Abstract
In a screen for zebrafish larval mutants with excessive liver lipid accumulation (hepatic steatosis), we identified harvest moon (hmn). Cytoplasmic lipid droplets, surrounded by multivesicular structures and mitochondria whose cristae appeared swollen, are seen in hmn mutant hepatocytes. Whole body triacylglycerol is increased in hmn mutant larvae. When we attempted to raise mutants, which were morphologically normal at the developmental stage that the screen was conducted, to adulthood, we observed that most hmn mutants do not survive to the juvenile period when raised. An arrest in growth occurs in the late larval period without obvious organ defects. Maternal zygotic mutants have no additional defects, suggesting that the mutation affects a late developmental process. The developmental window between embryogenesis and the metamorphosis remains under-studied, and hmn mutants might be useful for exploring the molecular and anatomic processes occurring during this transition period.
Keywords: genetic screen, hepatic steatosis, larval staging, zebrafish
Introduction
Evolutionarily conserved mechanisms are employed to defend against starvation. The liver is a central component of the infrastructure used to supply energy to vital organs during fasting. Orchestration of the metabolic processes during fasting involves multiple factors, including hormonal inputs, metabolite transport, altered gene expression, and large scale proteolysis and recycling of other macromolecules (Madrigal-Matute & Cuervo, 2016). To identify novel factors that participate in the metabolic transition from fed to fasted states, several laboratories have employed a zebrafish genetic system. This approach hinges on (i) the realization that master transcription factors, metabolic regulators, and multiple enzymes and transporters that control nutrient homeostasis in higher metazoa are present in zebrafish (Babin & Vernier, 1989; Ibabe et al. 2002; Schlombs et al. 2003; Matthews et al. 2009; Thakur et al. 2011; Hugo et al. 2012; Gu et al. 2014; Karanth et al. 2016); (ii) the feasibility of rapid screening for morphological and histological changes in the transparent livers of zebrafish larvae (Pack et al. 1996; Sadler et al. 2005), and (iii) the convenience of whole mount staining for select macromolecules. For our purposes, liver lipid stores are low in never-fed zebrafish larvae and excessive accumulation of triacylglycerol can be reliably and rapidly stained with conventional neutral lipid dyes, with a satisfactory spatio-temporal resolution (Schlombs et al. 2003; Schlegel & Stainier, 2006; Raldúa et al. 2008; Passeri et al. 2009; Imrie & Sadler, 2010; Progatzky et al. 2014; Chen et al. 2015; Levic et al. 2015; Liu et al. 2015).
Here we report the results of a genetic screen for mutants with hepatic steatosis, the excessive and inappropriate accumulation of neutral lipids in hepatocytes, that revealed a locus required for late larval growth and survival in zebrafish. The hmn mutant was found to have abnormal lipid accumulation in the liver. Furthermore, the accumulated lipids, which reflect an increase in triacylglycerol exclusively, are found in cytoplasmic droplets that are in close apposition to multivesicular structures suggestive of halted autophagosomal processing. Additionally, we observed that although never-fed hmn larvae are not more sensitive to starvation than wild-type (WT) larvae, they nevertheless fail to thrive when fed, dying in the late larval period at an arrested developmental stage. This study reveals a potential molecular handle on examining the developmental cues that are required for larval growth and transition through metamorphosis. Metabolic phenotyping in the emerging zebrafish model should consider the role of organ and whole-animal growth.
Materials and methods
Zebrafish mutagenesis
All animal studies were approved by the University of Utah Institutional Animal Care and Use Committee. Twelve male AB strain adults that were confirmed to be homozygous carriers of the WT slc16a6a allele (Hugo et al. 2012) were exposed to 3 cycles of N-ethylnitrosourea, and then bred to generate 200 F2 families. At least six sibling matings were performed per family. A specific locus test was not performed.
Oil Red O screening
The hmnz110 mutant was identified by fixing and staining 7-dpf larvae (n = 40–50) derived from in-crosses of F2 family members with 0.3% Oil Red O (ORO) dissolved in 60% propanol exactly as we described previously (Schlegel & Stainier, 2006). Only fully penetrant (25% of the larvae) and fully expressive (unambiguous, uniform increase in ORO liver-staining) mutants were analyzed further.
