Abstract
Quantitative hepatic perfusion parameters derived by fitting dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI) of liver to a pharmacokinetic model are prone to errors if the dynamic images are not corrected for respiratory motion by image registration. The contrast-induced intensity variations in pre- and postcontrast phases pose challenges for the accuracy of image registration. We propose an overdetermined system of transformation equations between the image volumes in the DCE-MRI series to achieve robust alignment. In this method, we register each volume to every other volume. From the transforms produced by all pairwise registrations, we constructed an overdetermined system of transform equations that was solved robustly by minimizing the L1/2-norm of the residuals. This method was evaluated on a set of 100 liver DCE-MRI examinations from 35 patients by examining the area under spikes appearing in the voxel time–intensity curves. The robust alignment procedure significantly reduced the area under intensity spikes compared with unregistered volumes (P<.001) and volumes registered to a single reference phase (P<.001). Our registration procedure provides a larger number of reliable time–intensity curve samples. The additional reliable samples in the precontrast baseline are important for calculating the postcontrast signal enhancement and thereby for converting intensity to contrast concentration. On the intensity ramp, retained samples help to better describe the uptake dynamics, providing a better foundation for parameter estimation. The presented method also simplifies the analysis of data sets with many patients by eliminating the need for manual intervention during registration.
Keywords: DCE-MRI, registration, contrast agent, outlier rejection, overdetermined, liver, perfusion, parameter estimation
INTRODUCTION
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) in the liver enables quantification of hepatic perfusion for assessment of tumor response to radiation therapy, as well as normal tissue damage (1–5). One of the challenges in liver DCE-MRI is respiratory motion during acquisition, which can cause blurring and artifacts in the images and intensity variations in the dynamic curves, and subsequently, errors in perfusion quantification using pharmacokinetic (PK) models. To reduce respiratory motion-induced degradation of liver images during DCE-MRI acquisition, a breathing control paradigm has been proposed, during which the patient is instructed to hold their breath multiple times (taking breaths in between) (5). The dynamic image volumes acquired during the 2–3 minutes of contrast uptake are still misaligned because of poor reproducibility of the liver position during multiple breath holds, loss of control during breath holding, and occasionally gross movement of the patient. Therefore, a voxel cannot be assumed to cover the same piece of tissue throughout the whole examination, but, instead, it covers different pieces at different points in time. Therefore, if not corrected for, breathing and gastrointestinal motions can induce artifacts, such as intensity spikes and wide bumps, in the contrast uptake curves. To estimate the arterial and portal venous perfusion in the liver, a sufficient number of reliable samples of the uptake curve must be collected to fully describe the contrast uptake dynamics. The shape of the precontrast plateau and the beginning of the intensity ramp following contrast agent administration can have a large effect on the estimated perfusion parameters, in particular on hepatic arterial perfusion. Unwanted artifacts introduced into the uptake curve by patient motion reduce the number of reliable samples, increasing the risk of inaccurate PK model parameter estimation.
To correct for image misalignment in the DCE series, image registration has been used to align the magnetic resonance (MR) images at different points in time or phases (1, 6–8). However, registration of the DCE-MRI phases has some unique challenges, for example, a large number of dynamic image volumes, low spatial resolution, and low contrast-to-noise ratio that are tradeoffs for the high temporal resolution. In addition, the image contrast is changed completely over the whole course from precontrast to rapid contrast uptake and to full contrast enhancement. Combined with all these effects, the dynamic contrast agent-induced intensity changes pose a difficult problem even to registration methods using multimodality image-to-image metrics such as mutual information (9–11).
A number of methods have been developed to tackle the DCE-MRI registration problem. Some methods attempt to register each phase to a reference phase selected among all phases. Such methods use either region-limited rigid registration (12) or deformable registration (9, 10). Deformable registration may yield incorrect volume changes around contrast-enhanced lesions unless the method is adapted to explicitly counteract such changes (11). As an alternative to reference phase-based registration, image intensity changes can be modeled using a PK model and the model incorporated into the registration algorithm (13–16). Such PK-based methods can be slow because of the need for repeated PK model parameter estimation. For a faster procedure, principal component analysis (PCA) can be used to generate reference images for registration (17). However, PCA may not be able to adequately describe the spatially varying time–intensity curves. Therefore, the phases could alternatively be decomposed into a low-rank and a sparse image component in a procedure called robust PCA that can be combined with image registration (18, 19). Robust PCA can also be used to combine registration with reconstruction, but this requires access to raw data from the MRI scanner (20). However, when comparing the 3 representative registration methods (21), 1 PK model-based, 1 PCA-based, and 1 sequentially registering each phase to its preceding phase, the sequential registration method performed better (average error 14.7% in estimated parameters) than the more complex iterative, PCA-based and PK model-based methods (average errors: 39.5% and 39.2%, respectively). Nevertheless, all image registration methods reduced errors in the estimated parameters in the tumors in comparison with without image registration methods.
The performance of the sequential method can be understood by considering that the changes between neighboring phases are small. Therefore, registration has a greater chance of success. However, misregistration errors are accumulated and propagated from the earlier phases to the later phases. If registration at the early phase fails, all following phases will be incorrectly aligned with the first phase.
