Abstract
The threat of a major coronavirus pandemic urges the development of suitable strategies to combat these pathogens. HCoV-NL63 is an α-coronavirus that can cause severe lower respiratory tract infections requiring hospitalization. We report here the 3.4 Å resolution cryo-electron microscopy reconstruction of the HCoV-NL63 coronavirus spike glycoprotein trimer, which is the conformational machine responsible for entry into host cells and the sole target of neutralizing antibodies during infection. The map resolves the extensive glycan shield obstructing the protein surface and, in combination with mass-spectrometry, provides a structural framework to understand accessibility to antibodies. The structure also reveals a remarkable modular architecture of the receptor-binding subunit and the complete architecture of the fusion machinery including the triggering loop and the C-terminal domains, which contribute to anchoring the trimer to the viral membrane. Our data further suggest that HCoV-NL63 and other coronaviruses use molecular trickery, based on masking of epitopes with glycans and activating conformational changes, to evade the immune system of infected hosts.
Coronaviruses are enveloped viruses, with large single-stranded positive-sense RNA genomes, classified in four genera (α, β, γ, and δ). In humans, coronaviruses are responsible for 30% of respiratory tract infections1. In addition, coronaviruses have fostered a lot of attention in the last decade due to the emergence of two deadly viruses with tremendous pandemic potential: severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle-East respiratory syndrome coronavirus (MERS-CoV)2. To date, there are no approved antiviral treatments or vaccines for any human coronavirus.
Coronaviruses are zoonotic viruses and surveillance studies suggest both SARS-CoV and MERS-CoV originated from bats; with camels likely acting as intermediate hosts for the latter virus3,4. Moreover, sequencing data demonstrated that bats constitute a reservoir of coronaviruses that have the potential to cross the species barrier and infect humans. This is illustrated by the fact that substitution of three amino acid residues in the spike (S) glycoprotein receptor-binding domain of the bat-infecting HKU4-CoV can enhance its affinity for human DPP4 (the MERS-CoV receptor) by two-orders of magnitude5,6. Also, substitution of two other residues enables processing by human proteases and allows the HKU4-CoV S protein to mediate entry into human cells7. As a result, cross-species transmission of coronaviruses poses an imminent and long-term threat to human health. Recombination with coronaviruses frequently involved in mild respiratory infections could potentially lead to the emergence of highly pathogenic viruses4. Understanding the pathogenesis, cross-species transmission and recombination of coronaviruses is key for evaluating long-term emerging disease potentials and for preventing and controlling the spread of coronaviruses in humans.
To date, α- and β-coronavirus genera have been implicated in human diseases and zoonoses. The human coronavirus NL63 (HCoV-NL63) is an α-coronavirus, which is genetically distinct from β-coronaviruses mouse hepatitis virus (MHV, the prototypical coronavirus), MERS-CoV and SARS-CoV, and was first isolated from a 7-month-old patient with a respiratory tract infection8,9. Further studies revealed HCoV-NL63 infections appear to be common in childhood, and most adult sera contain antibodies that neutralize the virus8,10. HCoV-NL63 is a major cause of bronchiolitis and pneumonia in newborns worldwide and can cause severe lower respiratory tract infections requiring hospitalization, especially of young children, the elderly, and immunocompromised adults11. HCoV-NL63 infections have been reported in 12 countries across Europe, Asia and North America, indicating that it is circulating among the human population worldwide. Other α-coronaviruses related to the human respiratory pathogen HCoV-229E have recently been identified in camels co-infected with MERS-CoV4, further underscoring the importance of characterizing this coronavirus genus. Additionally, the emergence of the highly lethal porcine epidemic diarrhea coronavirus (PEDV, α-genus) had devastating consequences for the US swine industry recently12.
Coronaviruses use S homotrimers to promote cell attachment and fusion of the viral and host membranes. As it is virtually the only antigen present at the virus surface, S is the main target of neutralizing antibodies during infection and a focus of vaccine design13. S is a class I viral fusion protein synthesized as a single chain precursor of about 1,300 amino acids which trimerizes upon folding14. It is composed of an N-terminal S1 subunit, containing the receptor-binding domain, and a C-terminal S2 subunit, driving membrane fusion. Upon virion uptake by target host cells, cleavage at the S2’ site (next to the putative fusion peptide) is required for fusion activation of all coronavirus S proteins such that they can subsequently transition to the post-fusion conformation15–17.