Nile Red staining
Larvae were placed in 5 mg L−1 Nile Red (Sigma) for 1 h at 28 °C, and then photographed under a GFP long-pass filter described previously (Schlegel & Stainier, 2006), except we used an AF6000 microscope (Leica Microsystems). Representative pictures were acquired with an attached DFC319FX camera (Cruz-Garcia & Schlegel, 2014).
Transmission electron microscopy
Larvae were fixed in 2.5% paraformaldehyde and 1% glutaraldehyde in 100 mm sodium cacodylate (pH 7.4), and counterstained with OsO4. Electron microscopy was performed using a JEOL JEM-1400 Plus transmission electron microscope at the University of Utah Electron Microscopy Core facility (Hugo et al. 2012; Karanth et al. 2013; Cruz-Garcia & Schlegel, 2014).
Lipid and glucose measurements
Free (not phosphorylated) glucose was measured in four individual pools of 10 larvae using a glucose oxidase kit (Biovision) exactly as described previously (Jurczyk et al. 2011). Glucose abundance was normalized to protein mass, which was assayed using a cuprous cation-bicinchoninic acid kit (ThermoFisher Pierce BCA) kit. Whole larval lipids were quantified as described previously (Hugo et al. 2012; Karanth et al. 2013).
Morphometric measurements
Standard length (SL), the distance from the snout to the caudle peduncle, and standardized standard length (SSL), a validated staging method for the larval and juvenile period that relies on the scoring of specific anatomic structures, were measured exactly as described (Parichy et al. 2009).
Survival experimental diets
Beginning at 7 dpf, 546 WT larvae and 547 hmn mutants were raised under normal laboratory conditions. Larvae were monitored for death every day by inspection for spontaneous movement. From days 7 to 10 dpf, 25 larvae were placed in 150 mL of saltwater (9 ppt) rotifers (Brachionus plicatilis, Reed Mariculture, San Jose, CA, USA; final rotifer concentration 250 individuals mL−1). These fish were fed 200 μL of diluted RotigrowPlus Algae (Reed Mariculture) at 9:00 and 16:00 h to support rotifer growth (final rotifer concentration was 250 mL−1). The diluted algae was made by mixing 10 mL RotigrowPlus (Reed Mariculture) and 40 mL of saltwater (6 ppt). From 11 to 17 dpf, system water was turned on to the tanks, and the fry were fed fresh filtered rotifers at 9:00 and 14:00 h (approximately 1500–2000 rotifers per fry per day), fresh-hatched Artemia at 9:00 and 16:00 h (approximately 800–900 napulii per fry per day), and Fry Hatch Encapsulation (Argent Aquaculture, Redmond, WA, USA) mixed 1 : 1 with Gemma micro-75 (Skretting, Westbrook, ME, USA) at 11:00, 13:00, and 17:00 h (approximately 3 mg per fry per day). From 18 to 24 dpf, the diet was the same as that for 11–17 dpf, except that Gemma micro-150 (Skretting) was used instead of the Gemma Micro-75. From 25 dpf onward, fish were fed fresh-hatched Artemia at 9:00 and 16:00 h (approximately 800–900 napulii per fry per day), and Gemma micro-300 (Skretting, Westbrook, ME) at 1:00 pm (approximately 5 mg per fry per day).
Statistical analysis
The two-tailed Student’s t-test was used to compare lipid and glucose levels, and for comparing SL and SSL. The SSL was treated as a continuous variable because intermediate stages were interpolated from the reference stages. For survival analysis, the log rank test was performed. Metabolites are reported as mean ± SEM.
