Abstract
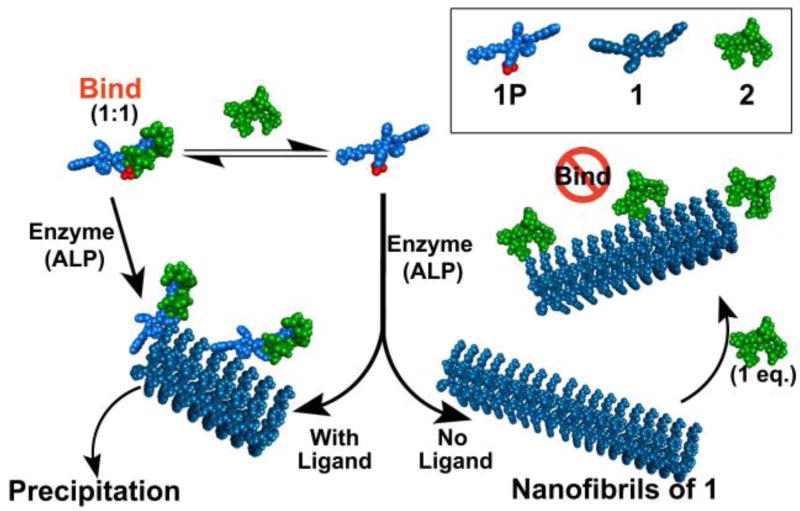
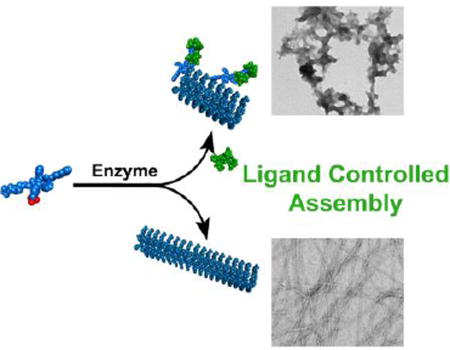
The concurrence of enzymatic reaction and ligand-receptor interactions is common for proteins, but rare for small molecules and has yet to be explored. Here we show that ligand-receptor interaction modulates the morphology of molecular assemblies formed by enzyme-instructed assembly of small molecules. While the absence of ligand-receptor interaction allows enzymatic dephosphorylation of a precursor to generate the hydrogelator that self-assembles to form long nanofibers, the presence of the ligand-receptor interaction biases the pathway to form precipitous aggregates containing short nanofibers. While the hydrogelators self-assemble to form nanofibers or nanoribbons that are unable to bind with the ligand (i.e., vancomycin), the addition of surfactant breaks up the assemblies to restore the ligand-receptor interaction. In addition, excess amount of the ligands can disrupt the nanofibers and result in the precipitates. As the first example of the use of ligand-receptor interaction to modulate the kinetics of enzymatic self-assembly, this work not only provides a solution to evaluate the interaction between aggregates and target molecules, but also offer new insight for understanding the emergent behavior of sophisticated molecular systems having multiple and parallel processes.
Table of Contents
Introduction
This article reports the first use of the ligand-receptor interaction to regulate enzymatic self-assembly and emergent properties of the assemblies of small molecules. Self-assembly of small molecules is a thermodynamically favorable process during which small molecule monomers assemble to form large supramolecular structures.1–9 Typically these supramolecular structures are static with properties dictated by their constituents.4 In nature, however, it is dynamic supramolecular structures and emergent properties of the assemblies which are the most prevalent.10–11 A prominent process is reversible protein phosphorylation and dephosphorylation that regulates many essential cellular functions.12 For example, tyrosine phosphorylation in VAV protein is a key mechanism in regulating the ligand-receptor interaction, thus further activating enzymes for immune responses.13 Additionally, the complex protein folding process has a well-established reliance on dephosphorylation of ATP by chaperone proteins.14–15 Meanwhile, immunological studies show that enzymatic transformation generates death ligands (e.g., TNF, TRAIL), which bind to cell death receptors to initiate oligomerization processes that control cell fates. A fundamental feature of these processes in living systems is the concurrence of enzymatic reaction and ligand-receptor interactions (e.g., enzymes or pseudo enzymes as molecular scaffolds for self-assembly),16 which results in sophisticated control of protein-protein interactions. This fact raises the possibility of employing small molecules to mimic this essential process for modulating protein-protein interactions, which would be a novel strategy for developing new therapeutics. Despite their significance, such an approach has received limited exploration because of the limited number of well-defined ligand-receptor systems of small molecules. Recently, we reported that enzymatic reaction is able to dimerize the ligand to mimic the activation of signal transduction.17 It would be highly desirable to use ligand-receptor interactions to modulate the outcome of enzyme-instructed self-assembly (EISA)18–23 of small molecules because morphological differences of the nanoscale assemblies may elicit different cellular responses.24–28 However, the use of ligand-receptor interactions for precisely controlling the kinetic behavior of small molecules remains challenging.
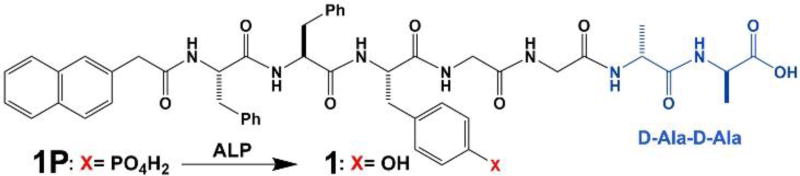
To understand the complex behavior of sophisticated molecular systems undergoing multiple and/or parallel processes, we choose to explore small molecules that are substrates of enzymes and are participants of ligand-receptor interactions. Specifically, we synthesized a small heptapeptide, Nap-FFYGGaa (1), which self-assembles to form nanofibers or nanosheets in aqueous solution. Phosphorylated 1 (i.e., 1P, phosphorylated at tyrosine residue) is a substrate of alkaline phosphatase (ALP) and a receptor of vancomycin (2). Our study reveals that the assemblies of 1 exhibit emergent properties of assembled molecules,29–30 which drastically affect the ligand-receptor interaction between the assemblies and the ligand, in effect “switching off” the ligand-receptor interaction between 2 and 1. On the other hand, the ligand-receptor interaction between 2 and 1P is able to bias EISA of 1 to generate aggregates containing short nanofibers, an observation that differs from EISA of 1 in the absence of 2. Additionally, during EISA of 1 in the presence of 2, short fibers emerge first, followed by aggregation and disruption of fibers, leading to formation of a precipitate. This transient fiber formation is coupled with a time-dependent change in the viscoelastic properties of the solution of 1, 2, and ALP, which is not observed when one of the components is missing, the hallmark of emergent behavior. As shown in Scheme 1, immediately after enzymatic dephosphorylation, 1 is ‘monomeric’ and is able to bind the ligand (2) or the enzyme can dephosphorylate the complex of 1P and 2. Thus, the binding between 1 and 2 favors an alternative pathway of assembly (i.e., different from the supramolecular polymerization of 1 observed without 2), leading to precipitation. While 2 shows no measurable binding to assemblies of 1, the addition of a surfactant Tween-80 (Tw-80) breaks up the assemblies of 1 and restores binding between 1 and 2. Isothermal titration calorimetry (ITC) measurement in the presence of surfactant, in fact, serves as a facile method to study interactions between ligands and receptors when either are prone to aggregate.