Our previously reported cryo-electron microscopy (cryoEM) reconstruction of the MHV S glycoprotein at 4.0 Å resolution revealed the pre-fusion architecture of the machinery mediating entry of β-coronaviruses into cells18. It also demonstrated that coronavirus S and paramyxovirus F proteins share a common evolutionary origin. Here, we report the atomic resolution structure of the pathogenic HCoV-NL63 S glycoprotein trimer which belongs to the α-coronavirus genus. The significant resolution improvement compared to earlier studies allows visualization of the S glycoprotein with an unprecedented level of details, which is a prerequisite for guiding drug and vaccine design, and reveals shared and unique features of the α-genus of human pathogens. Our results suggest that NL63 and other coronaviruses use molecular trickery, based on masking of epitopes with glycans and activating conformational changes, to evade the immune system of infected hosts similarly to what has been described for HIV-1.
RESULTS
Structure determination
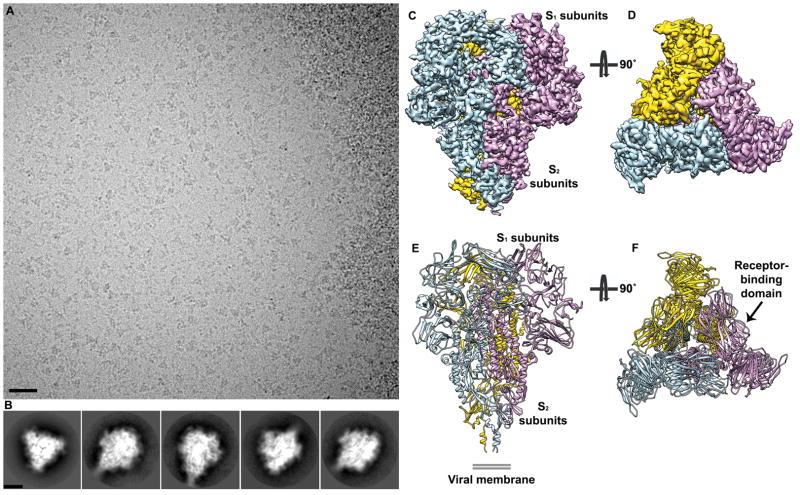
We produced the HCoV-NL63 S ectodomain N-terminally fused to a GCN4 trimerization motif in register with the heptad-repeat 2 (HR2) helix. Frozen-hydrated HCoV-NL63 spike ectodomain particles were imaged using an FEI Titan Krios electron microscope equipped with a Gatan Quantum GIF energy-filter operated in zero-loss mode with a slit width of 20 eV and a Gatan K2 Summit electron counting camera19. We determined a three-dimensional reconstruction of the HCoV-NL63 spike at 3.4 Å resolution using the gold-standard Fourier shell correlation (FSC) criterion of 0.14320,21 (Fig. 1 and Supplementary Fig. 1). The final model was built and refined using Coot22 and Rosetta23–25 and includes residues 28 to 1231 with an internal break between residues 994–998 (Supplementary Fig. 2 and Table 1). The HCoV-NL63 S ectodomain is a 160 Å long trimer with a triangular cross-section.
Figure 1. Cryo-electron microscopy structure of the HCoV-NL63 S trimer at atomic resolution.
a, Representative micrograph of frozen-hydrated HCoV-NL63 S particles (defocus 3.4 μm). Scale bar: 355 Å. b, Five selected class averages showing the particles along different orientations. Scale bar: 60 Å. c–d, 3D map filtered at 3.4 Å resolution colored by protomer. Two orthogonal views of the S trimer (from the side (c) and from the top, looking toward the viral membrane, (d)) are shown. e–f, Ribbon diagrams showing the HCoV-NL63 S atomic model oriented as in c–d.