Results
Isolation and initial characterization of hmn, a zebrafish mutant with hepatic steatosis
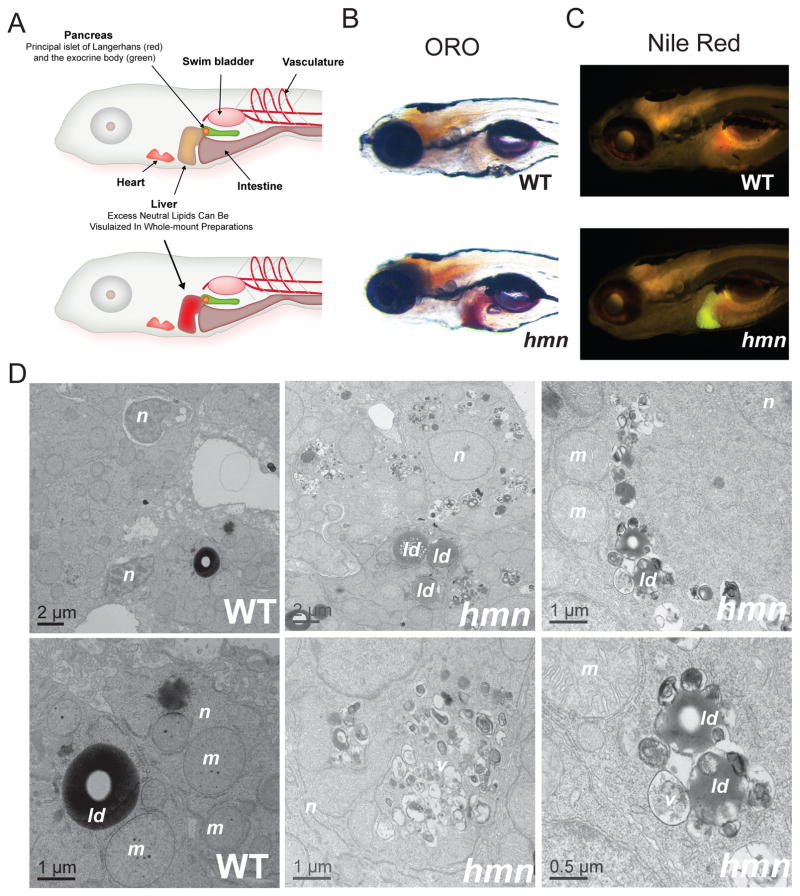
The hmn mutant was isolated using the same screening strategy we employed previously (Hugo et al. 2012). Namely, 7-dpf larvae derived from a conventional F2-recessive genetic screen were fixed and stained with ORO and examined for inappropriate hepatic lipid accumulation using whole-mount bright-field microscopic inspection (Fig. 1A,B). We also observed that staining of hmn mutants with the vital lipid stain Nile Red demonstrated excess lipid accumulation in the liver (Fig. 1C).
Fig. 1.
Identification of the hepatic steatosis mutant hmn. (A) Larval zebrafish anatomy and screening strategy for finding mutants with hepatic lipid accumulation. After exhaustion of the maternally deposited nutrients in the yolk, and commencement of feeding, larval zebrafish (shown here at 7 dpf, approximately) remain transparent, allowing evaluation of the development and function of the indicated metabolic organs. The genetic screening strategy used in this proposal relies on the ease of staining livers of never-fed larvae for excessive lipid accumulation. Modified from Schlegel & Gut (2015). (B,C) Whole-mount ORO and Nile Red staining of WT and hmn mutant larvae at 7 dpf revealed increased neutral lipid accumulation in hmn livers. (D) Transmission electron micrographs of WT and hmn mutant livers from 7-dpf larvae revealed the presence of numerous cytoplasmic lipid droplets (ld) in the latter, often in close proximity to multivesicular structures (v) and swollen mitochondria (m). The hepatocyte nuclei (n) appeared normal in hmn mutants. Note some nuclei labeled in the WT panel are of cholangiocytes and are more heterochromatic compared with the hepatocytes labeled for hmn mutants.
The accumulation of neutral lipids in hmn livers was confirmed with transmission electron microscopy (Fig. 1D). Numerous cytoplasmic lipid droplets were seen in hmn hepatocytes, but few lipid droplets were seen in the livers of WT siblings. Furthermore, the lipid droplets in hmn mutant livers were surrounded by multivesicular structures and mitochondria whose cristae appeared swollen. There was no evidence of inflammatory cell infiltration or excessive deposition of extracellular matrix.
