomal p-panTyr-IRS-1 (red) or B) lower pSer312-IRS-1 (green). Color bars represent t values. Voxel threshold was set to p < 0.001 (uncorrected) for visualization. Complete results are provided in Table 2. Brains are oriented in neurological space. Figure created in MRIcroGL version 1.150909.
Higher L1-exosome levels of p-panTyr-IRS-1 were associated with higher GM volume (less atrophy) in four clusters: The L angular gyrus, the R superior parietal lobule, the R fusiform gyrus, and the largest cluster at the R middle and superior temporal gyri (Table 2, Figure 1A). Higher L1-exosome levels of pSer312-IRS-1 were associated with lower GM volume (greater atrophy) in a very similar cluster at the R middle and superior temporal gyri with the same peak voxel as (Table 2, Figure 1B). Setting the statistical threshold to p < 0.001 (uncorrected) reveals an extensive overlap between the two contrast images (Figure 1). There were no significant positive volumetric associations for pSer312-IRS-1 or negative volumetric associations for p-panTyr-IRS-1. Fasting insulin yielded no significant cortical cluster-level associations with GM volume in the same model.
Alternative models including either HOMA-IR2 or CSF Aβ1-42 yielded no significant cortical cluster-level associations between these covariates and GM volumes; associations of pSer312-IRS-1 and p-panTyr-IRS-1 remained significant in these models (results reported as supplemental data sections 2 and 3).
A supplementary volumetric one-way t-test using a set of 26 age-matched control MRI control subjects confirmed that the images used above displayed the typical pattern of atrophy expected of AD patients (see supplemental data section 1).
3.2. Allen Atlas Expression-Volumetrics Correlation Results
For each of the six Allen Atlas brain specimens, IRS-1 expression levels (log2) were spatially correlated with the intensity of z-transformed SPM T-images (one depicting the whole-brain positive volumetric association with p-panTyr-IRS-1 and the other depicting the negative volumetric association with pSer312-IRS-1). The approximate random effects analysis for all six Allen specimens showed a significant effect of slope for p-panTyrIRS-1 (t[5] = 6.41 , p = 0.0014) and no overall significant slope effect for pSer312-IRS-1. The individual correlations were highly significant for the p-panTyrIRS-1 associated T-image across all six Allen specimens (see Table 3).
4. Discussion
We recently found that peripheral blood exosomes enriched by L1 immunoprecipitation show higher levels of pSer312-IRS-1 and lower levels of p-panTyr-IRS-1 in AD patients versus controls, replicating differences observed in post-mortem studies (Talbot et al., 2012). In the current report based on measurements in L1-enriched exosomes and MRIs in a cohort of well-characterized AD subjects, higher p-panTyr-IRS-1 levels were associated with less brain atrophy and pSer312-IRS-1 levels with greater brain atrophy, corroborating their respective protective and detrimental roles in AD pathogenesis. Moreover, these associations extended in a strikingly similar regional pattern. To identify the significance of this pattern, we examined the normal regional brain expression of IRS-1 and found a significant correlation with the volumetric associations. These results suggest that the recently introduced exosomal biomarkers of brain IR reflect molecular changes in the brains of AD patients in a regionally specific manner.
AD-related phosphorylation changes in IRS-1 are regionally diverse (Talbot & Wang, 2014; Talbot et al., 2012) rendering certain regions of the brain more insulin resistant than others. Although, to our knowledge, there has not been a systematic study of the whole brain, it is reasonable to assume that the regional density of phospho-IRS-1 follows, to a degree, the normal regional distribution of IRS-1. The regionally diverse brain IR in AD may result in regionally diverse neurodegeneration and atrophy through several mechanisms. First, decreased tyrosine and increased serine IRS-1 phosphorylation (Bomfim et al., 2012; Ma et al., 2009) results in uncoupling insulin receptors from downstream signaling pathways with cellular trophic actions, such as PI3K, and Akt/PKB (De Felice & Ferreira, 2014; Taniguchi et al., 2006). Second, IR may result in hyperactive mammalian target of rapamycin (mTOR) and Cdk5 activity, resulting in tau hyperphosphorylation and neurodegeneration (Caccamo et al., 2013; Orr, Salinas, Buffenstein, & Oddo, 2014). Third, insulin-dependent glucose uptake (via membrane translocation of the insulin-dependent glucose transporter GLUT4 (Taniguchi et al., 2006)) supplements insulin-independent neuronal glucose uptake in certain hippocampal and cortical areas (Choeiri, Staines, & Messier, 2002; de la Monte, 2012; Grillo, Piroli, Hendry, & Reagan, 2009; McEwen & Reagan, 2004; Sankar, Thamotharan, Shin, Moley, & Devaskar, 2002). Therefore, IR may enhance the AD-related “neuronal starvation” for glucose in these areas (Mamelak, 2012). Fourth, whereas in control brains IRS-1 is localized predominantly in the nucleus, in AD patients pSer312 and other Ser phosphorylated forms of IRS-1 are found diffusely in the cytosol within neuritic plaques and surrounding neurofibrillary lesions, suggesting a direct neurotoxic effect of these IRS-1 deposits (Talbot & Wang, 2014; Talbot et al., 2012).