The ordered glycan shield
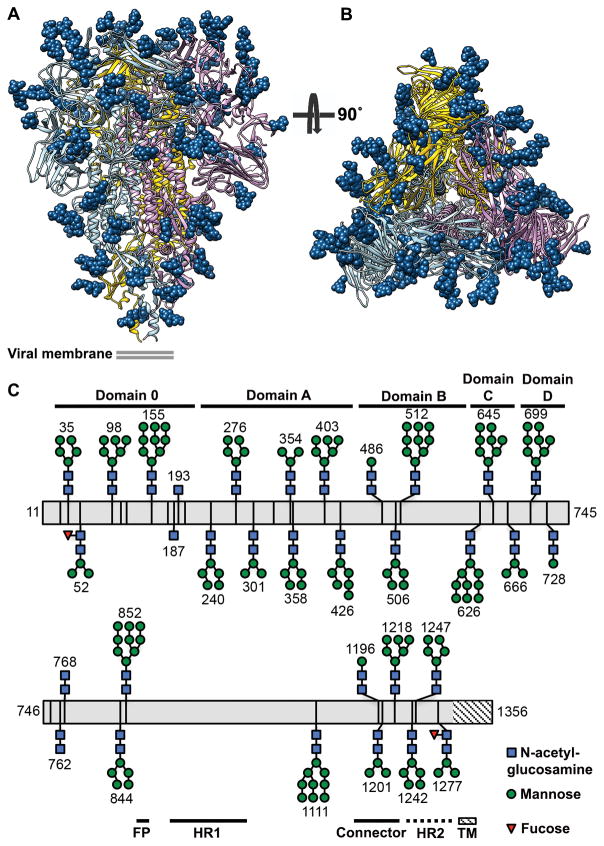
A striking feature of this structure is the extraordinary number of N-linked oligosaccharides that cover the spike trimer. In the cryoEM reconstruction, we could observe density for 30 N-linked glycans extending tangentially relative to the protein surface (Fig. 2A–B, Supplementary Fig. 2 and Supplementary Table 2). At least the two core N-acetyl glucosamine moieties are visible for the majority of glycosylation sites. Using on-line reversed phased liquid chromatography with electron transfer/high-energy collision-dissociation tandem mass-spectrometry26, we detected 24 N-linked glycosylation sites overlapping with those observed in the cryoEM map and identified 4 additional sites (Fig. 1C, Supplementary Fig 3 and Supplementary Table 2). These sites were identified from both intact glycopeptides as well as peptides with the glycan trimmed down to the N-linked core N-acetyl glucosamine moiety. The cryoEM and mass-spectrometry data combined provides evidence for glycosylation at 34 out of 39 possible NXS/T glycosylation sequons. The intact glycopeptides detected by MS/MS for HCoV-NL63 S expressed in Drosophila S2 cells corresponded to either paucimannosidic glycans containing 3 mannose residues (with or without core fucosylation) or high-mannose glycans containing 4 to 9 mannose residues. Previous reports suggested that several coronavirus S glycans are of the high-mannose type, as a result of direct budding from the endoplasmic reticulum-golgi intermediate compartment27,28, which underscores the biological relevance of the potential glycan structures identified. In the refined model, N-linked glycans significantly cover the accessible surface of the trimer (Fig. 1A–B). The higher glycan density per accessible surface area detected for the S2 subunits (865 Å2/glycan) compared to the S1 subunits (1350 Å2/glycan) could explain why most coronavirus neutralizing antibodies isolated to date target the latter region. As many of the observed glycosylation sites are topologically conserved among coronavirus S proteins, we put forward that the glycan footprint observed here could be representative of other S proteins. Besides potentially contributing to immune evasion as discussed below, S glycans have been proposed to play a role in host cell entry29 using L-SIGN lectin which is an alternative receptor for SARS-CoV30 and HCoV-229E27.
Figure 2. Organization of the HCoV-NL63 S protein glycan shield revealed by cryoEM and mass-spectrometry.
a–b, Ribbon diagrams showing two orthogonal views of the S trimer (from the side (a) and from the top, looking toward the viral membrane, (b)) with glycans shown as dark blue spheres. c, Residue-level schematic of N-linked glycans. The most extensive glycan structure detected by mass-spectrometry at each site is represented except for glycans observed only by cryoEM for which the resolved sugar moieties are shown. FP: fusion peptide, HR1: heptad-repeat 1 region, HR2: heptad-repeat 2 region (shown with a dashed line as it is not resolved in the map), TM: transmembrane domain (the striated texture indicates regions that are not part of the construct).
Structure of the S2’ trigger loop
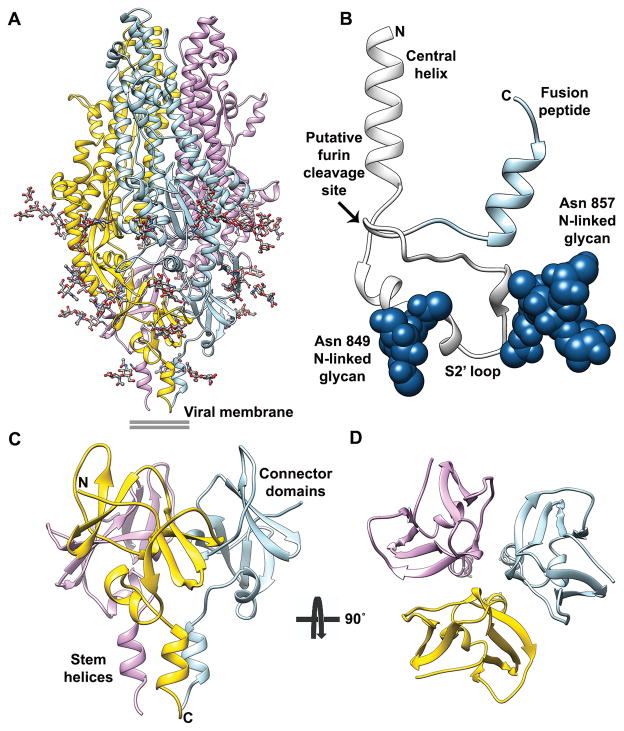
