Abstract
We have previously shown that commonly expressed miRNAs influenced tumor molecular phenotype in colorectal cancer. We hypothesize that infrequently expressed miRNAs, when showing higher levels of expression, help to define tumor molecular phenotype. In this study we examine 304 miRNAs expressed in at least 30 individuals but in less than 50% of the population and with a mean level of expression above 1.0 relative florescent unit. We examine associations in 1893 individuals who have tumor molecular phenotype data as well as miRNA expression levels for both carcinoma and normal colorectal tissue. We compare miRNAs uniquely associated with tumor molecular phenotype to RNAseq data to identify genes associated with these miRNAs. This information is used to further identify unique pathways associated with tumor molecular phenotypes of TP53-mutated, KRAS-mutated, CpG island methylator phenotype, and microsatellite instability tumors. Thirty-seven miRNAs were uniquely associated with TP53-mutated tumors; 30 of these miRNAs had higher level of expression in TP53-mutated tumors while seven had lower levels of expression. Of the 34 miRNAs associated with CpG island methylator phenotype-high tumors, 16 were more likely to have a CpG island methylator phenotype-high tumor and 19 were less likely to be CpG island methylator phenotype-high. For microsatellite instability, 13 of the 22 infrequently expressed miRNAs were significantly less likely to be expressed in microsatellite unstable tumors. KRAS-mutated tumors were not associated with any miRNAs after adjustment for multiple comparisons. Of the dysregulated miRNAs, 17 were more likely to be TP53-mutated tumors while simultaneously being less likely to be CpG island methylator phenotype-high and/or microsatellite instability tumors. Genes regulated by these miRNAs were involved in numerous functions and pathways that influence cancer risk and progression. In summary, some infrequently expressed miRNAs, when expressed at higher levels appear to have significant biological meaning in terms of tumor molecular phenotype and gene expression profiles.
Keywords: Colorectal Cancer, TP53, CIMP, MSI, miRNA
Introduction
Molecular pathological epidemiology is a growing field of study that utilizes molecular information from tumors to better understand disease processes and progression (1). Assessment of tumor molecular phenotype in colorectal cancer has led to a better understanding of lifestyle factors that are uniquely associated with specific tumor phenotype (2–10). Tumor markers also have been examined with survival in an effort to identify biomarkers that can be used to predict prognosis and provide individualized treatment (11–16). While most studies have focused on common tumor molecular phenotype, such as TP53-mutated and KRAS-mutated tumors, microsatellite instability, and CpG Island Methylator Phenotype, studies are now examining other characteristics of tumors, such as gene expression and miRNA expression that may be important in identifying key disease pathways (11, 14, 17, 18).
MiRNAs are small, non-protein-coding RNA molecules that regulate gene expression either by post-transcriptionally suppressing mRNA translation or by causing mRNA degradation (18–23). We have previously shown that commonly expressed miRNAs influence tumor molecular phenotype in colorectal cancer, with the greatest number of differentially expressed miRNAs being observed for microsatellite unstable tumors compared to microsatellite stable tumors (24). MiRNAs were less frequently differentially expressed for TP53-mutated tumors, KRAS-mutated tumors, and CpG island methylator phenotype-high tumors. Most research focusing on miRNAs and tumor phenotype have focused on microsatellite unstable and CpG island methylator phenotype-high tumors (25) and on targeted miRNAs. Most targeted miRNAs studied, such as miR-21, are commonly expressed in tumors. Examination of infrequently expressed miRNAs may provide insight into unique pathways associated with tumor molecular phenotype.
In this study we focus on miRNAs that are infrequently expressed in normal colorectal mucosa and carcinoma tissue. We have previously shown that 34.5% of miRNAs expressed in colon tumor tissue are expressed in fewer than 10% of the population (26). Almost half of the miRNAs expressed in colorectal cancer tissue are expressed in less than half of the population. This presents two interesting questions: first, are low levels of expression purely noise in the data representing background expression levels; second, are infrequently expressed miRNA meaningful when expressed at higher levels beyond what could be considered background noise? Since tumor molecular phenotype also varies in percentage of the population with a given phenotype, it is a logical question to determine if infrequently expressed miRNAs when expressed at higher levels are associated with unique tumor molecular phenotypes. In this study we examine associations between tumor molecular phenotype and infrequently expressed miRNA to determine if such associations exist. We further examine infrequently expressed miRNAs to determine genes they may be associated with gene expression when expressed at higher levels along with functions and pathways associated with those genes. The size and design of this study makes in uniquely powered to examine the role of infrequently expressed miRNAs as they relate to colorectal cancer.
Methods
Study Participants
Study participants were recruited as part of two population-based case-control studies that included all incident colon and rectal cancer between 30 to 79 years of age who resided in Utah or were from the Kaiser Permanente Medical Care Program in Northern California. Participants were white, Hispanic, or black for the colon cancer study and also included participants of Asian race for the rectal portion of the study (27, 28). Case diagnosis was verified by tumor registry data as a first primary adenocarcinoma of the colon or rectum and were diagnosed between October 1991 and September 1994 for the colon cancer study and between May 1997 and May 2001 for the rectal cancer study. Detailed study methods have been described (29). The study was approved by the Institutional Review Boards at the University of Utah and Kaiser Permanente Medical Care Program in Northern California.
RNA processing
Formalin-fixed paraffin embedded tissue from the initial biopsy or surgery was used to extract RNA. Both carcinoma tissue and adjacent normal mucosa were used. Tissue was micro-dissected from 1–4 sequential sections on aniline blue stained slides using an H&E slide for reference. Total RNA was extracted, isolated, and purified using the RecoverAll Total Nucleic Acid isolation kit (Ambion); NanoDrop spectrophotometer was used to determine RNA yields.
miRNA
The Agilent Human miRNA Microarray V19.0 containing probes for 2006 unique human miRNAs was used. Data were required to pass stringent quality control parameters established by Agilent to be included in the analyses. Quality control parameters included tests for excessive background fluorescence, excessive variation among probe sequence replicates on the array, and measures of the total gene signal on the array to assess low signal. If samples failed to meet quality standards for any of these parameters, the sample was re-labeled, hybridized to arrays, and re-scanned. If a sample failed quality control assessment a second time, the sample was deemed to be of poor quality and the sample was excluded from analysis. Our previous analysis has shown that the repeatability associated with this microarray was extremely high (r=0.98) (29), and that comparison of miRNA expression levels obtained from the Agilent microarray to those obtained from qPCR had an agreement of 100% in terms of directionality of findings and that the fold change calculated for the miRNA expression difference between carcinoma and normal colonic mucosa was almost identical (30). Of the 2006 unique human miRNAs assessed on the Agilent microarray, 1226 were expressed in colon carcinoma tissue and 1179 in normal colon mucosa.
To normalize differences in miRNA expression that could be attributed to the array, amount of RNA, location on array, or factors that could erroneously influence miRNA expression levels, total gene signal was normalized by multiplying each sample by a scaling factor (31), which was the median of the 75th percentiles of all the samples divided by the individual 75th percentile of each sample.
mRNA: RNA-Seq Sequencing Library Preparation and Data Processing
Total RNA was run on 245 carcinoma and normal mucosa pairs; of these 207 paired samples passed quality control and were used in analyses. Tissues samples taken from the study subjects at time of diagnosis were used for RNA extraction as previously described (32). For mRNA analysis, RNA library construction was done with the Illumina TruSeq Stranded Total RNA Sample Preparation Kit with Ribo-Zero. The samples were then fragmented and primed for cDNA synthesis, adapters were then ligated onto the cDNA, and the resulting samples were then amplified using PCR; the amplified library was then purified using Agencount AMPure XP beads. A more detailed description of the methods can be found in our previous work (33). Illumina TruSeq v3 single read flow cell and a 50 cycle single-read sequence run was performed on an Illumina HiSeq instrument. Reads were aligned to a sequence database containing the human genome (build GRCh37/hg19, February 2009 from genome.ucsc.edu) and alignment was performed using novoalign v2.08.01. Total gene counts were calculated for each exon and UTR of the genes using a list of gene coordinates obtained from http://genome.ucsc.edu. We dropped features that were not expressed in our RNA-Seq data or for which the expression was missing for the majority of samples, retaining 17,384 protein-coding genes (33).
Tumor Molecular Phenotype
We have previously assessed TP53 and KRAS mutations (4, 8, 34), the CpG island methylator phenotype using the classic panel that consisted of MLH1, CDKN2A, and MINT1, MINT2, and MINT31 (35), and microsatellite instability based on the mononucleotide repeats at BAT26 and TGFβR2 and a panel of 10 tetranucleotide repeats that were correlated highly with the Bethesda Panel (6); our original microsatellite instability studies were done prior to the development of the Bethesda Panel. Tumors were scored as CpG island methylator phenotype-high if two or more of the CpG islands were methylated for the five markers; otherwise they were classified as CpG island methylator phenotype -low/negative. This panel was run prior to the advent of more recent panels (36, 37).
Statistical Methods
The study focuses on infrequently expressed miRNAs which we define as being expressed in less than 50% of the study population for either normal mucosa or tumor. To be included in the analysis, miRNAs also had to have a mean level of expression of 1.0 Agilent Relative Florescent Unit (ARFU) in tumors or normal mucosa and be expressed in at least 30 individuals. Each infrequently expressed miRNA could be considered expressed or not in each tumor and normal, resulting in three primary dysregulation groups based on the tumor-normal expression differences: up-regulated (expressed more in tumor than in normal), down-regulated (expressed more in normal than in tumor), and referent (neither up- nor down-regulated at the 25%tile/75%tile cutpoints). Rather than forcing the same number of subjects to fall into these three groups for all infrequently expressed miRNAs, cutpoints were selected based on the upper 25% and lower 25% of the tumor-normal differences for all infrequently expressed miRNAs. The resulting three-level dysregulation group factor (up, down, or referent) was used as a predictor in a per-miRNA logistic regression model also adjusting for age, study center, and sex and standardized what was considered true expression for all miRNAs. A total of 304 miRNAs were analyzed that fit these criteria. We used paired carcinoma and normal mucosa miRNA expression, evaluating differential expression between the two tissue types to control for differences in expression by tumor site and other potential confounding factors. Analyses were run separately for overall colorectal cancer, colon cancer, and rectal cancer. We analyzed difference in association for infrequently expressed miRNAs by TP53-mutated versus non-TP53-mutated, KRAS-mutated versus non-KRAS-mutated, CpG island methylator phenotype-high relative to CpG island methylator phenotype-low/negative, and microsatellite unstable compared to microsatellite stable. Adjustment for multiple comparisons was done using the positive false discovery rate Q value (38); given the infrequent expression of these miRNA, we report any associations for which the Q value was less than 0.05.
