Abstract
The twin-supercoiled-domain model describes how transcription can drive DNA supercoiling, and how DNA supercoiling, in turn plays an important role in regulating gene transcription. In vivo and in vitro experiments have disclosed many details of the complex interactions in this relationship, and recently new insights have been gained with the help of genome-wide DNA supercoiling mapping techniques and single molecule methods. This review summarizes the general mechanisms of the interplay between DNA supercoiling and transcription, considers the biological implications, and focuses on recent important discoveries and technical advances in this field. We highlight the significant impact of DNA supercoiling in transcription, but also more broadly in all processes operating on DNA.
Keywords: transcription, gene regulation, supercoiling, chromatin, torque, mechanics
Introduction
DNA has a highly dynamic topology. In its relaxed B-DNA state, DNA forms a right-handed double helical structure with each helical turn containing ~ 10.5 base-pairs. When additional twists are introduced, DNA becomes overwound or underwound, resulting in (+) or (−) DNA supercoiling, respectively. DNA supercoiling is well known to be important for DNA compaction, and recently, the essential role of DNA supercoiling in gene regulation has also become prominent.
Transcription is an important source of DNA supercoiling in the cell. During transcription, RNA polymerase (RNAP) tracks the helical groove of DNA, over-twisting DNA downstream and under-twisting DNA upstream. Therefore, RNAP movement generates (+) DNA supercoiling in front of the RNAP, and (−) DNA supercoiling behind, as described in the twin-supercoiled-domain model (Liu and Wang 1987) (Fig. 1). The accumulation of this torsional stress may hinder transcription elongation and must be released in a timely fashion, either by topoisomerases or via DNA rotational relaxation.
Fig. 1.
A schematic of the twin-supercoiled domain model. During transcription, RNA polymerase must rotate relative to its helical DNA track. Owing to the size and typical confinement of the polymerase and associated machinery, the DNA is (+) supercoiled in the front and (−) supercoiled behind. Adapted from (Forth et al. 2013) with permission.
Early in vitro and in vivo biochemical studies have demonstrated that, in torsionally-constrained and topoisomerase-deficient systems, transcription can result in the accumulation of substantial torsional stress in DNA (Krasilnikov et al. 1999; Samul and Leng 2007; Tsao et al. 1989; Wu et al. 1988 ). Recent experiments show transcription-induced DNA supercoiling exists more broadly than previous thought and it plays highly dynamic roles. For example, Matsumoto et al. (Matsumoto and Hirose 2004) found that, in the presence of active topoisomerases, transcription coupled (−) DNA supercoiling impacted over 150 loci on polytene chromosomes in D. melanogaster. Additionally, Kouzine et al. (Kouzine et al. 2004; Kouzine et al. 2008) demonstrated that, even in a linear DNA system with a full complement of topoisomerases, transcription-induced DNA supercoiling and dynamic torsional stress can still exist and play an essential role in gene regulation. These results demonstrate that transcription and DNA supercoiling are coupled in a close and dynamic fashion. They also suggest that DNA supercoiling could potentially serve as an important feedback mediator to regulate transcription in real time.
Excess torsion can greatly alter DNA topology, including creating highly supercoiled DNA and fostering the formation of non-B DNA forms, such as R-loops (Leng et al. 2004), Z-DNA (Herbert and Rich 1996; Oberstrass et al. 2013), DNA cruciforms (Mizuuchi et al. 1982; Oussatcheva et al. 2004), and P-DNA (Allemand et al. 1998; Bryant et al. 2003; Deufel et al. 2007). These altered DNA structures may represent obstacles for transcription or targets for transcription factors to regulate transcription.
In addition, (−) DNA supercoiling and (+) DNA supercoiling have differential impacts on gene expression. (−) DNA supercoiling can facilitate DNA strand separation in promoter regions, thereby enabling open complex formation by RNAP. It can also help recruit transcription factors or other regulatory proteins. For example, it has been shown that (−) DNA supercoiling can facilitate recruitment of TATA binding proteins (TBPs), which are important factors for eukaryotic transcription initiation (Tabuchi et al. 1993). Kouzine et al. also showed that the transient torsional stress generated by transcription can melt the far upstream element (FUSE) of the human c-myc promoter (Kouzine et al. 2004; Kouzine et al. 2008), and hence regulate activator and repressor binding affinity to that region. In addition to localized effects, as a supercoiling wave propagates, transcription at one promoter could affect transcription at a distant promoter (Hanafi and Bossi 2000; Lilley and Higgins 1991), in the form of “topological promoter coupling”. The (+) DNA supercoiling generated by transcription can also play an important role in gene regulation. Gartenberg and Wang (Gartenberg and Wang 1992) have shown that, after preferential removal of transcription-generated negative supercoils, the accumulated (+) torsional stress in yeast greatly diminished mRNA synthesis. Transcription generated (+) supercoiling can also be used to disrupt, or remove, road-block proteins, such as destabilizing nucleosome structures (Sheinin et al. 2013) to make the DNA more accessible to RNAP. Bécavin et al (Bécavin et al. 2010) also theoretically demonstrated that transcription generated (+) torque could trigger the downstream nucleosomes to form reversomes stepwisely, making the nucleosomal barrier more permissive to RNAP processing. Moreover, the wavefront of reversomes can propagate ten times faster than RNAP progression, which explains the puzzling wavefront of nucleosome disruption observed during the transcription of the Drosophila Hsp70 locus, i.e. the nucleosome disruption observed downstream of the RNAP was much faster than the rate of elongation and was confined within the Hsp70 locus and stopped at its boundary (Petesch and Lis 2008; Zlatanova and Victor 2009). Together, all these highlight the important role of DNA supercoiling in regulating transcription.
Since the "twin supercoiled domain" model was proposed almost three decades ago, it has gained increasing support from both in vivo and in vitro studies. Particularly within the past decade, detailed and complex interactions between DNA supercoiling and transcription have begun to come to light, owing much to the advent of novel methodologies, such as genome-wide DNA supercoiling characterization and single molecule methods. This review will focus on these recent discoveries.