The HCoV-NL63 and MHV S2 fusion machineries are structurally similar and can be superimposed with excellent agreement (Fig. 3A, Supplementary Fig. 4, DALI31 Z-score 30.4, rmsd 2Å over 312 residues). In contrast to our previous MHV S structure18, most of the HCoV-NL63 S2’ trigger loop, which connects the upstream helix to the fusion peptide and participates in fusion activation, is resolved in the reconstruction (Fig. 3B). It runs almost perpendicularly to the long axis of the S2 subunit and forms three helical segments before looping back to connect to the fusion peptide. Multiple arginine residues, forming two putative furin cleavage sites, are present in the C-terminal region of the S2’ loop (863-RNIRSSR-870), which is characterized by weaker density as would be expected from a protease-sensitive polypeptide segment. These observations agree with previous studies suggesting fusion activation of the HCoV-NL63 S glycoprotein occurs upon S2’ proteolytic processing at the plasma membrane (by trypsin-like proteases such as TMPRS2) or in the endosomal pathway (by furin or cysteine proteases)15,32. The lack of strict amino acid sequence conservation at the S2’ cleavage site among coronavirus S proteins reflects the usage of different proteases found in distinct cellular compartments for fusion activation15,17. Similarly to the additional cleavage site present between the S1 and S2 subunits of MERS-CoV7, protease sensitivity is likely further influenced by the presence of multiple glycans in the vicinity of the S2’ loop (Fig. 3B). However, we emphasize that S2’ processing occurs at topologically equivalent positions for HCoV-NL63 S, MERS-CoV S, MHV S and probably most coronavirus S glycoproteins.
Figure 3. Architecture of the complete coronavirus fusion machinery.
a, Ribbon diagram of the S2 trimer colored by protomer with glycans rendered as sticks. b, Blow-up view of the S2’ trigger loop region which is comprised between the central helix and the fusion peptide (light blue). N-linked glycans are shown as dark blue spheres. c–d, Ribbon diagrams showing two orthogonal views of the S2’ C-terminal region which is assembled from the connector domains and stem helices. The N- and C- terminal extremities of the polypeptide segments shown are indicated.
The connector domain and stem helix contribute to anchoring the trimer to the viral membrane
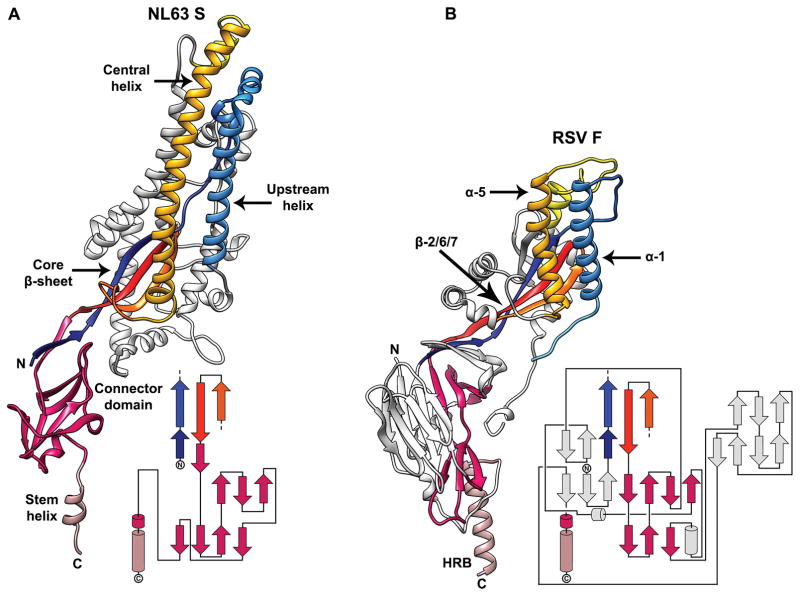
The HCoV-NL63 S reconstruction resolves a large part of the S2 C-terminal region that had not been observed in previous studies (Fig. 3A)18,33. Due to the significant resolution improvement, we were able to build an atomic model for the connector domain, so named as it connects to the HR2 region, and the stem helix. The connector folds as a β-rich domain decorated with one short α-helix. At its C-terminal end, the polypeptide chain folds as an α-helix (stem helix, Fig. 3A, C-D), aligned along the 3-fold molecular axis, which turns into the HR2 domain corresponding to 71 additional residues not resolved in our map. In the trimer, the connector domains assemble as a cup flanking the viral membrane proximal side of the ectodomain, and the stem helices form a bundle stabilized by hydrophobic interactions. The coronavirus S connector domain and the equivalent paramyxovirus F domain share a related topology although their tertiary structures are different and several structural motifs have been added to the latter domain throughout evolution34,35 (Fig. 4A–D). Moreover, the trimer of stem helices assemble as a helical bundle initiating the HR2 domain in a way reminiscent of the HRB region of paramyxovirus pre-fusion F structures34,35. These observations strengthen the evolutionary connection we previously proposed for the fusion machineries of these two viral families18. Comparison of the pre-fusion HCoV-NL63 S2 subunit with the structure of the post-fusion core implies that the C-terminal region of the connector domain and the stem helix must refold and/or change conformation to yield the canonical “trimer-of-hairpin” conformation that mediates fusion of the host and viral membrane in all class I fusion proteins18,36,37.