Inspired by the findings in Rajaraman S et al.’s study (21), and taking into account the aforementioned drawback of the sequential registration procedure, we introduce an overdetermined system for achieving robust registration for the DCE image series. In this procedure, rather than registering each phase to a reference phase or to a preceding phase, each phase is registered to every other phase. This produces an overdetermined system of transform equations that can be solved using robust statistical methods to reject outliers corresponding to failed registrations. This new method is evaluated on a set of 100 liver DCE-MRI examinations, and the results are compared with those of a conventional registration method where all phases are registered to 1 postcontrast reference phase. A robust registration method for liver DCE-MRI would greatly simplify the work-flow of clinical liver perfusion studies with a large number of participating patients by eliminating the need for manual intervention following faulty registration.
MATERIALS AND METHODS
Imaging
Under institutional review board approval, 100 DCE-MRI examinations of the liver from 35 patients were included in this study (women, 8; men, 27; age at examination, 51– 83 years; number of examinations per patient, 1– 4). The patients were imaged for about 3 minutes using a repeated breath-hold paradigm (5). For this paradigm, the patients were instructed to initially hold their breath for as long as they could and then hold their breath repeatedly with a single deep inhalation in between each breath hold. Images acquired during deep inhalations showed large liver movement and severe motion blur. Therefore, these were excluded from further analysis. Dynamic MR imaging was started at the beginning of the first breath hold and a gadobenate-dimeglumine-based contrast agent (MultiHance, Bracco S.p.A., Italy) was administered intravenously.
A 3 T MRI scanner (Magnetom Skyra, Siemens Medical Systems, Erlangen, Germany) was used for imaging. A 3-dimensional gradient echo sequence that speeds up acquisition using stochastic sampling with view sharing (time-resolved angiography with stochastic trajectories; TWIST) was used for 10 patients, and a sequence that enhances speed through partial Fourier reconstruction (volume interpolated breath hold examination; VIBE) was used for 25 patients. Sequence parameters varied among the patients (Table 1).
Table 1.
DCE-MRI Sequence Settings for TWIST and VIBE Sequences
| TWIST | VIBE | |
|---|---|---|
| Flip angle | 10–20° | 9° |
| Repetition time | 2.5–2.53 ms | 2.22 ms |
| Echo time | 0.84–0.86 ms | 0.78–0.79 ms |
| Temporal resolution | 2.3–4.3 s | 1.99–6.82 s |
| Voxel volume | 15.6–45.8 mm3 | 34.2–100 mm3 |
| Number of phases | 52–60 | 38–60 |
| Acquisition time | 2.0–3.7 min | 2.0–6.7 min |
| Number of examinations | 31 | 69 |
Abbreviations: DCE-MRI, Dynamic contrast-enhanced magnetic resonance imaging; TWIST, time-resolved angiography with stochastic trajectories; VIBE, volume interpolated breath hold examination.
During a breath hold, the liver should ideally be still. Unfortunately, partial inhalation during a breath hold, imperfect reproducibility of the exhale state, and changes to patient posture result in liver movement between images. These movements, however, are smaller than those observed during free breathing. For the spatial resolution, signal-to-noise ratio, and artifact level seen in this study, we, therefore, chose to use rigid registration to correct for the observed range of motions.
Preprocessing
Before registration, a number of preprocessing steps were performed. These were the same for the robust registration procedure (section Robust Registration) and the conventional procedure (section Conventional Registration) to which it was compared.
Reference Phase Selection
The conventional procedure involved alignment of all phases to 1 reference phase. For each examination, this reference phase was selected among all phases as the phase with the smallest sum of mean absolute distances to the other phases in intensity, which is calculated as follows:
where iref is the index of the reference phase among the nphases, phases, nvoxels is the number of voxels in a phase, and Ii(rk) is the image intensity of the ith phase at a spatial position of the kth voxel, rk. This step makes the selection of the reference phase deterministic and avoid manual intervention. The reference phase was also used for the robust registration procedure to define the coordinate system that the final transforms were given with respect to.
Region-Limited Image Registration
Because we wish to correct for the motion of the liver with a rigid transform and the rest of the body inside the field of view (FOV) does not move in the same way as the liver, region-limited registration was used. For this purpose, a region of interest (ROI) was drawn around the liver on the reference phase. This ROI was drawn snugly along the edge of the liver. For the region-limited registration, it is advantageous to include a margin outside the liver ROI. This margin covers the contrast-rich interface between the liver and the surrounding tissue, and it helps to drive the registration algorithm. The margin was set to 15 mm for all registrations.
Bias Field Correction
MR images commonly contain a spatial intensity variation that is not the result of intrinsic tissue properties but of the position of the tissue with respect to the transmission and receiver radiofrequency coils. This “bias field” can cause misregistration if it is the dominant feature in an image. Fortunately, the bias field typically varies slowly with respect to the spatial position. The effect of the bias field on the images used for registration was reduced by dividing each phase by a filtered version of itself. The modified phases used for registration were then given by the following equation:
where G(r, 0, ρ2 II) is a 3-dimensional Gaussian filter kernel with standard deviation ρ =30 mm in each dimension and * is the convolution operator. These bias-field-corrected images were only used for registration and not for the uptake curve extraction, as the uptake curves could be corrupted by the correction.