Genomic studies of transcription and DNA supercoiling
Although the interplay between transcription and DNA supercoiling is being increasingly established in vitro, details of these interactions at the genomic level remained, until recently, largely unknown. Generally, cellular (−) supercoiling can be estimated through the intercalation of psoralen, and its derivatives, into DNA, followed by DNA photo-cross-linking (Matsumoto and Hirose 2004). Psoralen is a planar, aromatic compound that can permeate cells and intercalate into DNA. The rate of psoralen photo-cross-linking to double-stranded DNA (dsDNA) is linearly related to the degree of (−) DNA supercoiling. Therefore, psoralen photo-binding can be used as a genome probe to detect in vivo DNA supercoiling.
In early studies, Matsumoto and Hirose (Matsumoto and Hirose 2004) introduced biotinylated psoralen into Drosophila salivary glands and visualized it, with fluorescent streptavidin, on polytene chromosomes. They observed bright psoralen signals at many transcriptionally active inter-bands and puffs, and demonstrated that transcription-coupled (−) supercoils of DNA exist widely and are co-localized to active transcribing sites in the genome. Subsequently, Bermúdez et al. (Bermúdez et al. 2010) combined a psoralen photo-binding method with micro-array analysis and mapped the distribution of DNA supercoils across the entire genome of S. cerevisiae. These studies provided a coarse-grain view of the distribution of torsional stress along chromosomes, although they did not have sufficient resolution to resolve supercoiling at the individual gene level.
Only very recently, several groups (Kouzine et al. 2013; Naughton et al. 2013; Teves and Henikoff 2014) have succeeded in generating high resolution maps of genome scale DNA helical tensions. Kouzine et al. (Kouzine et al. 2013) treated Raji human B cells in G1 phase with psoralen, and then used UV light to cross-link the psoralen with DNA. Subsequently, the fragments of DNA, with and without cross-linked psoralen, were separated, labeled with different dyes, and then hybridized to genomic oligo-nucleotide micro-arrays spanning the whole ENCODE regions. Information about DNA supercoiling around ENCODE promoters can thus be revealed through the log ratio (cross-linked/uncross-linked) of the fluorescent signals, defined as the cross-link level (CL). By treating the cell with inhibitors to different factors (e.g., Pol II, Topo I, or Topo II), the authors dissected the different roles of these factors in regulating DNA supercoiling. The results showed that transcription-dependent dynamic supercoiling is present genome-wide and transmits over ~1.5 kbp upstream of start sites of virtually every transcribed gene (Fig. 2a). Although (−) DNA supercoiling is strongly dependent on transcriptional activity, this dependence appears to be complex. Weakly expressed gene showed little DNA supercoiling, but interestingly, moderately expressed genes showed the highest DNA supercoiling, which was localized to the transcription start site. Highly expressed genes showed less DNA supercoiling than that of moderately expressed genes, however the supercoiling transmitted farther upstream. Inhibition of topoisomerases altered the pattern of DNA supercoiling, with Topo I preferentially recruited to moderately transcribed promoters and Topo II to highly transcribed promoters. The authors proposed that, with regard to regulating transcription-induced dynamic torsional stress, Topo I uses a "diffuse" mode of recruitment, whereas Topo II uses a "focal" mode (Fig. 2b).
Fig. 2.
Genome-wide coupling between transcription and DNA topology. (a) Dynamic (−) DNA supercoiling around transcription start sites (TSSs)for genes with low, medium, or high expression level. The vertical axis shows the ΔCL, which is the difference between CL values derived from transcription inhibited and uninhibited cells, and reflects DNA supercoiling induced by transcription. (b) A model for DNA supercoiling regulation by Topo I and Topo II. In this model, Topo I is diffusely recruited to the DNA supercoiling regions (diffuse mode), while Topo II is recruited in a focused manner to the most dynamic (−) supercoiling region (focal mode). For a moderately expressed gene, dynamic supercoiling is mainly managed by Topo I which is recruited to a broad range upstream of the TSSs. For a highly transcribed gene, dynamic supercoiling is resolved efficiently by Topo II which is recruited focally to the TSSs. Adapted, with permission, from (Kouzine et al. 2013).
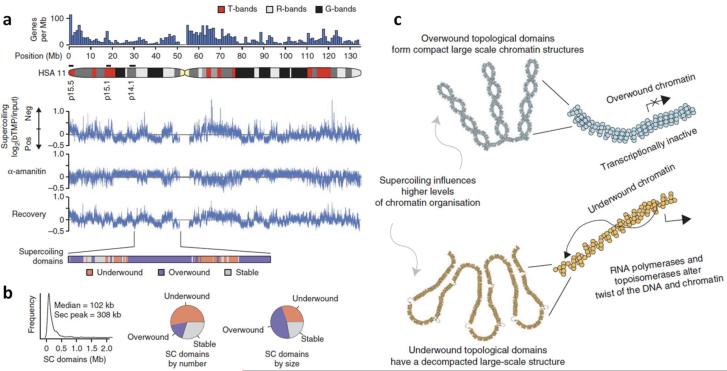
In another study, using biotin-tagged psoralen (bTMP), followed by UV cross-link and microarray analysis, Naughton et al. (Naughton et al. 2013) were able to map, with high resolution, supercoiling domains in the human genome. They utilized chromosome 11 as a representative, as it contains 5% of the genome and comprises regions of various gene densities and base compositions. As shown in Fig. 3, after examination of the chromosomal loci bound by bTMP, they identified hundreds of domains that are underwound, overwound, or stable, with an overall average size ~ 100 kb. Among them, more than half were underwound domains, but their sizes were generally smaller than those of the overwound domains. Further analysis indicated the supercoiling patterns of these domains were transcription and topoisomerase dependent, and the underwound domains were actively transcribed regions of "open" chromatin fibers, which were “GC” rich and enriched in Topo I and DNase I sites, but lacked Topo II. Their results support a model that transcription and topoisomerases are integral to the establishment and remodeling of supercoiling domains and influence the folding of large-scale chromatin structures.
Fig. 3.
High-resolution mapping of DNA supercoiling reveals hundreds of supercoiling domains in the human chromosome (HSA) 11. (a) Microarray data of biotin-tagged psoralen bTMP binding across HSA 11 to assay the level of DNA supercoiling. ά-amanitin is used to inhibit transcription. Supercoiling domains are categorized as underwound, overwound, or stable regions. (b) The distributions of supercoiling domains. (c) Model of the large-scale chromatin structures. An overwound domain corresponds to transcriptionally inactive chromatin and is compact over a large-scale; whereas an underwound domain corresponds to a transcriptionally active region and a decompacted chromatin structure. Adapted from (Naughton et al. 2013) with permission.