Scheme 1.
Illustration of ligand-receptor interaction of small molecules dictating the pathways of enzyme-instructed self-assembly.
While proteins usually change their conformation or shape upon enzymatic reaction to regulate ligand-receptor interaction for specific functions, as shown in the case of inherently disordered proteins,31–32 small molecules, in general, barely exhibit large conformational differences upon enzymatic reaction. As the first example illustrating reciprocal modulation between ligand-receptor interaction and enzymatic self-assembly, this work provides useful insights for developing nanoscale assemblies of small molecules for controlling biological and cellular processes, understanding the emergent behavior of sophisticated molecular systems undergoing multiple and parallel processes, and further offers a general approach to control the transformation of small molecules in the context of ligand-receptor interactions.
Results and Discussion
Molecular design and synthesis
We chose vancomycin (2) and a D-Ala-D-Ala derivative (1) as the ligand-receptor pair because of their well-established and specific interactions, as demonstrated by Walsh,33–34 Williams,35 Whitesides,36–37 as well as other groups.38–39 Recently, we have shown that ligand-receptor interaction modulates the cytotoxicity of molecular aggregates.40–41 Encouraged by these results, we designed a small molecule (Nap-FFpYGGaa (1P)) and the hydrogelator 1. The heptapeptide and various derivatives (Scheme 2) were synthesized by standard solid-phase peptide synthesis (SPPS) procedures (see Supporting Information) on a 2-chlorotrityl chloride resin,42 further purified by HPLC on a reverse phase C18 column, and lyophilized to give the peptides as fine white powders in approximately 80% yield. Phosphorylated tyrosine was synthesized following previously reported methods43 and the free amine further protected by an Fmoc group for SPPS. The overall yield of 1P is about 80%, based on resin loading.
Scheme 2.
Molecular structures of the heptapeptidic precursor 1P and its corresponding hydrogelator 1.
Ligand modulates enzymatic self-assembly
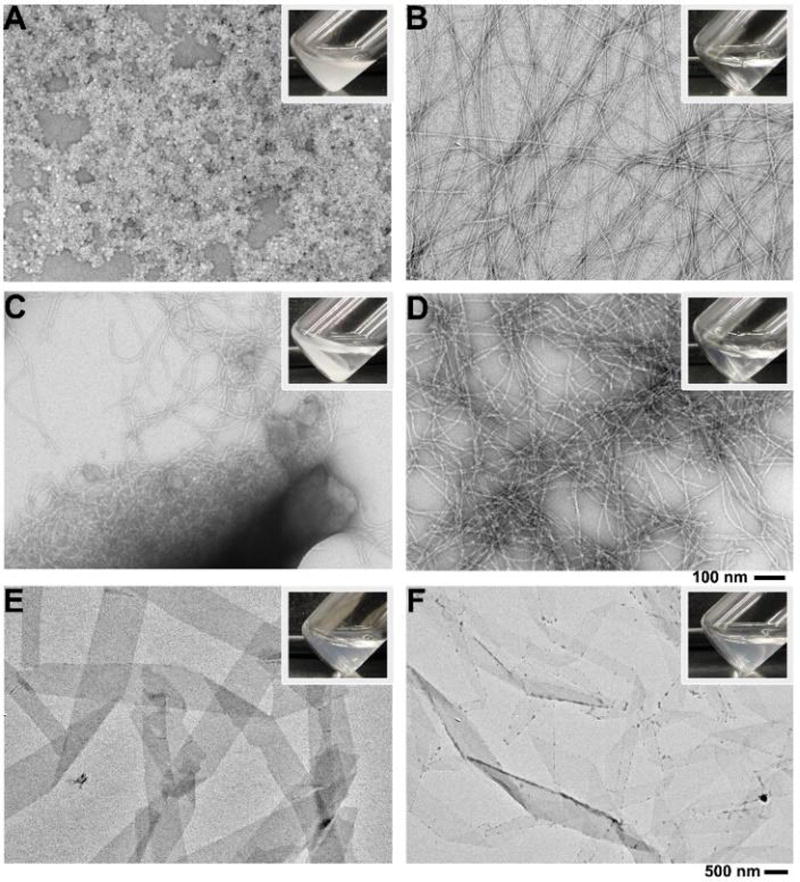
To understand how self-assembly affects the ligand-receptor interaction and how the ligand-receptor interaction modulates the process or behavior of enzymatic self-assembly, we used ALP to catalyze dephosphorylation of 1P without and with the presence of 2. As shown in Figure 1A, without ALP, the addition of one equivalent of 2 into the solution of 1P (500 µM) results in a colloidal suspension, which forms precipitates depending on pH and concentration, indicating strong intermolecular interaction between 1P and 2. Transmission electron microscopy (TEM) shows that the morphology of the aggregates is largely unstructured (Figure 1A). In the absence of 2, ALP (1.25 U/mL) converts 1P to 1, which, as expected, self-assembles to form long nanofibers (Figure 1B). When the concentration of 1P is 500 µM, the enzyme-induced formation of the nanofibers results in a viscous mixture. The simultaneous addition of 2 (1 eq.) and ALP (1.25 U/mL) into a solution of 1P (500 µM) induces formation of large aggregates, which cluster together and form precipitates (Figure 1C) over time. Although a self-supporting gel was only made upon changing the pH of a 4 mM solution of 1 to pH 6.4 (SI Figure S2), the addition of ALP into the solution of 1P (500 µM) and 2 (500 µM) also yield gel pieces that are sufficiently stable for rheology measurement (Figure S3&S4). These results confirm that 1 is a hydrogelator. The formation of precipitates over time agrees with an observed decrease in the storage modulus or critical strain of the mixture over 24 h (Figure S3). Furthermore, the time-dependence of the changes in storage and loss moduli are an emergent property of the combination of all three components. However, two days after using ALP to generate 1 from 1P, the addition of one equivalent of 2 to the solution of 1 hardly yields any precipitates after 24 hours. TEM reveals that the nanofibers (Figure 1D) are similar to those formed by mixing 1P and ALP without the post-self-assembly addition of 2. This result indicates that, being a kinetically trapped state following EISA of 1, the assemblies of 1 hardly favor binding with 2. Interestingly, while the direct addition of 1 in water results in a suspension consisting of nanoribbons (Figure 1E), TEM reveals small unstructured aggregates on the edge of the nanoribbon of 1 after the addition of 2 (Figure 1F). On the other hand, the addition of five equivalents of 2 into a 500 µM suspension of 1 almost completely destroys the nanoribbons formed by 1 and affords an opaque colloidal precipitate (Figure S5) 24–48h after the addition, indicating that high concentrations of 2 shift the equilibrium towards binding between 1 and 2.
Figure 1.
TEM images of (A) suspension of 1P and 2, (B) 1P and ALP, (C) 1P, 2, and ALP, (D) 1P treated with ALP for 2 days, then 2 was added. (E) 1, and (F) 1 and 2 in water. [1P]0 = [1]0 = [2]0 = 500µM, ALP = 1.25 U/mL, pH=7.4.