Robust Registration
The robust registration procedure for each DCE time series was divided into 5 stages—3 translational registration stages and 2 rigid registration stages with both translation and rotation. For the robust procedure, the workflow for each stage is illustrated in Figure 1. Either each possible pair of phases or each pair in a subset of all possible pairs was registered. Thus, each registered pair gave rise to a rigid transform describing the relative position of the 2 phases of the pair. For the registration of each pair, the first phase of the pair was used as a fixed image and the second as a moving image. The dilated liver ROI was applied as a mask to the fixed image to restrict the domain for which the registration metric was evaluated. Because of this asymmetry, the registration of pair (i, j), where i is the index of the fixed image and j is the index of the moving image, did not produce the same transform as the registration of pair (j,i).
Figure 1.
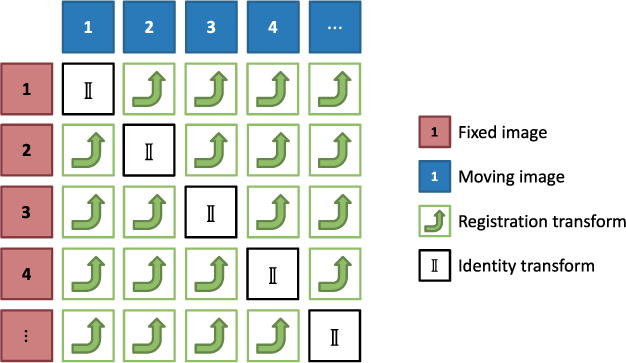
Registration of each possible pair of phases. The transforms produced by each registration transform any coordinate in the fixed image coordinate system into its corresponding point in the moving image coordinate system.
After the registration of all pairs of 1 of the steps, the resulting transforms were used to produce a set of consistent transforms as described in the next section. These transforms were then applied to the phases before proceeding to the next stage.
The registration of the 2 phases of a pair was performed using the open source registration software Plastimatch, version 1.6.0-beta (http://plastimatch.org). The settings of the translational and rigid registration steps are shown in Table 2. For the robust procedure, the first 2 translational steps only registered a subset with 10nphases pairs selected randomly among the nphases (nphases−1) possible pairs. These 2 steps helped roughly align the liver ROI for all the phases in preparation for the full registrations performed by the third through the fifth stage.
Table 2.
Plastimatch Parameters for Translational and Rigid Registration
| Plastimatch Parameter | Translational Registration | Rigid Registration |
|---|---|---|
| xform | translation | rigid |
| impl | itk | itk |
| optim | rsg | versor |
| metric | mattes | mattes |
| mi_histogram_bins | 48 | 48 |
| num_samples | 100 000 | 30 000 |
| max_step | 25 | 5 |
| min_step | 0.001 | 0.001 |
| max_its | 300 | 300 |
| min_its | 10 | 10 |
| translation_scale_factor | N/A | 10 000 |
| res | 1 1 1 | 1 1 1 |
Consistent Solution
The registration of a pair (i, j) of phases i and j results in a rigid transform described by a rotation matrix Bi,j and a translation vector bi,j. Thus, given a structure found in phase i at spatial coordinates r(i)=(x, y, z)T, the corresponding structure, as indicated by the registration, can be found at position r(j)=Bi,jr(i)+bi,j in phase j.
Let us assume that there is a transform given by rotation matrix Ai and translation vector ai, which transforms the coordinates of any structure in the reference phase iref into the coordinates of the same structure in phase i. If all of the coordinates for the voxels inside the liver ROI are inserted into the columns of a matrix R, the corresponding points in phase j are given by AjR + aj1, where . However, if the registration of phase pair (i, j) is successful, the same coordinates are also given by the application of the true transform from the reference phase to phase i followed by the registration transform from i to j such that: such that:
| (1) |
We then have npair of equations of the same form as equation (1), 1 for each registered phase pair, and up to nphases(nphases−1) are registered for a given stage. However, there are only nphases−1 true transforms given by Ai and ai. Therefore, the system in equation (1) is overdetermined if npair>nphases 1. Because the registration of a phase pair may not produce the true transform but only an estimate, equation (1) is not valid for all registered pairs (i, j) in general. Therefore, we must choose in what sense to solve the system in equation (1). If we define equation (2) as follows:
| (2) |
we can observe that is the sum of the squared distances between the liver voxel coordinates given by Bi,j(AiR+ai1)+bi,j1 and AjR+aj1. Finding the true transforms given by Ai and ai by minimizing the following equation:
| (3) |
where ║·║F is the Frobenius norm and would produce the least squares solution. If the error in the mapping of the liver voxel coordinates could be described by a normally distributed random variable, then minimizing equation (3) would produce a good solution for Ai and ai. However, because some of the phase pair registrations could fail and produce severely erroneous transforms, Bi,j and bi,j, the sum in equation (3) could be dominated by a few large terms that would thwart the accurate estimation of Ai and ai. Therefore, to lessen the impact of outlier registration transforms, Ai and ai can be robustly estimated by minimizing the following equation:
| (4) |
with p<2 (p=1⁄2 for this work). This can be done by using the iteratively reweighted least squares algorithm. For iteratively reweighted least squares, the following equation is solved repeatedly:
| (5) |
where
| (6) |
and β is the set of all transform parameters {Ai} and {ai}, and β(k) is the k th iteration of β. Equation (5) is iterated until convergence. To avoid division by 0 in equation (6), a nonzero regularization term E0 is needed. As is the root mean squared difference of the liver voxel coordinates produced by the left-hand side (LHS) and right-hand side (RHS) of equation (1), should be smaller than the best expected registration accuracy. For this work, was selected as 0.01 mm.
