Abstract
Two synthetic approaches that allow one to control PEG content within spherical nucleic acids (SNAs) have been developed. One approach begins with RNA-modified gold nanoparticles followed by a backfill of PEG 2K alkanethiols, and the other involves co-adsorption of the two entities on a gold nanoparticle template. These two methods have been used to explore the role of PEG density on the chemical and biological properties of RNA–SNAs. Such studies show that while increasing the extent of PEGylation within RNA–SNAs extends their blood circulation half-life in mice, it also results in decreased cellular uptake. Modified ELISA assays show that constructs, depending upon RNA and PEG content, have markedly different affinities for class A scavenger receptors, the entities responsible, in part, for cellular internalization of SNAs. In designing SNAs for therapeutic purposes, these competing factors must be considered and appropriately adjusted depending upon the desired use.
Graphical abstract
Introduction
Spherical nucleic acids (SNAs) are a unique class of nanoparticles that typically consist of a dense layer of highly oriented oligonucleotides surrounding a nanoparticle (NP) core,1,2 and they exhibit several properties that make them attractive for biological and medical applications. In contrast with their linear counterparts, SNAs are actively internalized by cells without the need for cytotoxic transfection reagents.3–5 This process is often mediated by class A scavenger receptors3,5 that have high affinities for SNAs but not the particle-free sequences. These structure-dependent properties have made SNAs promising single-entity agents for probing and regulating cellular processes.2,6–14 For example, RNA–SNAs are attractive agents for gene regulation applications, in which the design of the nucleic acid sequence of a SNA may be tailored to effect the knock-down of a genetic target.6 Indeed, small interfering RNA (siRNA) SNAs have been shown to penetrate the skin,11,13 when topically applied to mice, and cross the blood–brain barrier, when systemically administered. In fact, they have become lead compounds for treating diabetic wounds11 and glioblastoma multiforme.8
In addition to the nanoparticle core and oligonucleotide components that comprise the shell, RNA–SNAs typically include poly(ethylene glycol) (PEG) molecules interspersed within the shell (Scheme 1), which are intended to passivate the NP surface and improve particle stability.6 However, little is known about the effect of forming a mixed monolayer consisting of both RNA and PEG on the chemical and biological properties of SNAs. Indeed, PEG has been used as an agent to stabilize the RNA–SNAs and to increase their circulation times, although in the latter case, data has not yet been collected to evaluate the merit of such an approach. To further complicate matters, it is conceivable that the addition of PEG to the SNA could increase stability and circulation times but adversely affect cellular uptake because it could block the receptors that recognize the SNA and facilitate cellular internalization.
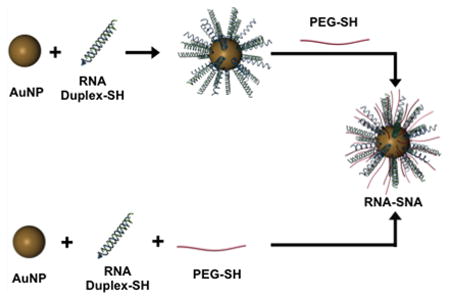
Scheme 1. Two Approaches to the Synthesis of RNA-SNAs, Which Consist of a NP Core, Duplexed siRNA (Thiolated Sense RNA Shown in Blue and Complementary Anti-Sense (AS) RNA Shown in Green), and a PEG-Thiol Passivating Molecule.
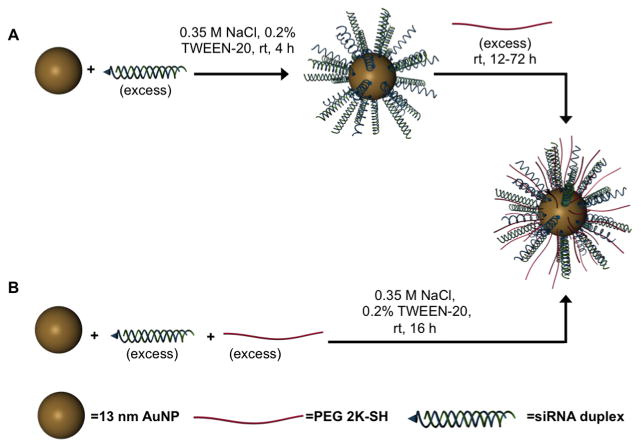
a(A) In the backfill method of synthesis, RNA is first adsorbed on the AuNP surface, and PEG is added subsequently. (B) Alternatively, RNA and PEG may be co-adsorbed to the AuNP.