ITC of binding and stability of the assembly
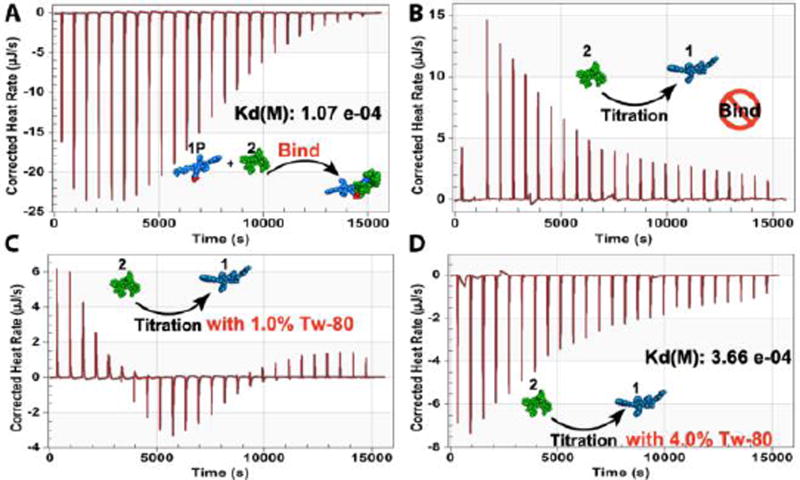
To investigate the interaction between 2 and the D-Ala-D-Ala derivatives (i.e., 1P and 1), we used ITC to estimate the dissociation constant (Kd) of the binding between 1P and 2. As shown in Figure 2A, 1P binds with 2 in a 1:1 ratio, with a Kd of 108 µM. This result agrees well with relatively tight binding between 2 and D-Ala-D-Ala. Titration in the presence of ALP appears to give a dissociation constant of 10 µM (Figure S6). This result suggests that the dephosphorylation process and subsequent self-assembly still permit the binding of 2 with 1P or 1. In contrast, assemblies of 1 hardly bind with 2. The heating profile (Figure 2B) of titration of 2 into a suspension of 1 is similar with that of the dissociation of the dimers of 2 (Figure S7), suggesting that the interaction between assembled 1 and 2 is too weak to be measured by ITC. In fact, this is the first case where 2 shows negligible binding with a D-Ala-D-Ala derivative.
Figure 2.
ITC of (A) 1P, and (B–D) 1 with 2 at different concentrations of Tw-80 for the determination of dissociation constant (Kd) and stoichiometry (n). Negative peaks indicate an exothermic release of heat.
Considering that the assemblies of a hydrogelator containing D-Ala-D-Ala,17,44 in a previous report, are able to bind with 2, the lack of binding between 2 and the assemblies of 1 is surprising. To understand this result, we synthesize control molecules Nap-ffpyGGaa (3P) and the corresponding self-assembling molecule, 3, by replacing the L-amino acid residues, FFY, in 1P or 1, with their D-amino acid enantiomers, ffy. Like 1P, 3P binds with 2 with a Kd of 82 µM. Similarly to the assemblies of 1, the assemblies of 3 barely bind with 2 (Figure S8). This control experiment excludes the possibility that the conformation of the receptor (i.e., 1 or 3) weakens the binding between 1 (or 3) and 2. Moreover, this result confirms that the assemblies of small molecules, indeed, differ considerably from their monomeric building blocks. As expected, upon the mutation of D-Ala-D-Ala to L-Ala-L-Ala, the resulting molecule, 4P (or 4), is unable to bind with 2 (Figure S8), which further confirms that the ligand-receptor interaction between 2 and 1P, 1, 3P or 3 still relies on the binding between vancomycin and D-Ala-D-Ala.
To compare the complex parallel processes occurring during the ITC experiments and to gain insights into the supramolecular behavior observed in the TEM in Figure 1, we examined the total heat released over the entire ITC experiment. We carried out three sets of titrations (in PBS buffer and at pH 7.4): 1P to (i) 2 alone, (ii) to ALP alone, and (iii) to a solution of 2 and ALP ([ALP]0 = 1.67 U/mL). Integrating the heat release profiles for each titration yields the total heat released for each titration, from which the total heat released by dilution of 1P is subtracted. The total heat release is −4261 µJ for titration of 1P to 2, −4653 µJ for titration of 1P to ALP, and −7186 µJ for titration of 1P to 2 and ALP (Figure S9), indicating that formation of nanofibers by dephosphorylation of 1P is more enthalpically favorable than binding of 1P and 2 alone. Importantly, the heat released from binding of 1P and 2 is comparable to heat released from dephosphorylation of 1P. This result agrees with hardly any disruption of fibers of 1 formed by dephosphorylation of 1P following the addition of 2 (Figure 1D). However, the fibers of nanosheets of 1, being kinetically trapped, can slowly transform over several weeks to months to a precipitate after the addition of five equivalent of 2. Furthermore, as the total heat released by titration of 1P to a solution of 2 and ALP is significantly larger than the heat released by simple dephosphorylation of 1P, 2 likely provides a low energy intermediate along the fibrilization pathway of 1, which drives the hydrogelator to form the unstructured aggregates that contain 1 and 2 (Figure 1C).
Addition of surfactant restores the ligand-receptor interaction
A series of studies by Shoichet et. al. have revealed that the aggregation of small molecules in water usually leads to false positives (up to 95%) in drug screening,45–46 which certainly is a form of abnormal binding. In addition to indicating the aggregates of small molecules is a rather general phenomenon, those results also imply that the assemblies can cause false negatives (i.e., the lack of ligand-receptor binding between 2 and 1). To verify that ‘monomeric’ 1 still is able to bind to 2, we employed a nonionic surfactant, Tw-80, to disrupt assemblies of 1. In the presence of 1.0 wt% of Tw-80, the appearance of negative peaks in the ITC heating profile indicates a release of heat (Figure 2C), likely due to molecular binding between 1 and 2. Upon increasing the Tw-80 concentration to 4.0 wt%, heating release dominates the whole titration process (Figure 2D), and fitting by an independent binding model gives a dissociation constant of 366 µM. This result confirms that surfactant restores the ligand-receptor interaction between 2 and 1. Moreover, this result suggests that using surfactant during ITC offers a facile method to study binding between ligands and aggregate-prone receptors. We further used ITC in the presence of Tw-80 to measure the binding of 2 and 3. Similar to the case of 1, the addition of 4.0 wt% Tw-80 recovers the ligand-receptor binding between 2 and 3 (Figure S10). In addition, the heating profile of the titration of 2 into a solution of 4 shows negligible change over various concentrations of Tw-80 (Figure S11). These results indicate that the surfactant itself has little contribution to heat released during the titrations shown in Figures 2C and 2D. The observed heat release likely originates from the interaction of 2 and the monomeric 1 after the surfactant disrupts the assemblies of 1.
