Abstract
Over the last several decades, it has become clear that epigenetic abnormalities may be one of the hallmarks of cancer. Post-translational modifications of histones, for example, may play a crucial role in cancer development and progression by modulating gene transcription, chromatin remodeling and nuclear architecture. Histone acetylation, a well-studied post-translational histone modification, is controlled by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs). By removing acetyl groups, HDACs reverse chromatin acetylation and alter transcription of oncogenes and tumor suppressor genes. In addition, HDACs deacetylate numerous non-histone cellular substrates that govern a wide array of biological processes including cancer initiation and progression. This review will discuss the role of HDACs in cancer and the therapeutic potential of HDAC inhibitors (HDACi) as emerging drugs in cancer treatment.
Introduction
Histone function is modulated by multiple post-translational modifications, including reversible acetylation of the N-terminal ε-group of lysines on histones. Histone acetylation is tightly controlled by a balance between the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs, also known as lysine deacetylases or KDACs). There are 18 potential human HDACs grouped into four classes. By removing the acetyl groups from the ε-amino lysine residues on histone tails HDACs may play a critical role in transcription regulation (Seto and Yoshida 2014).
Given that histone modification modulates chromatin structure and gene expression, it is not surprising that abnormal alterations in histone acetylation are associated with cancer development. For example, global loss of acetylation at lysine 16 and trimethylation at lysine 20 of histone H4 is reported to be a common abnormality in human cancer (Fraga et al. 2005), and a low level of histone H3 lysine 18 acetylation (H3K18ac) was found to be a predictor of poor survival in pancreatic, breast, prostate and lung cancers. In parallel, research increasingly shows aberrant expression of HDACs is frequently observed in various human cancers. Although it is not known whether the changes in histone modification are related to specific alterations in HDACs expression (there are obviously many other mechanisms that can explain why cancer cells might exploit HDACs to support tumorigenesis), they do nevertheless contribute to the overall principle of targeting HDACs for cancer therapy.
Because approximately equal numbers of genes are activated and repressed by HDAC inhibition, other mechanisms besides histone modification are involved in HDAC-mediated gene regulation. In addition to histones, HDACs also deacetylate a large number of non-histone proteins. This is consistent with the discovery of many acetylated non-histone proteins by global analysis in human cells (Choudhary et al. 2009). In tumorigenesis, the finely tuned acetylation status at the whole proteome level is greatly impaired by dysregulated deacetylases (Parbin et al. 2014). Through hyperacetylation of histone and non-histone targets, HDACi enable the re-establishment of cellular acetylation homeostasis and restore normal expression and function of numerous proteins that may reverse cancer initiation and progression. This article will describe recent advances in our understanding of the role of HDACs in cancer and the implications of HDACi in the treatment of cancer.
1. Dysregulation and mutation of HDACs in human cancer
Based on sequence homology to yeast, eighteen human HDACs are grouped into four classes. Class I Rpd3-like enzymes are comprised of HDAC1, 2, 3, and 8. Class II Hda1-like enzymes are further divided into two subclasses: IIa (HDAC4, 5, 6, 7, and 9) and IIb (HDAC6 and 10). Class III Sir2-like enzymes consist of seven sirtuins, which are NAD-dependent protein deacetylases and/or ADP ribosylases. Sirtuins have been shown to regulate many cellular processes including survival, aging, stress response, and metabolism. Class IV contains only HDAC11 which shares sequences similarity to both Class I and II proteins.
HDACs are involved in multiple different stages of cancer (Figure 1). Aberrant expression of Classical (Class I, II, IV) HDACs has been linked to a variety of malignancies, including solid and hematological tumors (Table 1). In most cases, a high level of HDACs is associated with advanced disease and poor outcomes in patients. For example, high expression of HDAC1, 2 and 3 are associated with poor outcomes in gastric and ovarian cancers (Weichert et al. 2008a; Weichert et al. 2008b; Sudo et al. 2011), and high expression of HDAC8 correlates with advanced-stage disease and poor survival in neuroblastoma (Oehme et al. 2009; Rettig et al. 2015). HDACs have also been found broadly dysregulated in multiple myeloma (MM). Overexpression of Class I HDACs, particularly HDAC1, is associated with inferior patient outcomes (Mithraprabhu et al. 2014).
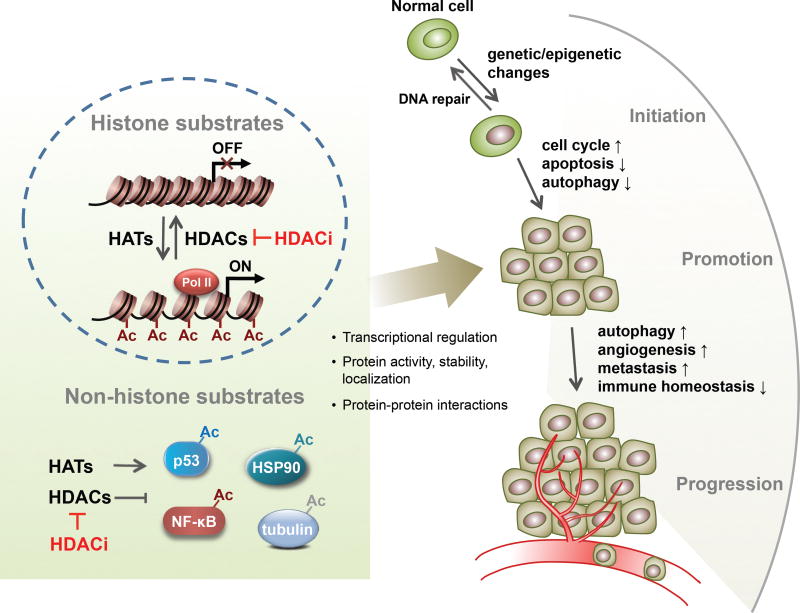
Figure 1.
A simplistic illustration of the diverse functions of HDACs and HDACi regulating different stages of cancer through multiple different mechanisms and changing different biological processes. Far right, ↑ indicates promotion or upregulation, ↓ indicates repression or downregulation.
Table 1.
Dysregulation and mutation of HDACs in human cancer.
| Cancer types | HDACs | Prognostic relevance | Genetic evidence | Molecular mechanism | References |
|---|---|---|---|---|---|
| Solid Tumours | |||||
| Neuroblastoma | HDAC8 | High transcript level correlates with advanced-stage disease and poor survival in neuroblastoma. | Knockdown and inhibition of HDAC8 promotes cell cycle arrest and differentiation, delays cell growth and induces cell death in vitro and in vivo. | HDAC8 inhibition induces p21WAF1/CIP1 and NTRK1/TrkA gene expression and enhances retinoic acid-mediated differentiation by regulating CREB phosphorylation. | (Oehme et al. 2009; Rettig et al. 2015) |
| HDAC10 | High expression correlates with poor overall patient survival in advanced INSS stage 4 neuroblastoma. | Knockdown and inhibition of HDAC10 in neuroblastoma cells interrupted autophagic flux resulting in an increase of sensitization to cytotoxic drug treatment. | HDAC10 controls autophagic processing and resistance to cytotoxic drugs via interaction with Hsp70 family proteins. | (Oehme et al. 2013) | |
| Medulloblastoma | HDAC2 | Overexpressed in medulloblastoma subgroups with poor prognosis. | HDAC2 depletion induces cell death and attenuates cell growth. MYC amplified and HDAC2 overexpressing cell lines are more sensitive to class I HDACi. | N/A | (Ecker et al. 2015) |
| HDAC5, 9 | Upregulated in high-risk medulloblastoma, and their expression is associated with poor survival. | Depletion of either HDAC5 or HDAC9 in MB cells resulted in a reduction of cell proliferation and increase in cell death. | N/A | (Milde et al. 2010) | |
| Lung | HDAC1 , 3 | High expression correlates with a poor prognosis in patients with lung adenocarcinoma. | N/A | N/A | (Minamiya et al. 2010; Minamiya et al. 2011) |
| HDAC2 | Abundant expression is observed in lung cancer tissues. | HDAC2 inactivation represses tumor cell growth in vitro and in vivo. | HDAC2 depletion activates apoptosis via p53 and Bax activation and Bcl2 suppression, induces cell cycle arrest by induction of p21 and suppression of cyclin E2, cyclin D1 and CDK2. | (Jung et al. 2012) | |
| HDAC5, 10 | Low expression is associated with poor prognosis in lung cancer patients. | N/A | N/A | (Osada et al. 2004) | |
| Gastric | HDAC1, 2, 3 | High expression is associated with nodal tumor spread and decreased overall patient survival. | N/A | N/A | (Weichert et al. 2008b; Sudo et al. 2011) |
| HDAC4 | Upregulated in gastric tumor cells compared with adjacent normal tissues. | HDAC4 inhibition has a synergistic effect with docetaxel treatment. | HDAC4 inhibition increased the level of cleaved caspases 3 and 9. | (Colarossi et al. 2014) | |
| HDAC10 | Low expression is a poor prognosis marker for gastric cancer patients. | N/A | N/A | (Jin et al. 2014) | |
| Liver | HDAC1 | Highly expressed in human HCCs and liver cancer cell lines. | HDAC1 inactivation impairs G1/S cell cycle transition and causes autophagic cell death. | Knockdown of HDAC1 induces p21 and p27 expression, and suppresses cyclin D1 and CDK2 expression. | (Xie et al. 2012) |
| HDAC1, 2, 3 | Upregulated in human HCCs and liver cancer cell lines. | The knockdown of HDAC1–3 leads to increased apoptosis and decreased proliferation. | Knockdown and inhibition of HDAC1–3 upregulates miR-449 and downregulates c-MET expression. Reduced c-MET dephosphorylates ERK1/2 and inhibits tumor growth. | (Buurman et al. 2012) | |
| HDAC1, 2, 3 | High expression is associated with poor survival in low-grade and early-stage tumors. | N/A | N/A | (Quint et al. 2011) | |
| HDAC3, 5 | Upregulation is correlated with DNA copy number gains. | N/A | N/A | (Lachenmayer et al. 2012) | |
| HDAC5 | Upregulated in HCC tissues. | Knockdown of HDAC5 promotes cell apoptosis and inhibits tumor cell growth in vitro and in vivo. | HDAC5 knockdown promotes apoptosis by upregulating cyto C, caspase 3, p53 and bax, and induces G1 phase cell cycle arrest by upregulating p21 and downregulating cyclin D1 and CDK2/4/6. | (Fan et al. 2014) | |
| HDAC5 | Upregulated in human HCC tissues. | Overexpression of HDAC5 promotes tumor cell proliferation, while knockdown of HDAC5 inhibits cell proliferation. | HDAC5 promotes cell proliferation by up-regulation of Six1. | (Feng et al. 2014) | |
| HDAC6 | Low expression is associated with poor prognosis in liver transplantation patients. | Knockdown of HDAC6 promotes HUVEC migration, proliferation, and tube formation in vitro, and suppress HCC cell apoptosis, and promote HCC cell proliferation in hypoxia. | HDAC6 knockdown promotes angiogenesis in HCC by HIF-1α/VEGFA axis. | (Lv et al. 2015) | |
| Pancreatic | HDAC2 | Highly expressed in PDAC. | HDAC2 confers resistance towards etoposide in PDAC cells. | HDAC2 knockdown upregulates NOXA expression which sensitize tumor cells towards etoposide-induced apoptosis. | (Fritsche et al. 2009) |
| HDAC6 | Highly expressed in human pancreatic cancer tissues. | Knockdown and inhibition of HDAC6 impairs the motility of cancer cells. | HDAC6 interacts with CLIP-170 and stimulates the migration of pancreatic cancer cells. | (Li et al. 2014) | |
| HDAC7 | Overexpression is associated with poor prognosis. | Knockdown of HDAC7 inhibits tumor cell growth. | N/A | (Ouaissi et al. 2008; Ouaissi et al. 2014) | |
| Colorectal | HDAC2 mutation | Truncating mutations are found in microsatellite unstable sporadic colorectal cancers, which lead to a loss of expression of the protein. | HDAC2 mutation renders cells more resistant to antiproliferative and proapoptotic effects of the HDAC inhibitor. | N/A | (Ropero et al. 2006) |
| HDAC1, 2, 3 | Highly expressed in a subset of colorectal carcinomas. HDAC2 is an independent prognostic factor in colorectal carcinoma. | Knockdown of HDAC1 and HDAC2 but not HDAC3 suppresses tumor cell growth. | N/A | (Weichert et al. 2008d) | |
| HDAC1, 2, 3, 5, 7 | Upregulated in human colorectal cancer. HDAC2 is an early biomarker of colon carcinogenesis. | N/A | N/A | (Stypula-Cyrus et al. 2013) | |
| Breast | HDAC1, 2, 3 | HDAC1 was highly expressed in hormone receptor positive tumors. HDAC2 and 3 are highly expressed in poorly differentiated and hormone receptor negative tumors. | N/A | N/A | (Zhang et al. 2005; Muller et al. 2013) |
| HDAC1, 6 | High expression is good prognostic factors for ER-positive invasive ductal carcinomas. | N/A | N/A | (Zhang et al. 2004; Seo et al. 2014) | |
| Ovarian | HDAC1, 2, 3 | High expression is associated with a poor outcome. | Knockdown of HDAC1 reduces cell proliferation via downregulating cyclin A expression, and knockdown of HDAC3 reduces the cell migration with elevated E-cadherin. | (Hayashi et al. 2010) | |
| HDAC1, 2, 3 | High-level expression is associated with a poor prognosis in ovarian endometrioid carcinomas. | N/A | N/A | (Weichert et al. 2008a). | |
| Cervical | HDAC10 | Low expression correlates with lymph node metastasis in cervical cancer. | Knockdown of HDAC10 promotes cervical cancer cell migration and invasion. | HDAC10 inhibits MMP2 and -9 expression. | (Song et al. 2013) |
| Prostate | HDAC1, 2, 3 | Highly expressed in prostate carcinomas. HDAC2 is an independent prognostic marker in prostate cancer cohort. | N/A | N/A | (Weichert et al. 2008c) |
| Renal | HDAC1, 2 | Highly expressed in renal cell cancer, but none of them are associated with patient survival. | N/A | N/A | (Fritzsche et al. 2008) |
| Bladder | HDAC1, 2, 3 | High expression is associated with higher tumor grades. High HDAC1 is a poor prognostic factor in urothelial bladder cancer. | N/A | N/A | (Poyet et al. 2014) |
| HDAC2, 4, 5, 7, 8 | Upregulation of HDAC2, 8 and downregulation of HDAC4, 5, and 7 mRNA are observed in urothelial cancer. | N/A | N/A | (Niegisch et al. 2013) | |
| Melanoma | HDAC3,8 | HDAC8 was increased in BRAF-mutated melanoma. HDAC8 and 3 over expression are associated with improved survival of patients with stage IV metastatic melanoma. | N/A | N/A | (Wilmott et al. 2015) |
| Hematological Tumors | |||||
| ALL | HDAC1-9 | HDAC2, 3, 6, 7, 8 are upregulated in ALL samples. HDAC1 and 4 show high expression in T-ALL and HDAC6 and 9 are highly expressed in B-lineage ALL. Higher expression of HDAC7 and 9 is associated with a poor prognosis in childhood ALL. | N/A | N/A | (Moreno et al. 2010) |
| HDAC4 | High expression is associated with high initial leukocyte count, T cell ALL and prednisone poor-response. | HDAC4 knockdown enhanced etoposide’s cytotoxic activity. | N/A | (Gruhn et al. 2013) | |
| CLL | HDAC1, 3, 6, 7, 9, 10, SIRT1 and 6 | Higher expressions are associated with poor prognosis and more advanced disease stage. | N/A | N/A | (Wang et al. 2011) |
| HDAC6, 7, 10 and SIRT2, 3, 6 | Overexpression of HDAC7 and 10 and underexpression of HDAC6 and SIRT3 are correlated with a poor prognosis. | N/A | N/A | (Van Damme et al. 2012) | |
| AML | HDAC5, 6, SIRT1 and 4. | HDAC6 and SIRT1 are overexpressed, and HDAC5 and SIRT4 are underexpressed in AML samples. | N/A | N/A | (Bradbury et al. 2005) |
| DLBCL | HDAC1 | Highly expressed in cases of DLBCL and correlated with a poor survival. | N/A | N/A | (Min et al. 2012) |
| HDAC2 | Highly expressed in nodal lymphomas, which is associated with shorter survival. | N/A | N/A | (Lee et al. 2014) | |
| HDAC1, 2, 6 | The expression is higher in cases of DLBCL or PTCL. High HDAC6 level is associated with favorable outcome in DLBCL, but with a negative outcome in PTCL. | N/A | N/A | (Marquard et al. 2009) | |
| HDAC3 | Overexpression is observed in phospho STAT3-positive ABC-type DLBCL. | HDAC3 knockdown inhibited survival of pSTAT3-positive DLBCL cells. | HDAC3 knockdown unregulated STAT3Lys685 acetylation but prevented STAT3Tyr705 Phosphorylation | (Gupta et al. 2012) | |
| CTCL | HDAC2, 6 | HDAC2 is highly expressed in aggressive rather than indolent CTCL. HDAC6 is associated with a favorable outcome independent of the subtype. | N/A | N/A | (Marquard et al. 2008) |
| HL | HDAC1, 2, 3 | Overexpressed in HL tissue samples. High HDAC1 expression is correlated with a worse outcome. | N/A | N/A | (Adams et al. 2010) |
| Myeloma | HDAC1 | Overexpression of class I HDAC, particularly HDAC1, is associated with poor prognosis in myeloma. | N/A | N/A | (Mithraprabhu et al. 2014) |
Human hepatocellular carcinoma (HCC), Pancreatic ductal adenocarcinoma (PDAC), Acute lymphoblastic leukemia (ALL), Chronic lymphocytic leukemia (CLL), Acute myelogenous leukemia (AML), Diffuse large B-cell lymphoma (DLBCL), Cutaneous T-cell lymphomas (CTCL), Hodgkin’s lymphoma (HL).