The pattern of atrophy associated with exosomal IRS-1 biomarkers represents a small but characteristic subset of the typical neocortical atrophy pattern observed in mild AD, in particular late-onset AD (for comparison, see figures 3 and 4 in Pini et al. (Pini et al., 2016)). Equally notable is the fact that we did not observe any IRS-1-associated atrophy in posteromedial regions such as the precuneus or the posterior cingulate cortex, or medial temporal and hippocampal regions. This suggests that AD-related pathways other than IRS-1 may be associated with atrophy in these areas. Our study contributes to the growing body of literature that shows that multiple genetic (e.g. APOE genotype), developmental, environmental, and syndromic factors, each with its own distinct associations, may converge to shape the overall pattern of atrophy observed in AD as well as its individual variations (Pini et al., 2016).
The overt similarity between regional IRS-1 expression and volumetric differences associated with an IRS-1 phosphotype is striking. The Allen atlas regional IRS-1 expression significantly aligns with the regionally distributed effect towards less atrophy associated with higher p-panTyrIRS-1 exosomal levels. This association serves to bolster the biological plausibility of the volumetric findings, but also the fidelity of a signal detected through the isolation and study of these exosomes from peripheral blood. Moreover, this signal is consistent with the physiological role of IRS-1 phosphotypes; p-panTyr-IRS-1 was previously shown to promote downstream insulin signaling (Gual et al., 2005) and we found it to be associated with less atrophy, whereas pSer312-IRS-1 was previously shown to impede downstream insulin signaling (Pederson et al., 2001; Sun et al., 1999) and we found it to be associated with more atrophy. Of the two GM patterns associated with IRS-1 phosphotypes, only the pattern associated with p-panTyr-IRS-1 showed spatial correlation with IRS-1 expression across all six Allen Brain Atlas brain specimens (individual correlations were highly significant, as well as the overall slope). This is not surprising considering the Allen Brain Atlas specimens came from normal controls. The p-panTyr-IRS-1 phosphotype is associated with normal downstream insulin signaling, whereas the pSer312-IRS-1 denotes insulin resistance, which may not have been present in these normal specimens. This highlights the importance of extending the Allen Brain Atlas project to include pathological specimens, such as specimens from patients with AD.
There are several limitations to this study that should be noted. The sample size was small, and no control group was included. Although the AD cohort studied here was largely the same as in the first report of exosomal IRS-1 biomarkers (Kapogiannis et al., 2015), MRI scans were not available for the control subjects included in that study. To alleviate concerns about the small sample size, we adopted a well-accepted statistical threshold and correction for multiple comparisons (Woo et al., 2014). Among the strengths of this study are the robustness of the volumetric associations, their meaningful regional pattern, and the direction of associations vis–à–vis the expected pathophysiological role of IRS-1 phosphotypes.
5. Conclusions
To our knowledge, this is the first biomarker study where a signal detected in the blood can be traced not only to the brain as a whole, but to specific brain regions where the relevant gene is preferentially expressed. These findings further support the search of exosome-based biomarkers for AD and other neurodegenerative diseases. Plasma exosomes enriched for neuronal origin may open a unique window for studying organ-specific cellular processes in living humans and may also be useful as thera-markers to follow response to experimental treatments.
Supplementary Material
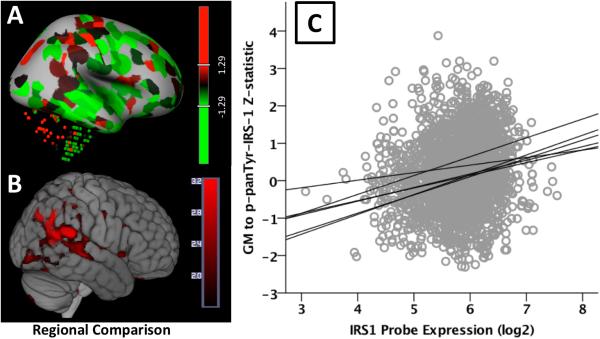
Figure 2. Allen Human Brain Atlas Expression Correlation with Volumetrics.