Increased triacylglycerol in hmn mutants
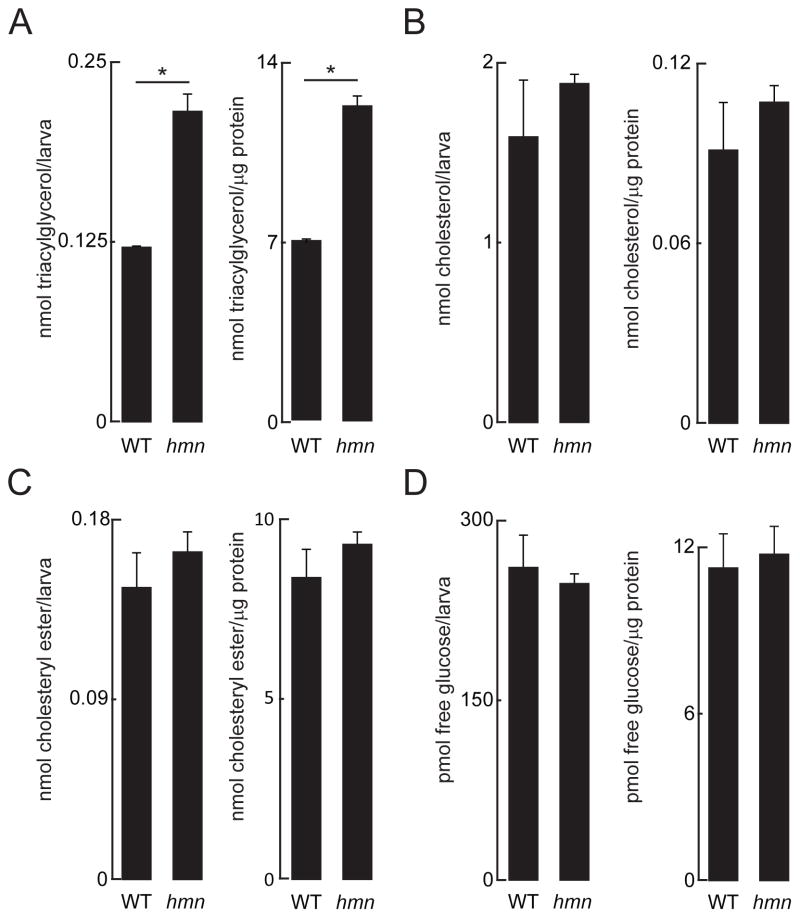
The hmn mutants showed increased whole-carcass triacylglycerol, and unchanged levels of cholesteryl ester and unesterified cholesterol (Fig. 2A–C). This pattern of elevated triacylglycerol and no changes in cholesteryl esters is commonly seen in human fatty liver disease (Cohen et al. 2011), and the lack of increase in cholesteryl esters argues against a lysosomal cholesteryl ester hydrolytic defect (Porto, 2014). Next we assessed whether mutation of hmn alters the amount of free glucose available in larvae. The abundance of free glucose in zebrafish embryos and larvae closely correlates with gluconeogenic transcriptional activation (Jurczyk et al. 2011; Gut et al. 2013); hmn mutant larvae did not show a change in free glucose compared with wild-type siblings at 7 dpf. The results of these analyses did not differ whether examined on a per-larva basis or when normalized to protein concentration, arguing that the size and mass the liver did not influence the accumulation of triacylglycerol (Tschop et al. 2012).
Fig. 2.
Increased triacylglycerol in hmn mutant larvae. (A) Triacylglycerol (B) unesterified cholesterol (C) cholesteryl esters and (D) free glucose were measured in lysates of 7-dpf larvae. *P < 0.01. Results are reported as mean ± SEM abundance of analyte per larva and abundance of analyte per mass of protein.
Arrested development and death in late larval period in hmn mutants
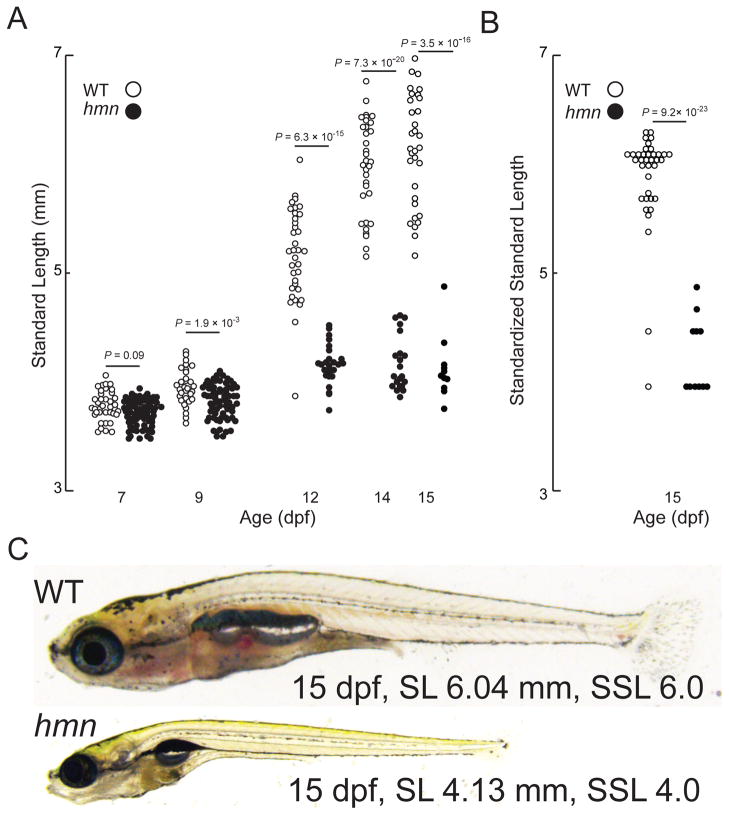
Next, we attempted to raise larvae to adulthood. We found that hmn mutant larvae showed a failure to grow, with a decrease in SL compared with WT manifesting by 9 dpf (Fig. 3A). To address whether there was a defect in linear growth or an arrest in progression through developmental stages, we evaluated the SSL of the surviving larvae at 15 dpf (Parichy et al. 2009). Compared with WT larvae, hmn mutants (sorted and raised separately beginning 7 dpf) showed an arrest in development, with a median SSL of 4.0 at 15 dpf compared with a median SSL of 6.0 in WT larvae (Fig. 3B). Finally, 15-dpf larvae all showed normal intestinal food content, suggesting that a there was no major defect in feeding per se (Fig. 3C).
