Abstract
Retinitis pigmentosa (RP) has a prevalence of approximately one in 4000; 25%–30% of these cases are autosomal dominant retinitis pigmentosa (adRP). Like other forms of inherited retinal disease, adRP is exceptionally heterogeneous. Mutations in more than 25 genes are known to cause adRP, more than 1000 mutations have been reported in these genes, clinical findings are highly variable, and there is considerable overlap with other types of inherited disease. Currently, it is possible to detect disease-causing mutations in 50%–75% of adRP families in select populations. Genetic diagnosis of adRP has advantages over other forms of RP because segregation of disease in families is a useful tool for identifying and confirming potentially pathogenic variants, but there are disadvantages too. In addition to identifying the cause of disease in the remaining 25% of adRP families, a central challenge is reconciling clinical diagnosis, family history, and molecular findings in patients and families.
Retinitis pigmentosa (RP) is an inherited dystrophic or degenerative disease of the retina with a prevalence of roughly one in 4000 (Haim 2002; Daiger et al. 2007). Typically, the disease progresses from the midperiphery of the retina into the central retina and, in many cases, into the macula and fovea (Heckenlively and Daiger 2007; Fahim et al. 2013). Clinical features include night blindness starting in adolescence, followed by progressive loss of peripheral vision—referred to as “tunnel vision”—culminating in legal blindness or complete blindness in adulthood. Characteristic retinal findings on examination include bone-spicule formations and attenuated blood vessels, reduced visual fields, reduced and/or abnormal electroretinograms (ERGs), changes in structure imaged by optical coherence tomography (OCT), and subjective changes in visual function (Fishman et al. 2005). However, features and findings are highly variable among patients, even among patients within the same family.
RP is exceptionally heterogeneous at a genetic level. All modes of inheritance are encountered: dominant, recessive, autosomal, X-linked, and even mitochondrial (Berger et al. 2010). It is assumed that each patient has a monogenic form of disease (or digenic in rare cases) but many different genes account for disease in RP patients as a group. Currently, mutations in more than 66 genes are known to cause nonsyndromic RP and an additional 31 genes cause syndromic forms such as Usher syndrome and Bardet–Biedl syndrome (Daiger et al. 2013; RetNet 2014). There are also many disease-causing mutations reported in these genes. More than 3100 mutations are listed in the Human Gene Mutation Database (HGMD) (HGMD 2014) in nonsyndromic RP genes alone (Daiger et al. 2013).
This review focuses on nonsyndromic autosomal dominant retinitis pigmentosa (adRP). This is partly to simplify the discussion but also because finding genes and mutations causing adRP entails unique opportunities and pitfalls. In addition, our laboratory has worked with adRP families for the past 25 years affording us insights into these issues. Despite the limited focus, many observations regarding genes and mutations causing adRP apply equally well to other forms of inherited retinopathy.
FINDING GENES AND MUTATIONS CAUSING adRP
The successes in finding adRP genes and disease-causing mutations over the past two decades are striking. Table 1 lists genes in which mutations are reported to cause adRP. References are in RetNet (RetNet 2014, see https://sph.uth.edu/retnet/). There are currently 25 reported adRP genes and an additional linked gene, RP63, which maps to human chromosome 6q23 (Kannabiran et al. 2012). The HGMD database (HGMD 2014) lists more than 1000 mutations in these genes, although not all are dominant-acting (and not all are pathogenic). To date, mutations in the known adRP genes account for 50%–75% of cases, depending on the tests performed and populations screened. Nonetheless, significant problems remain.
Table 1.
Genes reported to cause autosomal dominant retinitis pigmentosa (in chromosomal order)
| Symbol | Gene/protein | Chromosome | Alternate diseases | Function | |
|---|---|---|---|---|---|
| 1 | RPE65 | Retinal pigment epithelium-specific 65 kD protein | 1p31.2 | Recessive retinitis pigmentosa; recessive Leber congenital amaurosis | Visual cycle |
| 2 | PRPF3 | Pre-mRNA processing factor 3 | 1q21.2 | RNA processing | |
| 3 | SEMA4A | Semaphorin 4A | 1q22 | Dominant cone–rod dystrophy | Neuronal or immune response |
| 4 | SNRNP200 | Small nuclear ribonucleoprotein 200 kDa (U5) | 2q11.2 | RNA processing | |
| 5 | RHO | Rhodopsin | 3q22.1 | Dominant congenital stationary night blindness; recessive retinitis pigmentosa | Phototransduction, outer segment membrane structure |
| 6 | GUCA1B | Guanylate cyclase activating protein 1B | 6p21.1 | Dominant macular dystrophy | Phototransduction |
| 7 | PRPH2 | Peripherin 2 | 6p21.1 | Dominant macular dystrophy; digenic retinitis pigmentosa with ROM1; dominant adult vitelliform macular dystrophy; dominant cone–rod dystrophy; dominant central areolar choroidal dystrophy | Outer segment membrane structure |
| 8 | RP9 | RP9 protein or PIM1-kinase associated protein 1 | 7p14.3 | Unknown | |
| 9 | KLHL7 | Kelch-like 7 protein (Drosophila) | 7p15.3 | Ubiquitination pathway | |
| 10 | IMPDH1 | Inosine monophosphate dehydrogenase 1 | 7q32.1 | Dominant Leber congenital amaurosis | Purine or amino acid synthesis |
| 11 | RP1 | RP1 protein | 8q12.1 | Recessive retinitis pigmentosa | Ciliary structure/ function |
| 12 | TOPORS | Topoisomerase I binding arginine/ serine rich protein | 9p21.1 | Ciliary structure/ function | |
| 13 | PRPF4 | Pre-mRNA processing factor 4 | 9q32 | RNA processing | |
| 14 | HK1 | Hexokinase 1 | 10q22.1 | Glucose metabolism | |
| 15 | BEST1 | Bestrophin 1 | 11q12.3 | Dominant vitreoretinochoroidopathy; recessive bestrophinopathy; recessive retinitis pigmentosa; dominant macular dystrophy, Best type | Visual cycle and chloride channel |