We compared those miRNA with a Q value of <0.05 (58=7 miRNAs) to RNAseq data to identify genes whose expression was associated with these infrequently expressed miRNAs. To determine statistical significance between the miRNA∷mRNA associations, we ran a Fisher-Pitman Monte Carlo test with 10,000 permutation comparing differences in mean levels of gene expression across miRNA dysregulation groups of ≤75%ile vs >75%ile in R using the ‘coin’ package. RPKM (Reads Per Kilobase of transcript per Million mapped reads) mRNA expression level data were used in these analyses. Identification of networks and functions associated with genes whose mean expression was altered by miRNAs was done using Ingenuity Pathway Analysis®; adjustments for multiple comparisons were made using the Benjamini and Hochberg method (39). Both causal and interaction networks were generated. Interaction networks were limited to 35 molecules per network and 25 networks per analysis, and excluded endogenous chemicals. We focused on algorithmically derived interaction networks, which are assigned a score based on their relevance to the genes in the input dataset, the number of focus genes (i.e. dysregulated genes in our data that are in that network), and their connectivity (40). The score is calculated as −log10P, where P is generated using a Fisher’s exact test (41). Studies have found scores >3 to be significant, with a score of 3 indicating a 1/1000 chance that the focus genes are in a network due to random chance (42–44). Other studies have opted to utilize more stringent criteria and higher scores to ensure that their discovered networks are highly significant (45, 46); we utilized highly stringent criteria, only including networks with scores over 20.
Results
The study population is described in Table 1. Over half of the population were males. There were approximately equal numbers of individuals enrolled with proximal and distal colon tumors. Slightly less than half, 47.6%, of tumors had a TP53 mutation, 31.7% had a KRAS mutation, 21.2% were classified as CpG island methylator phenotype-high and 9.1% were microsatellite unstable.
Table 1.
Description of Study Population and miRNA expression
| Overall | Colon | Rectal | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Subject N | % | Subject N | % | Subject | % | ||
| Sex | |||||||
| Male | 1028 | 54.3 | 608 | 52.8 | 420 | 56.5 | |
| Female | 866 | 45.7 | 543 | 47.2 | 323 | 43.5 | |
| Center | |||||||
| Kaiser | 1144 | 60.4 | 740 | 64.3 | 404 | 54.4 | |
| Utah | 750 | 39.6 | 411 | 35.7 | 339 | 45.6 | |
| Site | |||||||
| Proximal Colon | 569 | 49.5 | 569 | 49.4 | 0 | 0.0 | |
| Distal Colon | 580 | 50.5 | 580 | 50.4 | 0 | 0.0 | |
| Study | |||||||
| Stage I | 559 | 30.0 | 259 | 22.7 | 300 | 41.5 | |
| Stage II | 489 | 26.3 | 350 | 30.7 | 139 | 19.2 | |
| Stage III | 548 | 29.4 | 340 | 29.9 | 208 | 28.8 | |
| Stage IV | 266 | 14.3 | 190 | 16.7 | 76 | 10.5 | |
| TP53 | |||||||
| Not Mutated | 953 | 52.4 | 597 | 54.4 | 356 | 49.4 | |
| Mutated | 864 | 47.6 | 500 | 45.6 | 364 | 50.6 | |
| KRAS | |||||||
| Not Mutated | 1240 | 68.5 | 724 | 67.6 | 516 | 69.9 | |
| Mutated | 569 | 31.5 | 347 | 32.4 | 222 | 30.1 | |
| CpG Island Methylator Phenotype | |||||||
| Low | 1312 | 78.8 | 700 | 71.8 | 612 | 88.6 | |
| High | 354 | 21.2 | 275 | 28.2 | 79 | 11.4 | |
| Microsatellite Instability | |||||||
| Stable | 1688 | 90.9 | 965 | 86.2 | 723 | 97.8 | |
| Unstable | 170 | 9.1 | 154 | 13.8 | 16 | 2.2 | |
|
|
|||||||
| Mean | STD | Mean | STD | Mean | STD | ||
|
|
|||||||
| Age | 64.2 | 10.2 | 65.4 | 9.5 | 62.3 | 11.0 | |
Assessment of TP53-mutated tumors associated with infrequently expressed miRNAs showed that 30 miRNAs were more likely to have a TP53 mutation if they were upregulated in tumors while seven miRNAs were associated with a lower likelihood of having a TP53 mutation if they were upregulated in tumors (Table 2). Most of the miRNAs (20 of the 37 miRNAs) were associated with a high level of differential expression in less than 20% of the. While some miRNAs were associated with a high level of differential expression in a large percentage of the population, these miRNAs were not expressed or extremely infrequently expressed in normal mucosa but were expressed to a greater degree in tumor tissue. There were no miRNAs more likely to have a TP53 mutation if down-regulated after adjusting for multiple comparisons. Site-specific associations for colon and rectal cancer generally had Q values of >0.05. However many of these miRNAs with a Q value of 0.03 to 0.04 overall had a Q value of 0.07 for colon cancer specifically, most likely reflecting the decrease in power when analyzing colon cancer specifically rather than colorectal cancer combined. The lowest Q values for miRNAs for rectal cancer were 0.083. There were no unique associations with KRAS-mutated tumors.
Table 2.
Associations between infrequently expressed miRNAs in colorectal cancer and TP53 mutations
| miRNA | Not-mutated | TP53-Mutated | P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | OR | (95% CI) | unadjusted | Q value | ||
| hsa-miR-1207-3p | Down-regulated1 | 68 | 7.1 | 80 | 9.3 | 1.31 | (0.94, 1.84) | 0.115 | 0.919 |
| Referent | 849 | 89.1 | 773 | 89.5 | 1.00 | ||||
| Up-regulated | 36 | 3.8 | 11 | 1.3 | 0.35 | (0.17, 0.69) | 0.002 | 0.030 | |
|
| |||||||||
| hsa-miR-1243 | Down-regulated | 161 | 16.9 | 140 | 16.2 | 0.92 | (0.72, 1.18) | 0.519 | 0.981 |
| Referent | 710 | 74.5 | 677 | 78.4 | 1.00 | ||||
| Up-Regulated | 82 | 8.6 | 47 | 5.4 | 0.60 | (0.41, 0.88) | 0.008 | 0.042 | |
|
| |||||||||
| hsa-miR-1296 | Down-regulated | 51 | 5.4 | 44 | 5.1 | 0.95 | (0.63, 1.45) | 0.824 | 0.981 |
| Referent | 878 | 92.1 | 777 | 89.9 | 1.00 | ||||
| Up-Regulated | 24 | 2.5 | 43 | 5.0 | 2.02 | (1.22, 3.37) | 0.007 | 0.039 | |
|
| |||||||||
| hsa-miR-133a | Down-regulated | 35 | 3.7 | 38 | 4.4 | 1.19 | (0.74, 1.91) | 0.469 | 0.981 |
| Referent | 910 | 95.5 | 805 | 93.2 | 1.00 | ||||
| Up-Regulated | 8 | 0.8 | 21 | 2.4 | 2.99 | (1.32, 6.80) | 0.009 | 0.043 | |
|
| |||||||||
| hsa-miR-133b | Down-regulated | 483 | 50.7 | 458 | 53.0 | 1.17 | (0.96, 1.43) | 0.116 | 0.919 |
| Referent | 415 | 43.5 | 326 | 37.7 | 1.00 | ||||
| Up-Regulated | 55 | 5.8 | 80 | 9.3 | 1.85 | (1.27, 2.69) | 0.001 | 0.030 | |
|
| |||||||||
| hsa-miR-151a-3p | Down-regulated | 43 | 4.5 | 34 | 3.9 | 1.03 | (0.64, 1.65) | 0.900 | 0.982 |
| Referent | 583 | 61.2 | 430 | 49.8 | 1.00 | ||||
| Up-Regulated | 327 | 34.3 | 400 | 46.3 | 1.63 | (1.35, 1.98) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-184 | Down-regulated | 197 | 20.7 | 204 | 23.6 | 1.29 | (1.02, 1.62) | 0.031 | 0.919 |
| Referent | 623 | 65.4 | 511 | 59.1 | 1.00 | ||||
| Up-Regulated | 133 | 14.0 | 149 | 17.2 | 1.40 | (1.08, 1.82) | 0.012 | 0.047 | |
|
| |||||||||
| hsa-miR-192-3p | Down-regulated | 222 | 23.3 | 189 | 21.9 | 0.97 | (0.77, 1.22) | 0.793 | 0.981 |
| Referent | 650 | 68.2 | 555 | 64.2 | 1.00 | ||||
| Up-Regulated | 81 | 8.5 | 120 | 13.9 | 1.73 | (1.27, 2.34) | 0.000 | 0.030 | |
|
| |||||||||
| hsa-miR-19a-3p | Down-regulated | 11 | 1.2 | 8 | 0.9 | 0.91 | (0.36, 2.27) | 0.836 | 0.981 |
| Referent | 745 | 78.2 | 597 | 69.1 | 1.00 | ||||
| Up-Regulated | 197 | 20.7 | 259 | 30.0 | 1.64 | (1.33, 2.04) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-224-5p | Down-regulated | 24 | 2.5 | 14 | 1.6 | 0.93 | (0.47, 1.84) | 0.843 | 0.981 |
| Referent | 364 | 38.2 | 222 | 25.7 | 1.00 | ||||
| Up-Regulated | 565 | 59.3 | 628 | 72.7 | 1.79 | (1.46, 2.19) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-3190-5p | Down-regulated | 11 | 1.2 | 11 | 1.3 | 1.14 | (0.49, 2.65) | 0.758 | 0.981 |