Fig. 3.
Most hmn mutants undergo an arrest of development during the late larval period. (A) The SL of 39 WT and 39 hmn mutant (MUT) larvae was measured on the indicated age (days post-fertilization, dpf). Note death of larvae is reflected by decreasing numbers of animals. (B) The SSL (a unitless index) was measured in the surviving 15 dpf larvae from panel B. (C) Representative (median SL and SSL for both) bright-field micrographs of 15-dpf WT and hmn mutant larvae, demonstrating food in the intestine of both WT and hmn mutant larvae.
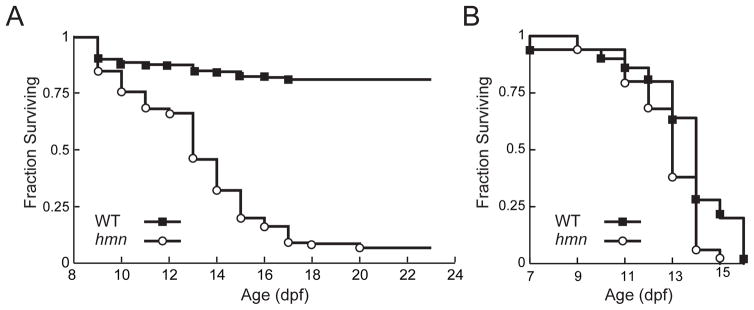
To quantify the effect of this failure to grow on survival, we attempted to raise 7-dpf larvae on the conventional diet used in our facility, and scored for death over the next 3 weeks. The median survival of hmn mutants was 13 dpf, and nearly 90% died by 21 dpf (Fig. 4A). In a replicate cohort, housing hmn mutant larvae individually did not affect their death during the larval period (not shown). Because larvae were co-cultured with live feed (Rofiera spp.), we asked whether hmn mutant larvae were also sensitive to death by deprivation of food. This experiment was motivated by our previous isolation of a zebrafish mutant with hepatic steatosis that was nutritionally suppressible (Hugo et al. 2012): mutation of the liver ketone body exporter gene slc16a6a causes accumulation of triacylglycerol in zebrafish larval livers during fasting, and renders animals susceptible to death by starvation. We found no difference in survival when larvae were never fed (Fig. 4B), suggesting that it is in an active growth (feeding state) that the loss of hmn is consequential.
Fig. 4.
A fraction of hmn mutants survive the late larval period. (A) In all, 546 WT larvae and 547 hmn mutant larvae were raised under normal laboratory conditions. Death was scored on the indicated days post-fertilization (dpf). For survival analysis, χ2 = 562, on 1 degree of freedom. P < 10−100. (B) In all, 100 WT larvae, and 100 hmn mutant larvae were scored for death on the indicated dpf. The median survival of WT larvae was 14.5 ± 2 dpf, and the median survival of hmn mutants was 14 ± 4 dpf. For survival analysis, χ2 = 2.4 on 1 degree of freedom. P = 0.123.