| 16 | ROM1 | Retinal outer segment membrane protein 1 | 11q12.3 | Digenic retinitis pigmentosa with PRPH2 | Outer segment membrane structure |
| 17 | NRL | Neural retina lucine zipper | 14q11.2 | Recessive retinitis pigmentosa | Transcription factor |
| 18 | RDH12 | Retinol dehydrogenase 12 | 14q24.1 | Recessive Leber congenital amaurosis with severe childhood retinal dystrophy | Visual cycle |
| 19 | NR2E3 | Nuclear receptor subfamily 2 group E3 | 15q23 | Recessive retinitis pigmentosa in Portuguese Crypto Jews; recessive Goldmann–Favre syndrome; recessive enhanced S-cone syndrome (ESC); combined dominant and recessive retinopathy | Transcription factor |
| 20 | PRPF8 | Pre-mRNA processing factor 8 | 17p13.3 | RNA processing | |
| 21 | CA4 | Carbonic anhydrase IV | 17q23.2 | Retinal capillary structure/function | |
| 22 | FSCN2 | Retinal fascin homolog 2, actin bundling protein | 17q25.3 | Dominant macular dystrophy | Ciliary structure/ function |
| 23 | CRX | Cone–rod otx-like photoreceptor homeobox transcription factor | 19q13.32 | Recessive, dominant and de novo Leber congenital amaurosis; dominant cone– rod dystrophy | Transcription factor |
| 24 | PRPF31 | Pre-mRNA processing factor 31 | 19q13.42 | RNA processing | |
| 25 | PRPF6 | Pre-mRNA processing factor 6 | 20q13.33 | RNA processing |
References as in RetNet, https://sph.uth.edu/retnet/, and Sullivan et al. 2014 (HK1)
For example, it should be easy to name genes known to cause adRP, but it is not. In recent years, the simple idea that one gene leads to one disease has proven highly inaccurate, especially for inherited retinal diseases. A gene first identified as the cause of one form of disease, for example, recessive RP, may harbor mutations that cause another form of retinal disease or mutations with a different mode of inheritance for example, adRP. Table 1 lists alternate forms of disease associated with adRP genes. One consequence of this overlap is that any simple count of “RP genes” is often arbitrary.
Likewise, there are complications in detecting disease-causing mutations in adRP genes. For one, the initial diagnosis in a family may be misleading. That is, because of exceptional clinical variation, affected family members may have different clinical diagnoses, or the mode of inheritance may be misconstrued. For example, families with X-linked RP and affected carrier females may be mistaken as adRP (Churchill et al. 2013). Further, a significant fraction of RP patients present as isolated cases, with no known affected relatives, so the mode of inheritance is undetermined. Also, because different mutations in the same gene can produce different clinical consequences, the actual mutation may be in a known gene not commonly associated with adRP. Examples include rare adRP mutations in RPE65 (Bowne et al. 2011a) and RDH12 (Fingert et al. 2008), genes more likely to cause recessive RP or recessive Leber congenital amaurosis, respectively.
Beyond these complications, there is the general problem of variants of unknown significance (VUS), that is, rare, potentially pathogenic variants in known adRP genes for which the evidence is equivocal. This is a fundamental problem in medical genetics given the extensive background of rare and apparently damaging variants in the human population (Marth et al. 2011). Suggestions for scoring potentially pathogenic variants are discussed in the next section.
Finally, there are substantial difficulties in identifying novel adRP genes. Three advanced technologies using next-generation sequencing (NGS) are in use: targeted retinal-gene capture NGS (Simpson et al. 2011; Audo et al. 2012; Neveling et al. 2012; O’Sullivan et al. 2012; Chen et al. 2013; Eisenberger et al. 2013; Glockle et al. 2013; Wang et al. 2014), whole-exome NGS (Bowne et al. 2011a; Ozgul et al. 2011; Tucker et al. 2011; Zeitz et al. 2013; El Shamieh et al. 2014; Jin et al. 2014; Sergouniotis et al. 2014), and whole-genome NGS (Nishiguchi et al. 2013). Each has advantages and disadvantages.
Because targeted retinal-capture NGS examines only a small portion of the human genome, that is, exons of known retinal genes and candidate genes, analysis of sequence data is simplified. Also, retinal capture panels are optimized for each exon and include retinal-specific splice variants. However, retinal-capture NGS is limited to known genes and detects a large number of VUS mutations (as do all NGS methods). In contrast, whole-exome sequencing is a powerful tool for identifying novel disease-causing genes but perforce is limited to exons, whereas whole genome NGS avoids this limitation but presents a very complex problem in data analysis. For autosomal dominant diseases, the problems are compounded by the need to detect a single, heterozygous mutation in a diploid organism, the proverbial needle-in-a-hay-stack.
Further, because NGS methods are based on short-read, paired-end, and shotgun sequencing, they are underpowered for detecting deletions, insertions, and variable-length repetitive elements. As examples, large deletions in the PRPF31 gene account for 2.5% of adRP cases (Sullivan et al. 2006b), but are generally not detectable by NGS, and the repetitive elements of the ORF15 region of the X-linked RP gene RPGR are refractory to short-read sequencing (Churchill et al. 2013).
Fortunately, segregation testing and linkage analysis in adRP families address many of these problems. Segregation testing—determining if a putative disease-causing variant tracks with disease in a family—can support or exclude pathogenicity. Complications are the late onset of disease and nonpenetrance. That is, a clinically unaffected family member may carry a disease allele, either because the disease has not manifested yet or because the mutation is non-penetrant in that individual. An example of the latter is mutations in PRPF31 for which non-penetrance may be as high as 20% (McGee et al. 1997; Venturini et al. 2012). However, nonpene-trance is not common among other adRP genes. Conversely, absence of a putative mutation in an affected family member is usually strong evidence against pathogenicity.