The mechanisms by which individual HDACs regulate tumorigenesis are quite diverse. Since HDACs induce a range of cellular and molecular effects through hyperacetylation of histone and non-histone substrates, HDACs could either repress tumor suppressor gene expression or regulate the oncogenic cell-signaling pathway via modification of key molecules. However, the contribution of HDACs to cancer may not necessarily be related to the level of HDAC expression, because aberrant activity of HDACs is also common in cancer development (West and Johnstone 2014). Certain HDACs function as the catalytic subunits of large corepressor complexes and could be aberrantly recruited to target genes by oncogenic proteins to drive tumorigenesis. For example, the aberrant recruitment of HDAC1, 2 or 3 by oncogenic fusion proteins AML1-ETO and PML-RAR contributes to the pathogenesis of acute myeloid leukemia (AML) (Hug and Lazar 2004; Falkenberg and Johnstone 2014).
While the broad anticancer effects of HDACi predict an oncogenic role of HDACs in tumor development, in some cancers it has been found that genetic inactivation of HDACs might have tumorigenic effects. HDAC1 somatic mutations were detected in 8.3% of dedifferentiated liposarcoma and HDAC4 homozygous deletions occurred in 4% of melanomas (Stark and Hayward 2007; Taylor et al. 2011). Truncating mutations of HDAC2 have been observed in human epithelial cancers with microsatellite instability, which causes a loss of HDAC2 protein expression and confers cells more resistant to HDACi (Ropero et al. 2006). Furthermore, the ectopic expression of HDAC2 in mutant cancer cells induces a reduction of tumor cell growth in vitro and in vivo, suggesting a putative tumor-suppressor role for HDAC2 in this cellular setting. Class II HDACs may also function as tumor suppressor in certain circumstances. Low expression of HDAC10 is associated with poor prognosis in lung and gastric cancer patients (Osada et al. 2004; Jin et al. 2014). HDAC6 is downregulated in human hepatocellular carcinoma (HCC) tissues, and low expression of HDAC6 is associated with poor prognosis in liver transplantation patients. Knockdown of HDAC6 promotes angiogenesis in HCC by HIF-1α/VEGFA axis (Lv et al. 2015). Also, a recent study revealed a dual role of HDAC1 in cancer initiation and maintenance. HDAC1 antagonizes the oncogenic activity of PML-RAR during the preleukemic stage of acute promyelocytic leukemia (APL), but favors the growth of APL cells at the leukemic stage (Santoro et al. 2013), indicating that elucidation of the role of HDACs at each step of tumorigenesis in different tumor cell types will provide a rationale for targeting HDACs in cancer therapy.
2. Possible mechanisms of HDACs in cancer development
Although identification of substrate specificity and biological function for individual HDACs still requires more comprehensive investigations, it is well known that HDACs play crucial roles in cancer by deacetylating histone and nonhistone proteins which are involved in the regulation of cell cycle, apoptosis, DNA damage response, metastasis, angiogenesis, autophagy and other cellular processes (Fig. 1).
2.1 Cell cycle
HDAC inhibition has been shown to have antiproliferative effects by inducing cell cycle arrest in G1 via upregulation of cyclin-dependent kinase (CDK) inhibitors or downregulation of cyclins and CDKs (Chun 2015). HDAC1 and 2 directly bind to the promoters of the p21WAF1/CIP1, p27KIP1, and p57KIP2 genes and negatively regulate their expression (Yamaguchi et al. 2010; Zupkovitz et al. 2010). Loss of HDAC1 and 2 induces expression of CDK inhibitors, leading to a cell cycle block in G1. Knockdown of HDAC5 leads to a significant up-regulation of p21 and down-regulation of cyclin D1 and CDK2/4/6, which results in G1-phase cell cycle arrest in human HCC cells (Fan et al. 2014). HDAC inhibition might block the cellular G1/S transition by reactivating Rb function by dephosphorylation and subsequently inhibiting E2F activities in the transcription of genes for G1 progression. Trichostatin A (TSA) suppressed retinal pigment epithelium (RPE) cell proliferation via a G1 phase arrest, caused through inhibition of Rb phosphorylation, reduction of cyclinD1/CDK4/6 complexes, and induction of p21 and p27 (Xiao et al. 2014). However, TSA also induced G1 phase arrest in malignant tumor cells with mutated Rb, indicating a Rb-independent G1 arrest (Tomosugi et al. 2012).
In addition to controlling the G1/S transition, HDAC1 knockdown in tumor cells impairs G2/M transition and inhibits cell growth as evidenced by a reduction of mitotic cells and an increased percentage of apoptotic cells (Senese et al. 2007). HDAC10 regulates the G2/M transition via modulation of cyclin A2 expression. The effect of HDAC10 on cyclin A2 transcription was dependent on let-7 and HMGA2 (Li et al. 2015b). Consistent with gene knockdown results, inhibition of HDACs by inhibitors including TSA, SAHA and VPA cause cell cycle arrest at the G2/M boundary in a variety of tumor cell lines, supporting pleiotropic roles of HDAC throughout the cell cycle (Juengel et al. 2014).
Besides transcriptional repression of cell cycle-related genes at the G1/S and G2/M cell cycle checkpoints, HDACs might also regulate cell cycle progression in a transcription-independent manner. During mitosis, A-Kinase-Anchoring Proteins AKAP95 and HA95 recruits HDAC3 along with Aurora B. In this context, HDAC3 deacetylates histone H3, which in turn allows maximal phosphorylation of Ser10 by Aurora B, leading to HP1β dissociation from mitotic chromosomes. The HDAC3-AKAP95/HA95-Aurora B pathway is required for normal mitotic progression (Li et al. 2006). Indeed, the histone deacetylase inhibitor LBH589 can induce G2-M cell cycle arrest and apoptosis in renal cancer cells through degradation of Aurora A and B kinases by targeting HDAC3 and HDAC6 (Cha et al. 2009). Moreover, HDAC3 directly interacts with cyclin A and regulates cyclin A stability by modulating its acetylation status. An abrupt loss of HDAC3 at metaphase facilitates cyclin A acetylation by PCAF/GCN5, which targets cyclin A for degradation. Given that cyclin A is crucial for S phase progression and entry into mitosis, HDAC3 knockdown causes cell accumulation in S and G2/M phases (Vidal-Laliena et al. 2013). Collectively, HDAC inhibition can arrest the cell cycle at either G1/S or G2/M phase, suggesting HDACs as therapeutic targets for abnormal cell growth and proliferation in cancer.
2.2 Apoptosis
HDACs have been shown to regulate apoptosis in a variety of cancer cells through changing expression of pro- and anti-apoptotic proteins. Treatment of tumor cells with HDACi can either directly activate apoptosis through the extrinsic (death receptor)/intrinsic (mitochondria) pathway, or enhance the susceptibility of tumor cells to apoptosis. The extrinsic apoptosis pathway induced by HDACi is via diverse mechanisms including upregulation of cell surface death receptors and/or ligands including FAS/APO1-FASL, TNF-TNF receptors and TRAIL-TRAIL receptors, reductions in the level of cytoplasmic FLICE-like inhibitory protein (c-FLIP), and enhanced recruitment of DISC formation (Zhang and Zhong 2014). Two non-selective HDACi, vorinostat and panobinostat, have been identified as modulators of FLIP expression in several pre-clinical cancer models (Bangert et al. 2012). Inhibition of HDAC1, 2 and/or 3, but not HDAC6, is necessary for efficient FLIP downregulation and caspase-8 activation in NSCLC. HDACi sensitize cancer cells for TRAIL-induced apoptosis in a FLIP- and caspase-8-dependent manner (Riley et al. 2013). Depletion of HDAC2 also synergizes pancreatic cancer cells towards TRAIL-induced apoptosis with an increased expression of TRAIL receptor DR5 (TRAIL-R2) (Schuler et al. 2010).
The intrinsic cell death pathway involves the interplay of the pro- and anti-apoptotic Bcl-2 superfamily of proteins, and HDAC inhibition could induce the intrinsic pathway by decreasing the expression of anti-apoptotic proteins, and increasing the expression of pro-apoptotic proteins (Zhang and Zhong 2014). HDAC2 depletion results in regression of tumor cell growth and activation of apoptosis via p53 and Bax activation and Bcl2 suppression in human lung cancer cells (Jung et al. 2012). In gastric cancer cells, HDAC2 knockdown selectively induced the expression of proapoptotic factors Bax, AIF, and Apaf-1, but repressed the expression of anti-apoptotic Bcl-2 (Kim et al. 2013). HDAC1 and HDAC8 cooperate to repress BMF (Zhang et al. 2006; Kang et al. 2014). Inhibition of HDAC8 by methylselenopyruvate (MSP), a competitive inhibitor of HDAC8, was sufficient to activate BMF transcription and promote BMF-mediated apoptosis in colon cancer cells (Kang et al. 2014). HDAC3 downregulates PUMA expression in gastric cancer cells and HDACi, like TSA, promotes PUMA expression through enhancing the binding of p53 to the PUMA promoter (Feng et al. 2013). TSA treatment can also effectively overcome resistance to DNA damage–induced cell death by reactivating PUMA expression in renal cell carcinoma cells (Zhou et al. 2014).
The relevance of p53 in HDACi-induced apoptosis is controversial. HDACi can activate p53, but does not necessarily require p53 for induction of anticancer action (Sonnemann et al. 2014). Most studies point to a p53-independent action of HDACi because the anticancer effect of HDACi is not influenced by the tumor’s p53 status (Ellis et al. 2009). Other studies, however, suggest an essential role of p53 in the response of tumor cells to HDACi treatment (Bajbouj et al. 2012). Using isogenic HCT-116 colon cancer cell lines with different p53 status, the antitumor effects of vorinostat, apicidin and VPA were largely independent of p53, while entinostat-induced cell death partially depends on p53 (Sonnemann et al. 2014), indicating HDACi may regulate apoptotic process via both p53-dependent and independent pathways.
2.3 DNA damage response
Numerous studies have shown that HDACs have important roles in DNA damage repair (DDR) responses since HDACs are critical in modulating chromatin remodeling and maintaining dynamic acetylation equilibrium of DNA damage-related proteins (Li and Zhu 2014). HDAC1 and HDAC2 are recruited to DNA damage sites to deacetylate histones H3K56 and H4K16, and facilitate non-homologous end-joining (NHEJ) (Miller et al. 2010), suggesting a direct role for these two enzymes during DNA replication and double-strand break (DSB) repair. HDAC3 is also associated with DNA damage control, although it is not localized to DSB DNA damage sites (Miller et al. 2010). Inactivation of HDAC3 causes genomic instability, and deletion of HDAC3 in the liver leads to hepatocellular carcinoma (Bhaskara et al. 2010). Besides altering histone acetylation status, Class I HDACs also regulate other proteins involved in the DNA damage response, including ATR, ATM, BRCA1 and FUS (Thurn et al. 2013). HDAC inhibition can repress DSB repair and render cancer cells more susceptible to ionizing radiation (IR) and DNA-damaging agents-induced cell death (Koprinarova et al. 2011).
Among the Class II HDACs, HDAC4, HDAC6, HDAC9 and HDAC10, have each been implicated in DNA damage repair processes. HDAC4 co-localizes with 53BP to nuclear foci after DSB. Depletion of HDAC4 reduces 53BP1 expression and abrogates the DNA damage-induced G2 checkpoint (Kao et al. 2003). HDAC9 and HDAC10 are reported to be required for homologous recombination (HR). Depletion of HDAC9 or HDAC10 inhibits HR and sensitizes cells to mitomycin C treatment (Kotian et al. 2011). The mismatch repair (MMR) system recognizes DNA mismatches that occur during DNA replication or recombination, and corrects these defects to maintain genomic integrity. MutS protein homolog 2 (MSH2), a key DNA mismatch repair protein, is regulated by Class IIb HDACs. HDAC6 sequentially deacetylates and ubiquitinates MSH2, causing a cellular tolerance to DNA damage and decreased cellular DNA mismatch repair activities by downregulation of MSH2 (Zhang et al. 2014). However, the deacetylation of MSH2 by HDAC10 might promote DNA mismatch repair activity (Radhakrishnan et al. 2015).
SIRT1 is a critical component of the DNA damage response pathway which regulates multiple steps of DDR, including damage sensing, signal transduction, DNA repair, and apoptosis (Gorospe and de Cabo 2008). SIRT1 interacts with and deacetylates several DDR proteins, including Ku70, NBS1, APE1, XPA, PARP-1, TopBP1, and KAP1 (Luna et al. 2013; Li and Zhu 2014; Wang et al. 2014; Lin et al. 2015). SIRT1 antagonizes p53 acetylation and facilitates cancer cells survival after DNA damage (Luo et al. 2001; Vaziri et al. 2001), making SIRT1 a promising target in cancer therapy. However, SIRT1 also plays an essential role in maintaining genome integrity and stability (Wang et al. 2008; Palacios et al. 2010), so the challenge still remains to modulate SIRT1 function in such a manner that it will be beneficial for cancer therapy. Recent research demonstrated that the role of SIRT1 in response to DNA damage requires posttranslational modifications such as site-specific phosphorylation and ubiquitination. HIPK2 interacts and phosphorylates SIRT1 at Ser682 after DNA damage. Phosphorylation of SIRT1 inhibits SIRT1 deacetylase activity on p53 which in turn potentiates apoptotic p53 target gene expression and DNA damage-induced apoptosis (Conrad et al. 2015). SIRT1 is also ubiquitinated by MDM2 during DDR, and ubiquitination of SIRT1 affects its function in cell death and survival in response to DNA damage (Peng et al. 2015).
SIRT6, also important in DNA repair, was first found to suppress genomic instability by regulating base excision DNA repair (BER) (Mostoslavsky et al. 2006). Recent studies have demonstrated that SIRT6 is involved in homologous recombination (HR) by deacetylating C-terminal binding protein (CtBP) and interacting protein (CtIP) (Kaidi et al. 2010). SIRT6 is rapidly recruited to DNA damage sites and stimulates DSB repair by mono-ADP-ribosylation of PARP1(Mao et al. 2011). SIRT6 also recruits the ISWI-chromatin remodeler SNF2H to DSBs, and deacetylates histone H3K56, preventing genomic instability through chromatin remodeling (Toiber et al. 2013). Together, results from these studies suggest that at least some of the Class III sirtuins (Sir2 proteins) have equally critical role in DNA damage response compared to the classical HDACs.
2.4 Metastasis
Epithelial-to-mesenchymal transition (EMT) is a major process in cancer cell invasion and metastasis, and emerging studies have demonstrated the key role of HDACs in EMT regulation in a variety of cancer contexts. EMT is characterized by the loss of epithelial cell markers, namely epithelial-cadherin (CDH1), and several transcriptional repressors of CDH1 have been identified, including Snail, Slug, Twist, ZEB1 and ZEB2. A mechanism of their action involves recruitment of HDACs to the CDH1 promoter resulting in deacetylation of H3 and H4 histones. Snail recruits HDAC1/2 and Sin3A complex to the CDH1 promoter for histone deacetylation, and the snail/HDAC1/HDAC2 repressor complex contributes to CDH1 silencing in the metastasis of pancreatic cancer (von Burstin et al. 2009). Moreover, the complex of snail/HDAC1/HDAC2 is required for EZH2-mediated CDH1 repression in nasopharyngeal carcinoma cells (Tong et al. 2012). Recruitment of HDACs to the CHD1 promoter is also regulated by ZEB1 in human pancreatic cancer cells (Aghdassi et al. 2012). Given that ZEB1 could also alter the splicing of CDH1 exon 11, a recent study revealed a dual effect of ZEB1 and its interacting Class I HDACs; the decrease in CDH1 is due to a combination of transcriptional inhibition and aberrant splicing (Liao et al. 2013). Treatment of cells with the HDACs inhibitor, LBH589 (panobinostat), induces CDH1 expression, and represses EMT and metastasis in triple-negative breast cancer (TNBC) cells. Besides preventing ZEB-mediated repression of CDH1 by inhibiting the Class I HDACs corepressors, the effect of LBH589 is partially mediated by inhibition of ZEB expression (Rhodes et al. 2014). The expression of ZEB1 is also reduced in pancreatic cancer cells after mocetinostat treatment that sensitizes the undifferentiated, ZEB1-expressing cancer cells for chemotherapy (Meidhof et al. 2015). These findings indicate the therapeutic potential of inhibition of Class I HDACs in targeting EMT and metastasis of cancer cells.
The role of SIRT1 in EMT regulation depends on the tumor type. In prostate cancer cells, SIRT1 induces cell migration in vitro and metastasis in vivo by cooperating with ZEB1 to suppress CDH1 transcription (Byles et al. 2012). A recent study reveals a vital role of MPP8-SIRT1 interaction in CDH1 silencing (Sun et al. 2015). The deacetylation of MPP8 at K439 by SIRT1 increases MPP8 protein stability, and MPP8 in turn facilitates SIRT1 recruitment to the CDH1 promoter for H4K16 deacetylation by regulating SIRT1-ZEB1 interaction. Thus disruption of MPP8-SIRT1 interaction de-represses CDH1 expression and reduces cell motility and invasiveness in prostate cancer cells (Sun et al. 2015). In breast cancer, SIRT1 overexpression is associated with decreased miR-200a. miR-200a negatively regulated SIRT1 expression and reduced EMT (Eades et al. 2011). miR-204 also inhibits EMT in gastric cancer and osteosarcoma cells by directly targeting and repressing SIRT1 at the post-transcriptional level (Shi et al. 2015). However, an opposite role of SIRT1 in cancer metastasis was indicated by the demonstration that SIRT1 reduces EMT in breast epithelial cells by deacetylating Smad4 and repressing the effect of TGF-β signaling on MMP7 (Simic et al. 2013). A similar mechanism is also found in oral squamous cell carcinoma (OSCC) cells, suggesting the key role of the SIRT1/Smad4/MMP7 pathway in EMT process. (Chen et al. 2014).