The hmn mutants that survived to adulthood were fertile, and the majority of their offspring had no additional defects when stained with ORO at 7 dpf: hepatic steatosis was seen in all larvae, and 10–15% showed cranial-facial defects that varied in frequency and severity from individual parental pair to pair. Thus, there did not seem to be additional, fully penetrant phenotypes revealed by examining maternal zygotic mutants.
Discussion
We conducted a genetic screen to identify new genes that participate in liver metabolism. We found that hmn mutant larvae accumulate excess neutral lipids in their livers. These stored lipids are in droplets surrounded by multivesicular structures and swollen mitochondria, suggesting that the isolated mutation perturbs normal autophagosomal processing, mitochondrial homeostasis, or both. In early zebrafish larvae, autophagy in heart and brain is observed (Hu et al. 2011; Lee et al. 2014). Nevertheless, the role of autophagy in fasting liver metabolism in an organism that can survive long bouts of fasting, such as zebrafish, has not been reported. Comprehensive examination of autophagic processes in older larvae and juveniles transitioning through metamorphosis would be of value if sufficient numbers of surviving hmn adults could be raised to perform the analyses. We plan to perform such a large-scale breeding effort to generate enough adult hmn mutants for such morphometric and biochemical studies. A more thorough examination of the body composition of these adults, including examination of the visceral and subcutaneous adipose stores, is also warranted (McMenamin et al. 2013).
Beyond the additional experiments to elucidate ultrastructural, biochemical, and molecular aspects of the hmn physiological phenotypes, the arrested development of these mutants requires further exploration. This developmental arrest was completely unexpected because we excluded mutants with morphological defects at the age of screening from analysis, positing that this approach would recover viable mutants, as we observed in a previous screen (Hugo et al. 2012). Since the arrested development in hmn mutants affected development of larvae in a uniform manner (i.e. there was not obvious, single-organ sparing or death), examining the role(s) of the hmn gene in individual cell types will require isolation of the molecular lesion. Crossing the isolated mutant allele with available null alleles will also allow us to leverage classical genetic approaches to elucidate hmn function (Kettleborough et al. 2013). If null alleles are not available, gene targeting with genome editing tools might be necessary to generate an allelic series (Reischauer et al. 2016). Finally, although the maternal zygotic mutants did not show additional phenotypes (i.e. it is likely that maternally deposited hmn mRNA is not required for oocyte and early embryo development), it is possible that generation of conditional hmn alleles would be of value to elucidate the role of the gene mutated in hmn mutant, should the gene be widely expressed.
Acknowledgments
Kazuyuki Hoshijima and David J. Grunwald provided guidance with chemical mutagenesis. Maurine Hobbs provided details on the diets used in the Centralized Zebrafish Animal Resource of the University of Utah. This work was supported by US NIH grant R01-DK096710 to A.S.
Footnotes
Author contributions
Conceptualization, A.S.; Methodology, S.E.H. and A.S.; Investigation, S.E.H. and A.S.; Writing – Original Draft, S.E.H. and A.S.; Writing – Reviewing & Editing, S.E.H. and A.S.; Funding Acquisition, A.S.; Resources, S.E.H. and A.S.; Supervision, A.S.
References
- Babin PJ, Vernier JM. Plasma lipoproteins in fish. J Lipid Res. 1989;30:467–489. [PubMed] [Google Scholar]
- Chen K, Wang CQ, Fan YQ, et al. Optimizing methods for the study of intravascular lipid metabolism in zebrafish. Mol Med Rep. 2015;11:1871–1876. doi: 10.3892/mmr.2014.2895. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Garcia L, Schlegel A. Lxr-driven enterocyte lipid droplet formation delays transport of ingested lipids. J Lipid Res. 2014;55:1944–1958. doi: 10.1194/jlr.M052845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Yang X, Lin L, et al. Genetic ablation of solute carrier family 7a3a leads to hepatic steatosis in zebrafish during fasting. Hepatology. 2014;60:1929–1941. doi: 10.1002/hep.27356. [DOI] [PubMed] [Google Scholar]
- Gut P, Baeza-Raja B, Andersson O, et al. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol. 2013;9:97–104. doi: 10.1038/nchembio.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhang J, Zhang Q. Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy. 2011;7:1514–1527. doi: 10.4161/auto.7.12.18040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo SE, Cruz-Garcia L, Karanth S, et al. A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 2012;26:282–293. doi: 10.1101/gad.180968.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibabe A, Grabenbauer M, Baumgart E, et al. Express ion of peroxisome proliferator-activated receptor’s in zebrafish. Histochem Cell Biol. 2002;118:231– 239. doi: 10.1007/s00418-002-0434-y. [DOI] [PubMed] [Google Scholar]