A). IRS-1 expression z-score map (red-high, green-low) from the Allen Human Brain atlas site for subject H0351.2001. Color bar is a z-statistic. (Image credit: Allen Institute). B). Volume to p-panTyrIRS-1 correlation statistical map thresholded at an uncorrected p value of 0.001. Color bar is a T-statistic. C). Scatterplot of IRS-1 log2 expression by strength of volumetric p-panTyrIRS-1 correlation. Regions showing significant volume increases with p-panTyrIRS-1 were also evident in the expected regional expression of IRS-1.
Acknowledgements
This research was supported entirely by the Intramural Research Program of the National Institute on Aging, NIH.
References
- Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Phelps CH. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. doi:10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. doi:10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. doi:S1053-8119(05)00110-2 [pii] 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Baker LD, Cross DJ, Minoshima S, Belongia D, Watson GS, Craft S. Insulin resistance and Alzheimer-like reductions in regional cerebral glucose metabolism for cognitively normal adults with prediabetes or early type 2 diabetes. Arch Neurol. 2011;68(1):51–57. doi: 10.1001/archneurol.2010.225. doi:archneurol.2010.225 [pii] 10.1001/archneurol.2010.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomfim TR, Forny-Germano L, Sathler LB, Brito-Moreira J, Houzel JC, Decker H, De Felice FG. An anti-diabetes agent protects the mouse brain from defective insulin signaling caused by Alzheimer's disease-associated Abeta oligomers. J Clin Invest. 2012;122(4):1339–1353. doi: 10.1172/JCI57256. doi:57256 [pii] 10.1172/JCI57256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Magri A, Medina DX, Wisely EV, Lopez-Aranda MF, Silva AJ, Oddo S. mTOR regulates tau phosphorylation and degradation: implications for Alzheimer's disease and other tauopathies. Aging Cell. 2013;12(3):370–380. doi: 10.1111/acel.12057. doi:10.1111/acel.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choeiri C, Staines W, Messier C. Immunohistochemical localization and quantification of glucose transporters in the mouse brain. Neuroscience. 2002;111(1):19–34. doi: 10.1016/s0306-4522(01)00619-4. doi:S0306452201006194 [pii] [DOI] [PubMed] [Google Scholar]
- Chua LM, Lim ML, Chong PR, Hu ZP, Cheung NS, Wong BS. Impaired neuronal insulin signaling precedes Abeta42 accumulation in female AbetaPPsw/PS1DeltaE9 mice. J Alzheimers Dis. 2012;29(4):783–791. doi: 10.3233/JAD-2012-111880. doi:10.3233/JAD-2012-111880. [DOI] [PubMed] [Google Scholar]
- Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55(10):2565–2582. doi: 10.1007/s00125-012-2644-8. doi:10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft S. Alzheimer disease: Insulin resistance and AD--extending the translational path. Nat Rev Neurol. 2012;8(7):360–362. doi: 10.1038/nrneurol.2012.112. doi:nrneurol.2012.112 [pii] 10.1038/nrneurol.2012.112. [DOI] [PubMed] [Google Scholar]
- Crane PK, Walker R, Hubbard RA, Li G, Nathan DM, Zheng H, Larson EB. Glucose levels and risk of dementia. N Engl J Med. 2013;369(6):540–548. doi: 10.1056/NEJMoa1215740. doi:10.1056/NEJMoa1215740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice FG, Ferreira ST. Inflammation, defective insulin signaling, and mitochondrial dysfunction as common molecular denominators connecting type 2 diabetes to Alzheimer disease. Diabetes. 2014;63(7):2262–2272. doi: 10.2337/db13-1954. doi:10.2337/db13-1954. [DOI] [PubMed] [Google Scholar]
- de la Monte SM. Insulin resistance and Alzheimer's disease. BMB Rep. 2009;42(8):475–481. doi: 10.5483/bmbrep.2009.42.8.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Monte SM. Therapeutic targets of brain insulin resistance in sporadic Alzheimer's disease. Front Biosci (Elite Ed) 2012;4:1582–1605. doi: 10.2741/482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiandaca MS, Kapogiannis D, Mapstone M, Boxer A, Eitan E, Schwartz JB, Goetzl EJ. Identification of preclinical Alzheimer's disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement. 2015;11(6):600–607. e601. doi: 10.1016/j.jalz.2014.06.008. doi:10.1016/j.jalz.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl EJ, Boxer A, Schwartz JB, Abner EL, Petersen RC, Miller BL, Kapogiannis D. Low neural exosomal levels of cellular survival factors in Alzheimer's disease. Ann Clin Transl Neurol. 2015;2(7):769–773. doi: 10.1002/acn3.211. doi:10.1002/acn3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl EJ, Boxer A, Schwartz JB, Abner EL, Petersen RC, Miller BL, Kapogiannis D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology. 2015;85(1):40–47. doi: 10.1212/WNL.0000000000001702. doi:10.1212/WNL.0000000000001702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Fox AS, Chang L, Schafter A, Arelin K, Burmann I, Margulies AD. Tight fitting genes: finding relations between statistical maps and gene expression patterns. 2014 Retrieved from http://f1000research.com/posters/1097120.