Herein, we evaluate methods for synthesizing RNA–SNAs with PEG as a stabilizing diluent. In general, two approaches are studied: (1) RNA–SNAs are initially prepared, followed by backfilling with PEG-alkanethiols (Scheme 1A); and (2) RNA duplexes, modified with alkanethiols and PEG-alkanethiols, are co-adsorbed onto the surface of a gold nanoparticle (Scheme 1B). The latter method has proven more efficient for preparing SNAs with more controllable PEG contents, and it has become the primary synthetic method for systematically controlling and studying RNA–SNA architecture and properties. We used this method to systematically study how RNA and PEG-loading affect uptake in C166 cells and circulation time in mice. This cell type was chosen because it is one of the most widely studied, and it has been the basis for the vast majority of SNA-based cellular uptake mechanistic work. The data show that the two primary components that define the RNA–SNA work against one another, and, depending upon intended use, the relative amounts of each will need to be carefully adjusted.
Results and Discussion
Synthesis of RNA–SNAs by Backfilling with PEG
PEGylated RNA–SNAs were first synthesized via the backfill method (Scheme 1A) using 2 kDa PEG thiol (PEG 2K) as a passivating molecule. This PEG was chosen because it is large enough to protrude past the RNA duplexes on the NP surface (16 nm for fully extended PEG 2K versus 8.3 nm for the RNA duplex; see the Methods section in the Supporting Information for a more detailed discussion on the determination of PEG and RNA length). A 200-fold excess of duplexed siRNA was first added to an aqueous solution of 13 nm gold nanoparticles (AuNPs), and after this mixture was incubated for 4 h to allow for the adsorption of RNA onto the NP surface through the formation of Au–S bonds, a 2000-fold excess of PEG 2K was added. The mixture was then incubated for 12 h to passivate the NP surface. Interestingly, the loading of RNA on the SNAs does not significantly change compared to that of SNAs that were not backfilled with PEG following functionalization with RNA (115 ± 5 sense RNA/NP, 43 ± 5 AS RNA/NP; 102 ± 8 sense RNA/NP, and 41 ± 2 AS RNA/NP, respectively), indicating that despite dense functionalization of the NP surface with negatively charged oligonucleotides, there is still significant room on the SNAs to load smaller, neutral molecules without displacing RNA. Thus, following a 12 h incubation with PEG 2K, PEG is loaded onto the RNA–SNAs (5 ± 1 PEG molecules per NP), corresponding to an increase in the hydrodynamic diameter from 34 ± 1 to 41 ± 1 nm due to the persistence length of PEG 2K, which extends past the RNA duplexes on the NP surface. In addition, a shift in the zeta potential from −33 ± 1 to −29 ± 2 mV is observed due to the shielding of the negatively charged RNA by neutral PEG molecules at the outermost surface of the SNA (Figure 1B).
Figure 1.