- Imrie D, Sadler KC. White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev Dyn. 2010;239:3013–3023. doi: 10.1002/dvdy.22443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyk A, Roy N, Bajwa R, et al. Dynamic glucoregulation and mammalian-like responses to metabolic and developmental disruption in zebrafish. Gen Comp Endocrinol. 2011;170:334–345. doi: 10.1016/j.ygcen.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanth S, Tran VM, Balagurunathan K, et al. Polyunsaturated fatty acyl-Coenzyme As are inhibitors of cholesterol biosynthesis. Dis Model Mech. 2013;6:1365–1377. doi: 10.1242/dmm.013425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanth S, Zinkhan EK, Hill JT, et al. FOXN3 regulates hepatic glucose utilization. Cell Rep. 2016;15:2745–2755. doi: 10.1016/j.celrep.2016.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettleborough RN, Busch-Nentwich EM, Harvey SA, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Koo Y, Ng A, et al. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy. 2014;10:572–587. doi: 10.4161/auto.27649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levic DS, Minkel JR, Wang WD, et al. Animal model of Sar1b deficiency presents lipid absorption deficits similar to Anderson disease. J Mol Med (Berl) 2015;93:165–176. doi: 10.1007/s00109-014-1247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Gates KP, Fang L, et al. Apoc2 loss-of-function zebrafish mutant as a genetic model of hyperlipidemia. Dis Model Mech. 2015;8:989–998. doi: 10.1242/dmm.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal-Matute J, Cuervo AM. Regulation of liver metabolism by autophagy. Gastroenterology. 2016;150:328–339. doi: 10.1053/j.gastro.2015.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews RP, Lorent K, Manoral-Mobias R, et al. TNFα–dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish s-adenosylhomocysteine hydrolase. Development. 2009;136:865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMenamin SK, Minchin JEN, Gordon TN, et al. Dwarfism and increased adiposity in the gh1 mutant zebrafish vizzini. Endocrinology. 2013;154:1476–1487. doi: 10.1210/en.2012-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack M, Solnica-Krezel L, Malicki J, et al. Mutations affecting development of zebrafish digestive organs. Development. 1996;123:321–328. doi: 10.1242/dev.123.1.321. [DOI] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, et al. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passeri MJ, Cinaroglu A, Gao C, et al. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49:443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porto AF. Lysosomal acid lipase deficiency: diagnosis and treatment of Wolman and Cholesteryl Ester Storage Diseases. Pediatr Endocrinol Rev. 2014;12(Suppl 1):125–132. [PubMed] [Google Scholar]
- Progatzky F, Sangha NJ, Yoshida N, et al. Dietary cholesterol directly induces acute inflammasome-dependent intestinal inflammation. Nat Commun. 2014;5:5864. doi: 10.1038/ncomms6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raldúa D, André M, Babin PJ. Clofibrate and gemfibrozil induce an embryonic malabsorption syndrome in zebrafish. Toxicol Appl Pharmacol. 2008;228:301–314. doi: 10.1016/j.taap.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Reischauer S, Stone OA, Villasenor A, et al. Cloche is a bHLH-PAS transcription factor that drives haemato-vascular specification. Nature. 2016;535:294–298. doi: 10.1038/nature18614. [DOI] [PubMed] [Google Scholar]
- Sadler KC, Amsterdam A, Soroka C, et al. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- Schlegel A, Gut P. Metabolic insights from zebrafish genetics, physiology, and chemical biology. Cell Mol Life Sci. 2015;72:2249–2260. doi: 10.1007/s00018-014-1816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, Stainier DY. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry. 2006;45:15179–15187. doi: 10.1021/bi0619268. [DOI] [PubMed] [Google Scholar]
- Schlombs K, Wagner T, Scheel J. Site-1 protease is required for cartilage development in zebrafish. Proc Natl Acad Sci USA. 2003;100:14024–14029. doi: 10.1073/pnas.2331794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur PC, Stuckenholz C, Rivera MR, et al. Lack of de novo phosphatidylinositol synthesis leads to endoplasmic reticulum stress and hepatic steatosis in cdipt-deficient zebrafish. Hepatology. 2011;54:452–462. doi: 10.1002/hep.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop MH, Speakman JR, Arch JRS, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2012;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]