Linkage analysis tests for cosegregation of polymorphic markers with disease in a family. Loosely, there are two contrasting approaches, linkage exclusion and linkage inclusion. Linkage exclusion is simply exclusion of a known adRP gene by recombination between polymorphic markers within or contiguous to the site. For example, small sets of highly variable STR (short-tandem repeat) polymorphisms can exclude many known adRP sites in families with five or more affected members available for testing (Sullivan et al. 2006b). In contrast, because of statistical issues, larger families are required for linkage inclusion, that is, mapping a disease gene by observing cosegregation of polymorphic markers. Since dominant retinal disease families may have many living, affected members, linkage mapping is an effective tool for localizing new adRP genes.
Contemporary marker sets for linkage mapping include hundreds of thousands of SNPs (single-nucleotide polymorphisms) such as markers in Affymetrix SNP Arrays (Friedman et al. 2009; Bowne et al. 2011a). Generally, in families with eight or more affected members available for testing, linkage mapping reduces the target region to one or a few chromosomal sites, each, 10 Mb in length. This substantially reduces the “space” to search for novel adRP genes using NGS approaches. As an alternative to using SNP marker arrays, data from whole-exome NGS testing of multiple family members can be combined for linkage mapping in RP families based on regions identical-by-descent and shared rare variants (Koboldt et al. 2014).
Each approach is useful: Most adRP genes in Table 1 were first localized by linkage mapping; NGS methods were used to find most of the recently identified genes; and retinal capture NGS is a rapid, efficient approach to finding disease-causing mutations in known adRP genes. Taken together, advanced DNA sequencing and mapping methods have revolutionized gene discovery and mutation detection for adRP.
DETERMINING PATHOGENEITY OF adRP MUTATIONS
Determining which of the many rare variants in our genomes are pathogenic and which are benign, with all possibilities in between, is one of the fundamental problems in medical genetics (MacArthur et al. 2014). Dominant Mendelian diseases such as adRP offer the advantage of segregation testing to evaluate pathogenicity. However, this is only one of multiple steps in establishing causality. Although there is no fully accepted protocol, each of the NGS publications cited above follows similar strategies. Note that establishing pathogenicity overlaps with, but is distinct from, the “pipeline” for sequence assembly and detection of variants. Also, criteria for establishing pathogenicity of variants that may cause autosomal dominant diseases are distinct from criteria for recessive and X-linked diseases. With these caveats in mind, the following are reasonable steps for determining pathogenicity of potential adRP mutations (not necessarily in order), using retinal capture NGS as a model.
Although the model assumes efficient retinal capture and a patient with autosomal inheritance, it is prudent to confirm coverage of the most common adRP genes, such as rhodopsin (RHO) and PRPH2, and to consider X-linked inheritance in the absence of male-to-male transmission.
The filtering pipeline should flag all variants with an allele frequency of <0.1% in publically available databases such as 1000 Genomes, ESP, and dbSNP (Abecasis et al. 2012; Fernandez-Suarez et al. 2014; NCBI 2014). Note that some recessive RP mutations have population frequencies ≥ 0.1%, whereas all reported adRP mutations are more rare.
Determine if any flagged variants are known, published disease-causing mutations. This requires searching, for example, relevant Leiden databases (Cremers et al. 2014), the HGMD database (HGMD 2014), and clin-VAR (NCBI 2014). In-house databases should be consulted too, of course. Caution: Even though a variant is listed in a disease database, it may not be pathogenic. For uncertain variants, it is essential to confirm pathogenicity based on original publications.
If there is no unequivocally pathogenic variant in a known adRP gene, consider deletion detection for PRPF31 (Sullivan et al. 2006b) and/or nested Sanger sequencing of ORF15 of RPGR (Churchill et al. 2013).
In the remaining cases, rank variants by pathogenicity scores using publically available services such as PolyPhen, SIFT, NNSplice, and Mutation Taster (Schwarz et al. 2010; Knecht and Krawczak 2014). Published functional data on specific genes may be useful too.
Confirm plausible mutations by conventional sequencing and test for segregation in available family members.
Finally, based on the totality of information available, including all of the above, plus clinical information, family history, and the biology and clinical phenotypes of possible disease genes, score putative mutations (one or more) for the likelihood of causing disease in the patient and family tested. Several scoring systems have been suggested, largely ranging from “recognized cause of disorder,” “expected cause of disorder,” “may or may not cause disorder” (VUS), to “probably not causative” (Richards et al. 2008).
What do these NGS strategies miss? Mutations in known retinal disease genes outside of exons, mutations in novel genes, and mutations not detectable by shotgun sequencing (e.g., variable repeats), among others.
PREVALENCE OF DISEASE-CAUSING MUTATIONS IN adRP FAMILIES
Our research has focused principally on genetic studies of adRP patients and families. For purposes of tracking progress in gene discovery and mutation detection, we have established a cohort of well-characterized adRP families (Sohocki et al. 2001; Sullivan et al. 2006a; Bowne et al. 2008, 2013). Families in the cohort have a clinical diagnosis of adRP by a knowledgeable clinician, three or more affected generations with affected females, or two or more generations with male-to-male transmission (to bias against X-linked RP). Currently, there are 270 families in the cohort (Table 2). The families are largely Americans of European origin, 75% white, 5% African American, 5% Hispanic, 3% Asian, and the remainder other or unknown.
Table 2.