Each iteration of equation (5) amounts to solving a constrained, weighted-least-squares problem, where the constraint is that all Ai must be rotation matrices. Because the minimization problem in equation (5) is a rigid-body problem, it can be broken up into 2 independent problems, 1 problem for the translation of the liver center of mass and 1 problem for the rotation of the liver about its center of mass. This can be seen by noting that:
| (7) |
where
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
| (13) |
and L is the lower triangular matrix in the Cholesky decomposition of Σ=LLT. Thus, the problem in equation (5) can be written as the following 2 problems:
| (14) |
| (15) |
where equation (14) is a problem to find the rotation matrices β1={Ai} only, whereas equation (15) is a problem to find the transformed translation vectors only and β={β1,β2}. The problem in equation (15) is a system of linear equations of the following form:
| (16) |
that can be solved in a least squares sense for using the Moore–Penrose pseudoinverse. When solving this system, the solution from equation (14) is used to fix to a constant by setting . The problem in equation (14) can similarly be written as a system of matrix equations as follows:
| (17) |
To find a solution for equation (17), a heuristic algorithm was used as described below. By summing over i, equation (17) can be rewritten as follows:
| (18) |
where the RHS is an approximation of the first term in the LHS, with an error given by the second term of the LHS. This formula can be solved recursively. However, {Ai} is a rotation matrix and needs to be orthogonal with det (Ai) = 1. To ensure this, the RHS of equation (18) can be decomposed into its singular value decomposition to produce a set of the following iteration equations:
| (19) |
| (20) |
| (21) |
where the sign of the last diagonal element in equation (20) is selected to make the determinant of positive and equation (21) is needed to ensure that . The stopping criteria for equations (19) to (21) are as follows:
| (22) |
Empirically, the outer recursive sequence given by equation (5) was observed to converge within 20 iterations for a few test cases. Therefore, 20 iterations of equation (5) were used for all examinations.
Conventional Registration
A conventional registration procedure was used as a benchmark for the robust registration. This conventional procedure registered each phase to the reference phase using the same 5 registration stages as for the robust procedure. However, phases were not registered to anything but the reference phase, and no processing of the resulting transforms was performed except the combination of the transforms from the 5 stages for each phase. The same ROI was used to limit the registration region, and the settings in Table 2 were used for alignment.
Evaluation
The effect of registration on the mean signal intensity versus time for a 1-cm3 cubic ROI placed 1 cm below the dome of the liver for 1 patient is illustrated in Figure 2. The spikes seen in the intensity for the nonregistered time series are caused by tissue moving in and out of the ROI due to breathing.
Figure 2.
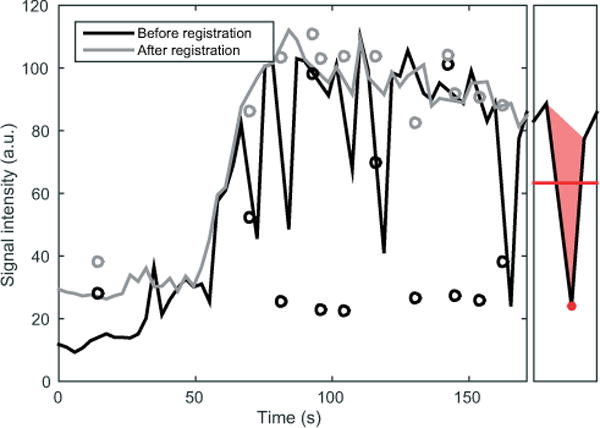
Signal intensity from an intrahepatic region of interest (ROI) from a dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) examination. The left side of the plot shows the time–intensity curves for a 1-cm3 cube placed 1 cm below the dome of the liver of a patient. The black curve shows the intensity before registration, and the gray curve shows the intensity after robust registration. Phases corresponding to deep inhalations during the examination are shown as circles and are not included in the main curves. The right box illustrates the local mean signal intensity (red line) and the local spike area (solid red triangle) around a sample (red dot).
Given the appearance of the signal variations seen in Figure 2, a suitable metric for the quality of the DCE-MRI time series should reflect how smooth the time–intensity curves are for the voxels inside the liver ROI. The temporal resolution of the time series as compared with the rise time of the time–intensity curve should also be reflected by the alignment quality metric. If only a few samples are collected during the rise time of a curve, the impact of 1 erroneous sample will be greater than that if a larger number of samples are collected. The variation of temporal sampling density across examinations can be taken into account by transforming the time of a given phase t into a normalized time τ as follows:
| (23) |
where t0 is the time at the center of the intensity ramp and T is half of the rise time. Thus, the rise time of the mean liver intensity for each examination is similar after transformation. The values of t0 and T were determined by fitting the time–intensity curve for the whole liver for each examination to the phenomenological function as follows:
| (24) |
where s0 is the baseline intensity, Δmax is the maximum signal enhancement, and α is the decay rate of the plateau. The function was fitted to the data using minimization of the L1-norm of the signal residuals.
Furthermore, errors in the baseline will be more troublesome for parameter estimation than had they appeared in the postcontrast plateau. In light of these considerations, we have chosen to use an approximation of the local spike area divided by the mean temporally local signal intensity to calculate the metric (see right panel of Figure 2 for illustration).