When Pol II transcribes a gene, it encounters an array of well-ordered nucleosomes, but it is not fully understood how Pol II traverses through this array in vivo. Teves and Henikoff (Teves and Henikoff 2014) investigated how transcriptional torsional stress affects Pol II kinetics and nucleosome turnover in the genome. They adapted a microarray method to next generation sequencing to achieve high resolution mapping of underwound DNA and then examined if the torsional stress generated by transcription could destabilize nucleosomes ahead of Pol II. They focused on the study of DNA torsional states around transcription start sites (TSSs) and transcription end sites (TESs). They found that inhibition of topoisomerases leads to rapid accumulation of torsional strain, accompanied by changes in Pol II kinetics and chromatin properties. The stalled Pol II accumulated immediately downstream of the TSSs after inhibition of either Topo I or Topo II, suggesting that both Topo I and Topo II can alter Pol II initiation kinetics (i.e. promoter-proximal pausing kinetics) (Core et al. 2008; Gilchrist et al. 2010; O'Brien and Lis 1991). However, as shown in Fig. 4a, Topo I and Topo II affect Pol II elongation differently. Topo I inhibition resulted in an overall increase in nascent RNA production near the 5′ end, while Topo II inhibition caused minimal overall changes in Pol II kinetics. Furthermore, Topo I inhibition primarily affected transcribed genes, whereas Topo II inhibition resulted in decreased nascent RNA production only for the highly expressed genes (Fig. 4b). These results support that Topo I acts within gene bodies and plays a main role in regulating both Pol II initiation and elongation kinetics, while Topo II acts primarily at the 5′ end to regulate Pol II initiation kinetics but plays only a secondary role in regulating Pol II elongation kinetics. The authors further used a CATCH-IT to measure nucleosome turnover, which utilizes covalent tags to capture histones and identify histone turnover. As shown in Fig. 4c and 4d, subsequent to the inhibition of topoisomerases, accumulated torsional stress destabilized histone-DNA interactions and increased nucleosome turnover within gene bodies genome-wide, but did not change the overall nucleosome occupancy. Their data support a model that the transient (+) torsional wave generated by Pol II can destabilize nucleosomes downstream allowing transcription to progress, and the transient (−) torsional wave generated by Pol II can facilitate nucleosome assembly upstream, thereby assisting in the maintenance of chromatin structure. In this way, the cell can achieve a delicate balance amidst nucleosomal destabilization, re-assembly, and Pol II progression.
Fig. 4.
Transcription-generated torsional stress can destabilize nucleosomes. (a) Altered Pol II elongation kinetics upon Topo I or Topo II inhibition. Top panel shows the average transcription profiles surrounding the transcription start sites (TSSs) and transcription end site (TESs) of all genes. Topo I inhibition increases the overall nascent-RNA production near TSSs, but not near TESs, suggesting Pol II stalling before the completion of the transcription; whereas Topo II inhibition showed no overall change relative to control. Bottom: Heat-map of the log ratio of nascent RNA for Topo I inhibited samples over control (left), and Topo II inhibited samples over control (right), with genes ordered by decreasing expression in control samples. Topo I inhibition primarily affects transcribed genes, whereas Topo II inhibition results in heterogeneous changes in nascent-RNA levels, suggesting Topo II plays only a secondary role in transcription. (b) Altered nucleosome turnover under torsion. Top: nucleosome turnover is measured by CATCH-IT followed by mapping of the positions of captured nucleosomes. Bottom: Heat maps showing changes in CATCH-IT signals, after Topo I (left) and Topo II (right) inhibition, from the controls. Adapted from (Teves and Henikoff 2014) with permission.
Single molecule studies of transcription under DNA supercoiling
Single molecule manipulation tools to study transcription under DNA supercoiling
Optical tweezers (OT) and magnetic tweezers (MT), as well as their variations and extensions, such as the rotor bead tracking (RBT) methods (Bryant et al. 2003; Oberstrass et al. 2013), angular optical trap (AOT) (La Porta and Wang 2004), magnetic torque tweezers (MTT) (Lipfert et al. 2010), and freely orbiting magnetic tweezers (FOMT) (Lipfert et al. 2011), are the primary single molecule manipulation tools used in the study of transcription under DNA supercoiling.
Optical tweezers (or optical traps) use a tightly focused laser beam to provide an attractive force on dielectric particles in three dimensions, enabling flexible and precise manipulation of small objects (Ashkin et al. 1986; Ashkin et al. 1987). Despite the capabilities of applying and measuring force, conventional optical tweezers cannot exert or detect torque, therefore this technique was initially limited to the studies of linear motion of transcription kinetics (Bai et al. 2007; Shundrovsky et al. 2004; Wang et al. 1998; Yin et al. 1995) and linear elastic properties of DNA (Wang et al. 1997). In contrast to the conventional optical trap, the angular optical trap (AOT) (Deufel et al. 2007; Inman et al. 2010; La Porta and Wang 2004) also termed the optical torque wrench (OTW), is a notable enhancement of an optical trap which enables full control over the angular orientation of a trapped particle and can directly, and simultaneously, exert and measure torque (Fig. 5a). Since its invention, the AOT has been successfully used to study transcription under DNA supercoiling, as well as torsional properties of various other biological systems (Forth et al. 2011; Forth et al. 2008; Forth et al. 2013; Ma et al. 2013; Sheinin et al. 2011; Sheinin et al. 2013; Sheinin and Wang 2009).
Fig. 5.