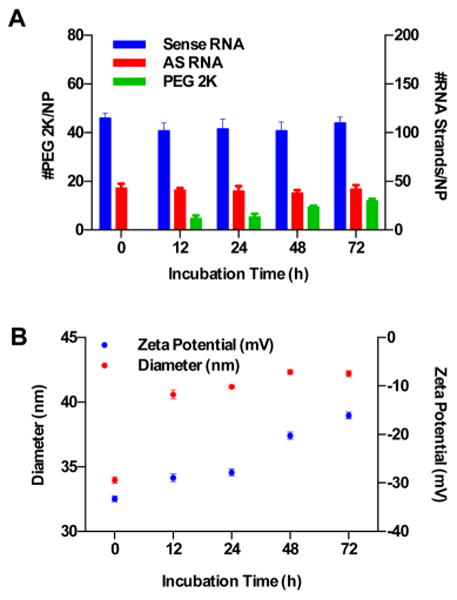
Effect of PEG 2K incubation time on the RNA and PEG loading of RNA–SNAs. (A) Antisense and sense RNA loading does not change significantly with increasing incubation time with PEG 2K; however, the loading of PEG does increase with longer incubation times. (B) The increase in PEG loading corresponds to an increase in the hydrodynamic diameter and the zeta potential of SNAs.
To increase the loading of PEG on the RNA–SNAs, the incubation time was varied from 12 to 72 h, and the resulting SNAs were purified and characterized. Correspondingly, an increase from 5 ± 1 to 12 ± 2 PEG molecules per NP was observed. Interestingly, however, the RNA loading (both sense and antisense strands) remains relatively constant, regardless of the duration of incubation of RNA–SNAs with PEG (Figure 1A). These changes in surface functionalization correlate with little change in the hydrodynamic diameter, but there is a shift in zeta potential from −29 ± 2 to −16 ± 1 mV for RNA–SNAs backfilled with PEG 2K for 12 and 72 h, respectively (Figure 1B). Despite the increase in PEG loading observed for longer PEG incubation times, the backfill method of synthesis generates a relatively small range of RNA–SNAs with respect to degrees of PEGylation. We hypothesize that by prefunction-alizing SNAs with RNA, the limited availability of surface sites prevents rapid adsorption of PEG onto the AuNP surface. To probe the relationships between the PEGylation of RNA–SNAs and cellular uptake and circulation half-life, we sought to synthesize SNAs with a larger range of PEG loadings. Although this may be achieved via the backfill method of synthesis by using an even greater excess of PEG or by further increasing the incubation time with PEG, these approaches are not desirable because they are inefficient in terms of materials use and synthesis time. Thus, we sought to synthesize RNA–SNAs through the coadsorption of RNA and PEG onto AuNPs, which we hypothesized could be used to make a range of SNAs with varying PEG loadings by changing the mole fraction of RNA and PEG present in solution.
Co-Adsorption of RNA and PEG to AuNPs in SNA Synthesis
To synthesize SNAs with a variety of RNA and PEG loadings, PEG 2K and duplexed siRNA were incubated with 13 nm gold nanoparticles (AuNPs) at different ratios (Figure 2A outlines nomenclature for the SNAs synthesized via this method). As the mole fraction of [RNA]/[PEG] incubated with AuNPs is increased, the number of sense RNA strands loaded per AuNP increases (Figure 2B). Interestingly, however, the antisense RNA loading is altered less-profoundly by the increase in PEG content. Thus, for SNAs more-densely functionalized with PEG, the percentage of sense RNA strands that are hybridized to antisense RNA strands is greater than SNAs with little or no PEG. In particular, on the SNA that we have labeled SNA 2-1 that was synthesized with a mole ratio of [RNA]/[PEG] = 0.1 and that has 19 ± 2 PEG 2K/SNA, 37 ± 2 sense RNA/SNA, and 32 ± 2 antisense RNA/SNA, ∼80% of sense strands are hybridized to antisense RNA. In contrast, on the SNA that we have labeled SNA 2-5 and that was synthesized with a mole ratio of [RNA]/[PEG] = 100 and that has 4 ± 1 PEG 2K/SNA, 125 ± 3 sense RNA/SNA, and 43 ± 3 antisense RNA/SNA, ∼30% of sense strands are hybridized to antisense RNA. This difference is likely due to reduced electrostatic repulsion in SNA 2-1 compared to that of SNAs with denser loading of negatively charged nucleic acids on the NP surface and correlates well with similar results observed for DNA-SNAs.15 This result is important because the functional moiety used to effect gene knockdown is the siRNA duplex.16 Thus, by coadsorbing RNA and PEG to the NP surface, one may effectively tailor the extent of PEGylation without significantly altering the loading of functional siRNA duplexes on the SNA's surface.