Figure 4. Structural conservation of coronavirus and paramyxovirus fusion machineries.
a–b, Ribbon diagrams of the HCoV-NL63 S2 subunit (a) and of the RSV F protein (b). The similar 3D organization of the core β-sheet, the upstream helix and the central helix among the two proteins are highlighted using identical colors. The topology diagrams underscore the fact that the HCoV-NL63 S connector domain and the equivalent RSV F domain also share a similar topology although their tertiary structures are different and several structural motifs have been added to the latter domain throughout evolution. The RSV F secondary structural elements are annotated according to34. The N- and C- terminal extremities of the polypeptide segments shown are indicated.
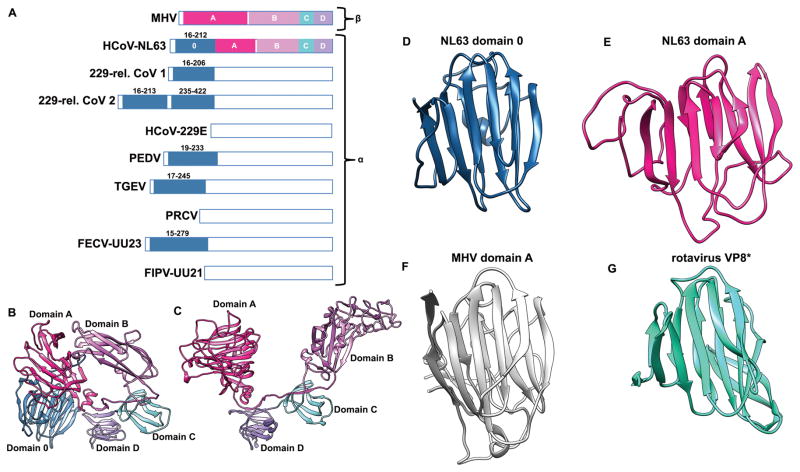
Duplication of the N-terminal domain in α-coronaviruses
The HCoV-NL63 S structure shows the presence of an additional N-terminal domain, relative to β-coronaviruses. Phylogenetic analyses suggest this is a canonical feature of most α-coronavirus S glycoproteins. (Fig. 5A–C). This domain, that we named domain 0, adopts a galectin-like β-sandwich fold supplemented with a three-stranded β-sheet similar to domain A (Fig. 5D–F, DALI Z-score 5.8, rmsd 4.1Å over 144 residues), suggesting a gene duplication event. Domain 0 interacts with the viral membrane proximal side of domain A and with domain D. We detected that domain 0 is also structurally similar to the VP8* sialic acid-binding domain of the rotavirus VP4 spike protein (Fig. 5G, PDB 1KQR, DALI Z-score 7.8, rmsd 3.1Å over 114 residues). In line with this finding, domain 0 of transmissible gastro-enteritis coronavirus (TGEV) and of PEDV bind to sialic acid and deletion of this domain in α-coronavirus S appears to correlate with loss of enteric tropism38. No sialic acid binding activity was detected for HCoV-NL63 domain 0 (Supplementary Fig. 5) and this might explain the strict respiratory tropism of this virus. Instead, host cell heparan sulfate proteoglycans have been shown to participate to HCoV-NL63 anchoring and infection39 and we detected binding of heparin sulfate to the HCoV-NL63 S protein using surface plasmon resonance (Supplementary Fig. 6 A). We hypothesize these interactions could be mediated either by domain 0, which exhibits several positively charged patches on its surface (Supplementary Fig. 6 B), or domain A that is known to bind carbohydrates in the case of bovine coronavirus40.