Surfactant breaks the assemblies and restores binding
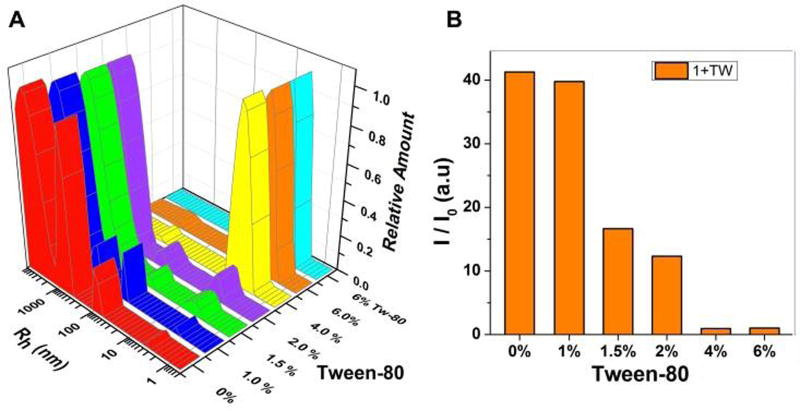
To verify the effect of Tw-80 on assemblies of 1, we used dynamic light scattering (DLS) to monitor the light scattering signal and hydrodynamic radius (Rh) of suspensions of 1 with different amounts of Tw-80 (Figure 3). As seen in Figure 3A, as the Tw-80 concentration increases from 1.0 wt% to 2.0 wt%, a peak representing species with an Rh ranging from 3–10 nm starts to grow. When the concentration of Tw-80 is increased to 4.0 wt%, this peak exhibits significant growth, accompanied by the disappearance of the peaks corresponding to the assemblies (larger than 100 nm), indicating that the addition of Tw-80, indeed, disrupts the assemblies of 1. Meanwhile, the light scattering signal of 1 decreases gradually with increasing Tw-80 concentration (Figure 3B). After the addition of 4.0 wt% Tw-80 into a suspension of 1, the light scattering signal decreases significantly, and is almost identical to a solution of 4.0 wt% Tw-80 alone (Figure S13). This result not only confirms that assemblies of 1 dissociate upon the addition of Tw-80, but also suggests that the dissociated species are too small to scatter light. TEM images (Figure 4) show that, at a higher Tw-80 concentrations, the long dense nanosheets of 1 become low density short nanosheets, with only a few small fibrils remaining. Finally, at 4.0 wt% Tw-80, nanoparticles dominate (Figure S15). These results confirm that Tw-80 disrupts the assemblies of 1 into oligomers.
Figure 3.
Dynamic light scattering measurements showing (A) hydrodynamic radii and (B) light scattering signals (I/I0) for the solution of 1 (800 µM) with various concentrations of Tw-80 (wt%).
Figure 4.
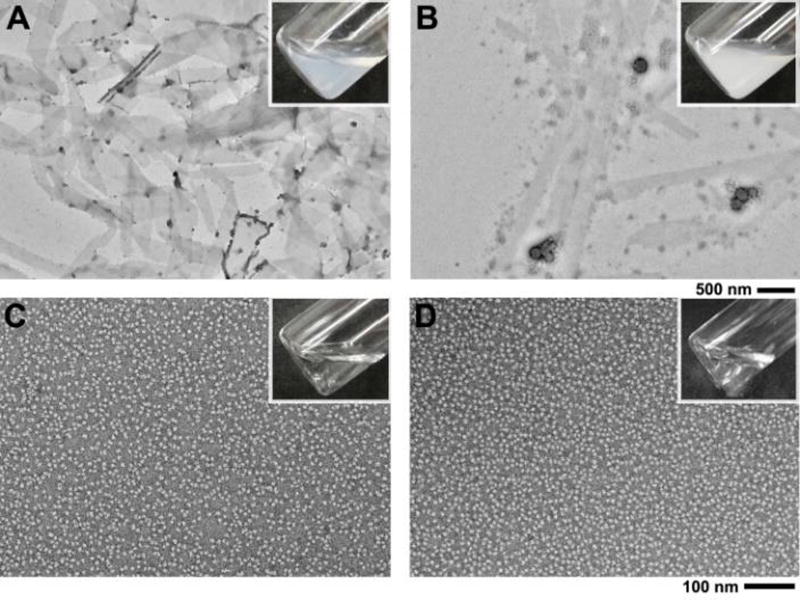
TEM images of suspensions of 1 and 2 with (A) 0 wt% (B) 1.0 wt% (C) 4.0 wt% of Tw-80 or TEM of (D) only 4.0 wt% of Tw-80. Inset are their optical images. [1]0 = [2]0 = 800 µM.
After the DLS study of the effect of Tw-80 on assemblies, we used TEM to examine the morphological properties of 1 and 2 under various amounts of Tw-80. TEM micrographs of the colloidal solution of 1 and 2 shows large amounts of nanosheets with dots along the edges, likely unstructured aggregates of 1 and 2. The presence of nanosheets and relatively few aggregates agrees well with no measurable ligand-receptor interaction between 2 and assembled 1. After adding 1.0 wt% Tw-80 into a suspension of 1 and 2, more unstructured aggregates form and nanosheets still remain (Figure 4B). Meanwhile, optical images clearly show the formation of precipitates. This result indicates that as 1.0 wt% Tw-80 breaks up assemblies of 1, the oligomers released are able to bind with 2 to form aggregates, which bind together to form precipitates. Such an observation is consistent with our previous results that as 1P is converted to 1, the forming hydrogelator binds with 2 to induce aggregation. At 4.0 wt% Tw-80, the aggregates disappear to give a clear solution, consisting of nanoparticles with a diameter of 12 ± 2 nm (Figure 4C), which is almost identical to a solution only containing 4.0 wt% Tw-80. This result indicates that, due to the strong dissolution of Tw-80, the complex of 1 and 2 is unable to form large aggregates. Together with DLS data, this result also confirms that 4.0 wt% Tw-80 completely breaks up assemblies of 1 to monomeric or oligomeric 1, which binds with 2 (i.e., restores the ligand-receptor interaction).
Morphological evolution
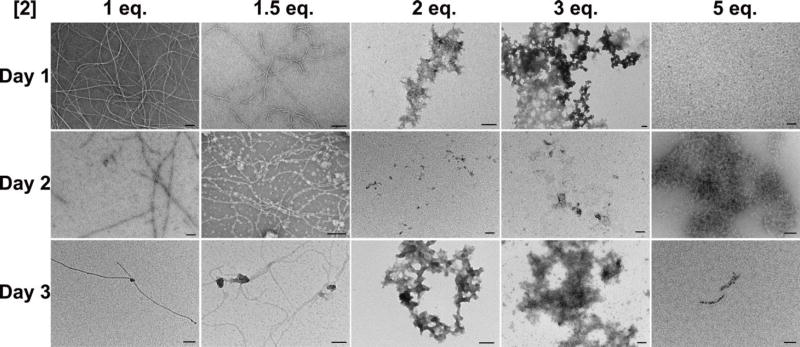
As demonstrated for both small molecule supramolecular polymerization41,47–49 and for assembly of amyloid proteins,50–52 aggregate morphology and even toxicity has a strong dependence on initial conditions and the aggregation pathway. A time-dependent study of the gelation and precipitation behavior of 1P in the presence of 2 and ALP reveals that the morphology and self-assembling behavior of 1 correlate with the concentration of 2. As shown in Figure 5, the morphologies of the fibers or amorphous precipitates formed after the addition of 2 to a solution of 1P and ALP exhibit a strong dependence on the concentration of 2. The addition of one equivalent of 2 is unable to prevent the formation of long nanofibers, but can turn the nanofibers into short fibers within 48 hours, accompanied by forming a precipitate identical to that in Figure 1C. While the addition of 1.5 equivalents of 2 decreases the nanofiber density, the addition of two or three equivalents of 2 results in precipitation within 24 hours. The addition of five equivalents of 2 completely prevents the formation of long nanofibers. These results indicate that, at pH 7.6, nanofibers are a metastable state along the precipitation pathway, suggesting that the interaction between 1 and 2 leading to precipitation is indeed energetically more favorable than self-assembly of 1 alone.