2.5 Angiogenesis
Tumor growth and metastasis depend on angiogenesis. Angiogenesis is triggered by hypoxia or hypoxic microenvironment, and the cellular response to hypoxia is primarily regulated by the transcription factor hypoxia-inducible factors 1 alpha (HIF-1α). Many HDACs are associated with HIF-1α activity as cell treatment with HDACi causes HIF-1α degradation and functional repression. HDAC1 and HDAC4 directly deacetylate HIF-1α and block degradation of the protein (Yoo et al. 2006; Geng et al. 2011). Instead of regulating HIF-1α acetylation, HDAC5 and HDAC6 facilitate HIF-1α maturation and stabilization by deacetylating its chaperones, HSP70 and HSP90 (Kong et al. 2006; Chen et al. 2015). Inhibition of HDAC5 and 6 results in hyperacetylation of these chaperones, accumulation of the immature HIF-1α complex, and degradation of HIF-1α by the 20S proteasome. HDAC4, HDAC5 and HDAC7 increased transcriptional activity of HIF-1α by promoting its association with p300 (Kato et al. 2004; Seo et al. 2009). In contrast, SIRT1-mediated deacetylation of HIF-1α at Lys674 inhibits HIF-1α activity by blocking p300 recruitment. The suppression of SIRT1 under hypoxic conditions provides a positive feedback loop that maintains a high level of HIF-1 activity (Lim et al. 2010).
Although the anti-angiogenic activity of HDAC inhibition has been demonstrated to be associated with decreased expression of pro-angiogenic genes, the specific effect of individual HDAC enzymes on angiogenic gene expression is controversial. KLF-4 recruits HDAC2 and HDAC3 at the VEGF promoter and represses its transcription. The upregulation of VEGF in cancer is associated with loss of KLF-4-HDAC-mediated transcriptional repression (Ray et al. 2013). HDAC5 is another negative regulator of angiogenesis by repressing proangiogenic gene expression, such as FGF2 or Slit2, in endothelial cells (Urbich et al. 2009). Recent studies demonstrated a dual role of HDAC6 in angiogenesis. HDAC6 promotes angiogenesis by deacetylating cortactin in endothelial cells, thereby regulating endothelial cell migration and sprouting (Kaluza et al. 2011). Another study reported the anti-angiogenic effect of HDAC6 in HCC, as depletion of HDAC6 facilitates angiogenesis by upregulating the expression of HIF-1α and VEGFA (Lv et al. 2015). HDAC7 is crucial in maintaining vascular integrity and endothelial angiogenic functions, such as tube formation, migration and proliferation (Turtoi et al. 2012). HDAC9 positively regulates endothelial cell sprouting and vascular growth by the repression of the miR-17–92 cluster, which reduces the expression of proangiogenic proteins (Kaluza et al. 2013). Taken together, HDACs play important roles in angiogenesis by modulating a multitude of pro- and anti-angiogenic factors, indicating that they are potential targets for anti-angiogenesis in cancer therapy.
2.6 Autophagy
The role of autophagy in cancer is complex. The failure to properly modulate autophagy in response to oncogenic stresses has been implicated both positively and negatively in tumorigenesis. On the one hand, autophagy functions as a surveillance mechanism to remove damaged organelles and cellular components, which might prevent normal cells from transforming to tumor cells. So the loss of autophagy proteins appears to promote cancer development. On the other hand, for established tumors, autophagy can help cancer cell survival under conditions of metabolic stress, and it might also confer resistance to anticancer therapies (Zhi and Zhong 2015). Consistent with the dual role of autophagy in cancer, many HDAC family members show both pro- and anti-autophagy activities (Koeneke et al. 2015). Depletion or inhibition of HDAC1 is reported to induce autophagy by promoting accumulation of the autophagosomal marker LC3-II (Xie et al. 2012). However, in mouse models, deletion of both HDAC1 and HDAC2 in skeletal muscle blocks autophagy flux (Moresi et al. 2012). Recent research indicates that the oncogenic role of Class IIa HDAC4 and HDAC5 in cancer cells would be derived at least partially via decreasing autophagic flux, but the detailed mechanism needs further investigations.
The key role of HDAC6 in autophagy was first established through the observation that HDAC6 provide an essential link between autophagy and the ubiquitin proteasome system (UPS) in neurodegenerative diseases. When the UPS is impaired, autophagy is strongly activated and acts as a compensatory degradation system in an HDAC6-dependent manner (Pandey et al. 2007). In HDAC6 knockout mouse embryonic fibroblasts (MEFs), HDAC6 appears to be important for ubiquitin-selective quality control (QC) autophagy, but not starvation-induced autophagy (Lee et al. 2010a). A similar mechanism is observed in mitophagy, a selective degradation of mitochondria. The parkin-mediated mitochondrial ubiquitination recruits HDAC6 and p62, which assemble the autophagy machinery and lead to mitochondrial clearance (Lee et al. 2010b). Besides Ub-based selective autophagy, HDAC6 is associated with autophagic clearance of IFN-induced ISG15-conjugated proteins. HDAC6 and p62 independently bind ISG15 and facilitate the autophagosome/lysosome degradation of ISG15 conjugates (Nakashima et al. 2015). The role of HDAC6 in the fusion event is to control acetylation of salt-inducible kinase 2 (SIK2). HDAC6-mediated deacetylation activates SIK2 kinase activity and promotes autophagosome processing (Yang et al. 2013). Although HDAC6 is dispensible for starvation-induced autophagy in MEFs (Lee et al. 2010a), another study demonstrated that HDAC6 is involved in this non-selective degradation by deacetylating LC3-II in HeLa cells (Liu et al. 2013). The acetylation level of LC3B–II is decreased upon serum deprivation and HDAC6 is at least partially responsible for deacetylating LC3-II. In neuroblastoma, depletion and inhibition of HDAC10 disables efficient autophagosome/lysosome fusion and interrupts autophagic flux, resulting in an increase of sensitization to cytotoxic drug treatment (Oehme et al. 2013). The deacetylation of Hsp70 protein families by HDAC10 might contribute to autophagy-mediated cell survival (Oehme et al. 2013). Overall, Class IIb HDACs seem to mainly regulate autophagic flux at the level of autophagosome-autolysosome fusion via deacetylation of cytoplasmic proteins (Koeneke et al. 2015).
Sirtuins also participate in regulating autophagy. Sirt1 activity is necessary for the induction of starvation-induced autophagy by directly deacetylating critical regulators of the autophagy machinery, incuding Atg5, Atg7, Atg8 and LC3 (Lee et al. 2008; Huang et al. 2015a). In embryonic stem cells (ESCs), SIRT1 mediates oxidative stress-induced autophagy at least in part by PI3K/Beclin 1 and mTOR pathways (Ou et al. 2014). SIRT1 and the PI3K/Akt/mTOR pathway are also found to be related to Plumbagin (PLB)-induced autophagy in prostate cancer cells (Zhou et al. 2015). SIRT3, a mitochondrial deacetylase, evokes mitophagy under oxidative stress or starvation conditions (Tseng et al. 2013; Webster et al. 2013). SIRT3 is found to confer cytoprotective effects by activating anti-oxidant defenses and mitophagy under mitochondrial proteotoxic stress and reestablishing homeostasis in breast cancer cells (Papa and Germain 2014). Two recent reports suggest a connection between autophagy with SIRT5 and SIRT6. SIRT5 reduces ammonia-induced autophagy and mitophagy by regulating glutamine metabolism (Papa and Germain 2014). SIRT6 is involved in autophagy activation during cigarette smoke-induced cellular senescence via attenuation of IGF-Akt-mTOR signaling (Takasaka et al. 2014). Collectively, a better understanding of the context-dependent effects of individual HDACs enzymes on autophagic process will give us an advantage to treat cancers by exploiting this area in a specifically-targeted manner.
3. Anticancer effect of HDAC inhibitors
The availability of HDACi has not only accelerated our understanding of HDAC functions and mechanism of actions, but also presented a promising new class of compounds for cancer treatment. To date, numerous synthetic or natural molecules that target Classes I, II and IV enzymes have been developed and characterized, although interest in the Class III Sirtuin family is increasing. Here we only describe the potential role of classical HDACi in cancer therapy. Since classical HDACs display Zn2+-dependent deacetylase activity, the binding of HDACi to the Zn2+ ion, which resides at the active site of HDACs, interferes with the activity of HDACs, thereby inhibiting their enzymatic function. On the bases of chemical structures, HDACi are classified into four groups, including hydroxamates, benzamides, short-chain fatty acids and cyclic peptides (Table 2). Most of these molecules have been developed as anticancer agents with varying specificity and efficiency, pharmacokinetic properties, and toxicological characteristics.
Table 2.
HDAC inhibitors currently under clinical investigations.
| HDACis | Specificity | Cancer Types | Clinical Trial | Reference |
|---|---|---|---|---|
| Hydroxamic acid | ||||
| Vorinostat (SAHA) | Classes I, II and IV | CTCL | FDA Approved in 2006 | (Mann et al. 2007) |
| Belinostat (Beleodaq/PXD101) | Classes I, II and IV | PTCL | FDA approved in 2014 | (McDermott and Jimeno 2014) |
| Panobinostat (LBH-589) | Classes I, II and IV | MM | FDA approved in 2015 | (Richardson et al. 2015) |
| Resminostat (4SC-201) | Classes I and II | Advanced colorectal and hepatocellular carcinoma; HL | Phase II trial | (Brunetto et al. 2013; Zhao and Lawless 2015) |
| Givinostat (ITF2357) | Classes I and II | CLL; MM; HL | Phase II trial | (Galli et al. 2010; Locatelli et al. 2014) |
| Pracinostat (SB939) | Classes I, II and IV | AML | Phase II trial | (Zorzi et al. 2013) |
| Abexinostat (PCI-24781) | Classes I and II | Metastatic solid tumors; HL; non-HL; CLL | Phase I trial | (Choy et al. 2015; Morschhauser et al. 2015) |
| Quisinostat (JNJ-26481585) | Class I and II HDACs | Advanced solid tumor; lymphoma; CTCL | Phase I and II trial | (Venugopal et al. 2013) |
| MPT0E028 | HDAC1, 2, 6 | Advanced solid tumor | Phase I trial | (Zwergel et al. 2015) |
| CHR-3996 | Class I | Solid tumor | Phase I trial | (Banerji et al. 2012) |
| CUDC-101 | Classes I and II HDAC,EGFR, HER2 | Solid tumor | Phase I trial | (Shimizu et al. 2015) |
| CUDC-907 | Classes I and II HDAC, PI3K | MM; lymphoma; solid tumor | Phase I trial | (Qian et al. 2012) |
| Benzamides | ||||
| Entinostat (MS-275) | Class I | Solid and hematological malignancies | Phase I and II trial | (Knipstein and Gore 2011) |
| Mocetinostat (MGCD0103) | Class I and IV | Solid and hematological malignancies | Phase I and II trial | (Younes et al. 2011) |
| Tacedinaline (CI-994) | Class I | MM; lung and pancreatic cancer | Phase II and III trial | (Pauer et al. 2004) |
| Ricolinostat (ACY-1215) | HDAC6 | MM; lymphoma | Phase I and II trial | (Santo et al. 2012) |
| Chidamide (CS055/HBI-8000) | HDAC1, 2, 3, and 10 | Breast cancer; NSCLC | Phase II and III trial | (Dong et al. 2012; Shi et al. 2015) |
| Cyclic peptides | ||||
| Romidepsin (Depsipeptide/FK228) | Class I | CTCL; PTCL | FDA approved in 2009 and 2011 | (Frye et al. 2012) |
| Aliphatic fatty acids | ||||
| Valproic acid (VPA) | Class I and II | Solid and hematological malignancies | Phase I and II trial | (Bilen et al. 2015) |
| Phenylbutyrate | Classes I and II | Solid and hematological malignancies | Phase I and II trial | (Iannitti and Palmieri 2011) |
| AR-42 | Class I and IIb | AML | Phase I trial | (Guzman et al. 2014) |
| Pivanex (AN-9) | Classes I and II | NSCLC; myeloma; CLL | Phase II trial | (Reid et al. 2004) |
AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CTCL, cutaneous T cell lymphoma; HL, Hodgkin’s lymphoma; MM, multiple myeloma; NSCLC, non-small-cell lung cancer; PTCL, peripheral T cell lymphoma.
The rationale for targeting HDACs in cancer therapy is that altered HDAC expression and/or function is frequently observed in a variety of cancer types. The disrupted acetylation homeostasis in cells might contribute to tumorigenesis, and the nature of reversible modulation by HDACs makes them attractive targets for cancer treatment. HDACs reversibly modify the acetylation status histones and non-histones and cause widespread changes in genes expression without a change in DNA sequence. HDACi can counteract the abnormal acetylation status of proteins found in cancer cells and can reactivate the expression of tumor suppressors, resulting in induction of cell cycle arrest, apoptosis, differentiation, and inhibition of angiogenesis and metastasis (Fig. 1). Furthermore, cancer cells are more sensitive to HDACi-induced apoptosis than normal cells (Ungerstedt et al. 2005), providing additional therapeutic potential of HDACi.
Currently, there are numerous HDACi under clinical development (Table 2), which can be divided into three groups based on their specificity: (1) nonselective HDACi, such as vorinostat, belinostat, and panobinostat; (2) selective HDACi, such as Class I HDACi (romidepsin and entinostat) and HDAC6 inhibitor (ricolinostat); and (3) multi-pharmacological HDACi, such as CUDC 101 and CUDC 907.
3.1 Non-selective Broad Spectrum HDAC inhibitors
The most extensively studied and commonly used HDACi are non-selective HDACi. For example, vorinostat (SAHA), a hydroxamate class agent, was the first histone deacetylase inhibitor approved by the Food and Drug Administration (FDA) to treat patients with cutaneous T-cell lymphoma (CTCL). Like TSA, SAHA inhibits all zinc-dependent HDACs in low nanomolar ranges, although more recent studies suggest that most hydroxamate-based HDACi have a weak effect on Class IIa enzymes (Bradner et al. 2010). Preclinical studies have demonstrated that SAHA induces apoptosis and cell cycle arrest, and reduces the proliferation and metastatic potential of tumor cells. SAHA also sensitized tumor cells to chemotherapy and/or radiotherapy (Shi et al. 2014; Xue et al. 2015). In addition to SAHA, two hydroxamate-based non-selective HDACi, belinostat (Beleodaq/PXD101) and panobinostat (LBH-589), were recently approved by the FDA to treat peripheral T-cell lymphomas (PTCL) and multiple myeloma, respectively. Both of these drugs are also under investigation in combination therapies for solid tumors.
3.2 Selective HDAC inhibitors
To date, there are relatively few highly selective HDACi, but compounds with proposed selectivity for several Class I and Class II HDACs have been developed (Table 3).
Table 3.
Specific HDAC inhibitors in cancer therapy.
| HDACis | Specificity | Cancer Types | Stage | References |
|---|---|---|---|---|
| Class I | ||||
| Romidepsin (Depsipeptide/FK228) | HDAC1, 2 | CTCL and PTCL | FDA approved | (Furumai et al. 2002; Frye et al. 2012) |
| BRD8430 | HDAC1, 2 | Neuroblastoma | Preclinical | (Frumm et al. 2013) |
| Compound 60 | HDAC1, 2 | Neuroblastoma | Preclinical | (Methot et al. 2008; Frumm et al. 2013; Schroeder et al. 2013) |
| MRLB-223 | HDAC1, 2 | Lymphomas | Preclinical | (Newbold et al. 2013) |
| Entinostat (MS-275) | HDAC1, 2, 3 | Multiple cancer cells | Clinical trial | (Hu et al. 2003; Khan et al. 2008; Knipstein and Gore 2011) |
| CHR-3996 | HDAC1, 2, 3 | Multiple cancer cells | Clinical trial | (Moffat et al. 2010; Banerji et al. 2012) |
| Tacedinaline (CI-994) | HDAC1, 2, 3 | Multiple myeloma; lung and pancreatic cancer | Clinical trial | (Kraker et al. 2003; Pauer et al. 2004) |
| Apicidin | HDAC1, 2, 3 | Multiple cancer cells | Preclinical | (Olsen, 2012 #1236; Ahn, 2015 #3269) |
| RGFP966 | HDAC3 | CTCL | Preclinical | (Wells et al. 2013) |
| BG45 | HDAC3 | Myeloma | Preclinical | (Minami et al. 2014) |
| T247 and T326 | HDAC3 | Colon and prostate cancer | Preclinical | (Suzuki et al. 2013) |
| PCI-34051 | HDAC8 | Lymphoma, neuroblastoma | Preclinical | (Balasubramanian et al. 2008; Oehme et al. 2009; Rettig et al. 2015) |
| Compound 2 (Cpd2) | HDAC8 | Neuroblastoma | Preclinical | (Krennhrubec et al. 2007; Oehme et al. 2009; Rettig et al. 2015) |
| C149 (NCC149) | HDAC8 | Multiple cancer cells | Preclinical | (Suzuki et al. 2012; Suzuki et al. 2014) |
| Compound 22 d | HDAC8 | Lung cancer | Preclinical | (Huang et al. 2012) |
| Class IIa | ||||
| TMP269 | HDAC4, 5, 7, 9 | Multiple myeloma | Preclinical | (Lobera et al. 2013; Kikuchi et al. 2015) |
| MC1568 | HDAC4, 5, 6, 7, 9 | Gastric, colorectal, pancreatic and breast cancer | Preclinical | (Mai et al. 2005; Duong et al. 2008; Wang et al. 2012; Colarossi et al. 2014; Ishikawa et al. 2014) |
| LMK235 | HDAC4, 5 | Multiple cancer cells | Preclinical | (Marek et al. 2013) |
| Class II b | ||||
| Ricolinostat (ACY-1215) | HDAC6 | Multiple myeloma, lymphoma, glioblastoma | Clinical trial | (Santo et al. 2012; Amengual et al. 2015; Li et al. 2015a; Mishima et al. 2015) |
| Tubacin | HDAC6 | Multiple cancer cells | Preclinical | (Haggarty et al. 2003; Aldana-Masangkay et al. 2011) |
| Tubastatin A | HDAC6 | Multiple cancer cells | Preclinical | (Butler et al. 2010) |
| C1A | HDAC6 | Multiple cancer cells | Preclinical | (Kaliszczak et al. 2013) |
| HPOB | HDAC6 | Multiple cancer cells | Preclinical | (Lee et al. 2013) |
| Nexturastat A (Compound 5g) | HDAC6 | Melanoma | Preclinical | (Bergman et al. 2012) |
| Compound 12 | HDAC6 | Colorectal cancer | Preclinical | (Lee et al. 2014) |
| Befexamac | HDAC6, 10 | Neuroblastoma, lung cancer | Preclinical | (Bantscheff et al. 2011; Oehme et al. 2013; Li et al. 2015b; Scholz et al. 2015) |
CTCL, cutaneous T cell lymphoma; PTCL, peripheral T cell lymphoma.