Prevalence of disease-causing mutations in adRP families (N = 270), with number of mutation types
| Gene | Families | Percent | Missense | Nonsense | Other |
|---|---|---|---|---|---|
| CRX | 2 | 0.7% | 1 | 0 | 1 |
| HK1 | 3 | 1.1% | 1 | 0 | 0 |
| IMPDH1 | 10 | 3.7% | 2 | 0 | 0 |
| KLHL7 | 3 | 1.1% | 3 | 0 | 0 |
| NR2E3 | 6 | 2.2% | 1 | 0 | 1 |
| NRL | 2 | 0.7% | 2 | 0 | 0 |
| PRPF3 | 4 | 1.5% | 2 | 0 | 0 |
| PRPF31 | 24 | 8.9% | 4 | 3 | 17 |
| PRPF8 | 7 | 2.6% | 4 | 0 | 1 |
| RDH12 | 1 | 0.4% | 0 | 0 | 1 |
| PRPH2 | 19 | 7.0% | 10 | 1 | 2 |
| PRPH2+ROM1 | 1 | 0.4% | 1 | 0 | 1 |
| RHO | 83 | 30.7% | 30 | 2 | 1 |
| RP1 | 13 | 4.8% | 0 | 3 | 1 |
| RP2 | 3 | 1.1% | 1 | 0 | 2 |
| RPE65 | 1 | 0.4% | 1 | 0 | 0 |
| RPGR | 22 | 8.1% | 4 | 2 | 13 |
| SNRNP200 | 4 | 1.5% | 4 | 0 | 0 |
| TOPORS | 4 | 1.5% | 3 | 0 | 1 |
| None | 58 | 21.5% | |||
| Total | 270 | 100% |
We have applied a variety of methods to detect disease-causing mutations in these families including linkage mapping (Sullivan et al. 1999; Bowne et al. 2002, 2011a), conventional Sanger sequencing (Bowne et al. 1999, 2006, 2008, 2013; Sullivan et al. 1999; Sohocki et al. 2001; Gire et al. 2007; Friedman et al. 2009), NGS (Bowne et al. 2011b; Wang et al. 2013), deletion detection (Sullivan et al. 2006b), and subclone sequencing of RPGR ORF15 (Churchill et al. 2013). Recently, we reported a novel adRP gene, hexokinase 1 (HK1), with a missense mutation in five independently ascertained families (Sullivan et al. 2014). This gene, too, was tested in the cohort.
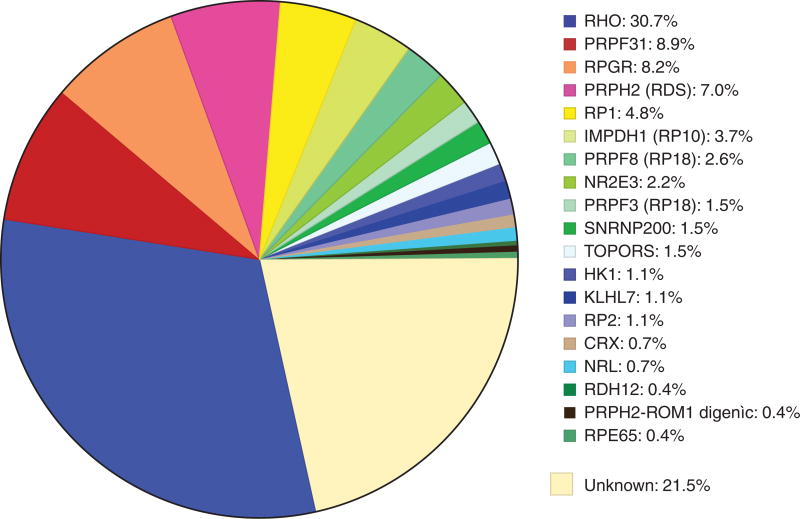
Table 2 summarizes our findings. To date we have identified the disease-causing mutation in 78% of the families, leaving 22% in which the known dominant RP genes have been excluded. Figure 1 shows the fraction of cases caused by mutations in each gene.
Figure 1.
Fraction of mutations causing dominant RP in a cohort of 270 “adRP” families.
Note that Table 2 includes mutations in two X-linked genes, RPGR and RP2, found in a total of 25 families (9%). Each of these families met reasonable criteria for inclusion in a cohort of “autosomal dominant” families, with multiple affected females and an initial diagnosis of adRP. On further review, most, but not all, affected females are more mildly affected than age-matched affected males (Wu et al. 2010; Churchill et al. 2013). Clinical symptoms are also more variable in carrier females, consistent with Lyonization or other factors modifying expression. This is an example of the power of molecular findings to dramatically change an initial diagnosis.
Based on the significant fraction of X-linked mutations among families with a diagnosis of adRP, mutations in RPGR are probably the most common cause of RP in the world. This is reinforced by the finding that RPGR mutations account for 13% of isolated RP in males (Branham et al. 2012).
The cohort families also have one instance of digenic inheritance of a PRPH2 Leu185Pro mutation combined with a ROM1 Leu114fs* 131 mutation (Kajiwara et al. 1994; Sullivan et al. 2006a). Technically, the inheritance pattern deviates from classical adRP because an affected child must inherit both alleles from an affected parent, or one allele from an unaffected parent. This is a rare condition but other ROM1 (rod outer-membrane protein 1) mutations are known to cause digenic RP in combination with the PRPH2 Leu185Pro mutation (Dryja et al. 1997).
Table 2 also indicates the distribution of mutation types in three broad classes, missense, nonsense, and “other.” From a functional prospective, missense mutations are likely to produce an abnormal protein and the mode of action is through partial loss-of-function, gain-of-function, dominant negative, or toxic effects. “Other” mutations most likely result in functional haploinsufficiency, that is, no protein or a nonfunctional protein. Nonsense mutations may or may not produce a protein depending on whether the premature stop is in the last exon, which escapes nonsense mediated decay, or earlier exons, which do not. Based on Table 2, most adRP mutations do not result in functional haploinsufficiency, except for PRPF31.