| Referent | 937 | 98.3 | 837 | 96.9 | 1.00 | ||||
| Up-Regulated | 5 | 0.5 | 16 | 1.9 | 3.62 | (1.32, 9.93) | 0.013 | 0.047 | |
|
| |||||||||
| hsa-miR-31-5p | Down-regulated | 14 | 1.5 | 11 | 1.3 | 0.83 | (0.37, 1.83) | 0.637 | 0.981 |
| Referent | 775 | 81.3 | 762 | 88.2 | 1.00 | ||||
| Up-Regulated | 164 | 17.2 | 91 | 10.5 | 0.57 | (0.43, 0.75) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-3607-3p | Down-regulated | 53 | 5.6 | 63 | 7.3 | 1.36 | (0.93, 1.99) | 0.111 | 0.919 |
| Referent | 835 | 87.6 | 715 | 82.8 | 1.00 | ||||
| Up-Regulated | 65 | 6.8 | 86 | 10.0 | 1.52 | (1.09, 2.14) | 0.014 | 0.0495 | |
|
| |||||||||
| hsa-miR-3609 | Down-regulated | 193 | 20.3 | 142 | 16.4 | 0.86 | (0.67, 1.11) | 0.247 | 0.981 |
| Referent | 588 | 61.7 | 492 | 56.9 | 1.00 | ||||
| Up-Regulated | 172 | 18.0 | 230 | 26.6 | 1.60 | (1.27, 2.02) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-3615 | Down-regulated | 207 | 21.7 | 201 | 23.3 | 1.04 | (0.83, 1.31) | 0.708 | 0.981 |
| Referent | 640 | 67.2 | 602 | 69.7 | 1.00 | ||||
| Up-Regulated | 106 | 11.1 | 61 | 7.1 | 0.63 | (0.45, 0.88) | 0.007 | 0.039 | |
|
| |||||||||
| hsa-miR-3622b-3p | Down-regulated | 22 | 2.3 | 26 | 3.0 | 1.48 | (0.83, 2.64) | 0.186 | 0.966 |
| Referent | 746 | 78.3 | 615 | 71.2 | 1.00 | ||||
| Up-Regulated | 185 | 19.4 | 223 | 25.8 | 1.48 | (1.19, 1.85) | 0.001 | 0.030 | |
|
| |||||||||
| hsa-miR-362-5p | Down-regulated | 26 | 2.7 | 13 | 1.5 | 0.60 | (0.31, 1.18) | 0.141 | 0.919 |
| Referent | 730 | 76.6 | 587 | 67.9 | 1.00 | ||||
| Up-Regulated | 197 | 20.7 | 264 | 30.6 | 1.66 | (1.34, 2.06) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-3687 | Down-regulated | 12 | 1.3 | 14 | 1.6 | 1.42 | (0.65, 3.11) | 0.376 | 0.981 |
| Referent | 726 | 76.2 | 580 | 67.1 | 1.00 | ||||
| Up-Regulated | 215 | 22.6 | 270 | 31.3 | 1.57 | (1.27, 1.93) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-374a-5p | Down-regulated | 15 | 1.6 | 7 | 0.8 | 0.55 | (0.22, 1.35) | 0.189 | 0.966 |
| Referent | 781 | 82.0 | 664 | 76.9 | 1.00 | ||||
| Up-Regulated | 157 | 16.5 | 193 | 22.3 | 1.43 | (1.13, 1.81) | 0.003 | 0.031 | |
|
| |||||||||
| hsa-miR-374b-5p | Down-regulated | 31 | 3.3 | 16 | 1.9 | 0.62 | (0.34, 1.15) | 0.131 | 0.919 |
| Referent | 711 | 74.6 | 563 | 65.2 | 1.00 | ||||
| Up-Regulated | 211 | 22.1 | 285 | 33.0 | 1.68 | (1.36, 2.08) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-424-5p | Down-regulated | 17 | 1.8 | 14 | 1.6 | 0.98 | (0.48, 2.00) | 0.950 | 0.997 |
| Referent | 701 | 73.6 | 572 | 66.2 | 1.00 | ||||
| Up-Regulated | 235 | 24.7 | 278 | 32.2 | 1.41 | (1.15, 1.74) | 0.001 | 0.030 | |
|
| |||||||||
| hsa-miR-4251 | Down-regulated | 72 | 7.6 | 76 | 8.8 | 1.35 | (0.95, 1.90) | 0.090 | 0.919 |
| Referent | 639 | 67.1 | 514 | 59.5 | 1.00 | ||||
| Up-Regulated | 242 | 25.4 | 274 | 31.7 | 1.42 | (1.15, 1.75) | 0.001 | 0.030 | |
|
| |||||||||
| hsa-miR-4296 | Down-regulated | 79 | 8.3 | 68 | 7.9 | 1.01 | (0.71, 1.42) | 0.975 | 0.997 |
| Referent | 728 | 76.4 | 625 | 72.3 | 1.00 | ||||
| Up-Regulated | 146 | 15.3 | 171 | 19.8 | 1.38 | (1.08, 1.76) | 0.011 | 0.047 | |
|
| |||||||||
| hsa-miR-4421 | Down-regulated | 216 | 22.7 | 179 | 20.7 | 0.95 | (0.75, 1.19) | 0.640 | 0.981 |
| Referent | 664 | 69.7 | 589 | 68.2 | 1.00 | ||||
| Up-Regulated | 73 | 7.7 | 96 | 11.1 | 1.51 | (1.09, 2.09) | 0.013 | 0.047 | |
|
| |||||||||
| hsa-miR-4654 | Down-regulated | 169 | 17.7 | 163 | 18.9 | 1.21 | (0.95, 1.56) | 0.128 | 0.919 |
| Referent | 569 | 59.7 | 450 | 52.1 | 1.00 | ||||
| Up-Regulated | 215 | 22.6 | 251 | 29.1 | 1.48 | (1.19, 1.85) | 0.001 | 0.030 | |
|
| |||||||||
| hsa-miR-4695-3p | Down-regulated | 25 | 2.6 | 25 | 2.9 | 1.14 | (0.65, 2.01) | 0.644 | 0.981 |
| Referent | 912 | 95.7 | 808 | 93.5 | 1.00 | ||||
| Up-Regulated | 16 | 1.7 | 31 | 3.6 | 2.20 | (1.19, 4.06) | 0.012 | 0.047 | |
|
| |||||||||
| hsa-miR-4711-5p | Down-regulated | 22 | 2.3 | 19 | 2.2 | 0.94 | (0.50, 1.75) | 0.849 | 0.981 |
| Referent | 905 | 95.0 | 840 | 97.2 | 1.00 | ||||
| Up-Regulated | 26 | 2.7 | 5 | 0.6 | 0.22 | (0.08, 0.57) | 0.002 | 0.030 | |
|
| |||||||||
| hsa-miR-484 | Down-regulated | 106 | 11.1 | 86 | 10.0 | 0.91 | (0.67, 1.24) | 0.550 | 0.981 |
| Referent | 763 | 80.1 | 659 | 76.3 | 1.00 | ||||
| Up-Regulated | 84 | 8.8 | 119 | 13.8 | 1.59 | (1.18, 2.15) | 0.002 | 0.030 | |
|
| |||||||||
| hsa-miR-5095 | Down-regulated | 134 | 14.1 | 105 | 12.2 | 0.89 | (0.67, 1.17) | 0.401 | 0.981 |
| Referent | 710 | 74.5 | 624 | 72.2 | 1.00 | ||||
| Up-Regulated | 109 | 11.4 | 135 | 15.6 | 1.42 | (1.08, 1.87) | 0.013 | 0.047 | |
|
| |||||||||
| hsa-miR-532-3p | Down-regulated | 61 | 6.4 | 44 | 5.1 | 0.83 | (0.55, 1.25) | 0.368 | 0.981 |
| Referent | 739 | 77.5 | 622 | 72.0 | 1.00 | ||||
| Up-Regulated | 153 | 16.1 | 198 | 22.9 | 1.51 | (1.19, 1.91) | 0.001 | 0.030 | |
|
| |||||||||
| hsa-miR-532-5p | Down-regulated | 48 | 5.0 | 22 | 2.5 | 0.60 | (0.35, 1.00) | 0.052 | 0.919 |
| Referent | 655 | 68.7 | 482 | 55.8 | 1.00 | ||||
| Up-Regulated | 250 | 26.2 | 360 | 41.7 | 1.94 | (1.59, 2.37) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-5685 | Down-regulated | 184 | 19.3 | 173 | 20.0 | 1.01 | (0.80, 1.28) | 0.922 | 0.993 |
| Referent | 683 | 71.7 | 645 | 74.7 | 1.00 | ||||
| Up-Regulated | 86 | 9.0 | 46 | 5.3 | 0.57 | (0.39, 0.83) | 0.003 | 0.031 | |
|
| |||||||||
| hsa-miR-625-5p | Down-regulated | 17 | 1.8 | 17 | 2.0 | 1.09 | (0.55, 2.15) | 0.806 | 0.981 |
| Referent | 898 | 94.2 | 831 | 96.2 | 1.00 | ||||
| Up-Regulated | 38 | 4.0 | 16 | 1.9 | 0.47 | (0.26, 0.84) | 0.012 | 0.047 | |
|
| |||||||||
| hsa-miR-652-3p | Down-regulated | 39 | 4.1 | 25 | 2.9 | 0.72 | (0.43, 1.20) | 0.209 | 0.966 |
| Referent | 824 | 86.5 | 702 | 81.3 | 1.00 | ||||
| Up-Regulated | 90 | 9.4 | 137 | 15.9 | 1.75 | (1.32, 2.33) | 0.000 | 0.030 | |
|
| |||||||||
| hsa-miR-664a-3p | Down-regulated | 201 | 21.1 | 186 | 21.5 | 1.13 | (0.89, 1.43) | 0.314 | 0.981 |
| Referent | 569 | 59.7 | 455 | 52.7 | 1.00 | ||||
| Up-Regulated | 183 | 19.2 | 223 | 25.8 | 1.50 | (1.19, 1.89) | 0.001 | 0.030 | |
|
| |||||||||
| hsa-miR-7-5p | Down-regulated | 14 | 1.5 | 10 | 1.2 | 0.98 | (0.43, 2.22) | 0.957 | 0.997 |
| Referent | 580 | 60.9 | 421 | 48.7 | 1.00 | ||||
| Up-Regulated | 359 | 37.7 | 433 | 50.1 | 1.67 | (1.38, 2.01) | <.0001 | 0.030 | |
|
| |||||||||
| hsa-miR-98-5p | Down-regulated | 26 | 2.7 | 18 | 2.1 | 0.82 | (0.44, 1.51) | 0.522 | 0.981 |
| Referent | 727 | 76.3 | 592 | 68.5 | 1.00 | ||||
| Up-Regulated | 200 | 21.0 | 254 | 29.4 | 1.54 | (1.24, 1.91) | <.0001 | 0.030 | |
Down-regulated have differential expression <-1.77; referent has differential expression between -1.77, 2.08; up- regulated has differential expression >2.08
Thirty-five infrequently expressed miRNAs were associated with CpG island methylator phenotype-high tumors (Table 3). Of these 35 miRNAs, 19 were less likely to be associated with a CpG island methylator phenotype-high tumor when up-regulated in tumor tissue, while 16 were more likely to have a CpG island methylator phenotype-high tumor if the miRNA was up-regulated in the tumor. Nine of these 35 miRNAs had over 20% of the population in the higher level of differential miRNA expression. As with TP53, many of these miRNAs had similar findings for colon cancer specifically as we observed for overall colorectal cancer, although the lowest FDR was 0.078 for colon cancer even when the raw p values were <0.0001 and comparable for both overall colorectal cancer and colon cancer specifically. Also like for TP53, after adjustment for multiple comparisons there were no significant findings between CIMP-high tumors and down-regulated miRNAs.