3.2.1 Class I HDACi
HDAC1 and HDAC2
Romidepsin (FK-228), the second histone deacetylase inhibitor approved for the treatment of CTCL and PTCL, exhibits a stronger inhibition towards HDAC1 and HDAC2 enzymes at low nanomolar levels (Furumai et al. 2002). Its antitumor efficacy has been demonstrated in different cancer models (McGraw 2013; Karthik et al. 2014). BRD8430, compound 60 and MRLB-223 are three novel HDAC1 and HDAC2 inhibitors under preclinical studies. The selective inhibition of HDAC1 and HDAC2 by BRD8430 and compound 60 induced differentiation and cell death in neuroblastoma cells, and synergistically activated retinoic acid signaling in combination treatment with 13-cis retinoic acid (Frumm et al. 2013). MRLB-223 induced tumor cell death via the intrinsic apoptotic pathway in a p53-independent manner. However, MRLB-223 had less effect on induction of apoptosis and therapeutic efficacy as seen using the broad-spectrum histone deacetylase inhibitor vorinostat (Newbold et al. 2013).
HDAC3
RGFP966 is an N-(o-aminophenyl) carboxamide HDAC3-selective inhibitor (Malvaez et al. 2013). RGFP966 decreased growth and increased apoptosis of refractory CTCL cells by targeting DNA replication (Wells et al. 2013). Consistent with previous research demonstrating the contribution of HDAC3 to the effects of SAHA on DNA replication (Conti et al. 2010), HDAC3 inhibition by RGFP966 reduces DNA replication fork velocity and causes replication stress in CTCL cells (Wells et al. 2013).
A recent preclinical study demonstrated that HDAC3 represents a promising therapeutic target in multiple myeloma (MM) (Minami et al. 2014). HDAC3 inhibition by BG45, a HDAC3-selective inhibitor, induces significant apoptosis in MM cells, without affecting normal donor PBMCs. BG45-induced MM cell toxicity might be associated with hyperacetylation and hypophosphorylation of STAT3. HDAC3 inhibition, but not HDAC1 or HDAC2, significantly enhances bortezomib-induced cell death in vitro and in vivo, providing the preclinical rationale for combination treatment of MM with HDAC3 and proteasome inhibitors.
T247 and T326 are identified as HDAC3-selective inhibitors by screening a series of compounds assembled using “click chemistry” (Suzuki et al. 2013). In cell-based assays, T247 and T326 selectively enhance the acetylation of NF-kB, a substrate of HDAC3, but did not regulate HDAC1 and HDAC6 substrates, suggesting they are HDAC3-selective inhibitors. T247 and T326 inhibited the growth of colon and prostate cancer cells (Suzuki et al. 2013). In TMEM16A–expressing cancer cells, T247 also exerts a suppressive effect on cancer cell viability via downregulatingTMEM16A (Matsuba et al. 2014).
HDAC8
HDAC8 has proven to be the most promising target to achieve selectivity. The unique features of HDAC8, such as conformational variability of the L1 and L2 loop segments (Dowling et al. 2008) and the presence of serine 39 phosphorylation near the active site (Lee et al. 2004), led to the design of higher selective inhibitors for HDAC8.
Modifications to the hydroxamic acid scaffold resulted in the discovery of PCI-34051, a potent HDAC8-specific inhibitor with a >200-fold selectivity over other HDACs. It induces caspase-dependent apoptosis in T-cell-derived malignant cells, but not in a panel of solid tumor cell lines or other hematopoietic cells. Mechanistically, PLCγ1-dependent calcium mobilization from the endoplasmic reticulum (ER) and in turn release of cytochrome c from mitochondria might contribute to PCI-34051-induced cell death (Balasubramanian et al. 2008). Besides T-cell leukemia and lymphoma, human and murine-derived malignant peripheral nerve sheath tumors (MPNST) cells also exhibited “sensitivity” to HDAC8 inhibitors: PCI-34051 and its variant PCI-48012 (Lopez et al. 2015). HDAC8 inhibition induced S-phase cell cycle arrest and apoptosis in MPNST cells, but the underlying mechanism remains unclear. Given that high HDAC8 expression is significantly correlated with advanced stage and poor outcome in neuroblastoma (Oehme et al. 2009), HDAC8 inhibition by selective inhibitors, Compound 2 (Cpd2) and PCI-34051, induced cell differentiation, cell cycle arrest and cell death in neuroblastoma cells, while untransformed cells were not affected (Rettig et al. 2015). PCI-48012, an in vivo stable variant of PCI-34051 with improved pharmacokinetic properties, displayed a significant antitumor activity without toxicity in xenograft mouse models. PCI-48012 in combination with retinoic acid further enhanced differentiation in neuroblastoma cells and delayed tumor growth in vivo (Rettig et al. 2015).
3.2.2 Class IIa HDACi
In contrast to Class I HDACs, much less is known about the molecular mechanisms and therapeutic potential of targeting Class IIa HDACs (HDAC4, 5, 7, and 9), and there is a lack of pharmacological tools to specifically probe Class IIa HDAC activities (Lobera et al. 2013). A high-throughput screen (HTS) identified trifluoromethyloxadiazole (TMFO) derivatives as inhibitors selective for Class IIa HDACs. Although TMP269, a compound in the TFMO series, has a modest growth inhibitory effect in multiple myeloma (MM) cell lines, it enhances protease inhibitor carfilzomib-induced apoptosis by activating ER stress signaling (Kikuchi et al. 2015), providing the combination of the inhibition of both proteasome and Class IIa HDACs as a novel treatment strategy in MM.
MC1568 and MC1575 are derivatives of aroyl-pyrrolyl–hydroxyamides (APHAs), showing selectivity towards Class IIa HDACs and HDAC6 (Mai et al. 2005; Fleming et al. 2014). Although Class IIa HDACs are mainly involved in tissue-specific growth and differentiation, rather than in cell proliferation, MC1568 and MC1575 treatment still displayed anti-proliferative effects in estrogen receptor (ER)–positive breast cancer cells (Duong et al. 2008) as well as human melanoma cells (Venza et al. 2013). MC1568 significantly enhanced MGCD0103-induced apoptosis and G2/M arrest in pancreatic cancer cells (Wang et al. 2012). The additional treatment with MC1568 to simvastatin led to further induction of p27 expression and displayed a considerable synergistic anti-proliferative effect in colorectal cancer cells (Ishikawa et al. 2014). MC1568 also had a synergistic effect with docetaxel treatment to increase cytotoxicity in gastric cancer cells (Colarossi et al. 2014).
Another new hydroxamate-based histone deacetylase inhibitor, LMK235, showed high selectivity for HDAC4 and HDAC5 (Marek et al. 2013). Compared with SAHA, LMK235 is less toxic and more suitable for the treatment of some cancers. Consistent with a recent study where silencing of HDAC4 was able to sensitize ovarian cancer cells to cisplatin (Stronach et al. 2011), the combination of LMK235 with cisplatin enhanced cisplatin sensitivity in resistant cells (Marek et al. 2013).
YK-4–272 and tasquinimod are two novel unconventional HDACi, which either target HDAC nuclear-cytoplasmic shuttling or alter the interaction of Class IIa HDACs with their partners. YK-4–272 represses the growth of human prostate cancer cells in vitro and in vivo (Kong et al. 2012). YK-4–272 binds HDAC4, and the localization of YK-4–272 in the cytoplasm traps and sequestrates HDAC4 in cytoplasm, resulting in increased acetylation of tubulin and nuclear histones in prostate cancer cells. However SAHA treatment also causes an accumulation of HDAC4 in cytoplasm similar to YK-4–272, which suggests the possibility that cytoplasmic restriction of Class II HDACs is an indirect effect of Class I inhibition. So far, the cytoplasmic functions of Class IIa HDACs is not well known and this uncertain function could be amplified by inhibition of HDAC nuclear transport, limiting the use of the HDAC shuttling inhibitor.
3.2.3 Class IIb HDACi
HDAC6 inhibition has been intensively studied and a number of HDAC6-selective inhibitors are developed, such as tubacin and tubastatin A; however, their poor pharmacokinetic properties prevented them from further clinical development. Among HDAC6-specific inhibitors available, ricolinostat (ACY-1215) was the first one entered in clinical studies of patients with relapsed/refractory multiple myeloma or lymphoma (Santo et al. 2012; Amengual et al. 2015). In multiple myeloma, the highly secretory antibody-producing cells are heavily reliant on protein handling pathways including the unfolded protein response (UPR), proteasome, aggresome and autophagy pathways. So targeting both of the proteasome and aggresome degradation pathways by proteasome and HDAC6 inhibitors, respectively, induces accumulation of polyubiquitinated proteins, followed by activation of apoptotic cascades and synergistic cytotoxicity. ACY-1215 in combination with bortezomib triggered synergistic anti-MM activity without significant adverse effects (Santo et al. 2012), and similar anti-MM effects were obtained by combination treatment of ACY-1215 with another proteasome inhibitor, carfilzomib (Mishima et al. 2015). Besides haematological tumors, recent research indicated that ACY-1215 also significantly inhibited glioblastoma multiforme (GBM) cell growth (Li et al. 2015a). C1A is another HDAC6-selective inhibitor, which modulates HDAC6 downstream targets (α-tubulin and HSP90) and induces growth inhibition of a panel of cancer cell lines. To date, HDAC10-specific inhibitors are not yet available. Like other selective HDAC inhibitors, development of HDAC10-selective inhibitors might help clarify the function and mechanism of action of HDAC10, and potentially provide additional anti-cancer drugs.
3.3 Multi-pharmacological HDAC inhibitors
Tumor heterogeneity requires a comprehensive approach to target multiple pathways underlying the initiation and progression of cancers. To enhance the therapeutic efficacy of HDACi, the combination with other anticancer agents have been explored and evaluated in preclinical and clinical studies. Another promising approach is to generate a single chemical compound that acts on multiple targets. CUDC-101, with a potent inhibitory activity against EGFR, HER2 and HDACs, is currently being evaluated in clinical trials as a treatment for advanced solid tumors, such as head and neck, gastric, breast, liver and non-small cell lung cancer tumors (Cai et al. 2010; Galloway et al. 2015). Recent research also indicates the antitumor effect of CUDC-101 in EGFR-overexpressing glioblastoma and anaplastic thyroid cancer (Liffers et al. 2015; Zhang et al. 2015a). CUDC-907 is another dual-acting agent developed by the same research group to inhibit both HDACs and phosphoinositide 3-kinase (PI3K) (Qian et al. 2012) and its clinical trials are underway for the treatment of lymphoma and multiple myeloma as well as advanced/relapsed solid tumors. Romidepsin (FK228, depsipeptide) is a potent Class I histone deacetylase inhibitor that has FDA approval for the treatment of cutaneous and peripheral T-cell lymphomas, and recent research demonstrated that FK228 and its analogs (FK-A5 and FK-A11) act as HDACs and PI3K dual inhibitors (Saijo et al. 2012; Saijo et al. 2015).
Numerous chemical hybrid molecules containing both HDACi activities and an additional anticancer module are under development, dual targeting HDACs and estrogen receptor (ER) (Tang et al. 2015), retinoid X receptor (RXR) (Wang et al. 2015), topoisomerase I/II (Guerrant et al. 2013), 1α, 25- vitamin D (Lamblin et al. 2010), oestrogen receptor (Gryder et al. 2013), receptor tyrosine kinases (RTK) (Zhang et al. 2013), tubulin (Zhang et al. 2015b) or DNA methyltransferase (Shukla et al. 2015), potentially leading to a rational efficacy in cancer therapy. Additionally, the hybrid of a nitric oxide (NO) donor and an histone deacetylase inhibitor has been developed and displayed outstanding antiproliferative activity in tumor cells (Duan et al. 2015).
4. Clinical landscape of HDAC inhibitors in cancer therapy
After Vorinostat (SAHA) was approved to treat CTCL in 2006, the other three HDACi, romidepsin, belinostat and panobinostat, have since been approved by the FDA for the treatment of cancer. Currently, more than 20 different HDACi are in different phases of clinical trials as single agents or in combination with chemotherapy or radiation therapy in patients with hematologic or solid tumors.
The efficacy of HDACi tested in clinicial trials has been largely restricted to hematological malignancies, with positive therapeutic responses in leukemias, lymphomas and multiple myeloma; however, the clinical outcomes in solid tumors are disappointing when used as monotherapy. It is not entirely clear why HDACi are more effective in hematological malignancies. One reason might be the poor pharmacokinetic properties of some HDACi, such as a short drug half-life that restricts them to distribute to solid tumors. Selective and accurate drug delivery of HDACi may help to overcome the issues associated with inefficient bioavailability. For example, HDACi conjugated to folic and pteroic acids selectively targets folate receptor (FR)-overexpressed solid tumors (Sodji et al. 2015). The other reason might be that HDACi do not target solid tumors. Identifying those cancers or patients where HDAC deregulation is important for tumor development might contribute to rational cancer therapy.
Another obstacle that limits the use of HDACi in patients is their side effects and toxicity displayed during early-phase clinical trials. The common toxicities related to vorinostat, romidepsin, and belinostat were nausea, vomiting, anorexia, and fatigue that are mostly manageable, but some may cause more serious adverse events. In general, acute toxicity of non-selective HDACi is mainly through HDAC1–3 inhibition, so these compounds from unrelated chemical classes have a similar toxicity profile. HDACi have a broad effect on chromatin and can reverse the aberrant epigenetic changes in cancers. However, while the inhibition of HDACs may re-activate some tumor suppressors, they can also affect numerous other genes (Guha 2015). Although the second-generation of HDACi have been developed with improved pharmacodynamic and pharmacokinetic values, given that these new agents possess similar specificity profiles as their parental compounds, it is unclear whether these newer agents will have improved and less toxic clinical outcomes. Currently, major efforts in therapeutic strategies are focused on developing selective inhibitors and studying combination therapies, with the aim of increasing potency against specific cancer types and overcoming drug toxicity and resistance.
HDACi are continuously explored for used in combination with other antitumor agents in order to optimize their efficacy and toxicity. Combining HDACi with primary chemotherapeutic agents that induce DNA damage or apoptosis has shown very promising results in preclinical research studies. HDAC inhibition might re-sensitize tumor cells to the primary agents and overcome therapy resistance. For example, hypoxia-induced cisplatin resistance in NSCLC can be overcome by combining cisplatin with panobinostat by increasing histone-acetylation and destabilization of HIF-1α (Fischer et al. 2015). ERCC1 and p53 were reported to have a predictive role for the efficacy of combined panobinostat and cisplatin treatment (Fischer et al. 2015). HDAC inhibitions could also overcome resistance to mTOR inhibitors (e.g., everolimus, temsirolimus, sirolimus and ridaforolimus) in advanced solid tumors or lymphoma (Dong et al. 2013; Beagle et al. 2015; Zibelman et al. 2015).
Given that cross talk exists between DNA methylation and histone deacetylation in gene expression, a combination of HDACi and DNA methyltransferases (DNMTs) have been shown to produce a synergistic effect on reactivation of tumor-suppressor genes and represent a promising future therapeutic approach. Large Phase I and II trials are currently underway to assess the efficacy of two chromatin-modifying agents, azacitidine and entinostat, for the treatment of chronic myelomonocytic leukemia, acute myeloid leukemia, NSCLC, advanced breast cancer and metastatic colorectal cancer (Juergens et al. 2011; Prebet et al. 2014). Recent research demonstrated that combined MS-275 and azacitidine treatment is more efficient and selectively targeted esophageal cancer cells by inducing DNA damage, cell viability loss, apoptosis and decreasing cell migration (Ahrens et al. 2015).
Preclinical studies also indicate a synergistic antitumor effect of HDACi with other epigenetic-targeted drugs, such as lysine-specific histone demethylase inhibitors (Vasilatos et al. 2013; Fiskus et al. 2014). These observations are consistent with recent findings that broad-acting HDAC inhibitors have minimal effect on promoter acetylation, but rather they promote H3K27 trimethylation, a silencing-associated histone modification (Halsall et al. 2015). These and other studies on the basic mechanisms of HDACs, HDACi, and their relationships with other histone modifications will no doubt guide the choice of future combination therapies.