If the normalized temporal spacing between phases is Δτ, an approximation of the time-normalized, intensity-normalized spike area (TISA) metric, m(r, i), at phase i and position r is then given by the following equation:
| (25) |
where the numerator is the spike area and the denominator is the mean signal intensity of the voxel for the phase and its 2 temporal neighbors. Because the preceding and the following phases are needed to evaluate equation (25) for a phase, equation (25) cannot be evaluated for phases adjacent to others omitted because of deep inhalations during the examination.
RESULTS
After the consistent registration solution has been found from the overdetermined system of transform equations by minimizing equation (4), the individual norms ║Ei,j║F show how well a particular transform {Bi,j, bi,j} agrees with the overall solution. This is illustrated in Figure 3 for the last registration stage of 1 examination. The small dark square in the lower left corner of the figure represents the registration errors among the precontrast phases. The larger dark square in the upper right corner represent the errors among the postcontrast phases. These 2 groups have relatively small deviations from the consistent solution compared with the errors found for registrations between pre- and postcontrast phases as represented by the brighter rectangles in the upper left and lower right corners. This pattern was seen for a majority of the examinations registered and indicates that registrations between pre- and postcontrast phases are more likely to produce transforms that do not agree with the consistent solution.
Figure 3.
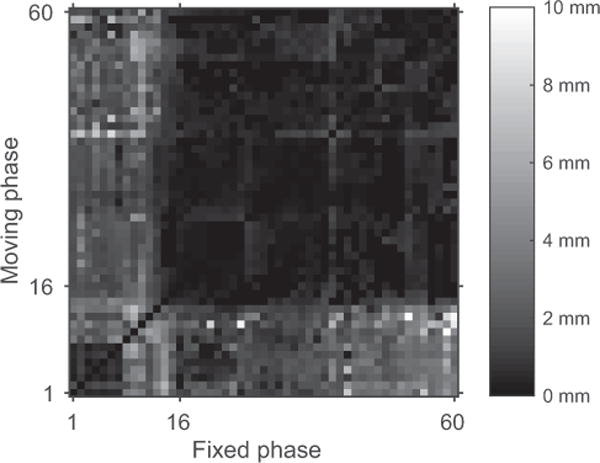
The value of ║Ei,j║F in millimeters for 1 examination following minimization of equation (4) is indicated by the color in the figure. The X-axis shows the phase index of the fixed registration phase and the Y-axis shows the index of the moving registration phase. Nine phases corresponding to deep inhalations have been omitted from the figure. Phase index 16 corresponds to the middle of the intensity ramp (τ=0). Indices <16 are precontrast phases and those >16 are postcontrast phases.
By sampling the intensity along a 1×1-cm-thick column extending 20 cm above and 20 cm below the liver dome, the effect of registration on the position of the liver can be illustrated. Figure 4 shows the intensity along such a column for the phases of 1 examination along with the time–intensity curves for a 1-cm3 cubic ROI placed 1 cm below the liver dome. The inferosuperior intensity profile exhibits motion before registration. After conventional registration, the motion is reduced for most phases, but it fails for a few precontrast phases. The intensity profile produced by the robust registration method has no such obviously failed registrations. Failed registrations like the one seen in Figure 4C–D were not found among any of the registrations produced by the robust method.
Figure 4.
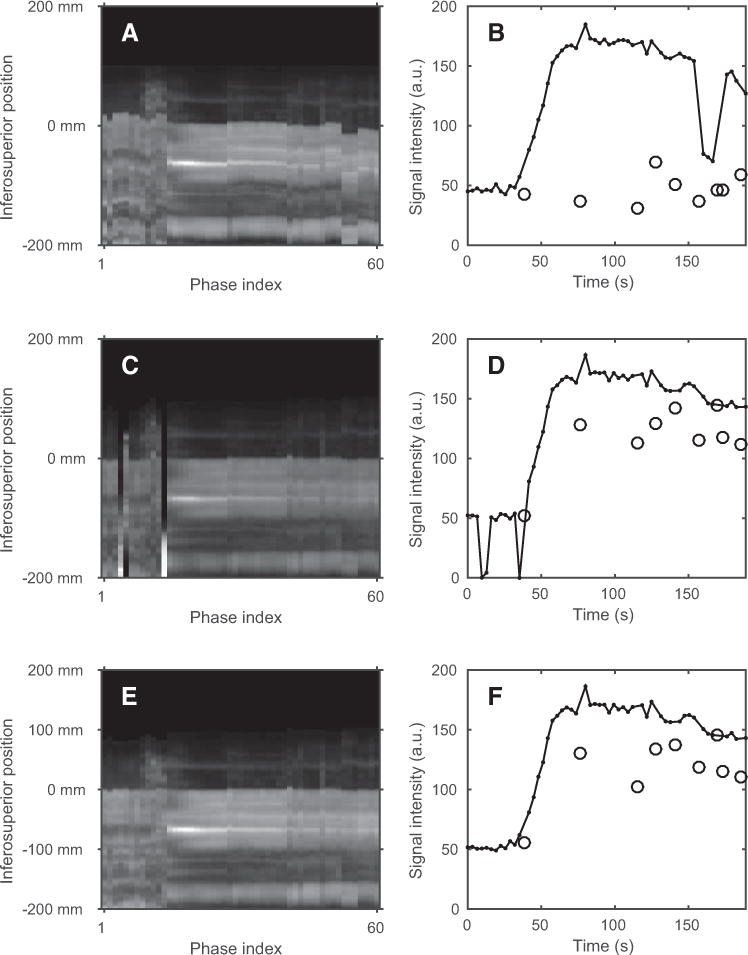
The intensity along a 40-cm-long column extending in the inferosuperior direction is seen for the phases before registration (A), after conventional registration (C), and after robust registration (E). The mean intensity of a 1-cm3 cubic ROI placed 1 cm below the liver dome is seen as a solid black curve for the phases before registration (B), after conventional registration (D), and after robust registration (F). The black circles indicate the intensities of phases excluded because of deep inhalations during examination. For each column of (A), (C), and (E), the intensity profile was divided by its mean to improve the visual contrast of the figure.