Table 3.
Overall associations between differential miRNA expression in infrequently expressed miRNA and CIMP High tumors
| miRNA | CIMP-Low/Negative | CIMP-High | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | OR | (95% CI) | P value | Q value | ||
| hsa-miR-151a-3p | Down-regulated1 | 45 | 3.4 | 20 | 5.6 | 1.46 | (0.84, 2.56) | 0.18 | 0.727 |
| Referent | 696 | 53.0 | 221 | 62.4 | 1.00 | ||||
| Up-Regulated | 571 | 43.5 | 113 | 31.9 | 0.64 | (0.50, 0.83) | 0.007 | 0.033 | |
|
| |||||||||
| hsa-miR-1915-5p | Down-regulated | 109 | 8.3 | 24 | 6.8 | 0.83 | (0.52, 1.31) | 0.42 | 0.773 |
| Referent | 1138 | 86.7 | 298 | 84.2 | 1.00 | ||||
| Up-Regulated | 65 | 5.0 | 32 | 9.0 | 1.90 | (1.21, 2.98) | 0.005 | 0.036 | |
|
| |||||||||
| hsa-miR-193a-3p | Down-regulated | 24 | 1.8 | 8 | 2.3 | 1.06 | (0.46, 2.42) | 0.90 | 0.895 |
| Referent | 908 | 69.2 | 273 | 77.1 | 1.00 | ||||
| Up-Regulated | 380 | 29.0 | 73 | 20.6 | 0.63 | (0.47, 0.84) | 0.002 | 0.033 | |
|
| |||||||||
| hsa-miR-199b-5p | Down-regulated | 80 | 6.1 | 29 | 8.2 | 1.25 | (0.79, 1.99) | 0.34 | 0.753 |
| Referent | 804 | 61.3 | 247 | 69.8 | 1.00 | ||||
| Up-Regulated | 428 | 32.6 | 78 | 22.0 | 0.61 | (0.46, 0.81) | 0.002 | 0.033 | |
|
| |||||||||
| hsa-miR-19a-3p | Down-regulated | 14 | 1.1 | 5 | 1.4 | 1.21 | (0.42, 3.43) | 0.73 | 0.803 |
| Referent | 924 | 70.4 | 281 | 79.4 | 1.00 | ||||
| Up-Regulated | 374 | 28.5 | 68 | 19.2 | 0.59 | (0.44, 0.79) | 0.0005 | 0.033 | |
|
| |||||||||
| hsa-miR-2110 | Down-regulated | 68 | 5.2 | 17 | 4.8 | 0.94 | (0.54, 1.63) | 0.83 | 0.826 |
| Referent | 1212 | 92.4 | 317 | 89.5 | 1.00 | ||||
| Up-Regulated | 32 | 2.4 | 20 | 5.6 | 2.48 | (1.39, 4.44) | 0.002 | 0.033 | |
|
| |||||||||
| hsa-miR-224-5p | Down-regulated | 27 | 2.1 | 5 | 1.4 | 0.38 | (0.14, 1.02) | 0.05 | 0.436 |
| Referent | 330 | 25.2 | 174 | 49.2 | 1.00 | ||||
| Up-Regulated | 955 | 72.8 | 175 | 49.4 | 0.35 | (0.27, 0.45) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-30e-5p | Down-regulated | 445 | 33.9 | 140 | 39.5 | 1.14 | (0.88, 1.48) | 0.31 | 0.753 |
| Referent | 644 | 49.1 | 177 | 50.0 | 1.00 | ||||
| Up-Regulated | 223 | 17.0 | 37 | 10.5 | 0.61 | (0.41, 0.89) | 0.01 | 0.041 | |
|
| |||||||||
| hsa-miR-31-5p | Down-regulated | 17 | 1.3 | 7 | 2.0 | 1.80 | (0.73, 4.44) | 0.20 | 0.727 |
| Referent | 1176 | 89.6 | 244 | 68.9 | 1.00 | ||||
| Up-Regulated | 119 | 9.1 | 103 | 29.1 | 4.17 | (3.08, 5.65) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-3609 | Down-regulated | 219 | 16.7 | 83 | 23.4 | 1.34 | (0.99, 1.80) | 0.05 | 0.436 |
| Referent | 757 | 57.7 | 228 | 64.4 | 1.00 | ||||
| Up-Regulated | 336 | 25.6 | 43 | 12.1 | 0.43 | (0.30, 0.61) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-3615 | Down-regulated | 295 | 22.5 | 88 | 24.9 | 1.22 | (0.92, 1.62) | 0.17 | 0.727 |
| Referent | 904 | 68.9 | 220 | 62.1 | 1.00 | ||||
| Up-Regulated | 113 | 8.6 | 46 | 13.0 | 1.65 | (1.13, 2.41) | 0.01 | 0.038 | |
|
| |||||||||
| hsa-miR-362-5p | Down-regulated | 27 | 2.1 | 5 | 1.4 | 0.63 | (0.24, 1.67) | 0.35 | 0.753 |
| Referent | 917 | 69.9 | 283 | 79.9 | 1.00 | ||||
| Up-Regulated | 368 | 28.0 | 66 | 18.6 | 0.58 | (0.43, 0.79) | 0.0004 | 0.033 | |
|
| |||||||||
| hsa-miR-3687 | Down-regulated | 20 | 1.5 | 5 | 1.4 | 0.95 | (0.35, 2.58) | 0.92 | 0.916 |
| Referent | 908 | 69.2 | 273 | 77.1 | 1.00 | ||||
| Up-Regulated | 384 | 29.3 | 76 | 21.5 | 0.66 | (0.50, 0.88) | 0.004 | 0.035 | |
|
| |||||||||
| hsa-miR-374a-5p | Down-regulated | 14 | 1.1 | 7 | 2.0 | 1.65 | (0.65, 4.19) | 0.29 | 0.753 |
| Referent | 1003 | 76.4 | 305 | 86.2 | 1.00 | ||||
| Up-Regulated | 295 | 22.5 | 42 | 11.9 | 0.48 | (0.34, 0.68) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-374b-5p | Down-regulated | 27 | 2.1 | 15 | 4.2 | 1.96 | (1.01, 3.80) | 0.05 | 0.436 |
| Referent | 878 | 66.9 | 272 | 76.8 | 1.00 | ||||
| Up-Regulated | 407 | 31.0 | 67 | 18.9 | 0.54 | (0.40, 0.72) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-3938 | Down-regulated | 44 | 3.4 | 12 | 3.4 | 1.03 | (0.54, 2.00) | 0.92 | 0.919 |
| Referent | 1239 | 94.4 | 324 | 91.5 | 1.00 | ||||
| Up-Regulated | 29 | 2.2 | 18 | 5.1 | 2.51 | (1.36, 4.65) | 0.003 | 0.033 | |
|
| |||||||||
| hsa-miR-3944-5p | Down-regulated | 308 | 23.5 | 68 | 19.2 | 0.85 | (0.62, 1.15) | 0.29 | 0.753 |
| Referent | 790 | 60.2 | 206 | 58.2 | 1.00 | ||||
| Up-Regulated | 214 | 16.3 | 80 | 22.6 | 1.47 | (1.08, 1.99) | 0.01 | 0.044 | |
|
| |||||||||
| hsa-miR-424-5p | Down-regulated | 21 | 1.6 | 8 | 2.3 | 1.33 | (0.58, 3.07) | 0.50 | 0.803 |
| Referent | 875 | 66.7 | 280 | 79.1 | 1.00 | ||||
| Up-Regulated | 416 | 31.7 | 66 | 18.6 | 0.52 | (0.39, 0.70) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-4492 | Down-regulated | 74 | 5.6 | 18 | 5.1 | 0.87 | (0.51, 1.49) | 0.62 | 0.803 |
| Referent | 1190 | 90.7 | 309 | 87.3 | 1.00 | ||||
| Up-Regulated | 48 | 3.7 | 27 | 7.6 | 2.18 | (1.33, 3.59) | 0.002 | 0.033 | |
|
| |||||||||
| hsa-miR-4533 | Down-regulated | 169 | 12.9 | 61 | 17.2 | 1.52 | (1.09, 2.11) | 0.01 | 0.184 |
| Referent | 1036 | 79.0 | 243 | 68.6 | 1.00 | ||||
| Up-Regulated | 107 | 8.2 | 50 | 14.1 | 2.07 | (1.43, 3.01) | 0.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-4709-3p | Down-regulated | 122 | 9.3 | 32 | 9.0 | 0.99 | (0.65, 1.50) | 0.96 | 0.965 |
| Referent | 1045 | 79.6 | 265 | 74.9 | 1.00 | ||||
| Up-Regulated | 145 | 11.1 | 57 | 16.1 | 1.55 | (1.10, 2.18) | 0.01 | 0.041 | |
|
| |||||||||
| hsa-miR-4722-5p | Down-regulated | 437 | 33.3 | 107 | 30.2 | 0.96 | (0.73, 1.26) | 0.75 | 0.803 |
| Referent | 683 | 52.1 | 173 | 48.9 | 1.00 | ||||
| Up-Regulated | 192 | 14.6 | 74 | 20.9 | 1.53 | (1.11, 2.10) | 0.01 | 0.038 | |
|
| |||||||||
| hsa-miR-484 | Down-regulated | 131 | 10.0 | 46 | 13.0 | 1.26 | (0.86, 1.83) | 0.23 | 0.753 |
| Referent | 1013 | 77.2 | 283 | 79.9 | 1.00 | ||||
| Up-Regulated | 168 | 12.8 | 25 | 7.1 | 0.55 | (0.35, 0.85) | 0.008 | 0.038 | |
|
| |||||||||
| hsa-miR-513a-3p | Down-regulated | 59 | 4.5 | 18 | 5.1 | 1.16 | (0.67, 2.01) | 0.58 | 0.803 |
| Referent | 1229 | 93.7 | 321 | 90.7 | 1.00 | ||||
| Up-Regulated | 24 | 1.8 | 15 | 4.2 | 2.42 | (1.24, 4.74) | 0.01 | 0.038 | |