Similarly, preclinical evidence from studies of HDACi together with proteasome inhibitors (e.g., bortezomib, carfilzomib and marizomib) provides a strong scientific rationale for combination therapy. Given that HDAC6 facilitates misfolded protein aggresome formation for proteosome-independent proteolysis, dual targeting of HDAC6 and proteasomes can produce synergistic effects in lymphoma and multiple myeloma (Amengual et al. 2015; Mishima et al. 2015). However, the combination of proteasome and Class I HDAC-specific inhibitors in nasopharyngeal carcinoma cells induced a significant apoptosis through an ROS-dependent and ER stress-induced mechanism, independent of HDAC6 inhibition (Hui and Chiang 2014). Because vorinostat and bortezomib are both FDA-approved drugs for the treatment of CTCL and multiple myeloma, respectively, the combination of these two agents has been tested in a variety of preclinical models and in clinical trials. Recent research explored the synergistic effect of vorinostat and bortezomib on host immune response and found co-treatment of HPV-expressing cervical cancer cells with bortezomib and vorinostat led to a tumor-specific immunity by rendering tumor cells more susceptible to killing by antigen-specific CD8+ T cells, suggesting that activated host immune surveillance contributes to antitumor effects (Huang et al. 2015b).
HDACi have also been evaluated in combination with a hormone antagonist for the treatment of patients whose tumors express hormone receptors. Three Phase II clinical trials are currently carried out with vorinostat and tamoxifen for the treatment of breast cancer (Munster et al. 2011). Although histone deacetylation plays a key role in estrogen receptor (ER) gene silencing, it remains unclear whether the addition of HDACi actually reactivate functional ERα expression (de Cremoux et al. 2015). A recent study demonstrates that Bcl-2 downregulation and induction of pro-apoptotic proteins by combined ER and HDAC inhibition leads to apoptotic cell death of tamoxifen-resistant cells (Raha et al. 2015).
HDACi have been shown to enhance the immunogenicity of cancer cells (Murakami et al. 2008; Christiansen et al. 2011; Jazirehi et al. 2014), and the antitumor efficacy of HDACi in vivo also relies on an intact immune system (West et al. 2013, 2014), so the combination of HDACi with immunotherapy is a promising strategy for the treatment of cancer. The efficacy of HDACi can be significantly enhanced by the concurrent administration of various immunotherapeutic approaches, such as cancer vaccines, adoptive T-cell transfer and immune checkpoint inhibitors (Park et al. 2015). For example, co-administration of HDACi with antibodies against cytotoxic T-lymphocyte antigen 4 (CTLA4) could further enhance the infiltration of CD4+ T cells and achieve a synergistic therapeutic effect on tumors by promoting antitumor immune responses (Cao et al. 2015). A recent preclinical study indicated that HDACi in combination with immunomodulatory drugs, such as lenalidomide and pomalidomide, showed a synergistic cytotoxicity in multiple myeloma by downregulating c-Myc expression. A Phase I trial is currently underway to assess the effect of ACY-241, a next generation selective inhibitor of HDAC6, with and without pomalidomide and low-dose dexamethasone for treatment of multiple myeloma. Another Phase II clinical trial evaluated the Class I histone deacetylase inhibitor romidepsin in combination with lenalidomide in patients with peripheral T-cell lymphoma.
There is also substantial evidence that HDACi such as vorinostat enhance radiation sensitization by inhibiting DNA damage repair, inducing apoptosis, inhibiting proliferation and angiogenesis, and enhancing immune surveillance for cancer.
Summary and Perspective
Studies over the past few decades have demonstrated that HDACs play a critical role in the development of cancer by reversibly modulating acetylation status of histone and non-histone proteins. As an eraser of histone acetylation and a key regulator of epigenetics, HDACs have been found to dysregulate and/or function incorrectly in cancer, providing a crucial attractive target against cancer. However, the precise function of HDACs as a central mediator of proliferation and tumorigenic capacity still remains a conundrum. Although genetic knockdown or knockout of HDACs in a variety of cancer cells induces cell cycle arrest and apoptosis, a putative tumor suppressor role of HDACs is also observed in certain circumstances. More studies are needed to systematically dissect the role of individual HDACs in different cancer types at different stages of tumorigenesis. Clearly, the development of HDACi, in particular selective inhibitors, could help clarify the function of distinct HDACs, and a better comprehension of HDACs in cancer will give us a mechanistic-based rationale for the clinical use of HDACi as antitumor agents. So far, the most common HDACi under preclinical and clinical evaluation are broad spectrum non-selective HDACi. The effectiveness of non-selective HDACi for the treatment of cancer relies on its broad-spectrum inhibition against HDACs, which is also the major reason for toxicity of these agents. Therefore, current emphasis is placed on developing HDACi with higher target specificity that might be more efficacious with less toxicity. In parallel, research is increasingly showing that combination therapy might be another important direction to enhance the therapeutic efficacy of HDACi. Further elucidation of the mechanisms of action of HDACs and HDACi will provide a bright future for the use of HDACi as one of many tools in the fight against cancer.
Acknowledgments
This work was supported in part by NIH grants R01CA169210 and R01CA187040 to E.S.
References
- Adams H, Fritzsche FR, Dirnhofer S, Kristiansen G, Tzankov A. Class I histone deacetylases 1, 2 and 3 are highly expressed in classical Hodgkin’s lymphoma. Expert Opin Ther Targets. 2010;14:577–584. doi: 10.1517/14728221003796609. [DOI] [PubMed] [Google Scholar]
- Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, Friess H, Buchler M, Evert M, Lerch MM, et al. Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut. 2012;61:439–448. doi: 10.1136/gutjnl-2011-300060. [DOI] [PubMed] [Google Scholar]
- Ahrens TD, Timme S, Hoeppner J, Ostendorp J, Hembach S, Follo M, Hopt UT, Werner M, Busch H, Boerries M, et al. Selective inhibition of esophageal cancer cells by combination of HDAC inhibitors and Azacytidine. Epigenetics. 2015;10:431–445. doi: 10.1080/15592294.2015.1039216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldana-Masangkay GI, Rodriguez-Gonzalez A, Lin T, Ikeda AK, Hsieh YT, Kim YM, Lomenick B, Okemoto K, Landaw EM, Wang D, et al. Tubacin suppresses proliferation and induces apoptosis of acute lymphoblastic leukemia cells. Leuk Lymphoma. 2011;52:1544–1555. doi: 10.3109/10428194.2011.570821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amengual JE, Johannet PM, Lombardo M, Zullo KM, Hoehn D, Bhagat G, Scotto L, Jirau-Serrano X, Radeski D, Heinen J, et al. Dual targeting of protein degradation pathways with the selective HDAC6 inhibitor, ACY-1215, and bortezomib is synergistic in lymphoma. Clin Cancer Res. 2015;21:4663–4675. doi: 10.1158/1078-0432.CCR-14-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajbouj K, Mawrin C, Hartig R, Schulze-Luehrmann J, Wilisch-Neumann A, Roessner A, Schneider-Stock R. P53-dependent antiproliferative and pro-apoptotic effects of trichostatin A (TSA) in glioblastoma cells. J Neurooncol. 2012;107:503–516. doi: 10.1007/s11060-011-0791-2. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- Banerji U, van Doorn L, Papadatos-Pastos D, Kristeleit R, Debnam P, Tall M, Stewart A, Raynaud F, Garrett MD, Toal M, et al. A phase I pharmacokinetic and pharmacodynamic study of CHR-3996, an oral class I selective histone deacetylase inhibitor in refractory solid tumors. Clin Cancer Res. 2012;18:2687–2694. doi: 10.1158/1078-0432.CCR-11-3165. [DOI] [PubMed] [Google Scholar]
- Bangert A, Cristofanon S, Eckhardt I, Abhari BA, Kolodziej S, Hacker S, Vellanki SH, Lausen J, Debatin KM, Fulda S. Histone deacetylase inhibitors sensitize glioblastoma cells to TRAIL-induced apoptosis by c-myc-mediated downregulation of cFLIP. Oncogene. 2012;31:4677–4688. doi: 10.1038/onc.2011.614. [DOI] [PubMed] [Google Scholar]
- Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- Beagle BR, Nguyen DM, Mallya S, Tang SS, Lu M, Zeng Z, Konopleva M, Vo TT, Fruman DA. mTOR kinase inhibitors synergize with histone deacetylase inhibitors to kill B-cell acute lymphoblastic leukemia cells. Oncotarget. 2015;6:2088–2100. doi: 10.18632/oncotarget.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman JA, Woan K, Perez-Villarroel P, Villagra A, Sotomayor EM, Kozikowski AP. Selective histone deacetylase 6 inhibitors bearing substituted urea linkers inhibit melanoma cell growth. J Med Chem. 2012;55:9891–9899. doi: 10.1021/jm301098e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilen MA, Fu S, Falchook GS, Ng CS, Wheler JJ, Abdelrahim M, Erguvan-Dogan B, Hong DS, Tsimberidou AM, Kurzrock R, et al. Phase I trial of valproic acid and lenalidomide in patients with advanced cancer. Cancer Chemother Pharmacol. 2015;75:869–874. doi: 10.1007/s00280-015-2695-x. [DOI] [PubMed] [Google Scholar]
- Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, Craddock C, Turner BM. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetto AT, Ang JE, Lal R, Olmos D, Molife LR, Kristeleit R, Parker A, Casamayor I, Olaleye M, Mais A, et al. First-in-human, pharmacokinetic and pharmacodynamic phase I study of Resminostat, an oral histone deacetylase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2013;19:5494–5504. doi: 10.1158/1078-0432.CCR-13-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buurman R, Gurlevik E, Schaffer V, Eilers M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kuhnel F, Skawran B. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012;143:811–820. doi: 10.1053/j.gastro.2012.05.033. e811-815. [DOI] [PubMed] [Google Scholar]
- Byles V, Zhu L, Lovaas JD, Chmilewski LK, Wang J, Faller DV, Dai Y. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene. 2012;31:4619–4629. doi: 10.1038/onc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Zhai HX, Wang J, Forrester J, Qu H, Yin L, Lai CJ, Bao R, Qian C. Discovery of 7-(4-(3-ethynylphenylamino)-7-methoxyquinazolin-6-yloxy)-N-hydroxyheptanamide (CUDc-101) as a potent multi-acting HDAC, EGFR, and HER2 inhibitor for the treatment of cancer. J Med Chem. 2010;53:2000–2009. doi: 10.1021/jm901453q. [DOI] [PubMed] [Google Scholar]
- Cao K, Wang G, Li W, Zhang L, Wang R, Huang Y, Du L, Jiang J, Wu C, He X, et al. Histone deacetylase inhibitors prevent activation-induced cell death and promote anti-tumor immunity. Oncogene. 2015 doi: 10.1038/onc.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha TL, Chuang MJ, Wu ST, Sun GH, Chang SY, Yu DS, Huang SM, Huan SK, Cheng TC, Chen TT, et al. Dual degradation of aurora A and B kinases by the histone deacetylase inhibitor LBH589 induces G2-M arrest and apoptosis of renal cancer cells. Clin Cancer Res. 2009;15:840–850. doi: 10.1158/1078-0432.CCR-08-1918. [DOI] [PubMed] [Google Scholar]
- Chen IC, Chiang WF, Huang HH, Chen PF, Shen YY, Chiang HC. Role of SIRT1 in regulation of epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis. Mol Cancer. 2014;13:254. doi: 10.1186/1476-4598-13-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yin C, Lao T, Liang D, He D, Wang C, Sang N. AMPK-HDAC5 pathway facilitates nuclear accumulation of HIF-1alpha and functional activation of HIF-1 by deacetylating Hsp70 in the cytosol. Cell Cycle. 2015;14:2520–2536. doi: 10.1080/15384101.2015.1055426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Choy E, Flamand Y, Balasubramanian S, Butrynski JE, Harmon DC, George S, Cote GM, Wagner AJ, Morgan JA, Sirisawad M, et al. Phase 1 study of oral abexinostat, a histone deacetylase inhibitor, in combination with doxorubicin in patients with metastatic sarcoma. Cancer. 2015;121:1223–1230. doi: 10.1002/cncr.29175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen AJ, West A, Banks KM, Haynes NM, Teng MW, Smyth MJ, Johnstone RW. Eradication of solid tumors using histone deacetylase inhibitors combined with immune-stimulating antibodies. Proc Natl Acad Sci. 2011;108:4141–4146. doi: 10.1073/pnas.1011037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch Pharm Res. 2015;38:933–949. doi: 10.1007/s12272-015-0571-1. [DOI] [PubMed] [Google Scholar]
- Colarossi L, Memeo L, Colarossi C, Aiello E, Iuppa A, Espina V, Liotta L, Mueller C. Inhibition of histone deacetylase 4 increases cytotoxicity of docetaxel in gastric cancer cells. Proteomics Clin Appl. 2014;8:924–931. doi: 10.1002/prca.201400058. [DOI] [PubMed] [Google Scholar]
- Conrad E, Polonio-Vallon T, Meister M, Matt S, Bitomsky N, Herbel C, Liebl M, Greiner V, Kriznik B, Schumacher S, et al. HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ. 2015 doi: 10.1038/cdd.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, Aladjem MI, Pommier Y. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cremoux P, Dalvai M, N’Doye O, Moutahir F, Rolland G, Chouchane-Mlik O, Assayag F, Lehmann-Che J, Kraus-Berthie L, Nicolas A, et al. HDAC inhibition does not induce estrogen receptor in human triple-negative breast cancer cell lines and patient-derived xenografts. Breast Cancer Res Treat. 2015;149:81–89. doi: 10.1007/s10549-014-3233-y. [DOI] [PubMed] [Google Scholar]
- Dong LH, Cheng S, Zheng Z, Wang L, Shen Y, Shen ZX, Chen SJ, Zhao WL. Histone deacetylase inhibitor potentiated the ability of MTOR inhibitor to induce autophagic cell death in Burkitt leukemia/lymphoma. J Hematol Oncol. 2013;6:53. doi: 10.1186/1756-8722-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Ning ZQ, Xing PY, Xu JL, Cao HX, Dou GF, Meng ZY, Shi YK, Lu XP, Feng FY. Phase I study of chidamide (CS055/HBI-8000), a new histone deacetylase inhibitor, in patients with advanced solid tumors and lymphomas. Cancer Chemother Pharmacol. 2012;69:1413–1422. doi: 10.1007/s00280-012-1847-5. [DOI] [PubMed] [Google Scholar]
- Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Hou J, Chu X, Li X, Zhang J, Li J, Xu W, Zhang Y. Synthesis and biological evaluation of novel histone deacetylases inhibitors with nitric oxide releasing activity. Bioorg Med Chem. 2015 doi: 10.1016/j.bmc.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Duong V, Bret C, Altucci L, Mai A, Duraffourd C, Loubersac J, Harmand PO, Bonnet S, Valente S, Maudelonde T, et al. Specific activity of class II histone deacetylases in human breast cancer cells. Mol Cancer Res. 2008;6:1908–1919. doi: 10.1158/1541-7786.MCR-08-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286:25992–26002. doi: 10.1074/jbc.M111.229401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker J, Oehme I, Mazitschek R, Korshunov A, Kool M, Hielscher T, Kiss J, Selt F, Konrad C, Lodrini M, et al. Targeting class I histone deacetylase 2 in MYC amplified group 3 medulloblastoma. Acta Neuropathol Commun. 2015;3:22. doi: 10.1186/s40478-015-0201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L, Bots M, Lindemann RK, Bolden JE, Newbold A, Cluse LA, Scott CL, Strasser A, Atadja P, Lowe SW, et al. The histone deacetylase inhibitors LAQ824 and LBH589 do not require death receptor signaling or a functional apoptosome to mediate tumor cell death or therapeutic efficacy. Blood. 2009;114:380–393. doi: 10.1182/blood-2008-10-182758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- Fan J, Lou B, Chen W, Zhang J, Lin S, Lv FF, Chen Y. Down-regulation of HDAC5 inhibits growth of human hepatocellular carcinoma by induction of apoptosis and cell cycle arrest. Tumour Biol. 2014;35:11523–11532. doi: 10.1007/s13277-014-2358-2. [DOI] [PubMed] [Google Scholar]
- Feng GW, Dong LD, Shang WJ, Pang XL, Li JF, Liu L, Wang Y. HDAC5 promotes cell proliferation in human hepatocellular carcinoma by up-regulating Six1 expression. Eur Rev Med Pharmacol Sci. 2014;18:811–816. [PubMed] [Google Scholar]
- Feng L, Pan M, Sun J, Lu H, Shen Q, Zhang S, Jiang T, Liu L, Jin W, Chen Y, et al. Histone deacetylase 3 inhibits expression of PUMA in gastric cancer cells. J Mol Med (Berl) 2013;91:49–58. doi: 10.1007/s00109-012-0932-x. [DOI] [PubMed] [Google Scholar]
- Fischer C, Leithner K, Wohlkoenig C, Quehenberger F, Bertsch A, Olschewski A, Olschewski H, Hrzenjak A. Panobinostat reduces hypoxia-induced cisplatin resistance of non-small cell lung carcinoma cells via HIF-1alpha destabilization. Mol Cancer. 2015;14:4. doi: 10.1186/1476-4598-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Shah B, Portier BP, Devaraj SG, Liu K, Iyer SP, Bearss D, Bhalla KN. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia. 2014;28:2155–2164. doi: 10.1038/leu.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming CL, Ashton TD, Gaur V, McGee SL, Pfeffer FM. Improved synthesis and structural reassignment of MC1568: A class IIa selective HDAC inhibitor. J Med Chem. 2014;57:1132–1135. doi: 10.1021/jm401945k. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Fritsche P, Seidler B, Schuler S, Schnieke A, Gottlicher M, Schmid RM, Saur D, Schneider G. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut. 2009;58:1399–1409. doi: 10.1136/gut.2009.180711. [DOI] [PubMed] [Google Scholar]