Figure 2.
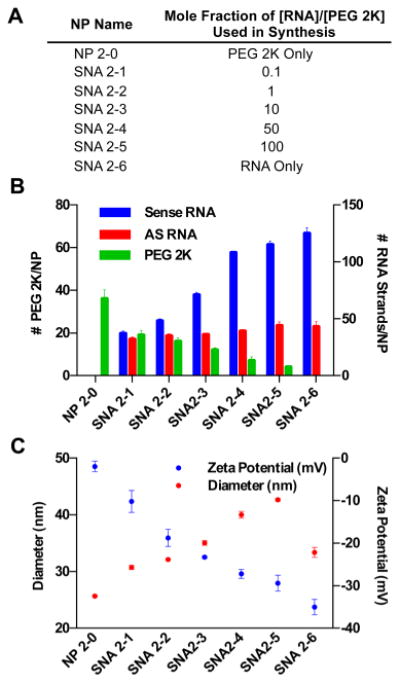
Co-adsorption of RNA and PEG to AuNPs to synthesize RNA-SNAs results in tunable RNA and PEG loading by varying the mole fraction of RNA/PEG used. (A) Nomenclature for RNA–SNAs, (B) RNA and PEG 2K loading, and (C) hydrodynamic diameter and zeta potential of RNA–SNAs synthesized using this method.
In addition, by coadsorbing RNA and PEG to AuNPs, a larger range of RNA–SNA PEG loadings was obtained than for the backfill method of synthesis. For example, SNA 2-1, the SNA with the lowest RNA loading and highest PEG 2K loading studied, has ∼60% more PEG 2K molecules per SNA than SNAs synthesized by backfilling with PEG 2K for 72 h. Collectively, these results show that the PEG content of RNA–SNAs synthesized via the coadsorption method is inversely related to the sense RNA loading. This may be the result of multiple factors: (1) the NP surface is more readily available for PEG 2K to bind because it has not been prefunctionalized with RNA, and (2) PEG 2K, which is lower in molecular weight than the RNA, may adsorb to the NP surface more rapidly.
To further understand the structure of the RNA–SNAs synthesized by coadsorption of RNA and PEG to AuNPs, the hydrodynamic diameters and zeta potentials of the SNAs were measured (Figure 2C). The sample of particles named NP 2-0, which is functionalized with PEG 2K only, is significantly smaller in size than the RNA–SNAs. This indicates that the PEG 2K on the NP surface is not fully extended when RNA is not present on the NP surface and that the presence of RNA on the NP surface facilitates the extension of PEG 2K to adopt a more linear conformation. When the RNA density is high enough (SNAs 2-4 and 2-5), the PEG 2K protrudes past the RNA surface, and the hydrodynamic diameter increases beyond the diameter of SNA 2-6 (40 ± 2 and 43 ± 3 nm for SNA 2-4 and SNA 2-5, respectively, versus 33 ± 1 nm for SNA-2-6), which is functionalized with RNA only. This correlates with the observation that the hydrodynamic diameter of SNAs backfilled with PEG 2K increases with longer incubation times because the dense loading of RNA on the NP surface remains relatively constant. In addition, the change in size with increasing RNA loading corresponds with increasingly negative surface potential. Due to the negative charge of the oligonucleotides, the zeta potential of the NPs approaches that of SNA 2-6, which consists of RNA only (−35 ± 2 mV), with increasing RNA loading. These results provide insight into the complicated structure of PEGylated RNA–SNAs. In particular, depending upon the extent of PEGylation, the RNA that is presented on the SNAs is shielded by PEG molecules, which extend past the RNA duplexes and alter the properties of the NP surface that is presented to proteins and cells in biological systems. This indicates that the extent of PEGylation of RNA–SNAs may impact their interactions with cell surface and serum proteins, an implication that must be considered in the design of therapeutic RNA–SNAs.