The mean TISA, m, for all examinations is shown with respect to the normalized time in Figure 5B. As a reference to help interpret the normalized time, τ, the mean of all liver time–intensity curves is shown in Figure 5A. This figure further illustrates where a particular value of τ corresponds to the precontrast region (τ < −1.5), the intensity ramp (−1.5< τ <1.5), or the postcontrast region t > 1.5). The TISA curves are consistently higher in the precontrast region compared with those in the postcontrast region. This is a result of the lower mean intensity of the phases in the precontrast region. In addition, at the beginning of the intensity ramp (around τ = −1), the conventional registration procedure performs almost as poorly as no registration, as seen by the close proximity of their TISA curves (green and red). Confidence intervals for the paired TISA differences between the robust method, the conventional method, and no registration are given in Table 3. The robust method is significantly better than no registration for the whole time interval (paired t test, P<.001). It is also significantly better than the conventional method for the precontrast and early contrast regions (P<.001).
Figure 5.
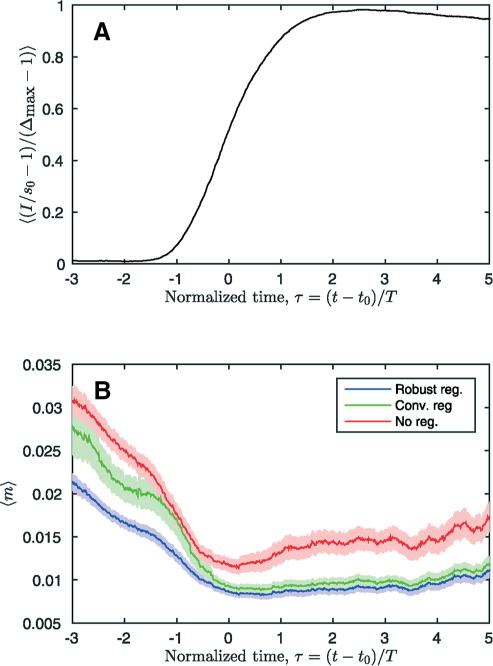
The mean time–intensity curve from all examinations is shown in (A). The mean time-normalized intensity-normalized spike area, m, is shown in (B). For (B), the mean of mwas calculated for all examinations and a time window that was one normalized time unit wide. The colored bands show the 95% confidence interval for the mean.
Table 3.
Paired TISA Difference for 3 Registration Methods and Time Intervals
| mconventional−mrobust | munregistered−mrobust | |||||
|---|---|---|---|---|---|---|
| Mean | SD | CI (95 %) | Mean | SD | CI (95 %) | |
| Pre-contrast (τ<−1.5) | 0.0046 | 0.0168 | [0.0034, 0.0057] | 0.0087 | 0.0087 | [0.0081, 0.0093] |
| Early contrast (−1.5<τ<0) | 0.0030 | 0.0140 | [0.0020, 0.0041] | 0.0043 | 0.0061 | [0.0038, 0.0048] |
| Whole time interval | 0.0022 | 0.0109 | [0.0018, 0.0026] | 0.0058 | 0.0076 | [0.0056, 0.0061] |
Abbreviations: TISA, time-normalized, intensity-normalized spike area; SD, standard deviation; CI, confidence interval.
DISCUSSION
In this study, we have implemented an overdetermined registration procedure for liver DCE-MRI examinations. We found that our procedure significantly reduced the image registration errors in the DCE time curves, particularly in the precontrast and early contrast uptake phases, compared with a conventional reference-phase-based registration method. Registration errors in the precontrast baseline could profoundly affect the calculation of signal intensity changes and the conversion to contrast concentration, whereas errors in the early contrast uptake curve propagate into derived parameters that are sensitive to the early curve dynamics, for example, hepatic arterial perfusion in liver DCE-MRI. The improved DCE curves describe the contrast agent uptake dynamics more correctly, and thereby, provide a better foundation for PK model parameter estimation.