Figure 5. Evolution of the α-coronavirus S glycoprotein fold appears to correlate wit tissue tropism.
a, Schematic representation of several α-coronavirus S glycoproteins S1 subunits highlighting the presence of one or several domains 0 (blue) compared to β-coronaviruses. HCoV-NL63 (GB: YP_003767.1), 229-rel CoV 1 (GB: ALK28775.1), 229-rel CoV 2 (GB: ALK28765.1), HCoV-229E (GB: NP_073551.1), PEDV: porcine epidemic diarrhea virus (GB: AAK38656.1), TGEV: transmissible gastro-enteritis virus strain Purdue P115 (GB: ABG89325.1), PRCV: porcine respiratory coronavirus strain ISU-1 (GB: ABG89317.1), FECV-UU23: feline enteric coronavirus strain UU23 (GB: ADC35472.1), and FIPV-UU21: feline infectious peritonitis coronavirus strain UU21 (GB: ADL71466.1). The β-coronavirus MHV S1 subunit is shown for comparison. Domains A–D are indicated for MHV and HCoV-NL63. b, Ribbon diagram of the HCoV-NL63 S1 subunit. c, Ribbon diagram of the MHV S1 subunit. d–g, Ribbon diagrams of HCoV-NL63 domain 0 (d), domain A (e), MHV domain A (f) and rotavirus VP8* (g) showing their structural similarity which points to a common ancestry. HCoV-NL63 domain 0 and A likely arose from a duplication event.
Glycan masking and conformational changes appear to participate in immune evasion
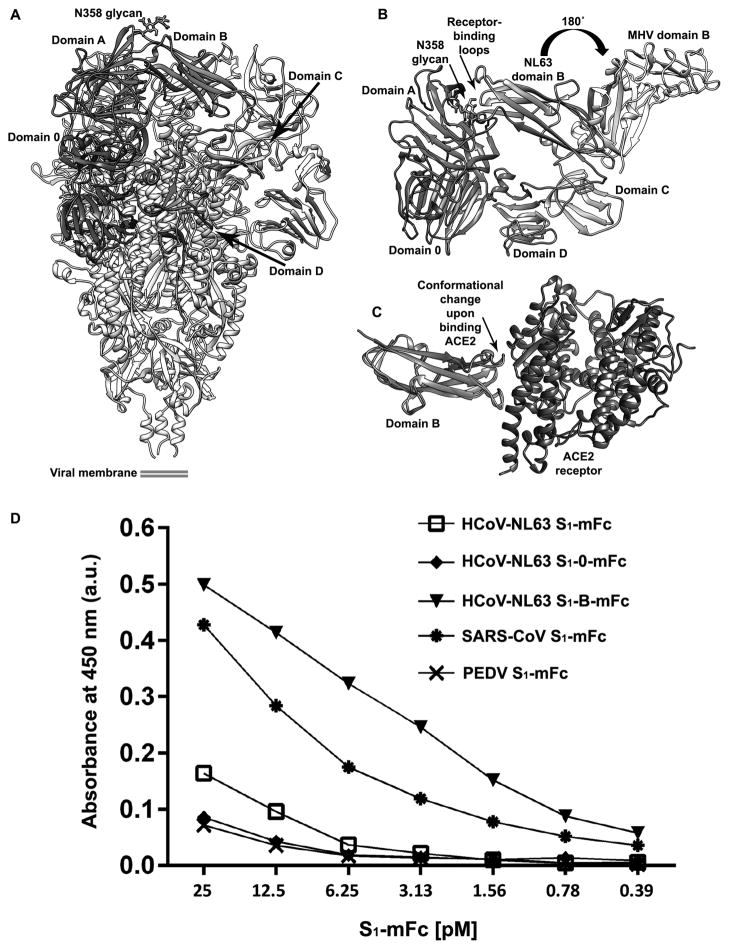
Domain B, which is the HCoV-NL63 receptor-binding domain, exhibits a structure distinct from β-coronavirus B domains although a topological relatedness could be detected among these β-rich domains41. Superimposition of the HCoV-NL63 and MHV S1 subunits highlights that their B domains feature opposite orientations related by a ~180° rotation (Fig. 6A–B). As a result, many of the HCoV-NL63 receptor-binding residues are either interacting with domain A belonging to the same protomer, including the glycan at residue asparagine 358, or are buried and not available to engage the host cell receptor (human angiotensin-converting enzyme 2, ACE2). Comparison of the HCoV-NL63 domain B structure in our cryoEM-derived model with the crystal structure of the same domain in complex with ACE242 reveals that the receptor-binding loop containing residues 531–539 significantly changes its conformation upon binding (Fig. 6C). These findings rationalize the markedly higher binding affinity of HCoV-NL63 domain B, compared to the full-length S1 domain, for ACE2 (Fig. 6D). Since the receptor-binding loops are known to elicit potent neutralizing antibodies in the case of TGEV43, MERS-CoV44 and SARS-CoV45–48, we speculate that HCoV-NL63 has evolved to limit exposure of this vulnerable site to B-cell receptors via protein-protein interactions and glycan masking. This mechanism is reminiscent of the HIV-1 immune evasion strategy which relies on a glycan shield and conformational changes triggered by binding of CD4 to expose the chemokine receptor-interacting motif (V3 loop)49,50.