A substantial fraction of mutations detected in the cohort are “common,” that is, they occur in three or more unrelated families. Table 3 lists the common mutations and the fraction of families with each. Twelve mutations in adRP genes account for nearly 30% of these families (i.e., excluding RPGR and RP2). Most of these mutations are ethnic-specific. Several arose by founder effect and have limited geographic distribution. That is, the mutation arose once only, increased in frequency by drift and population dynamics, and shares a distinct haplotype in cis to the mutation. For example, the rhodopsin Pro23His mutation (13%) and the peripherin 828+3A>T mutation (2%) arose by founder effect, are almost exclusive to the United States, and are found only in Americans of European origin (Dryja et al. 1990, 1991; Shankar et al. 2004). The hexokinase Glu847Lys mutation also arose from a common ancestor, but the mutation is found in North America and Europe (Sullivan et al. 2014). In contrast, the Asp226Asn mutation in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) arose independently on multiple occasions (Sullivan et al. 2006a).
Table 3.
Common mutations in adRP genes and RPGR (N = 270)
| Gene | Families | Mutation | Percent |
|---|---|---|---|
| HK1 | 3 | Glu847Lys | 1.1% |
| IMPDH1 | 6 | Asp226Asn | 2.2% |
| 4 | Asp311Asn | 1.5% | |
| NR2E3 | 5 | Gly56Arg | 1.9% |
| PRPF3 | 3 | Thr494Met | 1.1% |
| PRPH2 | 5 | 828+3A>T | 1.9% |
| 4 | Pro216Ser | 1.5% | |
| RHO | 5 | Arg135Leu | 1.9% |
| 5 | Arg135Trp | 1.9% | |
| 36 | Pro23His | 13.2% | |
| RP1 | 6 | Arg677X | 2.2% |
| 4 | Leu762Tyrfs*17 | 1.5% | |
| RPGR | 3 | Glu802Glyfs*32 | 1.1% |
| Total | 33.0% |
This is a general phenomenon for RP mutations: the prevalence of mutations in a given gene is often dominated by one or a few mutations that arose by founder effect. Thus, populations will have different prevalences as a result of founder mutations, even though the RP genes are universal and the fraction of new mutations is roughly the same worldwide. For example, rhodopsin mutations account for 30% of adRP cases among Americans of European origin but <10% in China, largely because the Pro23His mutation is absent from China (Li et al. 2010). In contrast, the prevalence of RPGR mutations, which are predominantly new mutations, is roughly the same worldwide (Bocquet et al. 2013; Xu et al. 2014).
DOMINANT-RECESSIVE ACTING adRP MUTATIONS
In most cases in which a gene in Table 1 has more than one disease association, this is the result of different mutations in the same gene causing different diseases. However, there are two instances in which the same mutation has different consequences depending on whether one or two copies are present, that is, whether the patient is heterozygous or homozygous. Technically, these mutations are dominant and recessive acting. In one large adRP family with the RP1 Arg677X mutation, two homozygous individuals are severely affected whereas heterozygous individuals are more mildly affected (Sullivan et al. 1999). In contrast, the consequences of this mutation are distinct from clinical expression of the several RP1 mutations, which are truly recessive, with unaffected carriers (Khaliq et al. 2005; Riazuddin et al. 2005). Similarly, in a large adRP family with the HK1 Glu847Lys mutation, a single homozygous individual is significantly more severely affected than heterozygotes (Sullivan et al. 2014). These cases may be rare, but because members of large, multigenerational adRP families occasionally live in the same community, this is more likely with adRP than with other dominant diseases.
NORMAL FUNCTION OF adRP GENES
Table 1 provides a brief phrase describing the functional class of each gene. Excellent, extensive reviews are also available (Berger et al. 2010; Wright et al. 2010). Even limiting discussion to genes that account for 1% or more of cases, genes causing adRP have diverse functional roles.
It is not surprising that mutations in genes coding for proteins that are abundant in rods or play fundamental roles in photoreceptor function may cause dominant RP. This includes rhodopsin, the most abundant rod protein; peripherin (PRPH2) and ROM1, which are essential structural components of rod outer membranes; and rod/cone expression factors such as CRX, NRL, and NR2E3. Mutations in these genes are frequent causes of adRP. On the other hand, the relationship between function and disease of other adRP genes is less clear.
For example, the protein products of four adRP genes, PRPF3, PRPF8, PRPF31, and SNRNP200, are essential components of the U4/U6-U5 tri-snRNP complex of the spliceo-some involved in mRNA splicing (Liu and Zack 2013). The products of two less frequent causes of adRP, PRPF4 and PRPF6, are also components of this complex. These genes are expressed in all tissues and are highly conserved in all eukaryotes. Why mutations in these genes cause dominant RP, and only RP, is not known. However, genes coding for other proteins in this complex are attractive candidates for novel adRP genes.
The protein products of the remaining common adRP genes, HK1, IMPDH1, KLHL7, RP1, and TOPORS, are enzymes found in all cells (HK1 and IMPDH1), have a universal role in protein degradation (KLHL7), or are components of sensory cilia (RP1). That is, their roles are diverse and not obviously predictive of dominant, degenerative disease limited to the retina.
RECONCILING CLINICAL DIAGNOSIS, FAMILY HISTORY AND MOLECULAR FINDINGS
An emerging realization in inherited retinopathies is apparent discrepancies between clinical findings, inheritance pattern, and results of genetic testing. Clinical findings of retinitis pigmentosa and evidence from family history of dominant inheritance are consistent with a diagnosis of adRP. Fortunately, in most cases, a diagnosis of adRP is supported by the results of genetic testing. However, given the complex relationship between genes, mutations, and inherited retinopathies, there are opportunities for conflicts, for instance, when an apparent disease-causing mutation is in a gene not known to cause adRP, or the clinical phenotype in the family is more variable than just RP, or the apparent mode of inheritance is misleading or not determined. For these reasons, clinicians, counselors, and geneticists must work closely with each other to resolve these conflicts. In some cases, a reconsideration of the clinical, family, and molecular findings, taken together, may substantially alter the predicted mode of inheritance and prognosis. Thus, in addition to all the other benefits of genetic testing of patients with RP, molecular findings may substantially alter diagnosis, prognosis, counseling, and treatment.