- Fritzsche FR, Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Scholman K, Denkert C, Dietel M, et al. Class I histone deacetylases 1, 2 and 3 are highly expressed in renal cell cancer. BMC Cancer. 2008;8:381. doi: 10.1186/1471-2407-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumm SM, Fan ZP, Ross KN, Duvall JR, Gupta S, VerPlank L, Suh BC, Holson E, Wagner FF, Smith WB, et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol. 2013;20:713–725. doi: 10.1016/j.chembiol.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R, Myers M, Axelrod KC, Ness EA, Piekarz RL, Bates SE, Booher S. Romidepsin: a new drug for the treatment of cutaneous T-cell lymphoma. Clin J Oncol Nurs. 2012;16:195–204. doi: 10.1188/12.CJON.195-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62:4916–4921. [PubMed] [Google Scholar]
- Galli M, Salmoiraghi S, Golay J, Gozzini A, Crippa C, Pescosta N, Rambaldi A. A phase II multiple dose clinical trial of histone deacetylase inhibitor ITF2357 in patients with relapsed or progressive multiple myeloma. Ann Hematol. 2010;89:185–190. doi: 10.1007/s00277-009-0793-8. [DOI] [PubMed] [Google Scholar]
- Galloway TJ, Wirth LJ, Colevas AD, Gilbert J, Bauman JE, Saba NF, Raben D, Mehra R, Ma AW, Atoyan R, et al. A Phase I Study of CUDC-101, a Multitarget Inhibitor of HDACs, EGFR, and HER2, in combination with chemoradiation in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2015;21:1566–1573. doi: 10.1158/1078-0432.CCR-14-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, Qian DZ. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286:38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M, de Cabo R. AsSIRTing the DNA damage response. Trends Cell Biol. 2008;18:77–83. doi: 10.1016/j.tcb.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn B, Naumann T, Gruner D, Walther M, Wittig S, Becker S, Beck JF, Sonnemann J. The expression of histone deacetylase 4 is associated with prednisone poor-response in childhood acute lymphoblastic leukemia. Leuk Res. 2013;37:1200–1207. doi: 10.1016/j.leukres.2013.07.016. [DOI] [PubMed] [Google Scholar]
- Gryder BE, Rood MK, Johnson KA, Patil V, Raftery ED, Yao LP, Rice M, Azizi B, Doyle DF, Oyelere AK. Histone deacetylase inhibitors equipped with estrogen receptor modulation activity. J Med Chem. 2013;56:5782–5796. doi: 10.1021/jm400467w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrant W, Patil V, Canzoneri JC, Yao LP, Hood R, Oyelere AK. Dual-acting histone deacetylase-topoisomerase I inhibitors. Bioorg Med Chem Lett. 2013;23:3283–3287. doi: 10.1016/j.bmcl.2013.03.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M. HDAC inhibitors still need a home run, despite recent approval. Nat Rev Drug Discov. 2015;14:225–226. doi: 10.1038/nrd4583. [DOI] [PubMed] [Google Scholar]
- Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia. 2012;26:1356–1364. doi: 10.1038/leu.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman ML, Yang N, Sharma KK, Balys M, Corbett CA, Jordan CT, Becker MW, Steidl U, Abdel-Wahab O, Levine RL, et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Mol Cancer Ther. 2014;13:1979–1990. doi: 10.1158/1535-7163.MCT-13-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsall JA, Turan N, Wiersma M, Turner BM. Cells adapt to the epigenomic disruption caused by histone deacetylase inhibitors through a coordinated, chromatin-mediated transcriptional response. Epigenetics Chromatin. 2015;8:29. doi: 10.1186/s13072-015-0021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, Horiuchi A, Kikuchi N, Hayashi T, Fuseya C, Suzuki A, Konishi I, Shiozawa T. Type-specific roles of histone deacetylase (HDAC) overexpression in ovarian carcinoma: HDAC1 enhances cell proliferation and HDAC3 stimulates cell migration with downregulation of E-cadherin. Int J Cancer. 2010;127:1332–1346. doi: 10.1002/ijc.25151. [DOI] [PubMed] [Google Scholar]
- Hu E, Dul E, Sung CM, Chen Z, Kirkpatrick R, Zhang GF, Johanson K, Liu R, Lago A, Hofmann G, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015a;57:456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Huang WJ, Wang YC, Chao SW, Yang CY, Chen LC, Lin MH, Hou WC, Chen MY, Lee TL, Yang P, et al. Synthesis and biological evaluation of ortho-aryl N-hydroxycinnamides as potent histone deacetylase (HDAC) 8 isoform-selective inhibitors. ChemMedChem. 2012;7:1815–1824. doi: 10.1002/cmdc.201200300. [DOI] [PubMed] [Google Scholar]
- Huang Z, Peng S, Knoff J, Lee SY, Yang B, Wu TC, Hung CF. Combination of proteasome and HDAC inhibitor enhances HPV16 E7-specific CD8+ T cell immune response and antitumor effects in a preclinical cervical cancer model. J Biomed Sci. 2015b;22:7. doi: 10.1186/s12929-014-0111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug BA, Lazar MA. ETO interacting proteins. Oncogene. 2004;23:4270–4274. doi: 10.1038/sj.onc.1207674. [DOI] [PubMed] [Google Scholar]
- Hui KF, Chiang AK. Combination of proteasome and class I HDAC inhibitors induces apoptosis of NPC cells through an HDAC6-independent ER stress-induced mechanism. Int J Cancer. 2014;135:2950–2961. doi: 10.1002/ijc.28924. [DOI] [PubMed] [Google Scholar]
- Iannitti T, Palmieri B. Clinical and experimental applications of sodium phenylbutyrate. Drugs R D. 2011;11:227–249. doi: 10.2165/11591280-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Hayashi H, Kinoshita K, Abe M, Kuroki H, Tokunaga R, Tomiyasu S, Tanaka H, Sugita H, Arita T, et al. Statins inhibit tumor progression via an enhancer of zeste homolog 2-mediated epigenetic alteration in colorectal cancer. Int J Cancer. 2014;135:2528–2536. doi: 10.1002/ijc.28672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazirehi AR, Kurdistani SK, Economou JS. Histone deacetylase inhibitor sensitizes apoptosis-resistant melanomas to cytotoxic human T lymphocytes through regulation of TRAIL/DR5 pathway. J Immunol. 2014;192:3981–3989. doi: 10.4049/jimmunol.1302532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Jiang W, Jiao F, Guo Z, Hu H, Wang L, Wang L. Decreased expression of histone deacetylase 10 predicts poor prognosis of gastric cancer patients. Int J Clin Exp Pathol. 2014;7:5872–5879. [PMC free article] [PubMed] [Google Scholar]
- Juengel E, Nowaz S, Makarevi J, Natsheh I, Werner I, Nelson K, Reiter M, Tsaur I, Mani J, Harder S, et al. HDAC-inhibition counteracts everolimus resistance in renal cell carcinoma in vitro by diminishing cdk2 and cyclin A. Mol Cancer. 2014;13:152. doi: 10.1186/1476-4598-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Xie HJ, Chang YG, Kim MG, Park H, Lee JY, et al. HDAC2 overexpression confers oncogenic potential to human lung cancer cells by deregulating expression of apoptosis and cell cycle proteins. J Cell Biochem. 2012;113:2167–2177. doi: 10.1002/jcb.24090. [DOI] [PubMed] [Google Scholar]
- Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kaliszczak M, Trousil S, Aberg O, Perumal M, Nguyen QD, Aboagye EO. A novel small molecule hydroxamate preferentially inhibits HDAC6 activity and tumour growth. Br J Cancer. 2013;108:342–350. doi: 10.1038/bjc.2012.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluza D, Kroll J, Gesierich S, Manavski Y, Boeckel JN, Doebele C, Zelent A, Rossig L, Zeiher AM, Augustin HG, et al. Histone deacetylase 9 promotes angiogenesis by targeting the antiangiogenic microRNA-17-92 cluster in endothelial cells. Arterioscler Thromb Vasc Biol. 2013;33:533–543. doi: 10.1161/ATVBAHA.112.300415. [DOI] [PubMed] [Google Scholar]
- Kaluza D, Kroll J, Gesierich S, Yao TP, Boon RA, Hergenreider E, Tjwa M, Rossig L, Seto E, Augustin HG, et al. Class IIb HDAC6 regulates endothelial cell migration and angiogenesis by deacetylation of cortactin. EMBO J. 2011;30:4142–4156. doi: 10.1038/emboj.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Nian H, Rajendran P, Kim E, Dashwood WM, Pinto JT, Boardman LA, Thibodeau SN, Limburg PJ, Lohr CV, et al. HDAC8 and STAT3 repress BMF gene activity in colon cancer cells. Cell Death Dis. 2014;5:e1476. doi: 10.1038/cddis.2014.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol. 2003;160:1017–1027. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik S, Sankar R, Varunkumar K, Ravikumar V. Romidepsin induces cell cycle arrest, apoptosis, histone hyperacetylation and reduces matrix metalloproteinases 2 and 9 expression in bortezomib sensitized non-small cell lung cancer cells. Biomed Pharmacother. 2014;68:327–334. doi: 10.1016/j.biopha.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Kato H, Tamamizu-Kato S, Shibasaki F. Histone deacetylase 7 associates with hypoxia-inducible factor 1alpha and increases transcriptional activity. J Biol Chem. 2004;279:41966–41974. doi: 10.1074/jbc.M406320200. [DOI] [PubMed] [Google Scholar]
- Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, Carey N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Suzuki R, Ohguchi H, Yoshida Y, Lu D, Cottini F, Jakubikova J, Bianchi G, Harada T, Gorgun G, et al. Class IIa HDAC inhibition enhances ER stress-mediated cell death in multiple myeloma. Leukemia. 2015;29:1918–1927. doi: 10.1038/leu.2015.83. [DOI] [PubMed] [Google Scholar]
- Kim JK, Noh JH, Eun JW, Jung KH, Bae HJ, Shen Q, Kim MG, Chang YG, Kim SJ, Park WS, et al. Targeted inactivation of HDAC2 restores p16INK4a activity and exerts antitumor effects on human gastric cancer. Mol Cancer Res. 2013;11:62–73. doi: 10.1158/1541-7786.MCR-12-0332. [DOI] [PubMed] [Google Scholar]
- Knipstein J, Gore L. Entinostat for treatment of solid tumors and hematologic malignancies. Expert Opin Investig Drugs. 2011;20:1455–1467. doi: 10.1517/13543784.2011.613822. [DOI] [PubMed] [Google Scholar]
- Koeneke E, Witt O, Oehme I. HDAC family members intertwined in the regulation of autophagy: A druggable vulnerability in aggressive tumor entities. Cells. 2015;4:135–168. doi: 10.3390/cells4020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HS, Tian S, Kong Y, Du G, Zhang L, Jung M, Dritschilo A, Brown ML. Preclinical studies of YK-4-272, an inhibitor of class II histone deacetylases by disruption of nucleocytoplasmic shuttling. Pharm Res. 2012;29:3373–3383. doi: 10.1007/s11095-012-0832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 2006;26:2019–2028. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprinarova M, Botev P, Russev G. Histone deacetylase inhibitor sodium butyrate enhances cellular radiosensitivity by inhibiting both DNA nonhomologous end joining and homologous recombination. DNA Repair (Amst) 2011;10:970–977. doi: 10.1016/j.dnarep.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Kotian S, Liyanarachchi S, Zelent A, Parvin JD. Histone deacetylases 9 and 10 are required for homologous recombination. J Biol Chem. 2011;286:7722–7726. doi: 10.1074/jbc.C110.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraker AJ, Mizzen CA, Hartl BG, Miin J, Allis CD, Merriman RL. Modulation of histone acetylation by [4-(acetylamino)-N-(2-amino-phenyl) benzamide] in HCT-8 colon carcinoma. Mol Cancer Ther. 2003;2:401–408. [PubMed] [Google Scholar]
- Krennhrubec K, Marshall BL, Hedglin M, Verdin E, Ulrich SM. Design and evaluation of ‘Linkerless’ hydroxamic acids as selective HDAC8 inhibitors. Bioorg Med Chem Lett. 2007;17:2874–2878. doi: 10.1016/j.bmcl.2007.02.064. [DOI] [PubMed] [Google Scholar]
- Lachenmayer A, Toffanin S, Cabellos L, Alsinet C, Hoshida Y, Villanueva A, Minguez B, Tsai HW, Ward SC, Thung S, et al. Combination therapy for hepatocellular carcinoma: additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J Hepatol. 2012;56:1343–1350. doi: 10.1016/j.jhep.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamblin M, Dabbas B, Spingarn R, Mendoza-Sanchez R, Wang TT, An BS, Huang DC, Kremer R, White JH, Gleason JL. Vitamin D receptor agonist/histone deacetylase inhibitor molecular hybrids. Bioorg Med Chem. 2010;18:4119–4137. doi: 10.1016/j.bmc.2010.03.078. [DOI] [PubMed] [Google Scholar]
- Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Tsai AC, Chen MC, Shen PJ, Cheng YC, Kuo CC, Pan SL, Liu YM, Liu JF, Yeh TK, et al. Azaindolylsulfonamides, with a more selective inhibitory effect on histone deacetylase 6 activity, exhibit antitumor activity in colorectal cancer HCT116 cells. J Med Chem. 2014;57:4009–4022. doi: 10.1021/jm401899x. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Mahendran A, Yao Y, Ngo L, Venta-Perez G, Choy ML, Kim N, Ham WS, Breslow R, Marks PA. Development of a histone deacetylase 6 inhibitor and its biological effects. Proc Natl Acad Sci. 2013;110:15704–15709. doi: 10.1073/pnas.1313893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010a;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010b;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yoo C, Im S, Jung JH, Choi HJ, Yoo J. Expression of histone deacetylases in diffuse large B-cell lymphoma and its clinical significance. Int J Med Sci. 2014;11:994–1000. doi: 10.7150/ijms.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Sun X, Zhang L, Yan B, Xie S, Liu R, Liu M, Zhou J. Histone deacetylase 6 and cytoplasmic linker protein 170 function together to regulate the motility of pancreatic cancer cells. Protein Cell. 2014;5:214–223. doi: 10.1007/s13238-013-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu X, Chen X, Zhang L, Wang X. Histone deacetylase 6 promotes growth of glioblastoma through inhibition of SMAD2 signaling. Tumour Biol. 2015a;36:9661–9665. doi: 10.1007/s13277-015-3747-x. [DOI] [PubMed] [Google Scholar]
- Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20:2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Peng L, Seto E. HDAC10 regulates cell cycle G2/M phase transition via a novel Let-7-HMGA2-Cyclin A2 pathway. Mol Cell Biol. 2015b;35:3547–3565. doi: 10.1128/MCB.00400-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhu WG. Targeting histone deacetylases for cancer therapy: from molecular mechanisms to clinical implications. Int J Biol Sci. 2014;10:757–770. doi: 10.7150/ijbs.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Jordaan G, Srivastava MK, Dubinett S, Sharma S, Sharma S. Effect of epigenetic histone modifications on E-cadherin splicing and expression in lung cancer. Am J Cancer Res. 2013;3:374–389. [PMC free article] [PubMed] [Google Scholar]
- Liffers K, Kolbe K, Westphal M, Lamszus K, Schulte A. Histone deacetylase inhibitors resensitize EGFR/EGFRvIII-overexpressing, Erlotinib-resistant glioblastoma cells to tyrosine kinase inhibition. Target Oncol. 2015;11:29–40. doi: 10.1007/s11523-015-0372-y. [DOI] [PubMed] [Google Scholar]
- Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Lin YH, Yuan J, Pei H, Liu T, Ann DK, Lou Z. KAP1 Deacetylation by SIRT1 Promotes Non-Homologous End-Joining Repair. PLoS One. 2015;10:e0123935. doi: 10.1371/journal.pone.0123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KP, Zhou D, Ouyang DY, Xu LH, Wang Y, Wang LX, Pan H, He XH. LC3B–II deacetylation by histone deacetylase 6 is involved in serum-starvation-induced autophagic degradation. Biochem Biophys Res Commun. 2013;441:970–975. doi: 10.1016/j.bbrc.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol. 2013;9:319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- Locatelli SL, Cleris L, Stirparo GG, Tartari S, Saba E, Pierdominici M, Malorni W, Carbone A, Anichini A, Carlo-Stella C. BIM upregulation and ROS-dependent necroptosis mediate the antitumor effects of the HDACi Givinostat and Sorafenib in Hodgkin lymphoma cell line xenografts. Leukemia. 2014;28:1861–1871. doi: 10.1038/leu.2014.81. [DOI] [PubMed] [Google Scholar]
- Lopez G, Bill KL, Bid HK, Braggio D, Constantino D, Prudner B, Zewdu A, Batte K, Lev D, Pollock RE. HDAC8, A potential therapeutic target for the treatment of malignant peripheral nerve sheath tumors (MPNST) PLoS One. 2015;10:e0133302. doi: 10.1371/journal.pone.0133302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Aladjem MI, Kohn KW. SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome Integr. 2013;4:6. doi: 10.1186/2041-9414-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Lv Z, Weng X, Du C, Zhang C, Xiao H, Cai X, Ye S, Cheng J, Ding C, Xie H, et al. Downregulation of HDAC6 promotes angiogenesis in hepatocellular carcinoma cells and predicts poor prognosis in liver transplantation patients. Mol Carcinog. 2015;55:1024–1033. doi: 10.1002/mc.22345. [DOI] [PubMed] [Google Scholar]