Figure 6. Immune evasion strategy employed by HCoV-NL63.
a, Ribbon diagram of the HCoV-NL63 S trimer highlighting the conformation of the S1 subunit. Domains 0, A, B, C and D are colored for one protomer. b, The HCoV-NL63 receptor-binding loops are buried via interactions with domain A belonging to the same protomer (including the glycan moiety harbored by residue Asn 358) and are not available to engage host cell receptors. Superimposition of the HCoV-NL63 (purple) and MHV (light grey) S1 subunits via their C domains highlights that their B domains feature opposite orientations related by a ~180° rotation. This suggests a putative trajectory for the conformational changes that have to occur to engage the host cell receptor. Only domain C is shown for MHV S. c, Comparison of the HCoV-NL63 domain B structure in our cryoEM derived model (purple) with the crystal structure of the same domain in complex with ACE2 (green/dark grey) shows that the receptor-binding loop containing residues 531–539 significantly changes its conformation upon binding. d, Isolated HCoV-NL63 domain B binds ACE2 with higher affinity than the full-length S1 domain. The ability of the HCoV-NL63 S1 subunit and its receptor binding domain (S1-B) to bind to the ACE2 receptor was compared using an ACE2-binding ELISA. ACE2-coated ELISA plates were incubated with serial dilutions of equimolar amounts of murine Fc-tagged (mFc) S1 proteins after which bound mFc-tagged S1 proteins were detected with anti-mFc conjugate. SARS-CoV S1-mFc was used as a positive control. HCoV-NL63 S1 domain 0 (S1-0-mFc) and PEDV S1-mFc (that do not bind ACE2) were used as negative controls. Mean values and errors bars of three independent experiments are shown.
DISCUSSION
Viruses have evolved several immune evasion strategies including rapid antigenic evolution, masking of epitopes, and exposure of non-neutralizing immune-dominant “decoy” epitopes. For instance, HIV-151, Lassa virus52, Hepatitis C virus53 and Epstein Barr virus54 exhibit extensive N-linked glycosylation, covering exposed protein surfaces, and whose mass can exceed that of the protein component. The HCoV-NL63 S trimer is covered by an extensive glycan shield consisting of 102 N-linked oligosaccharides obstructing the protein surface. This observation is reminiscent of what has been described for the HIV-1 envelope trimer51 although the glycan density is 30% higher in the latter case. Furthermore, our data suggests that, similarly to HIV-1, coronavirus S glycans are masking the protein surface to limit access to neutralizing antibodies and thwart the humoral immune response. This strategy is illustrated by the presence of a glycan linked to asparagine 358 in the HCoV-NL63 structure reported here. This glycan contributes, along with the proteinaceous moiety of domain A, to masking the receptor-binding loops, that have been shown to elicit potent neutralizing antibodies in vivo43 and in vitro44, and appear to represent a potential Achilles’ heel of coronaviruses. This hypothesis is further supported by the observation of three additional glycans directly protruding from the viral membrane distal side of domain B. As a result, conformational changes are required for the HCoV-NL63 S glycoprotein to be able to interact with ACE242. These rearrangements and/or receptor binding likely participate to initiate the fusion reaction by disrupting the interactions formed between domain B and the HR1 C-terminal region. It is tempting to speculate that interactions with heparan sulfate proteoglycans present at the host cell surface could contribute to activating HCoV-NL63 S and promote subsequent interactions with ACE2. A common theme arising from the analysis of α- and β-coronavirus S glycoprotein structures is that domain B-mediated host anchoring involves major structural rearrangements to expose the binding motifs18,33.
Visualization of the glycan shield obstructing access to the S surface and deciphering the molecular trickery employed by some coronaviruses provides a rational basis to understand accessibility to neutralizing antibodies and paves the way for guiding future immunogen and therapeutics design. We previously suggested that targeting the fusion machinery bears the promise of finding broadly neutralizing inhibitors of coronavirus infection18 and the high-density of glycans decorating this region will have to be taken into consideration to increase the likelihood of success.
METHODS PROCEDURES
Supplementary Material
Table S1. Data collection and refinement statistics.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under Award Number 1R01GM120553-01 (DV) and T32GM008268 (ACW). JS acknowledges support from the Netherlands Organization for Scientific Research (NWO, Rubicon 019.2015.2.310.006) and the European Molecular Biology Organisation (EMBO, ALTF 933-2015). MAT and FR acknowledge support from the Institut Pasteur and the CNRS. The authors are grateful to Jason Labonte for providing the Rosetta code handling glycans. The authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057 and 1S10OD018111), NSF (DBI-1338135) and CNSI at UCLA. This work was also partly supported by the University of Washington’s Proteomics Resource (UWPR95794). Part of this research was facilitated by the Hyak supercomputer system (including the STF initiative) at the University of Washington.