CONCLUDING REMARKS
The long-term objective of research on genes and mutations causing inherited retinopathies is to enable effective genetic testing of patients and families, to incorporate this information into clinical care and counseling, and to develop treatments and cures based on a fundamental understanding of disease pathology. Currently, the success rate for finding the causative mutation in adRP patients is 50%–75%, but this fraction will increase rapidly with widespread adaptation of NGS testing and development of “third-generation” sequencing (Laszlo et al. 2014). Gene-specific and mutation-specific treatment trials have begun already. In time the limitation will not be genetic testing per se, but interpretation of variants, reconciling diagnosis and molecular findings, and providing meaningful information to clinicians and useful counseling to patients. The realization of these goals requires close integration of retinal specialists, genetic counselors, molecular geneticists, and bioinformaticists.
Acknowledgments
Supported by Center and Individual Grants, and a Wynn-Gund TRAP Award, from the Foundation Fighting Blindness; by National Institutes of Health Grant EY007142; and by the Hermann Eye Fund.
Footnotes
Additional Perspectives on Retinal Disorders: Genetic Approaches to Diagnosis and Treatment available at www.perspectivesinmedicine.org
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audo I, Bujakowska KM, Leveillard T, Mohand-Said S, Lancelot ME, Germain A, Antonio A, Michiels C, Saraiva JP, Letexier M, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis. 2012;7:8. doi: 10.1186/1750-1172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29:335–375. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Bocquet B, Lacroux A, Surget MO, Baudoin C, Marquette V, Manes G, Hebrard M, Senechal A, Delettre C, Roux AF, et al. Relative frequencies of inherited retinal dystrophies and optic neuropathies in Southern France: Assessment of 21-year data management. Ophthalmic Epidemiol. 2013;20:13–25. doi: 10.3109/09286586.2012.737890. [DOI] [PubMed] [Google Scholar]
- Bowne SJ, Daiger SP, Hims MM, Sohocki MM, Malone KA, McKie AB, Heckenlively JR, Birch DG, Inglehearn CF, Bhattacharya SS, et al. Mutations in the RP1 gene causing autosomal dominant retinitis pigmentosa. Hum Mol Genet. 1999;8:2121–2128. doi: 10.1093/hmg/8.11.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Blanton SH, Cepko CL, Blackshaw S, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Daiger SP. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:559–568. doi: 10.1093/hmg/11.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Mortimer SE, Hedstrom L, Zhu J, Spellicy CJ, Gire AI, Hughbanks-Wheaton D, Birch DG, Lewis RA, et al. Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and Leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2006;47:34–42. doi: 10.1167/iovs.05-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Gire AI, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Daiger SP. Mutations in the TOPORS gene cause 1% of autosomal dominant retinitis pigmentosa. Mol Vis. 2008;14:922–927. [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Humphries MM, Sullivan LS, Kenna PF, Tam LCS, Kiang AS, Campbell M, Weinstock GM, Koboldt DC, Ding L, et al. A dominant mutation in RPE65 identified by whole-exome sequencing causes retinitis pigmentosa with choroidal involvement. Euro J Hum Genet. 2011a;19:1074–1081. doi: 10.1038/ejhg.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Koboldt DC, Ding L, Fulton R, Abbott RM, Sodergren EJ, Birch DG, Wheaton DH, Heck-enlively JR, et al. Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing. Invest Ophthalmol Vis Sci. 2011b;52:494–503. doi: 10.1167/iovs.10-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Avery CE, Sasser EM, Roorda A, Duncan JL, Wheaton DH, Birch DG, Branham KE, Heck-enlively JR, et al. Mutations in the SNRNP200 gene cause 1.6% of autosomal dominant retinitis pigmentosa. Mol Vis. 2013;19:2407–2417. [PMC free article] [PubMed] [Google Scholar]
- Branham K, Othman M, Brumm M, Karoukis AJ, Atmaca-Sonmez P, Yashar BM, Schwartz SB, Stover NB, Trzupek K, Wheaton D, et al. Mutations in RPGR and RP2 account for 15% of males with simplex retinal degenerative disease. Invest Ophthalmol Vis Sci. 2012;53:8232– 8237. doi: 10.1167/iovs.12-11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhao K, Sheng X, Li Y, Gao X, Zhang X, Kang X, Pan X, Liu Y, Jiang C, et al. Targeted sequencing of 179 genes associated with hereditary retinal dystrophies and 10 candidate genes identifies novel and known mutations in patients with various retinal diseases. Invest Ophthalmol Vis Sci. 2013;54:2186–2197. doi: 10.1167/iovs.12-10967. [DOI] [PubMed] [Google Scholar]
- Churchill JD, Bowne SJ, Sullivan LS, Lewis RA, Wheaton DK, Birch DG, Branham KE, Heckenlively JR, Daiger SP. Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2013;54:1411–1416. doi: 10.1167/iovs.12-11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers FP, den Dunnen JT, Ajmal M, Hussain A, Preising MN, Daiger SP, Qamar R. Comprehensive registration of DNA sequence variants associated with inherited retinal diseases in Leiden Open Variation Databases. Hum Mutat. 2014;35:147–148. doi: 10.1002/humu.22458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007;125:151–158. doi: 10.1001/archopht.125.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84:132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Hahn LB, Cowley GS, Olsson JE, Reichel E, Sandberg MA, Berson EL. Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med. 1990;323:1302–1307. doi: 10.1056/NEJM199011083231903. [DOI] [PubMed] [Google Scholar]