Figure 5.
TEM micrographs taken on three consecutive days of suspensions of 1P, 2, and ALP, with varying concentrations of 2. [1P]o = 500 µM, ALP = 1.25 U/mL, pH = 7.6, each scale bar is 100 nm.
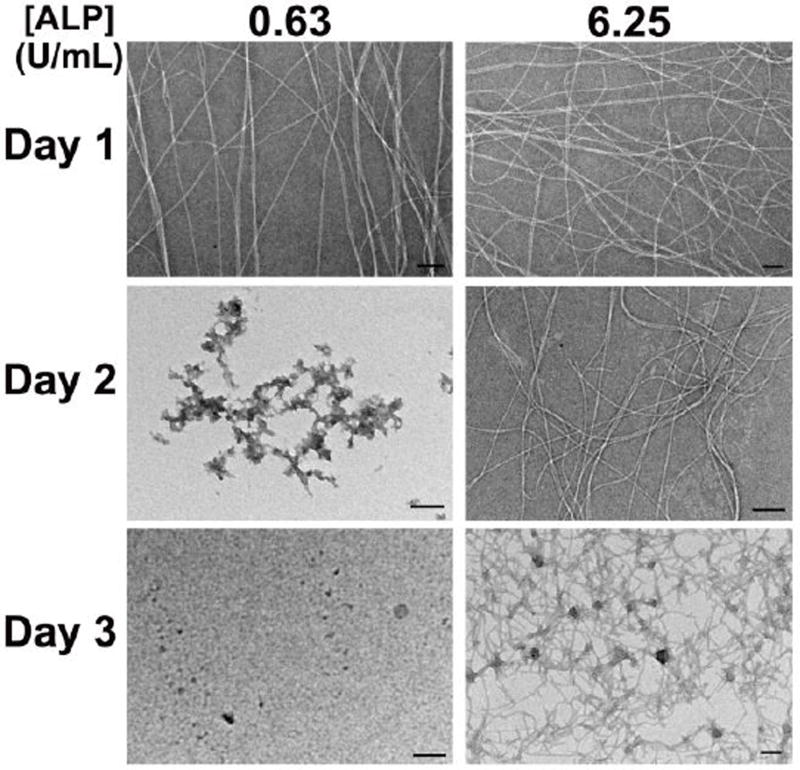
Variation of ALP concentration gives similar results, with higher ALP concentrations giving rise to more dense nanofibers prior to precipitate formation (Figure 6, and Figure S19). Surprisingly, at an ALP concentration of 6.25 U/ml, nanofibers remained on the third day, indicating increased order of the nanofibers of 1 formed at higher enzyme concentration, similar to enzyme-induced order of supramolecular polymerization reported by Ulijn et al.18,53 There also is an alternative explanation to the stability of the fibers formed at 6.25 U/mL ALP. Higher enzyme concentration likely gives higher concentrations of 1. Fibers may form with a combination of 1 and 1P. Hence higher relative concentrations of 1 would likely give fibers with a higher composition of 1 relative to 1P, leading to more stable structures.
Figure 6.
TEM micrographs taken on three consecutive days of suspensions of 1P, 2, and ALP with varying concentrations of ALP. [1P]0 = [2] = 500µM, pH = 7.6, each scale bar is 100nm.
In addition, dynamic oscillatory rheology confirms the disintegration of the gels formed by dephosphorylation of 1P in the presence of 2 and ALP. While both the storage and loss moduli are frequency independent, the storage and loss moduli of the gels formed by EISA of a solution of 500 µM 1P with 1.25 U/mL ALP in the presence of one equivalent of 2 decrease about an order of magnitude from 24 h to 48 h (Figures S3 and S4), indicating that 2 promotes the dissociation of the gel matrix. Hence both rheology and TEM confirm that the self-assembled fibers are a transient structure, indicative of a local energy minimum, with binding and subsequent precipitation of 1 and 2 being the global energy minimum.
Modulation of the Free Energy Landscape
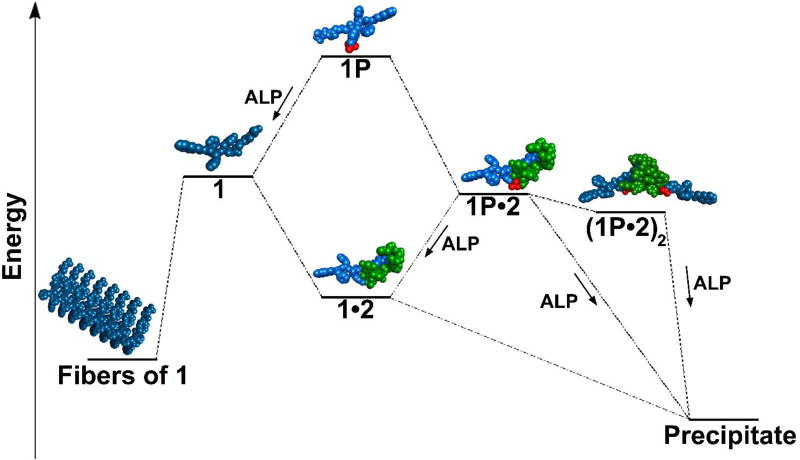
The transient formation of the nanofibers during EISA and the relative stability of assemblies of 1 against further precipitation after the addition of 2 indicate the presence of multiple structurally diverse intermediates along the fibril formation pathway. These intermediates likely interact with 2 to divert self-assembly away from fibril formation. Specifically, transient formation of nanofibers upon EISA of 1P and further deterioration of assembled 1 into a precipitate of 1 and 2, together with the observation that the addition of Tw-80 disrupts assemblies of 1 and restores binding between 1 and 2, indicates that either (i) a common di-, tri-, or oligomeric intermediates of 1 exist for both nanofiber assembly and precipitation or (ii) these pathways share only monomeric 1. Figure 7 illustrates the plausible energy landscape for EISA of 1P both in the presence and absence of 2. Containing an enzyme catalyzed step, EISA is inherently under kinetic control, and hence observations at thermodynamic equilibrium offer little information on perturbation of EISA by 2. However, analysis of time-dependent TEM, ITC, and the responses of the system to different concentrations of ligand provide insights into these kinetic pathways. Without 2, EISA of 1P follows a simple pathway illustrated on the left of Figure 7, whereby 1P (or oligomeric 1P) first undergoes dephosphorylation to provide 1, which further assembles to form nanofibrils. Although it is possible that 1P may form micelles before dephosphorylation,54 such a scenario is less likely in the presence of 2 because 2 binds to 1P and disfavor the formation of micelle (as shown in Figure 1A). However, in the presence of 2, the entire energy landscape appears to be available, yielding a complex mixture of species. 1P may bind with 2 in solution creating a lower energy species (1P•2) than simply ‘monomeric’ 1P, which can be further dephosphorylated to form 1•2, and may further assemble to yield precipitates. Additionally, as dimerization of 2 is well known,35 a dimeric complex (1P•2)2 of 2 and 1P and/or 1 is likely to form instead of the complex (1P•2), which further aggregates following dephosphorylation to form a precipitate. Aggregation is likely driven by the high local concentration of 2 caused by dimerization and perhaps also by the ability for 2 to promote dimerization of a peptide hydrogelator, as we have previously shown.44 The results in Figure 5 indicate that more than 1 equivalent of 2 is needed for the formation of the precipitate and 2 is part of the precipitate, thus 2 unlikely catalyzes the fiber to precipitate conversion. Importantly, when vancomycin aglycon55–56 replaces 2, no aggregates were observed (Figure S20); however, short fibers similar to those in Figure 5 were observed, indicating that vancomycin aglycon destabilizes assemblies of 1 in a similar fashion to 2. Because the glycogen of 2 is essential for dimerization in water, this result indicates that dimerization of 2 allows the formation of large structures, thereby promoting further aggregation. In addition, this observation supports formation of the dimeric complex (1P•2)2 as a key intermediate in the precipitation pathway. Therefore, during EISA of 1P, the presence of 2 allows for formation of lower energy complexes with 1P or 1, diverting the supramolecular aggregation by creating a lower energy pathway, similar to molecular catalysts or molecular chaperones.57
Figure 7.