- Gray SM, Meijer RI, Barrett EJ. Insulin regulates brain function, but how does it get there? Diabetes. 2014;63(12):3992–3997. doi: 10.2337/db14-0340. doi:10.2337/db14-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo CA, Piroli GG, Hendry RM, Reagan LP. Insulin-stimulated translocation of GLUT4 to the plasma membrane in rat hippocampus is PI3-kinase dependent. Brain Res. 2009;1296:35–45. doi: 10.1016/j.brainres.2009.08.005. doi:10.1016/j.brainres.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87(1):99–109. doi: 10.1016/j.biochi.2004.10.019. doi:10.1016/j.biochi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. doi:10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn KL, Hohnen-Behrens C, Cederberg A, Wu LE, Turner N, Yuasa T, James DE. IRS1-independent defects define major nodes of insulin resistance. Cell Metab. 2008;7(5):421–433. doi: 10.1016/j.cmet.2008.04.005. doi:10.1016/j.cmet.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapogiannis D, Boxer A, Schwartz JB, Abner EL, Biragyn A, Masharani U, Goetzl EJ. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical Alzheimer's disease. FASEB J. 2015;29(2):589–596. doi: 10.1096/fj.14-262048. doi:10.1096/fj.14-262048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JT, Ibrahimi S, Zhao L. Human ApoE Isoforms Differentially Modulate Glucose and Amyloid Metabolic Pathways in Female Brain: Evidence of the Mechanism of Neuroprotection by ApoE2 and Implications for Alzheimer's Disease Prevention and Early Intervention. J Alzheimers Dis. 2015;48(2):411–424. doi: 10.3233/JAD-150348. doi:10.3233/JAD-150348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QL, Yang F, Rosario ER, Ubeda OJ, Beech W, Gant DJ, Cole GM. Beta-amyloid oligomers induce phosphorylation of tau and inactivation of insulin receptor substrate via c-Jun N-terminal kinase signaling: suppression by omega-3 fatty acids and curcumin. J Neurosci. 2009;29(28):9078–9089. doi: 10.1523/JNEUROSCI.1071-09.2009. doi:10.1523/JNEUROSCI.1071-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamelak M. Sporadic Alzheimer's disease: the starving brain. J Alzheimers Dis. 2012;31(3):459–474. doi: 10.3233/JAD-2012-120370. doi:10.3233/JAD-2012-120370. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Reagan LP. Glucose transporter expression in the central nervous system: relationship to synaptic function. Eur J Pharmacol. 2004;490(1-3):13–24. doi: 10.1016/j.ejphar.2004.02.041. doi:10.1016/j.ejphar.2004.02.041 S0014299904001980 [pii] [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr., Kawas CH, Phelps CH. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. doi:10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr ME, Salinas A, Buffenstein R, Oddo S. Mammalian target of rapamycin hyperactivity mediates the detrimental effects of a high sucrose diet on Alzheimer's disease pathology. Neurobiol Aging. 2014;35(6):1233–1242. doi: 10.1016/j.neurobiolaging.2013.12.006. doi:10.1016/j.neurobiolaging.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott A, Stolk RP, van Harskamp F, Pols HA, Hofman A, Breteler MM. Diabetes mellitus and the risk of dementia: The Rotterdam Study. Neurology. 1999;53(9):1937–1942. doi: 10.1212/wnl.53.9.1937. [DOI] [PubMed] [Google Scholar]
- Pearson-Leary J, McNay EC. Intrahippocampal administration of amyloid-beta(1-42) oligomers acutely impairs spatial working memory, insulin signaling, and hippocampal metabolism. J Alzheimers Dis. 2012;30(2):413–422. doi: 10.3233/JAD-2012-112192. doi:10.3233/JAD-2012-112192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson TM, Kramer DL, Rondinone CM. Serine/threonine phosphorylation of IRS-1 triggers its degradation: possible regulation by tyrosine phosphorylation. Diabetes. 2001;50(1):24–31. doi: 10.2337/diabetes.50.1.24. [DOI] [PubMed] [Google Scholar]