|
| |||||||||
| hsa-miR-532-3p | Down-regulated | 70 | 5.3 | 20 | 5.6 | 0.93 | (0.55, 1.57) | 0.77 | 0.803 |
| Referent | 956 | 72.9 | 291 | 82.2 | 1.00 | ||||
| Up-Regulated | 286 | 21.8 | 43 | 12.1 | 0.51 | (0.36, 0.72) | 0.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-532-5p | Down-regulated | 38 | 2.9 | 17 | 4.8 | 1.38 | (0.76, 2.53) | 0.29 | 0.753 |
| Referent | 774 | 59.0 | 258 | 72.9 | 1.00 | ||||
| Up-Regulated | 500 | 38.1 | 79 | 22.3 | 0.47 | (0.36, 0.63) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-5685 | Down-regulated | 265 | 20.2 | 63 | 17.8 | 0.89 | (0.65, 1.22) | 0.48 | 0.798 |
| Referent | 969 | 73.9 | 252 | 71.2 | 1.00 | ||||
| Up-Regulated | 78 | 5.9 | 39 | 11.0 | 1.96 | (1.29, 2.97) | 0.001 | 0.033 | |
|
| |||||||||
| hsa-miR-590-5p | Down-regulated | 173 | 13.2 | 61 | 17.2 | 1.28 | (0.92, 1.77) | 0.14 | 0.727 |
| Referent | 983 | 74.9 | 269 | 76.0 | 1.00 | ||||
| Up-Regulated | 156 | 11.9 | 24 | 6.8 | 0.55 | (0.35, 0.87) | 0.01 | 0.038 | |
|
| |||||||||
| hsa-miR-6071 | Down-regulated | 151 | 11.5 | 37 | 10.5 | 0.89 | (0.60, 1.30) | 0.54 | 0.803 |
| Referent | 1135 | 86.5 | 301 | 85.0 | 1.00 | ||||
| Up-Regulated | 26 | 2.0 | 16 | 4.5 | 2.49 | (1.31, 4.75) | 0.006 | 0.036 | |
|
| |||||||||
| hsa-miR-625-5p | Down-regulated | 26 | 2.0 | 7 | 2.0 | 1.07 | (0.46, 2.51) | 0.88 | 0.876 |
| Referent | 1261 | 96.1 | 323 | 91.2 | 1.00 | ||||
| Up-Regulated | 25 | 1.9 | 24 | 6.8 | 3.55 | (1.98, 6.37) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-652-3p | Down-regulated | 41 | 3.1 | 15 | 4.2 | 1.35 | (0.72, 2.51) | 0.35 | 0.753 |
| Referent | 1085 | 82.7 | 310 | 87.6 | 1.00 | ||||
| Up-Regulated | 186 | 14.2 | 29 | 8.2 | 0.56 | (0.37, 0.86) | 0.007 | 0.038 | |
|
| |||||||||
| hsa-miR-664a-3p | Down-regulated | 301 | 22.9 | 63 | 17.8 | 0.64 | (0.47, 0.87) | <0.0001 | 0.184 |
| Referent | 687 | 52.4 | 234 | 66.1 | 1.00 | ||||
| Up-Regulated | 324 | 24.7 | 57 | 16.1 | 0.52 | (0.38, 0.72) | <.0001 | 0.033 | |
|
| |||||||||
| hsa-miR-873-3p | Down-regulated | 42 | 3.2 | 10 | 2.8 | 0.94 | (0.46, 1.90) | 0.86 | 0.858 |
| Referent | 1226 | 93.4 | 318 | 89.8 | 1.00 | ||||
| Up-Regulated | 44 | 3.4 | 26 | 7.3 | 2.17 | (1.30, 3.61) | 0.001 | 0.033 | |
|
| |||||||||
| hsa-miR-98-5p | Down-regulated | 25 | 1.9 | 16 | 4.5 | 2.44 | (1.26, 4.72) | 0.008 | 0.184 |
| Referent | 927 | 70.7 | 269 | 76.0 | 1.00 | ||||
| Up-Regulated | 360 | 27.4 | 69 | 19.5 | 0.68 | (0.51, 0.91) | 0.01 | 0.038 | |
MSI was associated with 22 infrequently expressed miRNAs (Table 4). Of these miRNAs, the majority (13 of 22) were less likely to be associated with a microsatellite unstable tumor if up-regulated in the tumor. Only two of the 22 miRNAs had over 20% of the population in the group of dysregulation. There were no significant associations with microsatellite unstable tumors and down-regulated infrequently expressed miRNAs.
Table 4.
Overall associations between differential miRNA expression in infrequently expressed miRNA and Microsatellite Instability tumors
| miRNA | Microsatellite Stable | Microsatellite Unstable | P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | OR | (95% CI) | unadjusted | Q value | ||
| hsa-miR-1207-3p | Down-regulated1 | 140 | 8.3 | 13 | 7.6 | 0.96 | (0.53, 1.75) | 0.90 | 0.90 |
| Referent | 1512 | 89.6 | 145 | 85.3 | 1.00 | ||||
| Up-Regulated | 35 | 2.1 | 12 | 7.1 | 3.28 | (1.65, 6.53) | 0.0007 | 0.04 | |
|
| |||||||||
| hsa-miR-133b | Down-regulated | 891 | 52.8 | 68 | 40.0 | 0.57 | (0.40, 0.79) | .001 | 0.19 |
| Referent | 664 | 39.4 | 96 | 56.5 | 1.00 | ||||
| Up-Regulated | 132 | 7.8 | 6 | 3.5 | 0.31 | (0.13, 0.74) | 0.08 | 0.04 | |
|
| |||||||||
| hsa-miR-151a-3p | Down-regulated | 70 | 4.1 | 8 | 4.7 | 0.85 | (0.39, 1.82) | 0.67 | 0.71 |
| Referent | 896 | 53.1 | 138 | 81.2 | 1.00 | ||||
| Up-Regulated | 721 | 42.7 | 24 | 14.1 | 0.22 | (0.14, 0.35) | <.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-192-3p | Down-regulated | 384 | 22.8 | 35 | 20.6 | 0.86 | (0.57, 1.29) | 0.47 | 0.67 |
| Referent | 1107 | 65.6 | 128 | 75.3 | 1.00 | ||||
| Up-Regulated | 196 | 11.6 | 7 | 4.1 | 0.31 | (0.14, 0.68) | 0.004 | 0.04 | |
|
| |||||||||
| hsa-miR-199b-5p | Down-regulated | 107 | 6.3 | 18 | 10.6 | 1.55 | (0.89, 2.71) | 0.12 | 0.41 |
| Referent | 1036 | 61.4 | 137 | 80.6 | 1.00 | ||||
| Up-Regulated | 544 | 32.2 | 15 | 8.8 | 0.22 | (0.13, 0.38) | <.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-203a | Down-regulated | 174 | 10.3 | 17 | 10.0 | 0.70 | (0.40, 1.23) | 0.21 | 0.52 |
| Referent | 474 | 28.1 | 71 | 41.8 | 1.00 | ||||
| Up-Regulated | 1039 | 61.6 | 82 | 48.2 | 0.52 | (0.37, 0.73) | 0.0002 | 0.04 | |
|
| |||||||||
| hsa-miR-28-3p | Down-regulated | 186 | 11.0 | 16 | 9.4 | 0.95 | (0.55, 1.64) | 0.85 | 0.85 |
| Referent | 1380 | 81.8 | 131 | 77.1 | 1.00 | ||||
| Up-Regulated | 121 | 7.2 | 23 | 13.5 | 1.88 | (1.16, 3.06) | 0.01 | 0.04 | |
|
| |||||||||
| hsa-miR-30a-5p | Down-regulated | 604 | 35.8 | 64 | 37.6 | 1.00 | (0.71, 1.41) | 1.00 | 1.00 |
| Referent | 900 | 53.3 | 101 | 59.4 | 1.00 | ||||
| Up-Regulated | 183 | 10.8 | 5 | 2.9 | 0.24 | (0.10, 0.61) | 0.003 | 0.04 | |
|
| |||||||||
| hsa-miR-30e-5p | Down-regulated | 578 | 34.3 | 70 | 41.2 | 1.22 | (0.87, 1.71) | 0.24 | 0.53 |
| Referent | 835 | 49.5 | 88 | 51.8 | 1.00 | ||||
| Up-Regulated | 274 | 16.2 | 12 | 7.1 | 0.43 | (0.23, 0.80) | 0.007 | 0.04 | |
|
| |||||||||
| hsa-miR-3609 | Down-regulated | 289 | 17.1 | 52 | 30.6 | 1.79 | (1.25, 2.58) | 0.002 | 0.19 |
| Referent | 1000 | 59.3 | 108 | 63.5 | 1.00 | ||||
| Up-Regulated | 398 | 23.6 | 10 | 5.9 | 0.23 | (0.12, 0.45) | <.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-3615 | Down-regulated | 378 | 22.4 | 41 | 24.1 | 1.24 | (0.84, 1.83) | 0.27 | 0.54 |
| Referent | 1173 | 69.5 | 99 | 58.2 | 1.00 | ||||
| Up-Regulated | 136 | 8.1 | 30 | 17.6 | 2.41 | (1.53, 3.79) | 0.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-374b-5p | Down-regulated | 44 | 2.6 | 5 | 2.9 | 0.97 | (0.37, 2.52) | 0.95 | 0.95 |
| Referent | 1146 | 67.9 | 160 | 94.1 | 1.00 | ||||