Qualitative energy landscape for the multiple paths of the assembly or precipitation of 1 and 2 based on thermodynamic data from ITC and relative stabilities of assemblies from TEM studies showing the role of 2 in stabilizing precipitation pathways. Oligomers of 1P and 1 which likely exist are left out for clarity.
In addition to 2 diverting assembly of 1 during EISA, TEM reveals that 2 destabilizes the assemblies of 1. The addition of one equivalent of 2 to the nanofibers of 1 is unable to lead to precipitation (Figure 1D), while the addition of five equivalents of 2 to nanosheets of 1 gives an opaque colloidal suspension paired with disruption of the nanosheets of 1 (Figure S4). Additionally, binding between 1 and 2 is restored upon the addition of Tw-80 that breaks up assemblies of 1, as evidenced by ITC (Figure 2d) and TEM (Figure 4), indicating that 1 and 2 can interact. Hence although assemblies of 1 barely revert to oligomers or monomers that can bind with 2, sufficiently high concentrations of 2 can perturb the energy landscape and pull the equilibrium towards binding of 1 and 2. Based on this, there are two plausible ‘mechanisms’ for 2 breaking the assemblies of 1. (i) Free 2 in solution can bind with monomeric or oligomeric 1 and initiate precipitation, thereby lowering the concentration of 1 in solution, leading to deterioration of large assemblies of 1. (ii) 2 may bind directly to assemblies of 1 leading to destruction of the assemblies and formation of an intermediate species of 1 and 2, followed by precipitate formation. Direct binding of 2 to the assemblies of 1, however, is unlikely as ITC showed little release of heat upon titration of 2 to assembled 1 (Figure 2b). Hence precipitation caused by 2 relies on either reversible supramolecular polymerization of 1 or else on the presence of intermediate species of 1 in solution. Therefore, transient formation of fibers of 1 is indicative of the dynamic nature of EISA, a process that dynamically evolves based on atomistic interactions between precursors (1P) of the self-assembling molecules (1) as well as ligands (2), eventually bringing the system to kinetic or thermal equilibrium dependence on both the initial and boundary conditions of the system.
Conclusion
The assembly or aggregation of proteins or peptides remains one of the most significant problems in biology and medicine, especially associated with diseases like Alzheimer’s disease (AD).58–63 The path taken, however, depends strongly on the initial conditions of the system, as well as intrinsic kinetic factors such as enzyme activation or critical nucleus formation. Recently, ionic strength was found to modulate the energy landscape of Aβ40.51 However, control over peptide concentration and initial state remains difficult.51 While it remains to prove that ligand-receptor interactions may modulate the aggregation of Aβ50 this study on how interaction with a ligand significantly alters supramolecular assembly of small molecules should provide useful insights. Because the formation of Aβ results from enzymatic reactions,64–65 the study of ligand-receptor interactions to modulate EISA of small molecules is more relevant to the disease condition than using hexafluoroisopropanol (HFIP) or dimethylsulfoxide (DMSO) to generate Aβ amyloids.66 In fact, we used HFIP to form the nanofibers of 1, and found that using HFIP leads to various different morphologies (Figure S21).
Moreover, it is well known that in drug screening small molecules hits with high aggregation potentials are poor candidates due to unpredictable efficacy of ligand-receptor interactions.45–46 This fact not only implies that self-assembly of small molecules should modulate specific ligand-receptor interactions, but also suggests a limited knowledge about the molecular interactions between aggregates and their target molecules. EISA creates multiple processes which run in parallel, while also providing control over aggregating peptide concentration. Additionally, this experimental system should be useful for the study of the kinetics of the interconversion of the molecular species, though one has to obtain accurate rate information of the reactions.
Although being extensively used by nature for controlling important cellular functions such as apoptosis67 and immune responses,68 exploration of EISA in the context of small molecules is at its infancy.26,69–77 Recently, EISA has found applications in selective inhibition of cancer cells22,78–82 or targeting tumors in an animal model,83 but enzymatic control over ligand-receptor interactions of small molecules has yet to be investigated. This work, thus, provides necessary understanding to develop EISA in sophisticated environments with prevailing ligand-receptor interactions. Hence this study demonstrates that perturbation of assembly can be accomplished through modulation of the relative energies of intermediate species. In a more general sense, the insights obtained in this work would contribute to the exploration of supramolecular chemistry in cellular milieu.
Supplementary Material
Acknowledgments
This work was partially supported by NIH (R01CA142746), the W.M. Keck Foundation.