Primary single molecule torsional manipulation tools for studies of DNA supercoiling. (a) A schematic of an angular optical trap (AOT) setup (Forth et al. 2011; Forth et al. 2008; Forth et al. 2013; Inman et al. 2010; La Porta and Wang 2004; Ma et al. 2013; Sheinin et al. 2011; Sheinin et al. 2013; Sheinin and Wang 2009). A DNA molecule is torsionally anchored at one end to the surface of a coverglass and at the other end to a nanofabricated quartz cylinder held in an optical trap of linear polarization. The optical trap exerts both a force and torque on the cylinder. Rotation of the cylinder via rotation of the laser polarization introduces supercoiling into DNA. During a measurement, torque, rotation, force, and DNA extension are simultaneously measured. (b) A schematic of a magnetic tweezers (MTs) setup. DNA is anchored in a similar fashion as in (a). To introduce DNA supercoiling, the magnetic bead is rotated via rotation of the magnetic field. In most magnetic tweezers, only force and DNA extension may be measured. Recent enhancement of magnetic tweezers also permits torque detection (Celedon et al. 2009; Janssen et al. 2012; Lipfert et al. 2010; Lipfert et al. 2011; Mosconi et al. 2011). (c) DNA force-torque phase diagram. Phase transitions between specific states of DNA are represented by solid black lines (Bryant et al. 2003; Marko 2007; Sarkar et al. 2001). Red points indicate torque values measured during phase transitions using an angular optical trap (Deufel et al. 2007; Forth et al. 2008; Sheinin et al. 2011; Sheinin and Wang 2009). Adapted from (Forth et al. 2013) with permission. (d) A cartoon depicting plectoneme migration via diffusion or hopping along DNA. Adapted from (Sheinin and Wang 2012) with permission.
Magnetic tweezers are the most well-known technique used to apply twist to a single biological molecule. They typically use a pair of permanent magnets to exert force and torque on a chemically functionalized super-paramagnetic bead linked to a biological molecule (Fig. 5b). As MTs are relatively simple to employ, this technique has been used to study transcription under DNA supercoiling (Harada et al. 2001; Revyakin et al. 2004; Revyakin et al. 2006), as well as many other torsional properties of DNA (Oberstrass et al. 2013; Strick et al. 1996), chromatin (Bancaud et al. 2006), and topoisomerases (Basu et al. 2012; Gore et al. 2006; Koster et al. 2005; Nollmann et al. 2007; Strick et al. 2000).
DNA supercoiling
Understanding DNA torsional mechanics is the first step towards deciphering the torsional mechanics of chromatin. A number of studies, using either optical tweezers or magnetic tweezers, have provided important characterizations of the torsional properties of a single DNA molecule. The torsional modulus of DNA has been measured to be ~400 pN·nm2 (Bryant et al. 2003; Forth et al. 2008). When (+) or (−) twists are introduced, DNA may buckle to form a plectoneme, and/or convert from BDNA to other structural forms of DNA. In particular, the critical torques were measured to be ~ +34 pN·nm for the B-DNA to P-DNA transition, ~ –3 pN·nm for the B-DNA to Z-DNA transition (at GC repeats), and ~ –10 pN·nm for the B-DNA to melted DNA (Bryant et al. 2003; Forth et al. 2008; Oberstrass et al. 2013; Sheinin et al. 2011). These studies have permitted the construction of a detailed phase diagram for DNA under force and torque (Fig. 5c).
Although the mechanical properties of plectonemes during DNA supercoiling have been characterized, the dynamics of plectoneme migration on DNA was more elusive. van Loenhout et al (van Loenhout et al. 2012) directly visualized the dynamics of individual plectonemes by combining MT with single molecule fluorescence. They found that a plectoneme can propagate along DNA via diffusion or hopping (Fig. 5d) and plectonemes showed preferential localization along DNA sequences. These results suggest that certain DNA sequences may be designed to “pin down” plectonemes and thus bring neighboring regulatory DNA elements into close proximity, while plectoneme hopping could provide a dramatic long-range rearrangement of the DNA conformation to permit fast searching during DNA recombination or enhancer-activated gene expression (Sheinin and Wang 2012).
Chromatin under twist
Transcription-induced supercoiling constantly perturbs the chromatin in the vicinity of an elongating RNAP. Using magnetic tweezers, Bancaud et al. (Bancaud et al. 2006) showed that a nucleosome array is extremely resilient to torsional stress and can reversibly accommodate a large amount of supercoiling. Chromatin may thus serve as a "powerful topological buffer", allowing RNAP to transcribe a longer gene under torsionally-constrained conditions, without any assistance from topoisomerases.
In addition, a canonical nucleosome has a distinctive chirality, with DNA wrapping around the histone octamer in a left-handed fashion, such that DNA in a nucleosome is (−) supercoiled. Using magnetic tweezers, Bancaud (Bancaud et al. 2007) examined the chiral transition of a single chromatin fiber under torsional stress (Figs. 6a and 6b). They found that the fiber can transiently trap positive twists at a rate of one turn per nucleosome, but does not trap negative twists. This hysteresis disappeared for a tetrasome fiber that lacked H2A-H2B dimers. These results suggest that, not only can a tetrasome readily fluctuate between left- and right-handed chiral conformations, a nucleosome can also undergo a similar chiral transition upon positive torsional stress. However, the energetic barrier for this nucleosome transition is high due to the presence of H2A-H2B dimers, leading to the observed hysteresis. More recently, Vlijm et al. (Vlijm et al. 2015) directly observed that a tetrasome can spontaneously fluctuate between left- and right-handed chirality and the addition of H2A and H2B converts a tetrasome into a stable left-handed nucleosome.
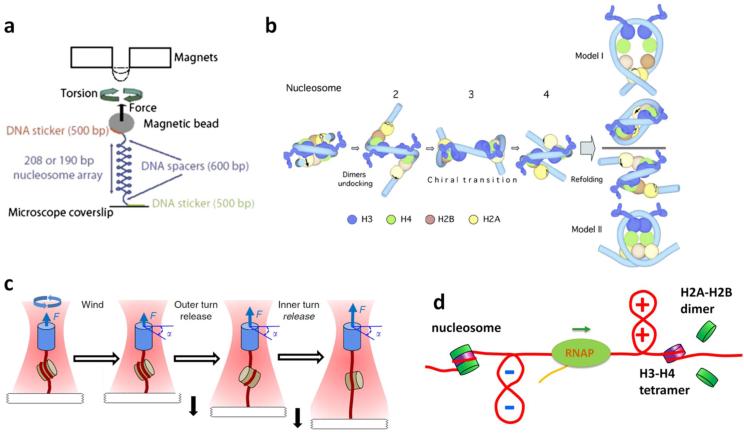
Fig. 6.