- Penny W, Holmes AP. Random Effects Analysis Human Brain Function. Elsevier; San Diego: 2004. [Google Scholar]
- Pini L, Pievani M, Bocchetta M, Altomare D, Bosco P, Cavedo E, Frisoni GB. Brain atrophy in Alzheimer's Disease and aging. Ageing Res Rev. 2016 doi: 10.1016/j.arr.2016.01.002. doi:10.1016/j.arr.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Sankar R, Thamotharan S, Shin D, Moley KH, Devaskar SU. Insulin-responsive glucose transporters-GLUT8 and GLUT4 are expressed in the developing mammalian brain. Brain Res Mol Brain Res. 2002;107(2):157–165. doi: 10.1016/s0169-328x(02)00487-4. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Maness PF. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr Opin Neurobiol. 2008;18(3):245–250. doi: 10.1016/j.conb.2008.07.015. doi:10.1016/j.conb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrijvers EM, Witteman JC, Sijbrands EJ, Hofman A, Koudstaal PJ, Breteler MM. Insulin metabolism and the risk of Alzheimer disease: the Rotterdam Study. Neurology. 2010;75(22):1982–1987. doi: 10.1212/WNL.0b013e3181ffe4f6. doi:10.1212/WNL.0b013e3181ffe4f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Goldberg JL, Qiao LY, Mitchell JJ. Insulin-induced insulin receptor substrate-1 degradation is mediated by the proteasome degradation pathway. Diabetes. 1999;48(7):1359–1364. doi: 10.2337/diabetes.48.7.1359. [DOI] [PubMed] [Google Scholar]
- Talbot K, Wang HY. The nature, significance, and glucagon-like peptide-1 analog treatment of brain insulin resistance in Alzheimer's disease. Alzheimers Dement. 2014;10(1 Suppl):S12–25. doi: 10.1016/j.jalz.2013.12.007. doi:10.1016/j.jalz.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Wang HY, Kazi H, Han LY, Bakshi KP, Stucky A, Arnold SE. Demonstrated brain insulin resistance in Alzheimer's disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J Clin Invest. 2012;122(4):1316–1338. doi: 10.1172/JCI59903. doi:59903 [pii] 10.1172/JCI59903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7(2):85–96. doi: 10.1038/nrm1837. doi:10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- Wang WY, Yu JT, Liu Y, Yin RH, Wang HF, Wang J, Tan L. Voxel-based meta-analysis of grey matter changes in Alzheimer's disease. Transl Neurodegener. 2015;4:6. doi: 10.1186/s40035-015-0027-z. doi:10.1186/s40035-015-0027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette AA, Bendlin BB, Starks EJ, Birdsill AC, Johnson SC, Christian BT, Asthana S. Association of Insulin Resistance With Cerebral Glucose Uptake in Late Middle-Aged Adults at Risk for Alzheimer Disease. JAMA Neurol. 2015;72(9):1013–1020. doi: 10.1001/jamaneurol.2015.0613. doi:10.1001/jamaneurol.2015.0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette AA, Johnson SC, Birdsill AC, Sager MA, Christian B, Baker LD, Bendlin BB. Insulin resistance predicts brain amyloid deposition in late middle-aged adults. Alzheimers Dement. 2015;11(5):504–510. e501. doi: 10.1016/j.jalz.2014.03.011. doi:10.1016/j.jalz.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette AA, Modanlo N, Kapogiannis D, Alzheimer's Disease Neuroimaging Initiative Insulin resistance predicts medial temporal hypermetabolism in mild cognitive impairment conversion to Alzheimer disease. Diabetes. 2015;64(6):1933–1940. doi: 10.2337/db14-1507. doi:10.2337/db14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette AA, Xu G, Johnson SC, Birdsill AC, Jonaitis EM, Sager MA, Bendlin BB. Insulin resistance, brain atrophy, and cognitive performance in late middle-aged adults. Diabetes Care. 2013;36(2):443–449. doi: 10.2337/dc12-0922. doi:10.2337/dc12-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Krishnan A, Wager TD. Cluster-extent based thresholding in fMRI analyses: pitfalls and recommendations. Neuroimage. 2014;91:412–419. doi: 10.1016/j.neuroimage.2013.12.058. doi:10.1016/j.neuroimage.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.