Footnotes
ASSOCIATED CONTENT
Details of the synthesis, TEM images, ITC. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.JL Atwood J-ML. Comprehensive Supramolecular Chemistry: Templating, self-assembly, and self-organization. Vol. 9 Pergamon; Oxford: 1996. [Google Scholar]
- 2.Fréchet JMJ. Proc. Natl. Acad.Sci., USA. 2002;99:4782. [Google Scholar]
- 3.Kato T. Science. 2002;295:2414. doi: 10.1126/science.1070967. [DOI] [PubMed] [Google Scholar]
- 4.Whitesides GM, Grzybowski B. Science. 2002;295:2418. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 5.Aida T, Meijer EW, Stupp SI. Science. 2012;335:813. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Morikawa M-a, Kimizuka N. J. Am. Chem. Soc. 2011;133:17370. doi: 10.1021/ja2057924. [DOI] [PubMed] [Google Scholar]
- 7.Wong KM-C, Yam VW-W. Acc. Chem. Res. 2011;44:424. doi: 10.1021/ar100130j. [DOI] [PubMed] [Google Scholar]
- 8.Po C, Tam AY-Y, Wong KM-C, Yam VW-W. J. Am. Chem. Soc. 2011;133:12136. doi: 10.1021/ja203920w. [DOI] [PubMed] [Google Scholar]
- 9.Kato T, Kihara H, Kumar U, Uryu T, Fréchet JMJ. Angew. Chem. Int. Ed. 1994;33:1644. [Google Scholar]
- 10.Boekhoven J, Hendriksen WE, Koper GJM, Eelkema R, van Esch JH. Science. 2015;349:1075. doi: 10.1126/science.aac6103. [DOI] [PubMed] [Google Scholar]
- 11.Pezzato C, Prins LJ. Nat Commun. 2015;6:7790. doi: 10.1038/ncomms8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves JD, Krebs EG. Pharm. Thera. 1999;82:111. doi: 10.1016/s0163-7258(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 13.Turner M, Billadeau DD. Nat. Rev. Immunol. 2002;2:476. doi: 10.1038/nri840. [DOI] [PubMed] [Google Scholar]
- 14.Craig EA, Weissman JS, Horwich AL. Cell. 1994;78:365. doi: 10.1016/0092-8674(94)90416-2. [DOI] [PubMed] [Google Scholar]
- 15.Horst R, Horwich AL, Wüthrich K. J. Am. Chem. Soc. 2011;133:16354. doi: 10.1021/ja206531c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, Wang X. Cell. 2012;148:213. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 17.Shi J, Du X, Yuan D, Haburcak R, Wu D, Zhou N, Xu B. Chem. Commun. 2015;51:4899. doi: 10.1039/c4cc10374b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams RJ, Smith AM, Collins R, Hodson N, Das AK, Ulijn RV. Nat Nano. 2009;4:19. doi: 10.1038/nnano.2008.378. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu H, Matsumoto S, Tamaru S-i, Kaneko K, Ikeda M, Hamachi I. J. Am. Chem. Soc. 2009;131:5580. doi: 10.1021/ja8098239. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Wang H, Wang L, Wang J, Kong D, Yang Z. J. Am. Chem. Soc. 2009;131:11286. doi: 10.1021/ja9042142. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Gu H, Yang Z, Xu B. J. Am. Chem. Soc. 2003;125:13680. doi: 10.1021/ja036817k. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka A, Fukuoka Y, Morimoto Y, Honjo T, Koda D, Goto M, Maruyama T. J. Am. Chem. Soc. 2015;137:770. doi: 10.1021/ja510156v. [DOI] [PubMed] [Google Scholar]
- 23.Zhou JDXW, Berciu C, He HJ, Shi JF, Nicastro D, Xu B. Chem. 2016;1:246. doi: 10.1016/j.chempr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuang Y, Gao Y, Xu B. Chem. Commun. 2011;47:12625. doi: 10.1039/c1cc15577f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye D, Shuhendler AJ, Cui L, Tong L, Tee SS, Tikhomirov G, Felsher DW, Rao J. Nat Chem. 2014;6:519. doi: 10.1038/nchem.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Gao Y, Kuang Y, Shi J, Du X, Zhou J, Wang H, Yang Z, Xu B. J. Am. Chem. Soc. 2013;135:9907. doi: 10.1021/ja404215g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Berciu C, Kuang Y, Shi J, Nicastro D, Xu B. ACS Nano. 2013;7:9055. doi: 10.1021/nn403664n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes M, Frederix PWJM, Raeburn J, Birchall LS, Sadownik J, Coomer FC, Lin IH, Cussen EJ, Hunt NT, Tuttle T, Webb SJ, Adams DJ, Ulijn RV. Soft Matter. 2012;8:5595. [Google Scholar]
- 29.Zhou J, Xu B. Bioconj. Chem. 2015;26:987. doi: 10.1021/acs.bioconjchem.5b00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J, Xu B. Nano Today. 2015;10:615. doi: 10.1016/j.nantod.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Nature. 2015;519:106. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 32.Fisher CK, Huang A, Stultz CM. J. Am. Chem. Soc. 2010;132:14919. doi: 10.1021/ja105832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh CT, Fisher SL, Park IS, Prahalad M, Wu Z. Chem. Biol. 1996;3:21. doi: 10.1016/s1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]
- 34.Walsh CT. Antibiotics: Actions, origins, resistance. ASM Press; Washongton DC: 2003. [Google Scholar]
- 35.Williams DH, Maguire AJ, Tsuzuki W, Westwell MS. Science. 1998;280:711. doi: 10.1126/science.280.5364.711. [DOI] [PubMed] [Google Scholar]
- 36.Rao JH, Lahiri J, Isaacs L, Weis RM, Whitesides GM. Science. 1998;280:708. doi: 10.1126/science.280.5364.708. [DOI] [PubMed] [Google Scholar]
- 37.Rao JH, Yan L, Xu B, Whitesides GM. J. Am. Chem. Soc. 1999;121:2629. [Google Scholar]
- 38.Sundram UN, Griffin JH, Nicas TI. J. Am. Chem. Soc. 1996;118:13107. [Google Scholar]
- 39.Leimkuhler C, Chen L, Barrett D, Panzone G, Sun BY, Falcone B, Oberthur M, Donadio S, Walker S, Kahne D. J. Am. Chem. Soc. 2005;127:3250. doi: 10.1021/ja043849e. [DOI] [PubMed] [Google Scholar]
- 40.Shi J, Du X, Huang Y, Zhou J, Yuan D, Wu D, Zhang Y, Haburcak R, Epstein IR, Xu B. J. Am. Chem. Soc. 2015;137:26. doi: 10.1021/ja5100417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi J, Du X, Yuan D, Haburcak R, Zhou N, Xu B. Bioconj. Chem. 2015;26:1879. doi: 10.1021/acs.bioconjchem.5b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan WCWPD, editor. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford University Press Inc.; 2000. [Google Scholar]
- 43.Alewood PF, Johns RB, Valerio RM, Kemp BE. Synthesis. 1983;1983:30. [Google Scholar]