Nucleosomes under DNA supercoiling. (a) A schematic of the magnetic tweezers experiments to study nucleosome chiral transition. A nucleosome array was twisted held under constant force while the DNA was supercoiled. (b) A model of the nucleosome chiral transition from left- to right-handed configuration. Two alternative routes for the refolding of the dimers are shown. (c) A cartoon depicting the nucleosome-stretching assay with an AOT. A DNA containing a single nucleosome was stretched under a defined torsion. (d) A model for nucleosome turn-over during transcription. (+) DNA supercoiling in front of a transcribing RNAP may induce H2A-H2B dimer loss, while (−) DNA supercoiling behind the RNAP facilitates nucleosome assembly and stabilizes assembled nucleosomes. (a) and (b) are adapted from (Bancaud et al. 2007), (c) from (Sheinin et al. 2013), all with permission.
Nucleosome chirality has been suggested to facilitate nucleosome disassembly in front of an elongating RNAP, where DNA is (+) supercoiled, and reassembly behind the RNAP, where DNA is (−) supercoiled. Sheinin et al. (Sheinin et al. 2013) examined the stability of a single nucleosome using an angular optical trap (Fig. 6c). They found that a (+) torque of ~19 pN·nm, or larger, can significantly disrupt nucleosome structures, leading to an almost complete (80%) loss of H2A-H2B dimer, whereas nucleosomes are stable against (−) torque. In a complementary study using scanning force microscopy and fluorescence correlation spectroscopy, Elbel and Langowski (Elbel and Langowski 2015) found that (+) supercoiling led to larger nucleosome opening angles than (−) supercoiled DNA, and also decreased nucleosome’s stability to high salt. These results corroborate those of Teves and Henikoff's genomic studies (Teves and Henikoff 2014), and support a model that Pol II generated (+) torque can disrupt nucleosomal roadblocks in vivo (Fig. 6d).
These single molecule studies provide significant quantitative information concerning how torsion affects both DNA and chromatin topology and structures, laying down the groundwork for understanding the interplay of DNA supercoiling and transcription.
Action over distance
DNA supercoiling can bring distal DNA elements into close proximity to permitn action over distance, for example, to allow enhancer-promoter communication over thousands of base pairs (Liu et al. 2001). Using magnetic tweezers, Ding et al. showed that (−) DNA supercoiling can encourage the formation of λ repressor (CI)-mediated loops in DNA (Ding et al. 2014), which in turn prevent over-expression of this repressor protein so that the cell can still preserve sensitivity to conditions that trigger virulence (lysis). Norregaard et al. (Norregaard et al. 2013) developed a unique peptide nucleic acid (PNA)-based assay with a tethered particle tracking method (Norregaard et al. 2014) and showed that the presence of DNA supercoils can greatly enhance the juxtaposition probability of λ repressor (CI) and, hence, significantly increase the efficiency and cooperativity of a λ epigenetic switch.
Transcription initiation under torsion
Transcription initiation requires promoter opening, which must be sensitively regulated by DNA supercoiling. Using magnetic tweezers, Revyakin et al. (Revyakin et al. 2004; Revyakin et al. 2006) developed an elegant method to study transcription initiation kinetics of E. coli RNAP on a supercoiled DNA template. In their assay, MTs were used to preset DNA into a plectonemic state by adding a number of (+) or (−) twists prior to transcription initiation. During initiation, RNAP unwound promoter DNA to form the transcription bubble of an RNAP open complex (RPo), thereby changing the linking number of DNA outside of the RNAP (Fig. 7). Because the DNA was in a plectonemic state, such a linking number change would have to be accommodated by a change in writhe (i.e. the number of loops in the plectonemic region). This resulted in a large change in DNA extension, and thus the detection sensitivity of an open complex formation was greatly enhanced. Using this assay, RNAP-dependent DNA unwinding was resolved to ~1 bp (spatial resolution) and ~1 s (temporal resolution). They show that DNA supercoiling can affect both the rate of the open complex formation and the lifetime of this complex. The authors compared DNA supercoiling impacts on abortive initiation at two canonical promoters, i.e. lacCONS (a consensus lac promoter) and rrnB P1 (one of the promoters driving transcription of ribosomal RNA) (Revyakin et al. 2004). At lacCONS, (−) supercoiling increases the lifetime of an RPo relative to that observed for (+) supercoiling. At rrnB P1, an RPo is significantly less stable under (−) supercoiling conditions, than observed at lacCONS, and is not even formed when DNA was (+) supercoiled. This work not only directly demonstrates the impact of DNA supercoiling on transcription initiation, but also establishes a novel methodology for studies of various protein-DNA interactions under supercoiling.
Fig. 7.
A schematic depicting the method of detection for promoter opening during transcription initiation using magnetic tweezers. Prior to a measurement, the DNA molecule was either (−) or (+) supercoiled under a small and constant force. Promoter opening by an RNAP increased or decreased the DNA extension for the (−) or (+) supercoiled DNA respectively. Adapted from (Revyakin et al. 2004) with permission.
Determination of RNAP generated torque
RNAP is a torsional motor; as it translocates along DNA, it must also rotate around the DNA helical axis. However, the determination of the torque that RNAP can generate is technically challenging and the torque value remained unknown until recently. An early study by Harada et al. (Harada et al. 2001) used a magnetic bead decorated with smaller fluorescent beads to visualize RNAP induced rotation and estimated that the lower limit of RNAP generated torque to be approximately ~ 5 pN·nm based on the rotational frictional drag of the magnetic bead.
Recently the maximum torque that RNAP can generate (i.e. the torque it takes to stall an elongating RNAP) was experimentally determined. Using an AOT, Ma et al. (Ma et al. 2013) developed a sophisticated single molecule assay to monitor transcription-generated DNA supercoiling and torque buildup, in real-time (Fig. 8a). With this method, the movement of an E. coli RNAP was followed under a defined torque, and the torque required to stall an elongating RNAP (“stall torque”) was measured. These experiments showed that the accumulated torsional stress, either in the (+) supercoiled DNA ahead of the RNAP or in the (−) supercoiled DNA behind the RNAP, could cause an elongating RNAP to stall. The measured average “stall torque” value, in either case, was ~11 pN·nm (Fig. 8b), which is twice that of the previously measured lower bound of RNAP generated torque. This torque nearly coincides with that required to melt DNA of arbitrary sequence (Bryant et al. 2003; Forth et al. 2008; Oberstrass et al. 2013) (Fig. 8c). Such melted DNA can serve as distinct targets for regulatory proteins in gene regulation. In addition, this torque is also sufficient to convert DNA into a plectonemic state (Bryant et al. 2003; Forth et al. 2008; Oberstrass et al. 2013), or to substantially modify chromatin topology (Bancaud et al. 2006). Thus, these results establish RNAP as a powerful torsional motor and suggest that the torque-generating capacity of RNAP may have been tuned to important transitions in DNA or chromatin structures. It also establishes the relevant physiological torque scale for DNA-based processes.