- Mai A, Massa S, Pezzi R, Simeoni S, Rotili D, Nebbioso A, Scognamiglio A, Altucci L, Loidl P, Brosch G. Class II (IIa)-selective histone deacetylase inhibitors. 1. Synthesis and biological evaluation of novel (aryloxopropenyl)pyrrolyl hydroxyamides. J Med Chem. 2005;48:3344–3353. doi: 10.1021/jm049002a. [DOI] [PubMed] [Google Scholar]
- Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek L, Hamacher A, Hansen FK, Kuna K, Gohlke H, Kassack MU, Kurz T. Histone deacetylase (HDAC) inhibitors with a novel connecting unit linker region reveal a selectivity profile for HDAC4 and HDAC5 with improved activity against chemoresistant cancer cells. J Med Chem. 2013;56:427–436. doi: 10.1021/jm301254q. [DOI] [PubMed] [Google Scholar]
- Marquard L, Gjerdrum LM, Christensen IJ, Jensen PB, Sehested M, Ralfkiaer E. Prognostic significance of the therapeutic targets histone deacetylase 1, 2, 6 and acetylated histone H4 in cutaneous T-cell lymphoma. Histopathology. 2008;53:267–277. doi: 10.1111/j.1365-2559.2008.03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquard L, Poulsen CB, Gjerdrum LM, de Nully Brown P, Christensen IJ, Jensen PB, Sehested M, Johansen P, Ralfkiaer E. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T-cell lymphomas. Histopathology. 2009;54:688–698. doi: 10.1111/j.1365-2559.2009.03290.x. [DOI] [PubMed] [Google Scholar]
- Matsuba S, Niwa S, Muraki K, Kanatsuka S, Nakazono Y, Hatano N, Fujii M, Zhan P, Suzuki T, Ohya S. Downregulation of Ca2+-activated Cl- channel TMEM16A by the inhibition of histone deacetylase in TMEM16A–expressing cancer cells. J Pharmacol Exp Ther. 2014;351:510–518. doi: 10.1124/jpet.114.217315. [DOI] [PubMed] [Google Scholar]
- McDermott J, Jimeno A. Belinostat for the treatment of peripheral T-cell lymphomas. Drugs Today (Barc) 2014;50:337–345. doi: 10.1358/dot.2014.50.5.2138703. [DOI] [PubMed] [Google Scholar]
- McGraw AL. Romidepsin for the treatment of T-cell lymphomas. Am J Health Syst Pharm. 2013;70:1115–1122. doi: 10.2146/ajhp120163. [DOI] [PubMed] [Google Scholar]
- Meidhof S, Brabletz S, Lehmann W, Preca BT, Mock K, Ruh M, Schuler J, Berthold M, Weber A, Burk U, et al. ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med. 2015;7:831–847. doi: 10.15252/emmm.201404396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE, Harrington P, et al. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2) Bioorg Med Chem Lett. 2008;18:973–978. doi: 10.1016/j.bmcl.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M, Taylor MD, von Deimling A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–3252. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SK, Koh YH, Park Y, Kim HJ, Seo J, Park HR, Cho SJ, Kim IS. Expression of HAT1 and HDAC1, 2, 3 in Diffuse Large B-Cell Lymphomas, Peripheral T-Cell Lymphomas, and NK/T-Cell Lymphomas. Korean J Pathol. 2012;46:142–150. doi: 10.4132/KoreanJPathol.2012.46.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami J, Suzuki R, Mazitschek R, Gorgun G, Ghosh B, Cirstea D, Hu Y, Mimura N, Ohguchi H, Cottini F, et al. Histone deacetylase 3 as a novel therapeutic target in multiple myeloma. Leukemia. 2014;28:680–689. doi: 10.1038/leu.2013.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamiya Y, Ono T, Saito H, Takahashi N, Ito M, Mitsui M, Motoyama S, Ogawa J. Expression of histone deacetylase 1 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Lung Cancer. 2011;74:300–304. doi: 10.1016/j.lungcan.2011.02.019. [DOI] [PubMed] [Google Scholar]
- Minamiya Y, Ono T, Saito H, Takahashi N, Ito M, Motoyama S, Ogawa J. Strong expression of HDAC3 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Tumour Biol. 2010;31:533–539. doi: 10.1007/s13277-010-0066-0. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Santo L, Eda H, Cirstea D, Nemani N, Yee AJ, O’Donnell E, Selig MK, Quayle SN, Arastu-Kapur S, et al. Ricolinostat (ACY-1215) induced inhibition of aggresome formation accelerates carfilzomib-induced multiple myeloma cell death. Br J Haematol. 2015;169:423–434. doi: 10.1111/bjh.13315. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Santo L, Eda H, Cirstea D, Nemani N, Yee AJ, O’Donnell E, Selig MK, Quayle SN, Arastu-Kapur S, et al. Ricolinostat (ACY-1215) induced inhibition of aggresome formation accelerates carfilzomib-induced multiple myeloma cell death. Br J Haematol. 2015;169:423–434. doi: 10.1111/bjh.13315. [DOI] [PubMed] [Google Scholar]
- Mithraprabhu S, Kalff A, Chow A, Khong T, Spencer A. Dysregulated Class I histone deacetylases are indicators of poor prognosis in multiple myeloma. Epigenetics. 2014;9:1511–1520. doi: 10.4161/15592294.2014.983367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat D, Patel S, Day F, Belfield A, Donald A, Rowlands M, Wibawa J, Brotherton D, Stimson L, Clark V, et al. Discovery of 2-(6-{[(6-fluoroquinolin-2-yl)methyl]amino}bicyclo[3.1.0]hex-3-yl)-N-hydroxypyrim idine-5-carboxamide (CHR-3996), a class I selective orally active histone deacetylase inhibitor. J Med Chem. 2010;53:8663–8678. doi: 10.1021/jm101177s. [DOI] [PubMed] [Google Scholar]
- Moreno DA, Scrideli CA, Cortez MA, de Paula Queiroz R, Valera ET, da Silva Silveira V, Yunes JA, Brandalise SR, Tone LG. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;150:665–673. doi: 10.1111/j.1365-2141.2010.08301.x. [DOI] [PubMed] [Google Scholar]
- Moresi V, Carrer M, Grueter CE, Rifki OF, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc Natl Acad Sci. 2012;109:1649–1654. doi: 10.1073/pnas.1121159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhauser F, Terriou L, Coiffier B, Bachy E, Varga A, Kloos I, Lelievre H, Sarry AL, Depil S, Ribrag V. Phase 1 study of the oral histone deacetylase inhibitor abexinostat in patients with Hodgkin lymphoma, non-Hodgkin lymphoma, or chronic lymphocytic leukaemia. Invest New Drugs. 2015;33:423–431. doi: 10.1007/s10637-015-0206-x. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Muller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W, Denkert C. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer--overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC Cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster PN, Thurn KT, Thomas S, Raha P, Lacevic M, Miller A, Melisko M, Ismail-Khan R, Rugo H, Moasser M, et al. A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer. 2011;104:1828–1835. doi: 10.1038/bjc.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Sato A, Chun NA, Hara M, Naito Y, Kobayashi Y, Kano Y, Ohtsuki M, Furukawa Y, Kobayashi E. Transcriptional modulation using HDACi depsipeptide promotes immune cell-mediated tumor destruction of murine B16 melanoma. J Invest Dermatol. 2008;128:1506–1516. doi: 10.1038/sj.jid.5701216. [DOI] [PubMed] [Google Scholar]
- Nakashima H, Nguyen T, Goins WF, Chiocca EA. Interferon-stimulated gene 15 (ISG15) and ISG15-linked proteins can associate with members of the selective autophagic process, histone deacetylase 6 (HDAC6) and SQSTM1/p62. J Biol Chem. 2015;290:1485–1495. doi: 10.1074/jbc.M114.593871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold A, Matthews GM, Bots M, Cluse LA, Clarke CJ, Banks KM, Cullinane C, Bolden JE, Christiansen AJ, Dickins RA, et al. Molecular and biologic analysis of histone deacetylase inhibitors with diverse specificities. Mol Cancer Ther. 2013;12:2709–2721. doi: 10.1158/1535-7163.MCT-13-0626. [DOI] [PubMed] [Google Scholar]
- Niegisch G, Knievel J, Koch A, Hader C, Fischer U, Albers P, Schulz WA. Changes in histone deacetylase (HDAC) expression patterns and activity of HDAC inhibitors in urothelial cancers. Urol Oncol. 2013;31:1770–1779. doi: 10.1016/j.urolonc.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Oehme I, Deubzer HE, Wegener D, Pickert D, Linke JP, Hero B, Kopp-Schneider A, Westermann F, Ulrich SM, von Deimling A, et al. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15:91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- Oehme I, Linke JP, Bock BC, Milde T, Lodrini M, Hartenstein B, Wiegand I, Eckert C, Roth W, Kool M, et al. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl Acad Sci. 2013;110:E2592–2601. doi: 10.1073/pnas.1300113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer. 2004;112:26–32. doi: 10.1002/ijc.20395. [DOI] [PubMed] [Google Scholar]
- Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 2014;32:1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouaissi M, Sielezneff I, Silvestre R, Sastre B, Bernard JP, Lafontaine JS, Payan MJ, Dahan L, Pirro N, Seitz JF, et al. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol. 2008;15:2318–2328. doi: 10.1245/s10434-008-9940-z. [DOI] [PubMed] [Google Scholar]
- Ouaissi M, Silvy F, Loncle C, Ferraz da Silva D, Martins Abreu C, Martinez E, Berthezene P, Cadra S, Le Treut YP, Hardwigsen J, et al. Further characterization of HDAC and SIRT gene expression patterns in pancreatic cancer and their relation to disease outcome. PLoS One. 2014;9:e108520. doi: 10.1371/journal.pone.0108520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34:699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parbin S, Kar S, Shilpi A, Sengupta D, Deb M, Rath SK, Patra SK. Histone deacetylases: a saga of perturbed acetylation homeostasis in cancer. J Histochem Cytochem. 2014;62:11–33. doi: 10.1369/0022155413506582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Thomas S, Munster PN. Epigenetic modulation with histone deacetylase inhibitors in combination with immunotherapy. Epigenomics. 2015;7:641–652. doi: 10.2217/epi.15.16. [DOI] [PubMed] [Google Scholar]
- Pauer LR, Olivares J, Cunningham C, Williams A, Grove W, Kraker A, Olson S, Nemunaitis J. Phase I study of oral CI-994 in combination with carboplatin and paclitaxel in the treatment of patients with advanced solid tumors. Cancer Invest. 2004;22:886–896. doi: 10.1081/cnv-200039852. [DOI] [PubMed] [Google Scholar]
- Peng L, Yuan Z, Li Y, Ling H, Izumi V, Fang B, Fukasawa K, Koomen J, Chen J, Seto E. Ubiquitinated sirtuin 1 (SIRT1) function is modulated during DNA damage-induced cell death and survival. J Biol Chem. 2015;290:8904–8912. doi: 10.1074/jbc.M114.612796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet C, Jentsch B, Hermanns T, Schweckendiek D, Seifert HH, Schmidtpeter M, Sulser T, Moch H, Wild PJ, Kristiansen G. Expression of histone deacetylases 1, 2 and 3 in urothelial bladder cancer. BMC Clin Pathol. 2014;14:10. doi: 10.1186/1472-6890-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prebet T, Sun Z, Figueroa ME, Ketterling R, Melnick A, Greenberg PL, Herman J, Juckett M, Smith MR, Malick L, et al. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US Leukemia Intergroup trial E1905. J Clin Oncol. 2014;32:1242–1248. doi: 10.1200/JCO.2013.50.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Lai CJ, Bao R, Wang DG, Wang J, Xu GX, Atoyan R, Qu H, Yin L, Samson M, et al. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clin Cancer Res. 2012;18:4104–4113. doi: 10.1158/1078-0432.CCR-12-0055. [DOI] [PubMed] [Google Scholar]
- Quint K, Agaimy A, Di Fazio P, Montalbano R, Steindorf C, Jung R, Hellerbrand C, Hartmann A, Sitter H, Neureiter D, et al. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch. 2011;459:129–139. doi: 10.1007/s00428-011-1103-0. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan R, Li Y, Xiang S, Yuan F, Yuan Z, Telles E, Fang J, Coppola D, Shibata D, Lane WS, et al. Histone Deacetylase 10 Regulates DNA Mismatch Repair and May Involve the Deacetylation of MutS Homolog 2. J Biol Chem. 2015;290:22795–22804. doi: 10.1074/jbc.M114.612945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha P, Thomas S, Thurn KT, Park J, Munster PN. Combined histone deacetylase inhibition and tamoxifen induces apoptosis in tamoxifen-resistant breast cancer models, by reversing Bcl-2 overexpression. Breast Cancer Res. 2015;17:26. doi: 10.1186/s13058-015-0533-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Alalem M, Ray BK. Loss of epigenetic Kruppel-like factor 4 histone deacetylase (KLF-4-HDAC)-mediated transcriptional suppression is crucial in increasing vascular endothelial growth factor (VEGF) expression in breast cancer. J Biol Chem. 2013;288:27232–27242. doi: 10.1074/jbc.M113.481184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T, Valone F, Lipera W, Irwin D, Paroly W, Natale R, Sreedharan S, Keer H, Lum B, Scappaticci F, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer. 2004;45:381–386. doi: 10.1016/j.lungcan.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Rettig I, Koeneke E, Trippel F, Mueller WC, Burhenne J, Kopp-Schneider A, Fabian J, Schober A, Fernekorn U, von Deimling A, et al. Selective inhibition of HDAC8 decreases neuroblastoma growth in vitro and in vivo and enhances retinoic acid-mediated differentiation. Cell Death Dis. 2015;6:e1657. doi: 10.1038/cddis.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes LV, Tate CR, Segar HC, Burks HE, Phamduy TB, Hoang V, Elliott S, Gilliam D, Pounder FN, Anbalagan M, et al. Suppression of triple-negative breast cancer metastasis by pan-DAC inhibitor panobinostat via inhibition of ZEB family of EMT master regulators. Breast Cancer Res Treat. 2014;145:593–604. doi: 10.1007/s10549-014-2979-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson PG, Laubach JP, Lonial S, Moreau P, Yoon SS, Hungria VT, Dimopoulos MA, Beksac M, Alsina M, San-Miguel JF. Panobinostat: a novel pan-deacetylase inhibitor for the treatment of relapsed or relapsed and refractory multiple myeloma. Expert Rev Anticancer Ther. 2015;15:737–748. doi: 10.1586/14737140.2015.1047770. [DOI] [PubMed] [Google Scholar]
- Riley JS, Hutchinson R, McArt DG, Crawford N, Holohan C, Paul I, Van Schaeybroeck S, Salto-Tellez M, Johnston PG, Fennell DA, et al. Prognostic and therapeutic relevance of FLIP and procaspase-8 overexpression in non-small cell lung cancer. Cell Death Dis. 2013;4:e951. doi: 10.1038/cddis.2013.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, Caballero R, Alaminos M, Setien F, Paz MF, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38:566–569. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- Saijo K, Imamura J, Narita K, Oda A, Shimodaira H, Katoh T, Ishioka C. Biochemical, biological and structural properties of romidepsin (FK228) and its analogs as novel HDAC/PI3K dual inhibitors. Cancer Sci. 2015;106:208–215. doi: 10.1111/cas.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo K, Katoh T, Shimodaira H, Oda A, Takahashi O, Ishioka C. Romidepsin (FK228) and its analogs directly inhibit phosphatidylinositol 3-kinase activity and potently induce apoptosis as histone deacetylase/phosphatidylinositol 3-kinase dual inhibitors. Cancer Sci. 2012;103:1994–2001. doi: 10.1111/cas.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo L, Hideshima T, Kung AL, Tseng JC, Tamang D, Yang M, Jarpe M, van Duzer JH, Mazitschek R, Ogier WC, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012;119:2579–2589. doi: 10.1182/blood-2011-10-387365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro F, Botrugno OA, Dal Zuffo R, Pallavicini I, Matthews GM, Cluse L, Barozzi I, Senese S, Fornasari L, Moretti S, et al. A dual role for Hdac1: oncosuppressor in tumorigenesis, oncogene in tumor maintenance. Blood. 2013;121:3459–3468. doi: 10.1182/blood-2012-10-461988. [DOI] [PubMed] [Google Scholar]
- Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, et al. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- Schroeder FA, Lewis MC, Fass DM, Wagner FF, Zhang YL, Hennig KM, Gale J, Zhao WN, Reis S, Barker DD, et al. A selective HDAC 1/2 inhibitor modulates chromatin and gene expression in brain and alters mouse behavior in two mood-related tests. PLoS One. 2013;8:e71323. doi: 10.1371/journal.pone.0071323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler S, Fritsche P, Diersch S, Arlt A, Schmid RM, Saur D, Schneider G. HDAC2 attenuates TRAIL-induced apoptosis of pancreatic cancer cells. Mol Cancer. 2010;9:80. doi: 10.1186/1476-4598-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, Bernard L, Draetta GF, Alcalay M, Seiser C, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HW, Kim EJ, Na H, Lee MO. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett. 2009;583:55–60. doi: 10.1016/j.febslet.2008.11.044. [DOI] [PubMed] [Google Scholar]
- Seo J, Min SK, Park HR, Kim DH, Kwon MJ, Kim LS, Ju YS. Expression of Histone Deacetylases HDAC1, HDAC2, HDAC3, and HDAC6 in Invasive Ductal Carcinomas of the Breast. J Breast Cancer. 2014;17:323–331. doi: 10.4048/jbc.2014.17.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Lawrence YR, Choy H, Werner-Wasik M, Andrews DW, Evans JJ, Judy KD, Farrell CJ, Moshel Y, Berger AC, et al. Vorinostat as a radiosensitizer for brain metastasis: a phase I clinical trial. J Neurooncol. 2014;118:313–319. doi: 10.1007/s11060-014-1433-2. [DOI] [PubMed] [Google Scholar]
- Shi Y, Dong M, Hong X, Zhang W, Feng J, Zhu J, Yu L, Ke X, Huang H, Shen Z, et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann Oncol. 2015;26:1766–1771. doi: 10.1093/annonc/mdv237. [DOI] [PubMed] [Google Scholar]