- 44.Shi J, Du X, Huang Y, Zhou J, Yuan D, Wu D, Zhang Y, Haburcak R, Epstein IR, Xu B. JJ. Am. Chem. Soc. 2015;137:26. doi: 10.1021/ja5100417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irwin JJ, Duan D, Torosyan H, Doak AK, Ziebart KT, Sterling T, Tumanian G, Shoichet BK. J. Med. Chem. 2015;58:7076. doi: 10.1021/acs.jmedchem.5b01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGovern SL, Helfand BT, Feng B, Shoichet BK. J. Med. Chem. 2003;46:4265. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 47.Teunissen AJP, Paffen TFE, Ercolani G, de Greef TFA, Meijer EW. J. Am. Chem. Soc. 2016;138:6852. doi: 10.1021/jacs.6b03421. [DOI] [PubMed] [Google Scholar]
- 48.van der Zwaag D, Pieters PA, Korevaar PA, Markvoort AJ, Spiering AJH, de Greef TFA, Meijer EW. J. Am. Chem. Soc. 2015;137:12677. doi: 10.1021/jacs.5b08138. [DOI] [PubMed] [Google Scholar]
- 49.Teunissen AJP, Nieuwenhuizen MML, Rodríguez-Llansola F, Palmans ARA, Meijer EW. Macromolecules. 2014;47:8429. [Google Scholar]
- 50.Garai K, Verghese PB, Baban B, Holtzman DM, Frieden C. Biochemistry. 2014;53:6323. doi: 10.1021/bi5008172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abelein A, Jarvet J, Barth A, Gräslund A, Danielsson J. J. Am. Chem. Soc. 2016;138:6893. doi: 10.1021/jacs.6b04511. [DOI] [PubMed] [Google Scholar]
- 52.Kurouski D, Lu X, Popova L, Wan W, Shanmugasundaram M, Stubbs G, Dukor RK, Lednev IK, Nafie LA. J. Am. Chem. Soc. 2014;136:2302. doi: 10.1021/ja407583r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirst AR, Roy S, Arora M, Das AK, Hodson N, Murray P, Marshall S, Javid N, Sefcik J, Boekhoven J, van Esch JH, Santabarbara S, Hunt NT, Ulijn RV. Nat Chem. 2010;2:1089. doi: 10.1038/nchem.861. [DOI] [PubMed] [Google Scholar]
- 54.Thornton K, Abul-Haija YM, Hodson N, Ulijn RV. Soft Matter. 2013;9:9430. [Google Scholar]
- 55.Xie J, Okano A, Pierce JG, James RC, Stamm S, Crane CM, Boger DL. J. Am. Chem. Soc. 2012;134:1284. doi: 10.1021/ja209937s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie J, Pierce JG, James RC, Okano A, Boger DL. J. Am. Chem. Soc. 2011;133:13946. doi: 10.1021/ja207142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartl FU. Nature. 1996;381:571. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 58.Wang J, Zhao C, Zhao A, Li M, Ren J, Qu X. J. Am. Chem. Soc. 2015;137:1213. doi: 10.1021/ja511030s. [DOI] [PubMed] [Google Scholar]
- 59.Hamley IW. Chemical Reviews. 2012;112:5147. doi: 10.1021/cr3000994. [DOI] [PubMed] [Google Scholar]
- 60.Economou NJ, Giammona MJ, Do TD, Zheng X, Teplow DB, Buratto SK, Bowers MT. J. Am. Chem. Soc. 2016;138:1772. doi: 10.1021/jacs.5b11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu L, Song Y, Cheng P-N, Moore JS. J. Am. Chem. Soc. 2015;137:8062. doi: 10.1021/jacs.5b01651. [DOI] [PubMed] [Google Scholar]
- 62.Colvin MT, Silvers R, Frohm B, Su Y, Linse S, Griffin RG. J. Am. Chem. Soc. 2015;137:7509. doi: 10.1021/jacs.5b03997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murphy MP, LeVine H. J. Alzheimer's disease : JAD. 2010;19:311. [Google Scholar]
- 64.Sisodia SS, St George-Hyslop PH. Nat Rev Neurosci. 2002;3:281. doi: 10.1038/nrn785. [DOI] [PubMed] [Google Scholar]
- 65.Chow VW, Mattson MP, Wong PC, Gleichmann M. NeuroMol. Med. 2010;12:1. doi: 10.1007/s12017-009-8104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stine WB, Jungbauer L, Yu C, LaDu MJ. In: Alzheimer's Disease and Frontotemporal Dementia: Methods and Protocols. Roberson DE, editor. Humana Press; Totowa, NJ: 2011. p. 13. [Google Scholar]
- 67.Ashkenazi A, Dixit VM. Science. 1998;281:1305. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 68.Lu A, Magupalli Venkat G, Ruan J, Yin Q, Atianand Maninjay K, Vos MR, Schröder Gunnar F, Fitzgerald Katherine A, Wu H, Egelman Edward H. Cell. 2014;156:1193. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toledano S, Williams RJ, Jayawarna V, Ulijn RV. J. Am. Chem. Soc. 2006;128:1070. doi: 10.1021/ja056549l. [DOI] [PubMed] [Google Scholar]
- 70.Yang Z, Liang G, Xu B. Acc. Chem. Res. 2008;41:315. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 71.Yang ZM, Ho PL, Liang GL, Chow KH, Wang QG, Cao Y, Guo ZH, Xu B. J. Am. Chem. Soc. 2007;129:266. doi: 10.1021/ja0675604. [DOI] [PubMed] [Google Scholar]
- 72.Zhou J, Du X, Gao Y, Shi J, Xu B. J. Am. Chem. Soc. 2014;136:2970. doi: 10.1021/ja4127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou J, Du X, Li J, Yamagata N, Xu B. J. Am. Chem. Soc. 2015;137:10040. doi: 10.1021/jacs.5b06181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li J, Kuang Y, Gao Y, Du X, Shi J, Xu B. J. Am. Chem. Soc. 2013;135:542. doi: 10.1021/ja310019x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao Y, Kuang Y, Guo Z-F, Guo Z, Krauss IJ, Xu B. J. Am. Chem. Soc. 2009;131:13576. doi: 10.1021/ja904411z. [DOI] [PubMed] [Google Scholar]
- 76.Yang ZM, Liang GL, Wang L, Xu B. J. Am. Chem. Soc. 2006;128:3038. doi: 10.1021/ja057412y. [DOI] [PubMed] [Google Scholar]
- 77.Yang Z, Gu H, Fu D, Gao P, Lam JK, Xu B. Adv Mater. 2006;18:545. [Google Scholar]
- 78.Kuang Y, Shi J, Li J, Yuan D, Alberti KA, Xu Q, Xu B. Angew. Chem. Int. Ed. 2014;53:8104. doi: 10.1002/anie.201402216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang Z, Liang G, Guo Z, Guo Z, Xu B. Angew. Chem. Int. Ed. 2007;46:8216. doi: 10.1002/anie.200701697. [DOI] [PubMed] [Google Scholar]
- 80.Shi J, Du X, Yuan D, Zhou J, Zhou N, Huang Y, Xu B. Biomacromolecules. 2014;15:3559. doi: 10.1021/bm5010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pires RA, Abul-Haija YM, Costa DS, Novoa-Carballal R, Reis RL, Ulijn RV, Pashkuleva I. J. Am. Chem. Soc. 2015;137:576. doi: 10.1021/ja5111893. [DOI] [PubMed] [Google Scholar]
- 82.Li J, Kuang Y, Shi J, Zhou J, Medina JE, Zhou R, Yuan D, Yang C, Wang H, Yang Z, Liu J, Dinulescu DM, Xu B. Angew. Chem. Int. Ed. 2015;54:13307. doi: 10.1002/anie.201507157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang P, Gao Y, Lin J, Hu H, Liao H-S, Yan X, Jin A, Tang Y, Song J, Niu G. ACS Nano. 2015;9:9517. doi: 10.1021/acsnano.5b03874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.