Footnotes
AUTHOR CONTRIBUTIONS
A.C.W., M.A.T., F.R., B.J.B. and D.V designed the experiments. B.J.B. designed and cloned the protein construct. M.A.T., W. L. and B.J.B. carried out protein expression and purification. A.C.W. and D.V. performed cryoEM sample preparation, data collection and processing. A.C.W., B.F., F.D. and D.V built the atomic model. A.C.W and J.S performed the MS experiments and analyzed the data. W.L. and B.J.B. performed the hemaglutination and binding assays. A.C.W., M.A.T., B.F., F.D., F.R., B.J.B. and D.V. analyzed the data. A.C.W., J.S., B.J.B. and D.V. prepared the manuscript with input from all authors.
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interest.
Accession codes
The cryoEM map and the atomic model have been deposited to the electron microscopy and protein data banks with accession codes EMD-XXX and XYZ, respectively. The mass-spectrometry data have been deposited to PRIDE with accession code PXDXXX and includes the raw data, COMET search results and annotated tandem MS spectra of all accepted glycopeptide identifications.
References
- 1.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–47. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vijay R, Perlman S. Middle East respiratory syndrome and severe acute respiratory syndrome. Curr Opin Virol. 2016;16:70–6. doi: 10.1016/j.coviro.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge XY, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–8. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabir JS, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–4. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A. 2014;111:12516–21. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–37. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, et al. Two Mutations Were Critical for Bat-to-Human Transmission of Middle East Respiratory Syndrome Coronavirus. J Virol. 2015;89:9119–23. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Hoek L, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouchier RA, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004;101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann H, et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A. 2005;102:7988–93. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu SS, et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–9. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mole B. Deadly pig virus slips through US borders. Nature. 2013;499:388. doi: 10.1038/499388a. [DOI] [PubMed] [Google Scholar]
- 13.Du L, et al. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–36. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–11. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burkard C, et al. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A. 2014;111:15214–9. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millet JK, Whittaker GR. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–34. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walls AC, et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–7. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–4. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–53. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiMaio F, et al. Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–5. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang RY, et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat Methods. 2015;12:335–8. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–42. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frese CK, et al. Unambiguous phosphosite localization using electron-transfer/higher-energy collision dissociation (EThcD) J Proteome Res. 2013;12:1520–5. doi: 10.1021/pr301130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffers SA, Hemmila EM, Holmes KV. Human coronavirus 229E can use CD209L (L-SIGN) to enter cells. Adv Exp Med Biol. 2006;581:265–9. doi: 10.1007/978-0-387-33012-9_44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie G, et al. Identification of N-linked carbohydrates from severe acute respiratory syndrome (SARS) spike glycoprotein. Virology. 2010;399:257–69. doi: 10.1016/j.virol.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, et al. A single asparagine-linked glycosylation site of the severe acute respiratory syndrome coronavirus spike glycoprotein facilitates inhibition by mannose-binding lectin through multiple mechanisms. J Virol. 2010;84:8753–64. doi: 10.1128/JVI.00554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeffers SA, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A. 2004;101:15748–53. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86:6537–45. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchdoerfer RN, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–21. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–7. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–8. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Q, et al. Core structure of S2 from the human coronavirus NL63 spike glycoprotein. Biochemistry. 2006;45:15205–15. doi: 10.1021/bi061686w. [DOI] [PubMed] [Google Scholar]
- 38.Krempl C, Schultze B, Laude H, Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J Virol. 1997;71:3285–7. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milewska A, et al. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J Virol. 2014;88:13221–30. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng G, et al. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem. 2012;287:41931–8. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F. Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits. J Virol. 2012;86:2856–8. doi: 10.1128/JVI.06882-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu K, Li W, Peng G, Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci U S A. 2009;106:19970–4. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reguera J, et al. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ying T, et al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun. 2015;6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prabakaran P, et al. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem. 2006;281:15829–36. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang WC, et al. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem. 2006;281:34610–6. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sui J, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101:2536–41. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Z, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A. 2007;104:12123–8. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–41. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 50.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart-Jones GB, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016 doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommerstein R, et al. Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection. PLoS Pathog. 2015;11:e1005276. doi: 10.1371/journal.ppat.1005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falkowska E, Kajumo F, Garcia E, Reinus J, Dragic T. Hepatitis C virus envelope glycoprotein E2 glycans modulate entry, CD81 binding, and neutralization. J Virol. 2007;81:8072–9. doi: 10.1128/JVI.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szakonyi G, et al. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat Struct Mol Biol. 2006;13:996–1001. doi: 10.1038/nsmb1161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Data collection and refinement statistics.