DCE images acquired at different parts of the dynamic curves have different sensitivities to image registration errors. We found that registering precontrast phases to postcontrast phases is more likely to result in registration errors than registering within the precontrast phases or within the postcontrast phases. The conventional reference-based registration method likely results in a higher misregistration rate for the precontrast phases, as well as the early contrast uptake phases, if the post-contrast phase is chosen as reference. As an effect of this phenomenon, spikes can appear in or around the baseline plateau in the time–intensity curves. The parameters of a PK model may still be possible to estimate with such spikes present by using robust regression methods. However, each registration failure reduces the number of reliable samples. Even worse, if sample points are lost on the intensity ramp by removing the misregistration points, the remaining samples may not be able to correctly describe the dynamics of the curve, resulting in faulty PK parameter estimates. Some parameters are highly sensitive to the shape of the initial ramp of the uptake curve, for instance, hepatic arterial perfusion in the liver. The robust registration procedure presented reduces the number of failed registrations in the precontrast region by rejecting them as outliers in an overdetermined system of transform equations and thereby provides a greater number of reliable samples for parameter estimation.
For the postcontrast plateau, the robust registration method and the conventional registration method perform almost equally well. This is a result of the higher signal-to-noise ratio and more stable contrast for the phases of this region. Both registration methods produce smoother curves than no registration. The robust method is seen to have a slightly lower TISA than the conventional method. This is because the robust method takes the mutual information of all pairs of phases into account when estimating the transform. Therefore, the robust method acts as a more efficient estimator for the true transforms than does the conventional method.
In this work, we performed each registration separately and then combined the resulting transforms using a method that rejects outliers. Another possible approach would be to maximize the sum of the mutual information metrics of all pairs of phases using strategies similar to those reported by others (22–24). Such an approach might improve the registration of DCE-MRI further. In addition, although the method presented in this paper is only applicable to rigid registration, a method based on the maximization of pairwise mutual information could aid in improving the robustness of deformable registration for DCE-MRI. Robust deformable registration could allow free breathing examinations, which would relieve subjects from the burden of repeatedly holding their breath.
The robust method produced a consistent registration quality across all examinations, without failed alignments such as those observed for the conventional method. It therefore reduces the need for manual intervention following registration and helps to simplify the workflow of clinical studies with many subjects.
CONCLUSIONS
A method has been presented that registers every possible pair of phases from a DCE-MRI examination and derives a final set of transforms from them using L1⁄2 minimization. The method is robust, insofar as to be able to reject such failed registrations that can appear when registering precontrast to postcontrast phases. The robust registration method improves the smoothness of the resulting time–intensity curves of voxels in the liver by eliminating spurious intensity spikes induced by motion or failed registration. Therefore, a baseline with reduced bias and a larger number of reliable samples in the intensity ramp are made available for the PK model parameter estimation. The consistent registration quality produced for all examinations studied shows that the method could simplify the workflow of clinical studies with many patients by eliminating the need for manual intervention following registration.
Acknowledgments
This work is supported, in part, by NIH/NCI RO1 CA132834 and PO1 CA059827.
Abbreviations
- DCE
Dynamic contrast-enhanced
- PK
pharmacokinetic
- MR
magnetic resonance
- PCA
principal component analysis
- ROI
region of interest
- RHS
right-hand side
- LHS
left-hand side
- TISA
time-normalized intensity-normalized spike area
Footnotes
Disclosure: No disclosures to report.
References
- 1.Wang H, Farjam R, Feng M, Hussain H, Ten Haken RK, Lawrence TS, Cao Y. Arterial perfusion imaging-defined subvolume of intrahepatic cancer. Int J Radiat Oncol Biol Phys. 2014;89(1):167–174. doi: 10.1016/j.ijrobp.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao Y, Platt JF, Francis IR, Balter JM, Pan C, Normolle D, Ben-Josef E, Haken RK, Lawrence TS. The prediction of radiation-induced liver dysfunction using a local dose and regional venous perfusion model. Med Phys. 2007;34(2):604–612. doi: 10.1118/1.2431081. [DOI] [PubMed] [Google Scholar]