| Up-Regulated | 497 | 29.5 | 5 | 2.9 | 0.08 | (0.03, 0.19) | <.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-3922-5p | Down-regulated | 192 | 11.4 | 8 | 4.7 | 0.45 | (0.22, 0.94) | 0.03 | 0.28 |
| Referent | 1318 | 78.1 | 127 | 74.7 | 1.00 | ||||
| Up-Regulated | 177 | 10.5 | 35 | 20.6 | 2.07 | (1.37, 3.13) | 0.0005 | 0.04 | |
|
| |||||||||
| hsa-miR-4492 | Down-regulated | 93 | 5.5 | 11 | 6.5 | 1.18 | (0.61, 2.26) | 0.63 | 0.71 |
| Referent | 1526 | 90.5 | 143 | 84.1 | 1.00 | ||||
| Up-Regulated | 68 | 4.0 | 16 | 9.4 | 2.29 | (1.28, 4.10) | 0.005 | 0.04 | |
|
| |||||||||
| hsa-miR-4533 | Down-regulated | 231 | 13.7 | 27 | 15.9 | 1.27 | (0.81, 1.98) | 0.30 | 0.56 |
| Referent | 1311 | 77.7 | 115 | 67.6 | 1.00 | ||||
| Up-Regulated | 145 | 8.6 | 28 | 16.5 | 2.09 | (1.32, 3.30) | 0.002 | 0.04 | |
|
| |||||||||
| hsa-miR-484 | Down-regulated | 182 | 10.8 | 11 | 6.5 | 0.57 | (0.30, 1.08) | 0.09 | 0.38 |
| Referent | 1307 | 77.5 | 151 | 88.8 | 1.00 | ||||
| Up-Regulated | 198 | 11.7 | 8 | 4.7 | 0.37 | (0.18, 0.78) | 0.008 | 0.04 | |
|
| |||||||||
| hsa-miR-513a-3p | Down-regulated | 81 | 4.8 | 7 | 4.1 | 0.84 | (0.38, 1.86) | 0.67 | 0.71 |
| Referent | 1575 | 93.4 | 154 | 90.6 | 1.00 | ||||
| Up-Regulated | 31 | 1.8 | 9 | 5.3 | 2.87 | (1.32, 6.24) | 0.01 | 0.04 | |
|
| |||||||||
| hsa-miR-520d-3p | Down-regulated | 124 | 7.4 | 16 | 9.4 | 1.33 | (0.76, 2.32) | 0.32 | 0.58 |
| Referent | 1495 | 88.6 | 140 | 82.4 | 1.00 | ||||
| Up-Regulated | 68 | 4.0 | 14 | 8.2 | 2.23 | (1.21, 4.11) | 0.008 | 0.04 | |
|
| |||||||||
| hsa-miR-532-5p | Down-regulated | 59 | 3.5 | 10 | 5.9 | 1.31 | (0.64, 2.65) | 0.46 | 0.67 |
| Referent | 1016 | 60.2 | 153 | 90.0 | 1.00 | ||||
| Up-Regulated | 612 | 36.3 | 7 | 4.1 | 0.08 | (0.04, 0.17) | <.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-5685 | Down-regulated | 337 | 20.0 | 27 | 15.9 | 0.83 | (0.53, 1.29) | 0.41 | 0.66 |
| Referent | 1243 | 73.7 | 116 | 68.2 | 1.00 | ||||
| Up-Regulated | 107 | 6.3 | 27 | 15.9 | 2.75 | (1.71, 4.40) | <.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-664a-3p | Down-regulated | 360 | 21.3 | 28 | 16.5 | 0.59 | (0.38, 0.91) | 0.02 | 0.26 |
| Referent | 923 | 54.7 | 130 | 76.5 | 1.00 | ||||
| Up-Regulated | 404 | 23.9 | 12 | 7.1 | 0.22 | (0.12, 0.40) | <.0001 | 0.04 | |
|
| |||||||||
| hsa-miR-98-5p | Down-regulated | 40 | 2.4 | 6 | 3.5 | 1.34 | (0.55, 3.26) | 0.52 | 0.67 |
| Referent | 1189 | 70.5 | 158 | 92.9 | 1.00 | ||||
| Up-Regulated | 458 | 27.1 | 6 | 3.5 | 0.10 | (0.04, 0.23) | <.0001 | 0.04 | |
We determined which genes were associated with each of the 57 miRNAs that had a Q value of <0.05 using our RNAseq data. Those associations for all genes whose expression was altered by significant miRNAs are summarized in Supplemental Table 1. There was considerable overlap in miRNAs associated with tumor molecular phenotype. For instance, 19 miRNAs were associated with both CpG island methylator phenotype-high tumors and TP53-mutated tumors; 9 of these miRNAs also were associated with microsatellite unstable tumors. For each miRNAs where a higher level of expression increased the likelihood of having a TP53-mutated tumor, there was a decreased the likelihood of having a CpG island methylator phenotype-high or microsatellite unstable tumor.
We have summarized the top three networks (Supplemental Table 2 has all networks with Scores of over 20) derived from genes linked to the 19 miRNAS that were associated with multiple tumor molecular phenotypes of TP53, CpG island methylator phenotype high, and/or microsatellite unstable (Figure 1). Network 1 (Immunological Disease, Inflammatory Disease, and Inflammatory Response) had a Score of 28 and 35 focus molecules including genes that were influenced by the miRNAs; Network 2 (Cell Cycle, Cancer, Cell-To-Cell Signaling and Interaction) had a Score of 25 and 34 Focus molecules influenced by the genes associated with these miRNAs; Network 3 (Amino Acid Metabolism, Small Molecule Biochemistry, Drug Metabolism) also had a Score of 25 and 34 Focus Molecules associated with genes linked to these miRNAs. The majority of genes in these networks were up-regulated (indicated in red) when the miRNAs were expressed at higher levels. The genes that were down-regulated (indicated in green NR3C1, TRPM6, GLP2R, ZFYVE28, FGD4, RNF112, TNFRSF17, TNFSF13, and CLEC3B) were all down-regulated in the presence of high levels of miR-224-5p. Higher levels of miR-224-5p were more likely to be present in TP53-mutated tumors and less likely to be present in CpG island methylator phenotype-high tumors. PHGDH was up-regulated at high levels of miR-19a-3p and KCND3 was up-regulated at high levels of miR-424-5p; high levels of miR-424-5p were more likely to have a TP53-mutated tumor and less likely to have a CpG island methylator phenotype-high tumor. MYC expression was associated with six miRNA, miR-151a-3p, miR-19a-3p, miR-3687, miR-374b-5p, miR-4533, and miR-7-5p. Higher levels of miRNA expression for all but miR-4533 were associated with TP53-mutated tumors, while miR-4533 was associated with tumors that were more likely to have microsatellite instability and CpG island methylator phenotype-high.
Figure 1.
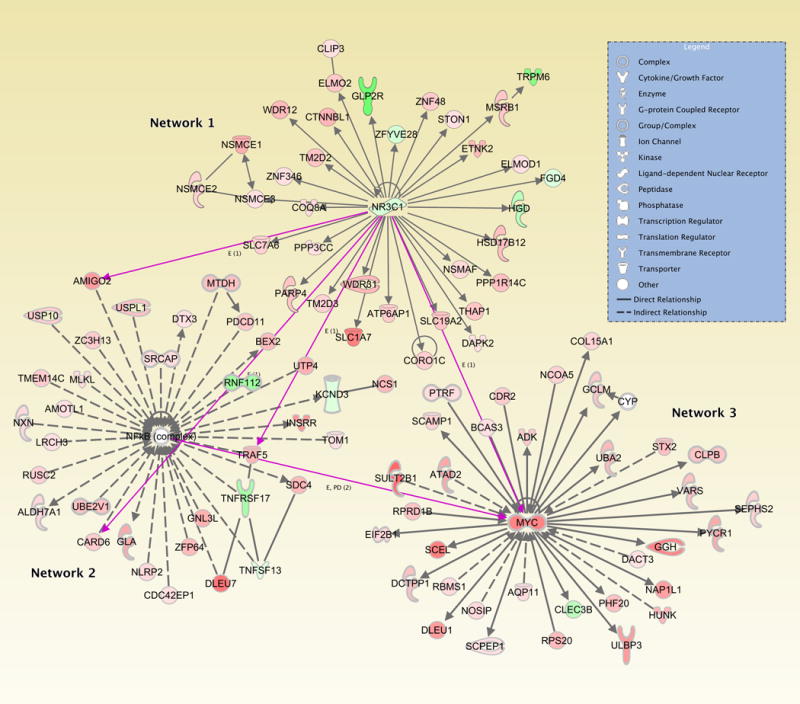
Top Ingenuity Pathway Analysis networks associated with genes whose expression is altered by high levels of miRNA expression associated with both TP53 and CpG island methylator phenotype-high and/or microsatellite instability
Discussion
Our data suggest that some miRNAs although infrequently expressed, when expressed at higher levels or up-regulated, are associated with specific tumor molecular phenotype. We did not have similar associations for down-regulated miRNAs. Of those infrequently expressed miRNAs significantly associated with tumor molecular phenotype when expressed at high levels were more likely to be highly expressed in TP53-mutated tumors and less likely to be associated with CpG island methylator phenotype-high or microsatellite unstable tumors. Many of these miRNAs were associated with altered gene mRNA expression in colorectal cancer tissue when expressed at high levels.