- Shi Y, Huang J, Zhou J, Liu Y, Fu X, Li Y, Yin G, Wen J. MicroRNA-204 inhibits proliferation, migration, invasion and epithelial-mesenchymal transition in osteosarcoma cells via targeting Sirtuin 1. Oncol Rep. 2015;34:399–406. doi: 10.3892/or.2015.3986. [DOI] [PubMed] [Google Scholar]
- Shimizu T, LoRusso PM, Papadopoulos KP, Patnaik A, Beeram M, Smith LS, Rasco DW, Mays TA, Chambers G, Ma A, et al. Phase I first-in-human study of CUDC-101, a multitargeted inhibitor of HDACs, EGFR, and HER2 in patients with advanced solid tumors. Clin Cancer Res. 2015;20:5032–5040. doi: 10.1158/1078-0432.CCR-14-0570. [DOI] [PubMed] [Google Scholar]
- Shukla S, Khan S, Kumar S, Sinha S, Farhan M, Bora HK, Maurya R, Meeran SM. Cucurbitacin B Alters the Expression of Tumor-Related Genes by Epigenetic Modifications in NSCLC and Inhibits NNK-Induced Lung Tumorigenesis. Cancer Prev Res (Phila) 2015;8:552–562. doi: 10.1158/1940-6207.CAPR-14-0286. [DOI] [PubMed] [Google Scholar]
- Simic P, Williams EO, Bell EL, Gong JJ, Bonkowski M, Guarente L. SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Rep. 2013;3:1175–1186. doi: 10.1016/j.celrep.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodji QH, Kornacki JR, McDonald JF, Mrksich M, Oyelere AK. Design and structure activity relationship of tumor-homing histone deacetylase inhibitors conjugated to folic and pteroic acids. Eur J Med Chem. 2015;96:340–359. doi: 10.1016/j.ejmech.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Zhu S, Wu C, Kang J. Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J Biol Chem. 2013;288:28021–28033. doi: 10.1074/jbc.M113.498758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnemann J, Marx C, Becker S, Wittig S, Palani CD, Kramer OH, Beck JF. p53-dependent and p53-independent anticancer effects of different histone deacetylase inhibitors. Br J Cancer. 2014;110:656–667. doi: 10.1038/bjc.2013.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- Stronach EA, Alfraidi A, Rama N, Datler C, Studd JB, Agarwal R, Guney TG, Gourley C, Hennessy BT, Mills GB, et al. HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 2011;71:4412–4422. doi: 10.1158/0008-5472.CAN-10-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stypula-Cyrus Y, Damania D, Kunte DP, Cruz MD, Subramanian H, Roy HK, Backman V. HDAC up-regulation in early colon field carcinogenesis is involved in cell tumorigenicity through regulation of chromatin structure. PLoS One. 2013;8:e64600. doi: 10.1371/journal.pone.0064600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo T, Mimori K, Nishida N, Kogo R, Iwaya T, Tanaka F, Shibata K, Fujita H, Shirouzu K, Mori M. Histone deacetylase 1 expression in gastric cancer. Oncol Rep. 2011;26:777–782. doi: 10.3892/or.2011.1361. [DOI] [PubMed] [Google Scholar]
- Sun L, Kokura K, Izumi V, Koomen JM, Seto E, Chen J, Fang J. MPP8 and SIRT1 crosstalk in E-cadherin gene silencing and epithelial-mesenchymal transition. EMBO Rep. 2015;16:689–699. doi: 10.15252/embr.201439792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kasuya Y, Itoh Y, Ota Y, Zhan P, Asamitsu K, Nakagawa H, Okamoto T, Miyata N. Identification of highly selective and potent histone deacetylase 3 inhibitors using click chemistry-based combinatorial fragment assembly. PLoS One. 2013;8:e68669. doi: 10.1371/journal.pone.0068669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Muto N, Bando M, Itoh Y, Masaki A, Ri M, Ota Y, Nakagawa H, Iida S, Shirahige K, et al. Design, synthesis, and biological activity of NCC149 derivatives as histone deacetylase 8-selective inhibitors. ChemMedChem. 2014;9:657–664. doi: 10.1002/cmdc.201300414. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Ota Y, Ri M, Bando M, Gotoh A, Itoh Y, Tsumoto H, Tatum PR, Mizukami T, Nakagawa H, et al. Rapid discovery of highly potent and selective inhibitors of histone deacetylase 8 using click chemistry to generate candidate libraries. J Med Chem. 2012;55:9562–9575. doi: 10.1021/jm300837y. [DOI] [PubMed] [Google Scholar]
- Takasaka N, Araya J, Hara H, Ito S, Kobayashi K, Kurita Y, Wakui H, Yoshii Y, Yumino Y, Fujii S, et al. Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J Immunol. 2014;192:958–968. doi: 10.4049/jimmunol.1302341. [DOI] [PubMed] [Google Scholar]
- Tang C, Li C, Zhang S, Hu Z, Wu J, Dong C, Huang J, Zhou HB. Novel bioactive hybrid compound dual targeting estrogen receptor and histone deacetylase for the treatment of breast cancer. J Med Chem. 2015;58:4550–4572. doi: 10.1021/acs.jmedchem.5b00099. [DOI] [PubMed] [Google Scholar]
- Taylor BS, DeCarolis PL, Angeles CV, Brenet F, Schultz N, Antonescu CR, Scandura JM, Sander C, Viale AJ, Socci ND, et al. Frequent alterations and epigenetic silencing of differentiation pathway genes in structurally rearranged liposarcomas. Cancer Discov. 2011;1:587–597. doi: 10.1158/2159-8290.CD-11-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurn KT, Thomas S, Raha P, Qureshi I, Munster PN. Histone deacetylase regulation of ATM-mediated DNA damage signaling. Mol Cancer Ther. 2013;12:2078–2087. doi: 10.1158/1535-7163.MCT-12-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomosugi M, Sowa Y, Yasuda S, Tanaka R, te Riele H, Ikawa H, Koyama M, Sakai T. Retinoblastoma gene-independent G1 phase arrest by flavone, phosphatidylinositol 3-kinase inhibitor, and histone deacetylase inhibitor. Cancer Sci. 2012;103:2139–2143. doi: 10.1111/cas.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH, Zhang HB, Liao YJ, Zheng F, Zhu W, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic Biol Med. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Turtoi A, Mottet D, Matheus N, Dumont B, Peixoto P, Hennequiere V, Deroanne C, Colige A, De Pauw E, Bellahcene A, et al. The angiogenesis suppressor gene AKAP12 is under the epigenetic control of HDAC7 in endothelial cells. Angiogenesis. 2012;15:543–554. doi: 10.1007/s10456-012-9279-8. [DOI] [PubMed] [Google Scholar]
- Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, Ngo L, Holmgren A, Jiang X, Marks PA. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C, Rossig L, Kaluza D, Potente M, Boeckel JN, Knau A, Diehl F, Geng JG, Hofmann WK, Zeiher AM, et al. HDAC5 is a repressor of angiogenesis and determines the angiogenic gene expression pattern of endothelial cells. Blood. 2009;113:5669–5679. doi: 10.1182/blood-2009-01-196485. [DOI] [PubMed] [Google Scholar]
- Van Damme M, Crompot E, Meuleman N, Mineur P, Bron D, Lagneaux L, Stamatopoulos B. HDAC isoenzyme expression is deregulated in chronic lymphocytic leukemia B-cells and has a complex prognostic significance. Epigenetics. 2012;7:1403–1412. doi: 10.4161/epi.22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y. Crosstalk between lysine-specific demethylase 1 (LSD1) and histone deacetylases mediates antineoplastic efficacy of HDAC inhibitors in human breast cancer cells. Carcinogenesis. 2013;34:1196–1207. doi: 10.1093/carcin/bgt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Venugopal B, Baird R, Kristeleit RS, Plummer R, Cowan R, Stewart A, Fourneau N, Hellemans P, Elsayed Y, McClue S, et al. A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors. Clin Cancer Res. 2013;19:4262–4272. doi: 10.1158/1078-0432.CCR-13-0312. [DOI] [PubMed] [Google Scholar]
- Venza I, Visalli M, Oteri R, Cucinotta M, Teti D, Venza M. Class II-specific histone deacetylase inhibitors MC1568 and MC1575 suppress IL-8 expression in human melanoma cells. Pigment Cell Melanoma Res. 2013;26:193–204. doi: 10.1111/pcmr.12049. [DOI] [PubMed] [Google Scholar]
- Vidal-Laliena M, Gallastegui E, Mateo F, Martinez-Balbas M, Pujol MJ, Bachs O. Histone deacetylase 3 regulates cyclin A stability. J Biol Chem. 2013;288:21096–21104. doi: 10.1074/jbc.M113.458323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, von Werder A, Schmidt A, Mages J, Pagel P, et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137:361–371. doi: 10.1053/j.gastro.2009.04.004. 371 e361-365. [DOI] [PubMed] [Google Scholar]
- Wang G, He J, Zhao J, Yun W, Xie C, Taub JW, Azmi A, Mohammad RM, Dong Y, Kong W, et al. Class I and class II histone deacetylases are potential therapeutic targets for treating pancreatic cancer. PLoS One. 2012;7:e52095. doi: 10.1371/journal.pone.0052095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Kafeel MI, Avezbakiyev B, Chen C, Sun Y, Rathnasabapathy C, Kalavar M, He Z, Burton J, Lichter S. Histone deacetylase in chronic lymphocytic leukemia. Oncology. 2011;81:325–329. doi: 10.1159/000334577. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen G, Chen K, Ren Y, Li H, Jiang X, Jia L, Fu S, Li Y, Liu X, et al. Dual targeting of retinoid X receptor and histone deacetylase with DW22 as a novel antitumor approach. Oncotarget. 2015;6:9740–9755. doi: 10.18632/oncotarget.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Lahusen TJ, Chen Q, Xu X, Jenkins LM, Leo E, Fu H, Aladjem M, Pommier Y, Appella E, et al. SIRT1 deacetylates TopBP1 and modulates intra-S-phase checkpoint and DNA replication origin firing. Int J Biol Sci. 2014;10:1193–1202. doi: 10.7150/ijbs.11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013;126:4843–4849. doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Denkert C, Noske A, Darb-Esfahani S, Dietel M, Kalloger SE, Huntsman DG, Kobel M. Expression of class I histone deacetylases indicates poor prognosis in endometrioid subtypes of ovarian and endometrial carcinomas. Neoplasia. 2008a;10:1021–1027. doi: 10.1593/neo.08474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Roske A, Gekeler V, Beckers T, Ebert MP, Pross M, Dietel M, Denkert C, Rocken C. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol. 2008b;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008c;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Roske A, Niesporek S, Noske A, Buckendahl AC, Dietel M, Gekeler V, Boehm M, Beckers T, Denkert C. Class I histone deacetylase expression has independent prognostic impact in human colorectal cancer: Specific role of class I histone deacetylases in vitro and in vivo. Clin Cancer Res. 2008d;14:1669–1677. doi: 10.1158/1078-0432.CCR-07-0990. [DOI] [PubMed] [Google Scholar]
- Wells CE, Bhaskara S, Stengel KR, Zhao Y, Sirbu B, Chagot B, Cortez D, Khabele D, Chazin WJ, Cooper A, et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PLoS One. 2013;8:e68915. doi: 10.1371/journal.pone.0068915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AC, Mattarollo SR, Shortt J, Cluse LA, Christiansen AJ, Smyth MJ, Johnstone RW. An intact immune system is required for the anticancer activities of histone deacetylase inhibitors. Cancer Res. 2013;73:7265–7276. doi: 10.1158/0008-5472.CAN-13-0890. [DOI] [PubMed] [Google Scholar]
- West AC, Smyth MJ, Johnstone RW. The anticancer effects of HDAC inhibitors require the immune system. Oncoimmunology. 2014;3:e27414. doi: 10.4161/onci.27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmott JS, Colebatch AJ, Kakavand H, Shang P, Carlino MS, Thompson JF, Long GV, Scolyer RA, Hersey P. Expression of the class 1 histone deacetylases HDAC8 and 3 are associated with improved survival of patients with metastatic melanoma. Mod Pathol. 2015;28:884–894. doi: 10.1038/modpathol.2015.34. [DOI] [PubMed] [Google Scholar]
- Xiao W, Chen X, Liu X, Luo L, Ye S, Liu Y. Trichostatin A, a histone deacetylase inhibitor, suppresses proliferation and epithelial-mesenchymal transition in retinal pigment epithelium cells. J Cell Mol Med. 2014;18:646–655. doi: 10.1111/jcmm.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HJ, Noh JH, Kim JK, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Lee JY, Park H, et al. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PLoS One. 2012;7:e34265. doi: 10.1371/journal.pone.0034265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K, Gu JJ, Zhang Q, Mavis C, Hernandez-Ilizaliturri FJ, Czuczman MS, Guo Y. Vorinostat, a histone deacetylase (HDAC) inhibitor, promotes cell cycle arrest and re-sensitizes rituximab- and chemo-resistant lymphoma cells to chemotherapy agents. J Cancer Res Clin Oncol. 2015 doi: 10.1007/s00432-015-2026-y. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Cubizolles F, Zhang Y, Reichert N, Kohler H, Seiser C, Matthias P. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang FC, Tan BC, Chen WH, Lin YH, Huang JY, Chang HY, Sun HY, Hsu PH, Liou GG, Shen J, et al. Reversible acetylation regulates salt-inducible kinase (SIK2) and its function in autophagy. J Biol Chem. 2013;288:6227–6237. doi: 10.1074/jbc.M112.431239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO J. 2006;25:1231–1241. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes A, Oki Y, Bociek RG, Kuruvilla J, Fanale M, Neelapu S, Copeland A, Buglio D, Galal A, Besterman J, et al. Mocetinostat for relapsed classical Hodgkin’s lymphoma: an open-label, single-arm, phase 2 trial. Lancet Oncol. 2011;12:1222–1228. doi: 10.1016/S1470-2045(11)70265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhong Q. Histone deacetylase inhibitors and cell death. Cell Mol Life Sci. 2014;71:3885–3901. doi: 10.1007/s00018-014-1656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Mehta A, Boufraqech M, Davis S, Wang J, Tian Z, Yu Z, Boxer MB, Kiefer JA, et al. Dual inhibition of HDAC and EGFR signaling with CUDC-101 induces potent suppression of tumor growth and metastasis in anaplastic thyroid cancer. Oncotarget. 2015a;6:9073–9085. doi: 10.18632/oncotarget.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xiang S, Joo HY, Wang L, Williams KA, Liu W, Hu C, Tong D, Haakenson J, Wang C, et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSalpha. Mol Cell. 2014;55:31–46. doi: 10.1016/j.molcel.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kong Y, Zhang J, Su M, Zhou Y, Zang Y, Li J, Chen Y, Fang Y, Zhang X, et al. Design, synthesis and biological evaluation of colchicine derivatives as novel tubulin and histone deacetylase dual inhibitors. Eur J Med Chem. 2015b;95:127–135. doi: 10.1016/j.ejmech.2015.03.035. [DOI] [PubMed] [Google Scholar]
- Zhang X, Su M, Chen Y, Li J, Lu W. The design and synthesis of a new class of RTK/HDAC dual-targeted inhibitors. Molecules. 2013;18:6491–6503. doi: 10.3390/molecules18066491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Adachi M, Kawamura R, Imai K. Bmf is a possible mediator in histone deacetylase inhibitors FK228 and CBHA-induced apoptosis. Cell Death Differ. 2006;13:129–140. doi: 10.1038/sj.cdd.4401686. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat. 2005;94:11–16. doi: 10.1007/s10549-005-6001-1. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y, Mita K, Hamaguchi M, Hayashi S, Iwase H. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res. 2004;10:6962–6968. doi: 10.1158/1078-0432.CCR-04-0455. [DOI] [PubMed] [Google Scholar]
- Zhao J, Lawless MW. Resminostat: Opening the door on epigenetic treatments for liver cancer. Hepatology. 2015 doi: 10.1002/hep.27853. [DOI] [PubMed] [Google Scholar]
- Zhi X, Zhong Q. Autophagy in cancer. F1000Prime Rep. 2015;7:18. doi: 10.12703/P7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Tolstov Y, Arslan A, Roth W, Grullich C, Pahernik S, Hohenfellner M, Duensing S. Harnessing the p53-PUMA axis to overcome DNA damage resistance in renal cell carcinoma. Neoplasia. 2014;16:1028–1035. doi: 10.1016/j.neo.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZW, Li XX, He ZX, Pan ST, Yang Y, Zhang X, Chow K, Yang T, Qiu JX, Zhou Q, et al. Induction of apoptosis and autophagy via sirtuin1- and PI3K/Akt/mTOR-mediated pathways by plumbagin in human prostate cancer cells. Drug Des Devel Ther. 2015;9:1511–1554. doi: 10.2147/DDDT.S75976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zibelman M, Wong YN, Devarajan K, Malizzia L, Corrigan A, Olszanski AJ, Denlinger CS, Roethke SK, Tetzlaff CH, Plimack ER. Phase I study of the mTOR inhibitor ridaforolimus and the HDAC inhibitor vorinostat in advanced renal cell carcinoma and other solid tumors. Invest New Drugs. 2015 doi: 10.1007/s10637-015-0261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzi AP, Bernstein M, Samson Y, Wall DA, Desai S, Nicksy D, Wainman N, Eisenhauer E, Baruchel S. A phase I study of histone deacetylase inhibitor, pracinostat (SB939), in pediatric patients with refractory solid tumors: IND203 a trial of the NCIC IND program/C17 pediatric phase I consortium. Pediatr Blood Cancer. 2013;60:1868–1874. doi: 10.1002/pbc.24694. [DOI] [PubMed] [Google Scholar]
- Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J, Rembold M, Meunier D, Egger G, Lagger S, et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol. 2010;30:1171–1181. doi: 10.1128/MCB.01500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwergel C, Valente S, Jacob C, Mai A. Emerging approaches for histone deacetylase inhibitor drug discovery. Expert Opin Drug Discov. 2015;10:599–613. doi: 10.1517/17460441.2015.1038236. [DOI] [PubMed] [Google Scholar]