Fig. 8.
Determination of the torque generated by RNAP during transcription. (a) A cartoon depicting the "twin supercoiled domain" model and experimental configuration to measure transcription against (−) supercoiling upstream (behind the RNAP) or (+) supercoiling downstream (in front of the RNAP). E. coli RNAP was torsionally anchored to the surface of a coverglass while either the downstream end or upstream end of the DNA template was torsionally constrained to a quartz cylinder held in an AOT. The AOT monitored the translocation of the RNAP along DNA and the torque RNAP generated in real time. RNAP elongation accumulated (+) or (−) DNA supercoiling respectively. As torque increased, RNAP was eventually stalled and the AOT reported the value of the stall torque. (b) Distributions of the measured downstream (left) and upstream (right) stall torques of RNAP. Adapted from (Ma et al. 2013) with permission.
Ma et al. also found that upon torque release, a significant fraction of stalled RNAPs resumed transcription in a short time. This suggests that, in vivo, stalled RNAPs can be rescued if torsional stress is released in a timely fashion, either by topoisomerases or via DNA and/or RNAP rotation, thereby preventing RNAPs from becoming roadblocks to hinder other vital cellular processes such as replication (Dutta et al. 2011). In addition, they also demonstrated that torque can directly affect transcription speed and pausing, providing direct evidence that DNA supercoiling can be an effective regulator of transcription elongation.
DNA supercoiling induced transcriptional bursting
Transcription has been found to be highly stochastic in a diverse array of organisms, from bacteria to mammals. At many highly transcribed genes, expression happens in the form of transcriptional bursting, during which transcription occurs as stochastic “bursts” of RNA synthesis interspersed with long periods of inactivity (Golding et al. 2005; Levens and Larson 2014; So et al. 2011; Suter et al. 2011; Taniguchi et al. 2010; Zong et al. 2010). However, the origin of such a ubiquitous phenomenon was not well understood. Recently, using single molecule fluorescence techniques, Chong et al. (Chong et al. 2014) directly monitored RNA synthesis in vitro when a T7 RNAP was transcribing on a torsionally-constrained template. They observed that, in the presence of Topo I which preferentially removes (−) DNA supercoiling, transcription initiation and elongation slowed down and eventually halted due to an accumulation of (+) supercoiling built up by transcription, but once gyrase was introduced, transcription was fully recovered (Fig. 9). Since there are more chromosomal DNA loops than the number of gyrase molecules in E. coli, these results support that, in a chromosomal DNA loop containing a highly expressed gene, the accumulation and removal of positive supercoiling can switch genes off and on. They further used quantitative RT-PCR to examine whether the chromosomal supercoiling level affects transcription elongation in live E. coli cells. The results were consistent with the in vitro experimental results, and confirmed that (+) supercoiling buildup can slow down transcription in vivo. These data together led them to an insightful explanation for transcriptional bursting in bacteria. The E coli chromosome is organized into hundreds of topologically constrained loops. During active transcription, transient DNA supercoiling can be generated locally. (−) DNA supercoiling is rapidly removed by Topo I, whereas the activity of gyrase is not sufficient to keep up with transcription, leading to (+) DNA supercoiling accumulation within the DNA loops. The torsional stress eventually inhibits transcription initiation and switches genes off. Subsequent gyrase binding to DNA releases the (+) DNA supercoiling and switches genes back to the "on" state. Thus, genes are stochastically switched between on and off states due to the release and accumulation of (+) DNA supercoiling. Chong et al.’s experiments provide a simple, but elegant, demonstration that supercoiling dynamics are the primary origin of transcriptional bursting in bacteria. It is worth noting that in eukaryotic cells, topoisomerases are different from that in prokaryotic cells (Champoux 2001). For example, there is no DNA gyrase in eukaryotic cells. In addition, in contrast to Topo I in E. coli which is a type IA topoisomerase and can only relax (−) supercoils via a “single-strand break and passage” mechanism, eukaryotic topoisomerase I is a type IB topoisomerase and can operate on both (+) and (−) supercoils using a “single-strand break and rotation” mechanism (Dekker et al. 2002; Koster et al. 2005). Eukaryotic topoisomerase II, using a “double-strand break and passage” mechanism, can also release DNA supercoils of both signs. (Berger et al. 1996; Champoux 2001; Strick et al. 2000). These dissimilarities may result in different patterns of transcription and noise in eukaryotes versus prokaryotes.
Fig. 9.
Single molecule observations of transcription bursting. (a) A cartoon depicting how multiple rounds of transcription on a circular template led to an accumulation of significant (+) torsional stress and inhibition of transcription in the presence of Topo I and absence of gyrase. The subsequent addition of gyrase recovered transcription. (b) Time dependence of T7 transcription initiation rate (blue) under the conditions shown in (a). Transcription initiation was inhibited by (+) supercoiling, but then recovered after the addition of gyrase. Adapted from (Chong et al. 2014) with permission.
Some open questions
Despite important insights gained from recent experiments, many fundamental questions are left unanswered. First, single molecule studies have established E. coli RNAP as a powerful torsional motor with the ability to significantly alter DNA topology. However, the torque that a eukaryotic RNA polymerase, such as Pol II, can generate remains unknown. Is the torque generated by Pol II large enough to significantly disrupt nucleosome structures? Genomic studies (Teves and Henikoff 2014) have shown that topoisomerase inhibition results in increased nucleosome turnover within gene bodies. This observation suggests that Pol II transiently introduces torsion into DNA, which in turn alters nucleosome stability. However, direct and quantitative evidence is lacking to show that Pol II generated torque is sufficient to cause nucleosome turnover. In addition, in vivo RNAP works closely with various transcription factors, and how these factors regulate the capability of RNAP in driving DNA supercoiling needs to be examined. It is intriguing to examine whether inhibiting some transcription factors could result in a measurable effect on DNA supercoiling at a genomic scale. Furthermore, it would be interesting to see how DNA supercoiling induced by transcription could affect other vital cellular processes, such as replication and DNA recombination. Finally, single molecule and single cell studies have provided insight into transcriptional bursting, showing DNA supercoiling dynamics play a key role, in E. coli, in turning genes "on" or "off". As transcriptional bursting is a phenomenon across species, it will be interesting to see if DNA supercoiling also plays important roles in the determination of gene "noise" in eukaryotic systems.