Many miRNAs are expressed infrequently in the population and often have low levels of expression when detected (29). Many of the miRNAs that have levels of expression around 0 could be considered background noise from slight differences in RNA samples despite high quality control. Additionally, although the data were normalized, picking a scale to normalize on is arbitrary and a different scale could have slightly altered what was considered background levels of expression. The Agilent Platform that we used to collect miRNA data in this study has been noted as being able to detect low levels of expression (47, 48). Based on our findings, it appears that very low levels of expression are similar to no expression for most miRNAs, and that distinct associations for specific tumor molecular phenotype can only be seen when examining expression of these miRNAs at higher levels. These higher levels of expression are less likely to be the result of background expression, especially considering associations with tumor molecular phenotype.
To gain insight into pathways and functions of infrequently expressed miRNAs, we utilized our colorectal gene expression data from RNAseq. We assessed which genes were associated with miRNAs when miRNAs were more highly expressed. Since most of these miRNAs are infrequently expressed, there is less information regarding gene associations in existing databases, and even less information for colorectal tissue-specific expression, thus making use of our data imperative. Examining gene expression provided some insight into how these infrequently expressed miRNAs could be associated with various disease pathways. A limitation of RNAseq data, although a common method to determine miRNA∷mRNA associations (49), is that miRNA targeted genes could be missed since gene expression studies more likely capture associations with transcription better than translation. However, we believe that our having RNAseq data in conjunction with miRNA data provides insight into colon-specific direct and indirect functions and pathways associated with these infrequently expressed miRNAs.
Given their infrequent expression, many of the miRNAs evaluated in our study have no known association with colorectal tumor molecular phenotype in the literature. However, our findings suggest that some infrequently expressed miRNAs, when they have high levels of expression in a tumor, may play an important role in tumorigenesis and the development of specific tumor phenotype. For instance, miR-19a-3p, which had about 25% to 30% of the population with high differential expression, was included previously in a miRNA cluster that functioned alongside Epstein-Barr Virus to control gene expression in human B cells through a TP53-induced mechanism (50). While we could find no reported association between this miRNA, or the others evaluated in this study, and colorectal cancer-specific tumor molecular phenotype, these findings are consistent with our finding that high levels of miR-19a-3p is associated with a TP53 phenotype in colorectal cancer.
It has been shown that TP53 mutations are inversely related to CpG island methylator phenotype-high and microsatellite unstable in colorectal cancer; TP53 mutations are present in higher rates in microsatellite stable tumors while CpG island methylator phenotype-high tumors also are frequently microsatellite unstable tumors (34, 51). Our findings support this pattern by demonstrating that certain infrequently expressed miRNAs when upregulated in TP53-mutated tumors are simultaneously more likely to be down-regulated in CpG island methylator phenotype-high and microsatellite unstable tumors.
To further put these findings in perspective, we identified three major networks that represented the genes associated with those miRNAs that were up-regulated in TP53-mutated tumors and down-regulated in CpG island methylator phenotype-high and microsatellite unstable tumors. The first network has NR3C1 as one of its central components (See Figure 1). NR3C1 is a glucocorticoid receptor that induces apoptotic cell death, via decreased expression of anti-apoptotic proteins, such as BCL2 and MCL1, and induces expression of pro-apoptotic proteins like BCL2-like apoptosis initiator 11 (52). In earlier studies, NR3C1 has been associated with proximal microsatellite unstable tumors, with hypermethylation of NR3C1 being identified as a marker for microsatellite unstable tumors and a marker to differentiate between CpG island methylator phenotype-high and CpG island methylator phenotype-low/negative phenotypes (53). These findings correlate with our identified association between the NR3C1 pathway and tumor phenotype; NR3C1 was down-regulated in our data, suggesting less likely association with CpG island methylator phenotype-high and microsatellite unstable tumors. Our findings suggest that differential methylation of NR3C1, and its subsequent role in tumorigenesis and phenotype, may be in part due to the dysregulation of previously unstudied, infrequently expressed miRNAs.
The NFkB complex is central in our second Ingenuity Pathway Analysis network and is well known in literature for up-regulating and promoting various pro-inflammatory cytokines and linking various gastrointestinal conditions such as inflammatory bowel disease, diabetes mellitus, and colorectal cancer (54). The classical NFkB pathway plays a major role in linking inflammation to the onset and progression of malignancy in various tissues (55). One pro-inflammatory stimulus includes red meat consumption which has been linked to colon cancer and TP53-mutated tumors specifically (56, 57). A prospective study in Denmark has shown that the combination of polymorphisms in NFκB that down-regulate its expression, and high red meat consumption increases the likelihood of developing colorectal cancer (58). They proposed that lower NFkB activity leads to higher loads of reactive oxygen species secondary to heme degradation, contributing to colorectal carcinogenesis. Moreover, other studies have found that the NFκB pathway to be linked with the TP53 pathway in hepatocellular carcinoma; the crosstalk between these two pathways is critical for the survival of HCC cells in the setting of reactive oxygen species (59). These previous findings further support an association between the NFkB complex and a TP53 molecular phenotype in certain cancers, especially in the setting of pro-inflammatory stimuli. Here we suggest that the up-regulation of infrequently expressed miRNAs may provide an important link between NFκB and its related genes and TP53 phenotype in colorectal cancer.
In our third Ingenuity Pathway Analysis network, MYC encodes for c-myc, a transcription factor often constitutively amplified leading to tumor progression of many cancers. In colorectal cancer, aberrant WNT/b-catenin pathway influences the amplification of MYC, leading to increased cellular proliferation (60). In our data MYC was up-regulated in conjunction with miRNAs that were up-regulated in TP53-mutated tumors. Furthermore, the consensus molecular subtype 2 subtype of colorectal cancer is canonically known to have strong WNT/MYC activation in microsatellite stable tumors; this subtype was also found to be highly correlated with TP53-mutated tumors (61). This suggests that miRNA dysregulation from infrequently expressed miRNAs, may play an important role in MYC’s function in TP53-mutated molecular phenotype.
The study has several strengths and weaknesses. First, given the size of the study and the Agilent Platform used, we can identify and examine the impact of infrequently expressed miRNAs. Many studies are too small to be able to determine associations with infrequently expressed miRNAs. Our dataset is rich, in that we have information on tumor molecular phenotype as well as RNAseq for a subset of these samples to improve our understanding of how miRNAs alter specific genes in colorectal tissue. One of the limitations of the study, which applies to the field of miRNA research, is the difficulty in understanding the pathways and genes associated with miRNA expression, especially when miRNAs alter multiple genes and genes are modified by multiple miRNAs. We have attempted to address this weakness in part by using our colorectal RNAseq data in conjunction with our miRNA data to identify genes that are up- or down-regulated by infrequently expressed miRNAs. In this study we have used adjacent tissue to the tumor as our comparison tissue. However there are limitation that the “normal” tissue is not true normal, although the best tissue available for comparison.
In summary, our data suggest that a large percentage of miRNAs expressed in colorectal tissue are infrequently expressed. However, some of the infrequently expressed miRNAs, when expressed at higher levels influence tumor molecular phenotype. This information is important for consideration pathways associated with cancer as well as examining lifestyle and environmental factors that may alter those pathways. Genes associated with these infrequently expressed miRNAs are involved in a variety of functions that may impact cancer development and prognosis.
Supplementary Material
Supplemental Table 1. Associations with genes whose expression was associated with miRNAs significantly different by tumor molecular phenotype.
Supplemental Table 2. Summary of Ingenuity Pathway Analysis networks with scores over 20 for genes associated with TP53-mutated tumors, CpG island methylator phenotype-high/or microsatellite instability.
Acknowledgments
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute. We acknowledge Sandra Edwards for data oversight and study management, and Michael Hoffman and Erica Wolff for miRNA analysis. We acknowledge Dr. Bette Caan and the staff at the Kaiser Permanente Medical Research Program for sample and data collection.
Financial Support:
This study was supported by NCI grants CA163683 and CA48998.
Footnotes
Potential Competing Interests:
None
Specific Author Contributions:
MS obtained funding, planned study, oversaw study data collection and analysis, and wrote the manuscript.
AP and FL helped interpret findings and helped write the manuscript.
JS provided input into the statistical analysis
LM conducted bioinformatics analysis and helped write manuscript
RW oversaw laboratory analysis.
WS reviewed manuscript and did pathology overview for the study
JH conducted statistical analysis and managed data.