Effects of RNA and PEG Content on the Cellular Uptake of RNA–SNAs
The effects of RNA and PEG loading on the cellular uptake of RNA–SNAs was then studied within the context of their recognition by class A scavenger receptors, which are known to facilitate their uptake by a variety of cell types.3 Previous work has shown that PEG-NPs exhibit almost no measurable binding to class A scavenger receptors3 because they are not recognized by the protein. Thus, it was hypothesized that RNA–SNAs with higher PEG loading and lower RNA loading would suffer from poorer uptake into certain cell types due to decreased recognition by class A scavenger receptors. To probe this hypothesis, we conducted a modified ELISA assay to measure the relative affinity of RNA–SNAs for class A scavenger receptors (Figure 3A), where wells were coated with protein and treated with increasing concentrations of NPs and the amounts of the NPs that bound the protein in each well were quantified using inductively coupled plasma mass spectrometry (ICP-MS). Correlating with previous studies, NP 2-0 (which consists only of PEG 2K on its NP core) has a relatively low affinity for class A scavenger receptors, as evidenced by the minimal binding of NP 2-0 to the protein in this assay. On the basis of the results from previous studies that showed that class A scavenger receptors recognize the nucleobases of oligonucleo-tides,17 we hypothesized that the affinity of the protein for RNA–SNAs would increase with increasing RNA loading. As expected, more NPs bind class A scavenger receptors as RNA loading increases and PEG loading decreases (∼150 times more SNA 2-6 than NP 2-0 bound per well treated with 100 nM NP) due to the increased recognition of the RNA shell by the protein.
Figure 3.
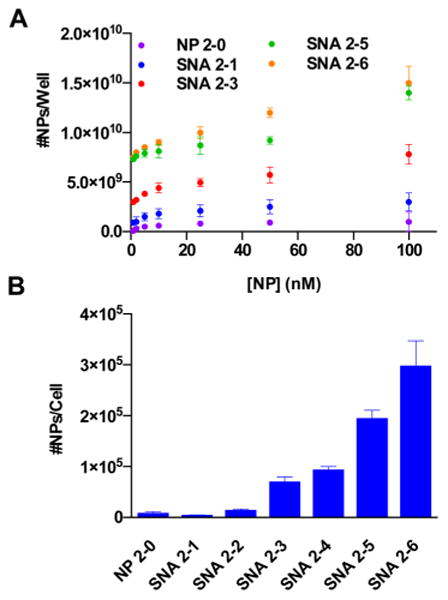
Effects of RNA and PEG content on the recognition of RNA–SNAs by cell surface receptors. (A) A modified ELISA assay measuring the relative affinity of class A scavenger receptors for RNA–SNAs with varying PEG content. (B) ICP-MS determination of RNA–SNA uptake by C166 cells.
On the basis of these results, we hypothesized that the RNA and PEG content of RNA–SNAs would impact their uptake into C166 mouse endothelial cells, which have been widely used to study the mechanism of uptake of SNAs.3 C166 cells were treated with RNA–SNAs functionalized with increasing amounts of PEG 2K, and their uptake was measured using ICP-MS to quantify the Au content in cells (Figure 3B). The results show that corresponding to their relative affinities for class A scavenger receptors, the uptake of RNA–SNAs into C166 cells is dependent upon both their RNA and PEG content, where SNAs containing more RNA are taken up into C166 cells to a greater extent than SNAs that contain less RNA and more PEG. This correlates well with previous work that has shown a decrease in cellular uptake of SNAs with lower oligonucleotide density4 and the relative lack of affinity for PEG-NPs by class A scavenger receptors.3 Taken together, these results indicate that the RNA and PEG components of the SNA inversely affect recognition by cell surface receptors and may be used to modulate the cellular uptake of SNAs.