- Dryja TP, Hahn LB, Cowley GS, McGee TL, Berson EL. Mutation spectrum of the rhodopsin gene among patients with autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci. 1991;88:9370–9374. doi: 10.1073/pnas.88.20.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Hahn LB, Kajiwara K, Berson EL. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1997;38:1972–1982. [PubMed] [Google Scholar]
- Eisenberger T, Neuhaus C, Khan AO, Decker C, Preising MN, Friedburg C, Bieg A, Gliem M, Charbel Issa P, Holz FG, et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: The example of retinal dystrophies. PLoS ONE. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Shamieh S, Neuille M, Terray A, Orhan E, Condroyer C, Demontant V, Michiels C, Antonio A, Boyard F, Lancelot ME, et al. Whole-exome sequencing identifies KIZ as a ciliary gene associated with autosomal-recessive rod-cone dystrophy. Am J Hum Genet. 2014;94:625–633. doi: 10.1016/j.ajhg.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahim AT, Daiger SP, Weleber RG. Retinitis pigmentosa overview. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews. University of Washington; Seattle: 2013. [Google Scholar]
- Fernandez-Suarez XM, Rigden DJ, Galperin MY. The 2014 Nucleic Acids Research Database Issue and an updated NAR online Molecular Biology Database Collection. Nucleic Acids Res. 2014;42:D1–D6. doi: 10.1093/nar/gkt1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingert JH, Oh K, Chung M, Scheetz TE, Andorf JL, Johnson RM, Sheffield VC, Stone EM. Association of a novel mutation in the retinol dehydrogenase 12 (RDH12) gene with autosomal dominant retinitis pigmentosa. Arch Ophthalmol. 2008;126:1301–1307. doi: 10.1001/archopht.126.9.1301. [DOI] [PubMed] [Google Scholar]
- Fishman GA, Jacobson SG, Alexander KR, Cideciyan AV, Birch DG, Weleber RG, Hood DC. Outcome measures and their application in clinical trials for retinal degenerative diseases: Outline, review, and perspective. Retina. 2005;25:772–777. doi: 10.1097/00006982-200509000-00014. [DOI] [PubMed] [Google Scholar]
- Friedman JS, Ray JW, Waseem N, Johnson K, Brooks MJ, Hugosson T, Breuer D, Branham KE, Krauth DS, Bowne SJ, et al. Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2009;84:792–800. doi: 10.1016/j.ajhg.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gire AI, Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Daiger SP. The Gly56Arg mutation in NR2E3 accounts for 1–2% of autosomal dominant retinitis pigmentosa. Mol Vis. 2007;13:1970–1975. [PubMed] [Google Scholar]
- Glockle N, Kohl S, Mohr J, Scheurenbrand T, Sprecher A, Weisschuh N, Bernd A, Rudolph G, Schubach M, Polo-schek C, et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur J Hum Genet. 2013;22:99–104. doi: 10.1038/ejhg.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim M. Epidemiology of retinitis pigmentosa in Denmark. Acta Ophthalmol Scand Suppl. 2002;233:1–34. doi: 10.1046/j.1395-3907.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- Heckenlively JR, Daiger SP. Emery and Rimoin’s principles and practice of medical genetics, Chap 147. Churchill Livingston; London: 2007. Hereditary retinal and choroidal degenerations; pp. 3197–3227. [Google Scholar]
- HGMD. Human Gene Mutation Database. 2014 http://www.hgmd.cf.ac.uk/. Biobase Biological Databases.
- Jin ZB, Huang XF, Lv JN, Xiang L, Li DQ, Chen J, Huang C, Wu J, Lu F, Qu J. SLC7A14 linked to autosomal recessive retinitis pigmentosa. Nat Commun. 2014;5:3517. doi: 10.1038/ncomms4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- Kannabiran C, Singh HP, Jalali S. Mapping of locus for autosomal dominant retinitis pigmentosa on chromosome 6q23. Hum Genet. 2012;131:717–723. doi: 10.1007/s00439-011-1115-5. [DOI] [PubMed] [Google Scholar]
- Khaliq S, Abid A, Ismail M, Hameed A, Mohyuddin A, Lall P, Aziz A, Anwar K, Mehdi SQ. Novel association of RP1 gene mutations with autosomal recessive retinitis pigmentosa. J Med Genet. 2005;42:436–438. doi: 10.1136/jmg.2004.024281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht C, Krawczak M. Molecular genetic epidemiology of human diseases: From patterns to predictions. Hum Genet. 2014;133:425–430. doi: 10.1007/s00439-013-1396-y. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Larson DE, Sullivan LS, Bowne SJ, Steinberg KM, Churchill JD, Buhr AC, Nutter N, Pierce EA, Blanton SH, et al. Exome-based mapping and variant prioritization for inherited Mendelian disorders. Am J Hum Genet. 2014;94:373–384. doi: 10.1016/j.ajhg.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, Craig JM, Langford KW, Samson JM, Daza R, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32:829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xiao X, Wang P, Guo X, Zhang Q. Mutation spectrum and frequency of the RHO gene in 248 Chinese families with retinitis pigmentosa. Biochem Biophys Res Commun. 2010;401:42–47. doi: 10.1016/j.bbrc.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Liu MM, Zack DJ. Alternative splicing and retinal degeneration. Clin Genet. 2013;84:142–149. doi: 10.1111/cge.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth GT, Yu F, Indap AR, Garimella K, Gravel S, Leong WF, Tyler-Smith C, Bainbridge M, Blackwell T, Zheng-Bradley X, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee TL, Devoto M, Ott J, Berson EL, Dryja TP. Evidence that the penetrance of mutations at the RP11 locus causing dominant retinitis pigmentosa is influenced by a gene linked to the homologous RP11 allele. Am J Hum Genet. 1997;61:1059–1066. doi: 10.1086/301614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Collin RW, Gilissen C, van Huet RA, Visser L, Kwint MP, Gijsen SJ, Zonneveld MN, Wieskamp N, de Ligt J, et al. Next-generation genetic testing for retinitis pigmentosa. Hum Mutat. 2012;33:963–972. doi: 10.1002/humu.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi KM, Tearle RG, Liu YP, Oh EC, Miyake N, Benaglio P, Harper S, Koskiniemi-Kuendig H, Venturini G, Sharon D, et al. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc Natl Acad Sci. 2013;110:16139–16144. doi: 10.1073/pnas.1308243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan J, Mullaney BG, Bhaskar SS, Dickerson JE, Hall G, O’Grady A, Webster A, Ramsden SC, Black GC. A paradigm shift in the delivery of services for diagnosis of inherited retinal disease. J Med Genet. 2012;49:322–326. doi: 10.1136/jmedgenet-2012-100847. [DOI] [PubMed] [Google Scholar]