- 3.Cao Y, Pan C, Balter JM, Platt JF, Francis IR, Knol JA, Normolle D, Ben-Josef E, Ten Haken RK, Lawrence TS. Liver function after irradiation based on computed tomographic portal vein perfusion imaging. Int J Radiat Oncol Biol Phys. 2008;70(1):154–160. doi: 10.1016/j.ijrobp.2007.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao Y. The promise of dynamic contrast-enhanced imaging in radiation therapy. Semin Radiat Oncol. 2011;21(2):147–156. doi: 10.1016/j.semradonc.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y, Wang H, Johnson TD, Pan C, Hussain H, Balter JM, Normolle D, Ben-Josef E, Ten Haken RK, Lawrence TS, Feng M. Prediction of liver function by using magnetic resonance-based portal venous perfusion imaging. Int J Radiat Oncol Biol Phys. 2013;85(1):258–263. doi: 10.1016/j.ijrobp.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thng CH, Koh TS, Collins DJ, Koh DM. Perfusion magnetic resonance imaging of the liver. World J Gastroenterol. 2010;16(13):1598–1609. doi: 10.3748/wjg.v16.i13.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y, Tsien CI, Sundgren PC, Nagesh V, Normolle D, Buchtel H, Junck L, Lawrence TS. Dynamic contrast-enhanced magnetic resonance imaging as a biomarker for prediction of radiation-induced neurocognitive dysfunction. Clin Cancer Res. 2009;15(5):1747–1754. doi: 10.1158/1078-0432.CCR-08-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farjam R, Tsien CI, Feng FY, Gomez-Hassan D, Hayman JA, Lawrence TS, Lawrence TS, Cao Y. Physiological imaging-defined, response-driven subvolumes of a tumor. Int J Radiat Oncol Biol Phys. 2013;85(5):1383–1390. doi: 10.1016/j.ijrobp.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanner C, Schnabel JA, Chung D, Clarkson MJ, Rueckert D, Hill DLG, Hawkes DJ. Volume and shape preservation of enhancing lesions when applying non-rigid registration to a time series of contrast enhancing MR breast images. Lect Notes Comput Sci. 2000;1935:327–337. [Google Scholar]
- 10.Tanner C, Schnabel JA, Degenhard A, Castellano-Smith AD, Hayes C, Leach MO, Hose DR, Hill DLG, Hawkes DJ. Validation of volume-preserving non-rigid registration: application to contrast-enhanced MR-mammography. Lect Notes Comput Sci. 2002;2488:307–14. [Google Scholar]
- 11.Rohlfing T, Maurer CR, Bluemke DA, Jacobs MA. Volume-preserving nonrigid registration of MR breast images using free-form deformation with an incompressibility constraint. IEEE Trans Med Imaging. 2003;22(6):730–741. doi: 10.1109/TMI.2003.814791. [DOI] [PubMed] [Google Scholar]
- 12.Zuo CS, Jiang A, Buff BL, Mahon TG, Wong TZ. Automatic motion correction for breast MR imaging. Radiology. 1996;198(3):903–906. doi: 10.1148/radiology.198.3.8628891. [DOI] [PubMed] [Google Scholar]
- 13.Hayton P, Brady M, Tarassenko L, Moore N. Analysis of dynamic MR breast images using a model of contrast enhancement. Med Image Anal. 1997;1(3):207–224. doi: 10.1016/s1361-8415(97)85011-6. [DOI] [PubMed] [Google Scholar]
- 14.Adluru G, DiBella EVR, Schabel MC. Model-based registration for dynamic cardiac perfusion MRI. J Magn Reson Imaging. 2006;24(5):1062–1070. doi: 10.1002/jmri.20756. [DOI] [PubMed] [Google Scholar]
- 15.Buonaccorsi GA, Roberts C, Cheung S, Watson Y, O’Connor JPB, Davies K, Jackson A, Jayson GC, Parker GJ. Comparison of the performance of tracer kinetic model-driven registration for dynamic contrast enhanced MRI using different models of contrast enhancement. Acad Radiol. 2006;13(9):1112–1123. doi: 10.1016/j.acra.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Buonaccorsi GA, O’Connor JPB, Caunce A, Roberts C, Cheung S, Watson Y, Davies K, Hope L, Jackson A, Jayson GC, Parker GJ. Tracer kinetic model-driven registration for dynamic contrast-enhanced MRI time-series data. Magn Reson Med. 2007;58(5):1010–1019. doi: 10.1002/mrm.21405. [DOI] [PubMed] [Google Scholar]
- 17.Melbourne A, Atkinson D, White MJ, Collins D, Leach M, Hawkes D. Registration of dynamic contrast-enhanced MRI using a progressive principal component registration (PPCR) Phys Med Biol. 2007;52(17):5147–5156. doi: 10.1088/0031-9155/52/17/003. [DOI] [PubMed] [Google Scholar]
- 18.Hamy V, Dikaios N, Punwani S, Melbourne A, Latifoltojar A, Makanyanga J, Chouhan M, Helbren E, Menys A, Taylor S, Atkinson D. Respiratory motion correction in dynamic MRI using robust data decomposition registration - application to DCE-MRI. Med Image Anal. 2014;18(2):301–313. doi: 10.1016/j.media.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Menys A, Hamy V, Makanyanga J, Hoad C, Gowland P, Odille F, Taylor SA, Atkinson D. Dual registration of abdominal motion for motility assessment in free-breathing data sets acquired using dynamic MRI. Phys Med Biol. 2014;59(16):4603–4619. doi: 10.1088/0031-9155/59/16/4603. [DOI] [PubMed] [Google Scholar]
- 20.Otazo R, Koesters T, Candès EJ, Sodickson DK. Motion-guided low-rank plus sparse (L+S) reconstruction for free-breathing dynamic MRI. Proc Intl Soc Mag Reson Med. 2014;22:0742. [Google Scholar]
- 21.Rajaraman S, Rodriguez JJ, Graff C, Altbach MI, Dragovich T, Sirlin CB, Korn RL, Raghunand N. Automated registration of sequential breath-hold dynamic contrast-enhanced MR images: a comparison of three techniques. Magn Reson Imaging. 2011;29(5):668–682. doi: 10.1016/j.mri.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lok-Chuen Lo J, Brady M, Moore N. Simultaneous multiple image registration method for T1 estimation in breast MRI images. Med Image Comput Assist Interv. 2006;9(Pt 1):865–872. doi: 10.1007/11866565_106. [DOI] [PubMed] [Google Scholar]
- 23.Wachinger C, Navab N. Simultaneous registration of multiple images: similarity metrics and efficient optimization. IEEE Trans Pattern Anal Mach Intell. 2013;35(5):1221–1233. doi: 10.1109/TPAMI.2012.196. [DOI] [PubMed] [Google Scholar]
- 24.Yigitsoy M, Wachinger C, Navab N. Temporal groupwise registration for motion modeling. Inf Process Med Imaging. 2011;22:648–659. doi: 10.1007/978-3-642-22092-0_53. [DOI] [PubMed] [Google Scholar]