All authors approved final manuscript
References
- 1.Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol. 2016 doi: 10.1007/s00535-016-1272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishihara R, Lochhead P, Kuchiba A, Jung S, Yamauchi M, Liao X, et al. Aspirin use and risk of colorectal cancer according to BRAF mutation status. JAMA. 2013;309(24):2563–71. doi: 10.1001/jama.2013.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishihara R, Wang M, Qian ZR, Baba Y, Yamauchi M, Mima K, et al. Alcohol, one-carbon nutrient intake, and risk of colorectal cancer according to tumor methylation level of IGF2 differentially methylated region. Am J Clin Nutr. 2014;100(6):1479–88. doi: 10.3945/ajcn.114.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slattery ML, Anderson K, Curtin K, Ma K, Schaffer D, Edwards S, et al. Lifestyle factors and Ki-ras mutations in colon cancer tumors. Mutat Res. 2001;483(1–2):73–81. doi: 10.1016/s0027-5107(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 5.Slattery ML, Anderson K, Curtin K, Ma KN, Schaffer D, Samowitz W. Dietary intake and microsatellite instability in colon tumors. Int J Cancer. 2001;93(4):601–7. doi: 10.1002/ijc.1370. [DOI] [PubMed] [Google Scholar]
- 6.Slattery ML, Curtin K, Anderson K, Ma KN, Ballard L, Edwards S, et al. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J Natl Cancer Inst. 2000;92(22):1831–6. doi: 10.1093/jnci/92.22.1831. [DOI] [PubMed] [Google Scholar]
- 7.Slattery ML, Curtin K, Anderson K, Ma KN, Edwards S, Leppert M, et al. Associations between dietary intake and Ki-ras mutations in colon tumors: a population-based study. Cancer Res. 2000;60(24):6935–41. [PubMed] [Google Scholar]
- 8.Slattery ML, Curtin K, Ma K, Edwards S, Schaffer D, Anderson K, et al. Diet activity, and lifestyle associations with p53 mutations in colon tumors. Cancer Epidemiol Biomarkers Prev. 2002:541–8. [PubMed] [Google Scholar]
- 9.Slattery ML, Curtin K, Sweeney C, Levin TR, Potter J, Wolff RK, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120(3):656–63. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein AM, Song M, Zhang X, Pan A, Wang M, Fuchs CS, et al. Processed and Unprocessed Red Meat and Risk of Colorectal Cancer: Analysis by Tumor Location and Modification by Time. PLoS One. 2015;10(8):e0135959. doi: 10.1371/journal.pone.0135959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slattery ML, Herrick JS, Mullany LE, Gertz J, Wolff RK. Improved survival among colon cancer patients with increased differentially expressed pathways. BMC Med. 2015;13:75. doi: 10.1186/s12916-015-0292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samowitz WS, Curtin K, Ma KN, Schaffer D, Coleman LW, Leppert M, et al. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev. 2001;10(9):917–23. [PubMed] [Google Scholar]
- 13.Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9(11):1193–7. [PubMed] [Google Scholar]
- 14.Dou R, Nishihara R, Cao Y, Hamada T, Mima K, Masuda A, et al. MicroRNA let-7, T Cells, and Patient Survival in Colorectal Cancer. Cancer Immunol Res. 2016;4(11):927–35. doi: 10.1158/2326-6066.CIR-16-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanyuda A, Kim SA, Martinez-Fernandez A, Qian ZR, Yamauchi M, Nishihara R, et al. Survival Benefit of Exercise Differs by Tumor IRS1 Expression Status in Colorectal Cancer. Ann Surg Oncol. 2016;23(3):908–17. doi: 10.1245/s10434-015-4967-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–68. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino S, Meyerhardt JA, Kawasaki T, Clark JW, Ryan DP, Kulke MH, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450(5):529–37. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 18.Drusco A, Nuovo GJ, Zanesi N, Di Leva G, Pichiorri F, Volinia S, et al. MicroRNA profiles discriminate among colon cancer metastasis. PLoS One. 2014;9(6):e96670. doi: 10.1371/journal.pone.0096670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Murray BS, Choe SE, Woods M, Ryan TE, Liu W. An in silico analysis of microRNAs: mining the miRNAome. Mol Biosyst. 2010;6(10):1853–62. doi: 10.1039/c003961f. [DOI] [PubMed] [Google Scholar]
- 21.Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288(3–4):77–87. doi: 10.1007/s00438-013-0734-z. [DOI] [PubMed] [Google Scholar]
- 22.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18(2):103–10. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–62. doi: 10.1093/nar/gkp294. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slattery ML, Herrick JS, Mullany LE, Wolff E, Hoffman MD, Pellatt DF, et al. Colorectal tumor molecular phenotype and miRNA: expression profiles and prognosis. Mod Pathol. 2016;29(8):915–27. doi: 10.1038/modpathol.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valeri N, Vannini I, Fanini F, Calore F, Adair B, Fabbri M. Epigenetics, miRNAs, and human cancer: a new chapter in human gene regulation. Mamm Genome. 2009;20(9–10):573–80. doi: 10.1007/s00335-009-9206-5. [DOI] [PubMed] [Google Scholar]
- 26.Slattery ML, Herrick JS, Pellatt DF, Stevens JR, Mullany LE, Wolff E, et al. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: variations in miRNA expression and disease progression. Carcinogenesis. 2016 doi: 10.1093/carcin/bgv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slattery ML, Potter J, Caan B, Edwards S, Coates A, Ma KN, et al. Energy balance and colon cancer–beyond physical activity. Cancer research. 1997;57(1):75–80. [PubMed] [Google Scholar]
- 28.Slattery ML, Caan BJ, Benson J, Murtaugh M. Energy balance and rectal cancer: an evaluation of energy intake, energy expenditure, and body mass index. Nutr Cancer. 2003;46(2):166–71. doi: 10.1207/S15327914NC4602_09. [DOI] [PubMed] [Google Scholar]
- 29.Slattery ML, Herrick JS, Pellatt DF, Stevens JR, Mullany LE, Wolff E, et al. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: variations in miRNA expression and disease progression. Carcinogenesis. 2016;37(3):245–61. doi: 10.1093/carcin/bgv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellatt DF, Stevens JR, Wolff RK, Mullany LE, Herrick JS, Samowitz W, et al. Expression Profiles of miRNA Subsets Distinguish Human Colorectal Carcinoma and Normal Colonic Mucosa. Clin Transl Gastroenterol. 2016;7:e152. doi: 10.1038/ctg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agilent Technologies I. Agilent GeneSpring User Manual. Santa Clara, CA: Aglient Technologies Inc; 2013. cited 2015 July 16. [Google Scholar]
- 32.Slattery ML, Herrick JS, Mullany LE, Valeri N, Stevens J, Caan BJ, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer. 2015;137(2):428–38. doi: 10.1002/ijc.29384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slattery ML, Pellatt DF, Mullany LE, Wolff RK, Herrick JS. Gene expression in colon cancer: A focus on tumor site and molecular phenotype. Genes, chromosomes & cancer. 2015;54(9):527–41. doi: 10.1002/gcc.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, et al. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158(4):1517–24. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129(3):837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 36.Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22(2):271–82. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9(3):305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B. 2002;64S(8):479–98. [Google Scholar]
- 39.Benjamini YH,Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 40.Savli H, Szendroi A, Romics I, Nagy B. Gene network and canonical pathway analysis in prostate cancer: a microarray study. Experimental & molecular medicine. 2008;40(2):176–85. doi: 10.3858/emm.2008.40.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Carrillo JA, Ding Y, He Y, Zhao C, Zan L, et al. Ruminal Transcriptomic Analysis of Grass-Fed and Grain-Fed Angus Beef Cattle. PLoS One. 2015;10(6):e0116437. doi: 10.1371/journal.pone.0116437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Human molecular genetics. 2009;18(11):2078–90. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan-Fang T, Dong W, Li P, Wen-Li Z, Jun L, Na W, et al. Analyzing the gene expression profile of pediatric acute myeloid leukemia with real-time PCR arrays. Cancer cell international. 2012;12(1):40. doi: 10.1186/1475-2867-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naito Y, Kuroda M, Mizushima K, Takagi T, Handa O, Kokura S, et al. Transcriptome Analysis for Cytoprotective Actions of Rebamipide against Indomethacin-Induced Gastric Mucosal Injury in Rats. Journal of clinical biochemistry and nutrition. 2007;41(3):202–10. doi: 10.3164/jcbn.2007029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reyes-Gibby CC, Yuan C, Wang J, Yeung SC, Shete S. Gene network analysis shows immune-signaling and ERK1/2 as novel genetic markers for multiple addiction phenotypes: alcohol, smoking and opioid addiction. BMC systems biology. 2015;9:25. doi: 10.1186/s12918-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia P, Kao CF, Kuo PH, Zhao Z. A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC Syst Biol. 2011;5(Suppl 3):S12. doi: 10.1186/1752-0509-5-S3-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sah S, McCall MN, Eveleigh D, Wilson M, Irizarry RA. Performance evaluation of commercial miRNA expression array platforms. BMC Res Notes. 3:80. doi: 10.1186/1756-0500-3-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–15. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 49.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–47. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31(9):2207–21. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bond CE, Umapathy A, Ramsnes I, Greco SA, Zhen Zhao Z, Mallitt KA, et al. p53 mutation is common in microsatellite stable, BRAF mutant colorectal cancers. Int J Cancer. 2012;130(7):1567–76. doi: 10.1002/ijc.26175. [DOI] [PubMed] [Google Scholar]
- 52.Wei TT, Lin YT, Chen WS, Luo P, Lin YC, Shun CT, et al. Dual Targeting of 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase and Histone Deacetylase as a Therapy for Colorectal Cancer. EBioMedicine. 2016;10:124–36. doi: 10.1016/j.ebiom.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahlquist T, Lind GE, Costa VL, Meling GI, Vatn M, Hoff GS, et al. Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol Cancer. 2008;7:94. doi: 10.1186/1476-4598-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jurjus A, Eid A, Al Kattar S, Zeenny MN, Gerges-Geagea A, Haydar H, et al. Inflammatory bowel disease, colorectal cancer and type 2 diabetes mellitus: The links. BBA Clin. 2016;5:16–24. doi: 10.1016/j.bbacli.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Pinard M, Wang Y, Yang L, Lin R, Hiscott J, et al. Crosstalk between the TNF and IGF pathways enhances NF-kappaB activation and signaling in cancer cells. Growth Horm IGF Res. 2015;25(5):253–61. doi: 10.1016/j.ghir.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Pellatt AJ, Slattery ML, Mullany LE, Wolff RK, Pellatt DF. Dietary intake alters gene expression in colon tissue: possible underlying mechanism for the influence of diet on disease. Pharmacogenet Genomics. 2016;26(6):294–306. doi: 10.1097/FPC.0000000000000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slattery ML, Curtin K, Ma K, Edwards S, Schaffer D, Anderson K, et al. Diet activity, and lifestyle associations with p53 mutations in colon tumors. Cancer Epidemiol Biomarkers Prev. 2002;11(6):541–8. [PubMed] [Google Scholar]
- 58.Andersen V, Christensen J, Overvad K, Tjonneland A, Vogel U. Polymorphisms in NFkB, PXR, LXR and risk of colorectal cancer in a prospective study of Danes. BMC Cancer. 2010;10:484. doi: 10.1186/1471-2407-10-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Q, Zhan L, Cao H, Li J, Lyu Y, Guo X, et al. Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NFKB and TP53 pathways. Autophagy. 2016;12(6):999–1014. doi: 10.1080/15548627.2016.1166318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rennoll S, Yochum G. Regulation of MYC gene expression by aberrant Wnt/beta-catenin signaling in colorectal cancer. World J Biol Chem. 2015;6(4):290–300. doi: 10.4331/wjbc.v6.i4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Salas N, Dominguez G, Barderas R, Mendiola M, Garcia-Albeniz X, Maurel J, et al. Clinical relevance of colorectal cancer molecular subtypes. Crit Rev Oncol Hematol. 2017;109:9–19. doi: 10.1016/j.critrevonc.2016.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Associations with genes whose expression was associated with miRNAs significantly different by tumor molecular phenotype.
Supplemental Table 2. Summary of Ingenuity Pathway Analysis networks with scores over 20 for genes associated with TP53-mutated tumors, CpG island methylator phenotype-high/or microsatellite instability.