- Ozgul RK, Siemiatkowska AM, Yucel D, Myers CA, Collin RW, Zonneveld MN, Beryozkin A, Banin E, Hoyng CB, van den Born LI, et al. Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet. 2011;89:253–264. doi: 10.1016/j.ajhg.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RetNet. The Retinal Information Network. Stephen P. Daiger, Ph.D., Administrator, The University of Texas Health Science Center at Houston; 2014. http://www.sph.uth.edu/RetNet/ [Google Scholar]
- Riazuddin SA, Zulfiqar F, Zhang Q, Sergeev YV, Qazi ZA, Husnain T, Caruso R, Riazuddin S, Sieving PA, Hejtman-cik JF. Autosomal recessive retinitis pigmentosa is associated with mutations in RP1 in three consanguineous Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2264–2670. doi: 10.1167/iovs.04-1280. [DOI] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. Mutation Taster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Sergouniotis PI, Chakarova C, Murphy C, Becker M, Lenassi E, Arno G, Lek M, MacArthur DG, Bhattacharya SS, Moore AT, et al. Biallelic variants in TTLL5, encoding a tubulin glutamylase, cause retinal dystrophy. Am J Hum Genet. 2014;94:760–769. doi: 10.1016/j.ajhg.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar SP, Kozma P, Birch DG, Hughbanks-Wheaton D, Locke KG, Ruiz RS, Sullivan LS, Bowne SJ, Stone EM, Daiger SP. Analysis of protein haplotypes in trans as factors modifying phenotypic variation of retinal dystrophies caused by a splice site mutation in the peripherin/RDS gene. Invest Ophthalmol Vis Sci. 2004;45:3719. [Google Scholar]
- Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE. Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J Med Genet. 2011;48:145–151. doi: 10.1136/jmg.2010.083568. [DOI] [PubMed] [Google Scholar]
- Sohocki MM, Daiger SP, Bowne SJ, Rodriquez JA, Northrup H, Heckenlively JR, Birch DG, Mintz-Hittner H, Ruiz RS, Lewis RA, et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001;17:42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LS, Heckenlively JR, Bowne SJ, Zuo J, Hide WA, Gal A, Denton M, Inglehearn CF, Blanton SH, Daiger SP. Mutations in a novel retina-specific gene cause autosomal dominant retinitis pigmentosa. Nat Genet. 1999;22:255–259. doi: 10.1038/10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, Garcia CA, Ruiz RS, Blanton SH, Northrup H, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: A screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006a;47:3052–3064. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LS, Bowne SJ, Seaman CR, Blanton SH, Lewis RA, Heckenlively JR, Birch DG, Hughbanks-Wheaton D, Daiger SP. Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006b;47:4579–4588. doi: 10.1167/iovs.06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LS, Koboldt DC, Bowne SJ, Lang S, Blanton SH, Cadena EL, Avery CE, Lewis RA, Webb-Jones K, Wheaton DK, et al. A dominant mutation in hexokinase 1 (HK1) causes retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014 doi: 10.1167/iovs.14-15419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Scheetz TE, Mullins RF, DeLuca AP, Hoffmann JM, Johnston RM, Jacobson SG, Sheffield VC, Stone EM. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci. 2011;108:E569–576. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini G, Rose AM, Shah AZ, Bhattacharya SS, Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012;8:e1003040. doi: 10.1371/journal.pgen.1003040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang H, Tuan HF, Nguyen DH, Sun V, Keser V, Bowne SJ, Sullivan LS, Luo H, Zhao L, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: Identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2013;133:331–345. doi: 10.1007/s00439-013-1381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang VW, Feng Y, Tian X, Li FY, Truong C, Wang G, Chiang PW, Lewis RA, Wong LJ. Dependable and efficient clinical utility of target capture based deep sequencing in molecular diagnosis of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014 doi: 10.1167/iovs.14-14936. [DOI] [PubMed] [Google Scholar]
- Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: Genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11:273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- Wu DM, Khanna H, Atmaca-Sonmez P, Sieving PA, Bran-ham K, Othman M, Swaroop A, Daiger SP, Hecken-lively JR. Long-term follow-up of a family with dominant X-linked retinitis pigmentosa. Eye. 2010;24:764–774. doi: 10.1038/eye.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Guan L, Shen T, Zhang J, Xiao X, Jiang H, Li S, Yang J, Jia X, Yin Y, et al. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Hum Genet. 2014 doi: 10.1007/s00439-014-1460-2. [DOI] [PubMed] [Google Scholar]
- Zeitz C, Jacobson SG, Hamel CP, Bujakowska K, Neuille M, Orhan E, Zanlonghi X, Lancelot ME, Michiels C, Schwartz SB, et al. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2013;92:67–75. doi: 10.1016/j.ajhg.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]