Effects of RNA and PEG Content on the Circulation Half-Life of RNA–SNAs in Mice
The uptake of SNAs into cells via recognition by cell surface receptors is only one of many SNA-protein interactions that affect SNA function. In addition, the recognition of SNAs by serum proteins and the subsequent recognition by macrophages18 are important processes that can result in the clearance of NPs from the bloodstream upon sequestration by macrophage-rich tissues, such as the liver and spleen.19–29 PEG has been used widely to minimize the adsorption of serum proteins to NP surfaces and extend their circulation half-lives in animal models.21–23,29–33 To study the effects of RNA and PEG loading on the adsorption of serum proteins on RNA–SNAs, we incubated SNAs containing increasing amounts of PEG 2K in 10% fetal bovine serum for 0–4 h at physiological temperature (37 °C) and measured the amount of protein adsorbed to the NP surface qualitatively using dynamic light scattering (DLS) (Figure S1), and quantitatively using a bicinchoninic acid (BCA) assay (Figure 4A). The DLS and BCA results show that, indeed, SNAs with increasing PEG content and decreasing RNA content adsorb less serum protein than SNAs with little or no PEG.
Figure 4.
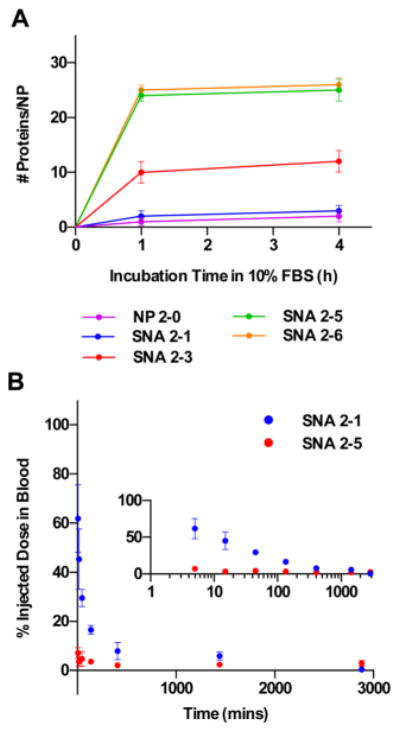
Effect of RNA and PEG content on the blood circulation time of RNA–SNAs in mice. (A) Increasing PEG content on RNA–SNAs minimizes the adsorption of serum proteins. (B) Circulation of PEGylated RNA–SNAs in mice is extended by increasing PEG loading. Inset shows data plotted on a log scale to highlight the differences in SNA content in the blood at earlier time points.
To study the effect of PEG loading and serum protein adsorption on the blood circulation half-life of RNA–SNAs, two SNAs (one with high RNA loading but low PEG loading, SNA 2-5; and one with low RNA loading but high PEG loading, SNA 2-1) were injected into mice intravenously via the tail vein. Blood samples were collected at various time points following the injection, and the samples were analyzed for Au content by ICP-MS (Figure 4B). The results indicate that SNA 2-5 is cleared from the blood more rapidly and has about a 10 times shorter circulation half-life than SNA 2-1 (∼1 min for SNA 2-5 and ∼10 min for SNA 2-1). Though the exact mechanism responsible for the decreased clearance of SNA 2-1 observed is not known, we hypothesize that SNAs designed for intravenous use may benefit from dense PEGylation because this would extend their circulation time in the bloodstream and could lead to enhanced accumulation in the targeted tissue. This may be of importance when designing SNAs to target cancer, where leaky vasculature often results in higher accumulation of NPs in tumor tissue through the enhanced permeability and retention effect,34,35 and NPs with long circulation half-lives exhibit higher accumulation in tumor tissue.36–39
Conclusions
This work establishes the role of PEG in the design and synthesis of efficacious RNA–SNAs. In particular, SNA synthesis parameters, including incubation time and order of addition of PEG, are presented as a means to tune the extent of SNA PEG content. The addition of PEG as a “backfill” molecule to RNA-functionalized AuNPs was shown to have little effect on the RNA loading, indicating that despite dense functionalization with negatively charged oligonucleotides, there is significant room to add neutral molecules to the NP surface. To further increase the density of PEG on the SNA surface, one may choose to co-adsorb RNA and PEG onto the AuNP surface, which enables facile tuning of the loading of RNA and PEG on SNAs by varying the mole fraction of each component. Doing so at high ratios of PEG to RNA enables one to load more PEG than is possible by “backfilling” RNA–SNAs with PEG by reducing the number of sense RNA strands loaded on the NP while maintaining a roughly constant loading of AS RNA.