In summary, genomic sequencing and micro-array analysis studies have painted a very dynamic picture of the complicated interplay between transcription and DNA supercoiling in vivo, and also disclosed the entangled roles of RNAPs, topoisomerases, and DNA supercoiling in gene regulation. Single molecule studies complement these approaches by providing invaluable quantitative information about the torsional characteristics of DNA, chromatin, and RNAP, and thus new insights into cellular mechanisms. With continued technological innovation, we expect that unprecedented knowledge will be gained about how cells use DNA supercoiling as an important regulator for many cellular processes.
Acknowledgements
This work was supported by National Science Foundation grant (MCB- 1517764 to M.D.W.) and the Fundamental Research Funds for the Central Universities, China (15lgjc15 to J.M.)
Footnotes
Conflict of interest
None
Compliance with Ethical Standards
Ethical approval
This article does not contain any studies with human or animal subjects performed by the authors.
References
- Allemand JF, Bensimon D, Lavery R, Croquette V. Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc Natl Acad Sci USA. 1998;95:14152–14157. doi: 10.1073/pnas.95.24.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett. 1986;11:288–290. doi: 10.1364/OL.11.000288. [DOI] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic JM, Yamane T. Optical trapping and manipulation of single cells using infrared laser beams. Nature. 1987;330:769–771. doi: 10.1038/330769a0. [DOI] [PubMed] [Google Scholar]
- Bécavin C, Barbi M, Victor J-M, Lesne A. Transcription within Condensed Chromatin: Steric Hindrance Facilitates Elongation. Biophys J. 2010;98:824–833. doi: 10.1016/j.bpj.2009.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Fulbright RM, Wang MD. Mechanochemical Kinetics of Transcription Elongation. Phys Rev Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- Bancaud A, et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol. 2006;13:444–450. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- Bancaud A, et al. Nucleosome Chiral Transition under Positive Torsional Stress in Single Chromatin Fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- Basu A, Schoeffler AJ, Berger JM, Bryant Z. ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat Struct Mol Biol. 2012;19:538–546. doi: 10.1038/nsmb.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- Bermúdez I, García-Martínez J, Pérez-Ortín JE, Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen–DNA photobinding. Nucl Acids Res. 2010;38:e182–e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- Celedon A, et al. Magnetic Tweezers Measurement of Single Molecule Torque. Nano lett. 2009;9:1720–1725. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. DNA Topoisomerases: Structure, Function, and Mechanism. Annual Review of Biochemistry. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chong S, Chen C, Ge H, Xie XS. Mechanism of Transcriptional Bursting in Bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker NH, Rybenkov VV, Duguet M, Crisona NJ, Cozzarelli NR, Bensimon D, Croquette V. The mechanism of type IA topoisomerases. Proc Natl Acad Sci USA. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat Meth. 2007;4:223–225. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- Ding Y, Manzo C, Fulcrand G, Leng F, Dunlap D, Finzi L. DNA supercoiling: A regulatory signal for the λ repressor. Proc Natl Acad Sci USA. 2014;111:15402–15407. doi: 10.1073/pnas.1320644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA Polymerase Backtracking to Genome Instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbel T, Langowski J. The effect of DNA supercoiling on nucleosome structure and stability. J Phys: Condens Matter. 2015;27:064105. doi: 10.1088/0953-8984/27/6/064105. [DOI] [PubMed] [Google Scholar]
- Forth S, Deufel C, Patel Smita S, Wang Michelle D. Direct Measurements of Torque During Holliday Junction Migration. Biophys J. 2011;101:L5–L7. doi: 10.1016/j.bpj.2011.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth S, Deufel C, Sheinin MY, Daniels B, Sethna JP, Wang MD. Abrupt Buckling Transition Observed during the Plectoneme Formation of Individual DNA Molecules. Phys Rev Lett. 2008;100:148301–148304. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forth S, Sheinin MY, Inman J, Wang MD. Torque Measurement at the Single-Molecule Level. Annu Rev Biophys. 2013;42:583–604. doi: 10.1146/annurev-biophys-083012-130412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci USA. 1992;89:11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Santos GD, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II Disrupts DNA-specified Nucleosome Organization to Enable Precise Gene Regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-Time Kinetics of Gene Activity in Individual Bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Gore J, Bryant Z, Stone MD, Nöllmann M, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–104. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafi DE, Bossi L. Activation and silencing of leu-500 promoter by transcription-induced DNA supercoiling in the Salmonella chromosome. Mol Microbiol. 2000;37:583–594. doi: 10.1046/j.1365-2958.2000.02015.x. [DOI] [PubMed] [Google Scholar]
- Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, Kinosita K. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- Herbert A, Rich A. The Biology of Left-handed Z-DNA. J Biol Chem. 1996;271:11595–11598. doi: 10.1074/jbc.271.20.11595. [DOI] [PubMed] [Google Scholar]
- Inman J, Forth S, Wang MD. Passive torque wrench and angular position detection using a single-beam optical trap. Opt Lett. 2010;35:2949–2951. doi: 10.1364/OL.35.002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen XJA, Lipfert J, Jager T, Daudey R, Beekman J, Dekker NH. Electromagnetic Torque Tweezers: A Versatile Approach for Measurement of Single-Molecule Twist and Torque. Nano Lett. 2012;12:3634–3639. doi: 10.1021/nl301330h. [DOI] [PubMed] [Google Scholar]
- Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- Kouzine F, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Liu J, Sanford S, Chung H-J, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in Vivo1. J Mol Biol. 1999;292:1149–1160. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- La Porta A, Wang MD. Optical Torque Wrench: Angular Trapping, Rotation, and Torque Detection of Quartz Microparticles. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- Leng F, Amado L, McMacken R. Coupling DNA Supercoiling to Transcription in Defined Protein Systems. J Biol Chem. 2004;279:47564–47571. doi: 10.1074/jbc.M403798200. [DOI] [PubMed] [Google Scholar]