In addition, the effect of PEG content on the biological fate of RNA-SNAs was established. While increased PEG loading reduces the formation of the SNA protein corona, thereby enhancing the blood circulation half-life, SNAs with high densities of PEG also exhibit reduced cellular uptake in other cell lines, most likely due to decreased interactions with class A scavenger receptors. While PEG may be used to modulate the interactions of SNAs with biological entities, including cell surface receptors and serum proteins, the optimal extent of PEG content will depend upon each application. For example, a SNA intended for topical application to the skin may not benefit from dense PEG loading as much as an SNA that requires intravenous delivery. Contrary to the conventional thought that one must densely PEGylate a nanoparticle for optimal function in vivo, in the case of SNAs, doing so could compromise function by sacrificing the high cellular uptake that is typically afforded by the SNA's architecture and dense oligonucleotide shell. Ultimately, it will be important to identify the ideal amount of PEG needed for SNAs for each specific application with the understanding that there is an inverse relationship between circulation time and cellular uptake that must be considered. A SNA architecture with an exposed PEG shell that enables longer circulation times but is shed by an external stimulus upon reaching target tissue to expose the oligonucleotide layer and facilitate cellular uptake may be valuable for mitigating these competing effects.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers U54CA151880 and U54CA199091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C.A.M. also gratefully acknowledges support from the NTU-NU Institute for NanoMedicine located at the International Institute for Nanotechnology, Northwestern University (Evanston, IL) and the Nanyang Technological University (Singapore). A.B.C. acknowledges a National Defense Science and Engineering Graduate Fellowship. Research reported in this publication was supported by the National Institute Of General Medical Sciences of the National Institutes of Health under award number T32GM105538 and F31GM119392 awarded to J.R.F. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. T.J.M. acknowledges an IBNAM-Baxter Early Career Development Award in Bioengineering and support from the International Institute for Nanotechnology of Northwestern University.
Footnotes
Tables showing RNA sequences used and properties of PEG 2K used. A figure showing the effect of PEGylation of the biological properties of RNA–SNAs. Additional details on materials and methods. (PDF)
Notes: The authors declare the following competing financial interest(s): C.A.M. has financial interests in/relative to Exicure, Inc. and AuraSense LLC, companies developing SNA therapeutic platforms.
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconj-chem.6b00483.
References
- 1.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 2.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 3.Choi CH, Hao L, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc Natl Acad Sci U S A. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halo TL, McMahon KM, Angeloni NL, Xu Y, Wang W, Chinen AB, Malin D, Strekalova E, Cryns VL, Cheng C, et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci U S A. 2014;111:17104–17109. doi: 10.1073/pnas.1418637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et al. Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3006839. 209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radovic-Moreno AF, Chernyak N, Mader CC, Nallagatla S, Kang RS, Hao L, Walker DA, Halo TL, Merkel TJ, Rische CH, et al. Immunomodulatory spherical nucleic acids. Proc Natl Acad Sci U S A. 2015;112:3892–3897. doi: 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randeria PS, Briley WE, Chinen AB, Guan CM, Petrosko SH, Mirkin CA. Nanoflares as probes for cancer diagnostics. Cancer Treat Res. 2015;166:1–22. doi: 10.1007/978-3-319-16555-4_1. [DOI] [PubMed] [Google Scholar]
- 11.Randeria PS, Seeger MA, Wang XQ, Wilson H, Shipp D, Mirkin CA, Paller AS. siRNA-based spherical nucleic acids reverse impaired wound healing in diabetic mice by ganglioside GM3 synthase knockdown. Proc Natl Acad Sci U S A. 2015;112:5573–5578. doi: 10.1073/pnas.1505951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: Probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng D, Giljohann DA, Chen DL, Massich MD, Wang XQ, Iordanov H, Mirkin CA, Paller AS. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc Natl Acad Sci U S A. 2012;109:11975–11980. doi: 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Nano-flares for mRNA Regulation and Detection. ACS Nano. 2009;3:2147–2152. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randeria PS, Jones MR, Kohlstedt KL, Banga RJ, Olvera de la Cruz M, Schatz GC, Mirkin CA. What Controls the Hybridization Thermodynamics of Spherical Nucleic Acids? J Am Chem Soc. 2015;137:3486–3489. doi: 10.1021/jacs.5b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayan SP, Choi CH, Hao L, Calabrese CM, Auyeung E, Zhang C, Goor OJ, Mirkin CA. The Sequence-Specific Cellular Uptake of Spherical Nucleic Acid Nanoparticle Conjugates. Small. 2015;11:4173–4182. doi: 10.1002/smll.201500027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinen AB, Guan CM, Mirkin CA. Spherical Nucleic Acid Nanoparticle Conjugates Enhance G-Quadruplex Formation and Increase Serum Protein Interactions. Angew Chem, Int Ed. 2014;54:527–531. doi: 10.1002/anie.201409211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv Drug Delivery Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cedervall T, Lynch I, Foy M, Berggad T, Donnelly SC, Cagney G, Linse S, Dawson KA. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew Chem, Int Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 21.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci U S A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K. The Evolution of the Protein Corona around Nanoparticles: A Test Study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 23.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci U S A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moghimi SM, Patel HM. Serum-mediated recognition of liposomes by phagocytic cells of the reticuloendothelial system - The concept of tissue specificity. Adv Drug Delivery Rev. 1998;32:45–60. doi: 10.1016/s0169-409x(97)00131-2. [DOI] [PubMed] [Google Scholar]
- 25.Nagayama S, Ogawara K, Fukuoka Y, Higaki K, Kimura T. Time-dependent changes in opsonin amount associated on nanoparticles alter their hepatic uptake characteristics. Int J Pharm. 2007;342:215–221. doi: 10.1016/j.ijpharm.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 26.Owens DE, III, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 28.Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J Am Chem Soc. 2012;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 29.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DW, Cohen Y, Emili A, Chan WC. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano. 2014;8:2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 30.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time evolution of the nanoparticle protein corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 31.Palchetti S, Colapicchioni V, Digiacomo L, Caracciolo G, Pozzi D, Capriotti AL, La Barbera G, Lagana A. The protein corona of circulating PEGylated liposomes. Biochim Biophys Acta, Biomembr. 2016;1858:189–196. doi: 10.1016/j.bbamem.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Runa S, Hill A, Cochran VL, Payne CK. PEGylated Nanoparticles: Protein Corona and Secondary Structure. Proc SPIE 9165,91651F10.1117/12.2062767 2014 [Google Scholar]
- 33.Venerando R, Miotto G, Magro M, Dallan M, Baratella D, Bonaiuto E, Zboril R, Vianello F. Magnetic Nano-particles with Covalently Bound Self-Assembled Protein Corona for Advanced Biomedical Applications. J Phys Chem C. 2013;117:20320–20331. [Google Scholar]
- 34.Jain RK. Transport of Molecules across Tumor Vasculature. Cancer Metastasis Rev. 1987;6:559–593. doi: 10.1007/BF00047468. [DOI] [PubMed] [Google Scholar]
- 35.Matsumura Y, Maeda H. A New Concept for Macromolecular Therapeutics in Cancer-Chemotherapy - Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 36.Fang J, Nakamura H, Maeda H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv Drug Delivery Rev. 2011;63:136–151. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Iyer AK, Khaled G, Fang J, Maeda H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discovery Today. 2006;11:812–818. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Controlled Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 39.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Challenges and Key Considerations of the Enhanced Permeability and Retention Effect for Nanomedicine Drug Delivery in Oncology. Cancer Res. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