- Levens D, Larson Daniel R. A New Twist on Transcriptional Bursting. Cell. 2014;158:241–242. doi: 10.1016/j.cell.2014.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ, Higgins CF. Local DNA topology and gene expression: the case of the leu-500 promoter. Mol Microbiol. 1991;5:779–783. doi: 10.1111/j.1365-2958.1991.tb00749.x. [DOI] [PubMed] [Google Scholar]
- Lipfert J, Kerssemakers JWJ, Jager T, Dekker NH. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat Meth. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- Lipfert J, Wiggin M, Kerssemakers JWJ, Pedaci F, Dekker NH. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nat Commun. 2011;2:439. doi: 10.1038/ncomms1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bondarenko V, Ninfa A, Studitsky VM. DNA supercoiling allows enhancer action over a large distance. Proc Natl Acad Sci USA. 2001;98:14883–14888. doi: 10.1073/pnas.261477898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bai L, Wang MD. Transcription Under Torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev E. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci. 2004;117:3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K, Mizuuchi M, Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982;156:229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Mosconi F, Allemand JF, Croquette V. Soft magnetic tweezers: A proof of principle. Rev Sci Instrum. 2011;82:034302. doi: 10.1063/1.3531959. [DOI] [PubMed] [Google Scholar]
- Naughton C, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollmann M, et al. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat Struct Mol Biol. 2007;14:264–271. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- Norregaard K, Andersson M, Nielsen PE, Brown S, Oddershede LB. Tethered particle analysis of supercoiled circular DNA using peptide nucleic acid handles. Nat Protoc. 2014;9:2206–2223. doi: 10.1038/nprot.2014.152. [DOI] [PubMed] [Google Scholar]
- Norregaard K, Andersson M, Sneppen K, Nielsen PE, Brown S, Oddershede LB. DNA supercoiling enhances cooperativity and efficiency of an epigenetic switch. Proc Natl Acad Sci USA. 2013;110:17386–17391. doi: 10.1073/pnas.1215907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Lis JT. RNA polymerase II pauses at the 5' end of the transcriptionally induced Drosophila hsp70 gene. Mol Cell Biol. 1991;11:5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Fernandes LE, Lebel P, Bryant Z. Torque spectroscopy of DNA: base-pair stability, boundary effects, backbending, and breathing dynamics. Phys Rev Lett. 2013;110:178103–178103. doi: 10.1103/PhysRevLett.110.178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oussatcheva EA, Pavlicek J, Sankey OF, Sinden RR, Lyubchenko YL, Potaman VN. Influence of Global DNA Topology on Cruciform Formation in Supercoiled DNA. J Mol Biol. 2004;338:735–743. doi: 10.1016/j.jmb.2004.02.075. [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT. Rapid, Transcription-Independent Loss of Nucleosomes over a Large Chromatin Domain at Hsp70 Loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: Detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A, Liu C, Ebright RH, Strick TR. Abortive Initiation and Productive Initiation by RNA Polymerase Involve DNA Scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samul R, Leng F. Transcription-coupled Hypernegative Supercoiling of Plasmid DNA by T7 RNA Polymerase in Escherichia coli Topoisomerase I-Deficient Strains. J Mol Biol. 2007;374:925–935. doi: 10.1016/j.jmb.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Léger J-F, Chatenay D, Marko JF. Structural transitions in DNA driven by external force and torque. Phys Rev E. 2001;63:051903. doi: 10.1103/PhysRevE.63.051903. [DOI] [PubMed] [Google Scholar]
- Sheinin MY, Forth S, Marko JF, Wang MD. Underwound DNA under Tension: Structure, Elasticity, and Sequence-Dependent Behaviors. Phys Rev Lett. 2011;107:108102. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin MY, Li M, Soltani M, Luger K, Wang MD. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun. 2013;4 doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin MY, Wang MD. Twist-Stretch Coupling and Phase Transition During DNA Supercoiling. Phys Chem Chem Phys. 2009;11:4800–4803. doi: 10.1039/b901646e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinin MY, Wang MD. A DNA Twist Diffuses and Hops. Science. 2012;338:56–57. doi: 10.1126/science.1228656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shundrovsky A, Santangelo TJ, Roberts JW, Wang MD. A Single-Molecule Technique to Study Sequence-Dependent Transcription Pausing. Biophys J. 2004;87:3945–3953. doi: 10.1529/biophysj.104.044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So L-h, Ghosh A, Zong C, Sepúlveda LA, Segev R, Golding I. GENERAL PROPERTIES OF THE TRANSCRIPTIONAL TIME-SERIES IN ESCHERICHIA COLI. Nat Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick TR, Allemand J-F, Bensimon D, Bensimon A, Croquette V. The Elasticity of a Single Supercoiled DNA Molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian Genes Are Transcribed with Widely Different Bursting Kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Tabuchi H, Handa H, Hirose S. Underwinding of DNA on Binding of Yeast TFIID to the TATA Element. Biochem Biophys Res Commun. 1993;192:1432–1438. doi: 10.1006/bbrc.1993.1576. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao Y-P, Wu H-Y, Liu LF. Transcription-driven supercoiling of DNA: Direct biochemical evidence from in vitro studies. Cell. 1989;56:111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- van Loenhout MTJ, de Gruntb MV, Dekker C. Dynamics of DNA Supercoils. Science. 2012;338:94–97. doi: 10.1126/science.1225810. [DOI] [PubMed] [Google Scholar]
- Vlijm R, Lee M, Lipfert J, Lusser A, Dekker C, Dekker Nynke H. Nucleosome Assembly Dynamics Involve Spontaneous Fluctuations in the Handedness of Tetrasomes. Cell Rep. 2015;10:216–225. doi: 10.1016/j.celrep.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-Y, Shyy S, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yin H, Wang MD, Svoboda K, Landick R, Block SM, Gelles J. Transcription Against an Applied Force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Victor J-M. How are nucleosomes disrupted during transcription elongation? HFSP J. 2009;3:373–378. doi: 10.2976/1.3249971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C, So Lh, Sepúlveda LA, Skinner SO, Golding I. Lysogen stability is determined by the frequency of activity bursts from the fate-determining gene. Molecular Systems Biology. 2010;6 doi: 10.1038/msb.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]