Abstract
The tumor suppressor p53 is lost or mutated in approximately half of human cancers. Mutant p53 not only loses its anti-tumor transcriptional activity, but often acquires oncogenic functions to promote tumor proliferation, invasion, and drug resistance. Traditional strategies have been taken to directly target p53 mutants through identifying small molecular compounds to deplete mutant p53, or to restore its tumor suppressive function. Accumulating evidence suggest that cancer cells with mutated p53 often exhibit specific functional dependencies on secondary genes or pathways to survive, providing alternative targets to indirectly treat p53-mutant cancers. Targeting these genes or pathways, critical for survival in the presence of p53 mutations, holds great promise for cancer treatment. In addition, mutant p53 often exhibits novel gain-of-functions to promote tumor growth and metastasis. Here, we review and discuss strategies targeting mutant p53, with focus on targeting the mutant p53 protein directly, and on the progress of identifying genes and pathways required in p53-mutant cells.
Keywords: p53 mutation, targeted therapies, survival pathway, gain of function
INTRODUCTION
The p53 protein is a tumor suppressor that serves as a genomic guardian to maintain a dynamic balance between cell growth and cell arrest in response to genomic stress (1–4). As a transcription factor, the p53 gene contains a core DNA binding domain, a transcriptional activation domain, and a tetramerization domain (5, 6). p53 is normally expressed at low levels, and its expression is stabilized and increased upon various genotoxic and cellular stress signals, such as DNA damage, hypoxia, oncogene activation, and nutritional deprivation (7). These stress signals stimulate the binding of p53 to a specific DNA sequence, activating or repressing a set of downstream targeting genes that can regulate cell cycle arrest, apoptosis, or DNA repair. Thus, dysfunction of p53 will disrupt the balance of cell growth and arrest, allowing abnormal cells to proliferate and develop into cancer.
In contrast to traditional tumor suppressors that are often reduced or deleted in human cancers, p53 is more commonly mutated in most human cancers (8, 9). Genomic sequencing data across different human cancers reveal that p53 gene is the most frequently and commonly mutated tumor suppressor gene in human cancers, with over 50% of cancers harboring somatic p53 mutations (10–12). The mutation rate is even higher in certain cancer subtypes, such as ovarian serous carcinomas, squamous lung cancer, and triple-negative breast cancer (13–15). The presence of mutated p53 has been associated with poor prognosis in various tumor types (16, 17). In general, over 75% of p53 mutations occur as missense mutations with a single amino acid change in the core DNA binding domain, resulting in loss of its transcriptional activity and accumulation of dysfunction p53 protein (18). Of these mutations, there are several hotspots that occur with a higher frequency across cancer types. These hotspots include amino acids R175, Y220, R248, and R273. Additionally, two distinct types of p53 point mutations are frequently observed in cancers: conformational mutations, and DNA contact mutations. Conformational mutations of p53 disrupt the structure of p53 and abolish DNA-binding ability, while DNA contact mutants alter amino acids that directly bind DNA (19, 20). Both mutations cause p53 to lose its transcription activity and gain dominate-negative (DN) activity over the remaining wild-type allele, through hetero-oligomerization with wild-type p53 (21). Drugs have been developed to induce the degradation of mutant p53 or restore its wild-type function. Furthermore, increasing evidence suggests that mutant p53 also acquires new oncogenic functions (gain-of-function, GOF mutations) to increase tumor progression, metastasis, and drug resistance (22–26). Therefore, p53 mutations cause loss of tumor suppressive activity, as well as gain of oncogenic activity to promote tumor development, posing an attractive druggable target for cancer therapy.
COMPOUNDS THAT DIRECTLY TARGET MUTANT P53
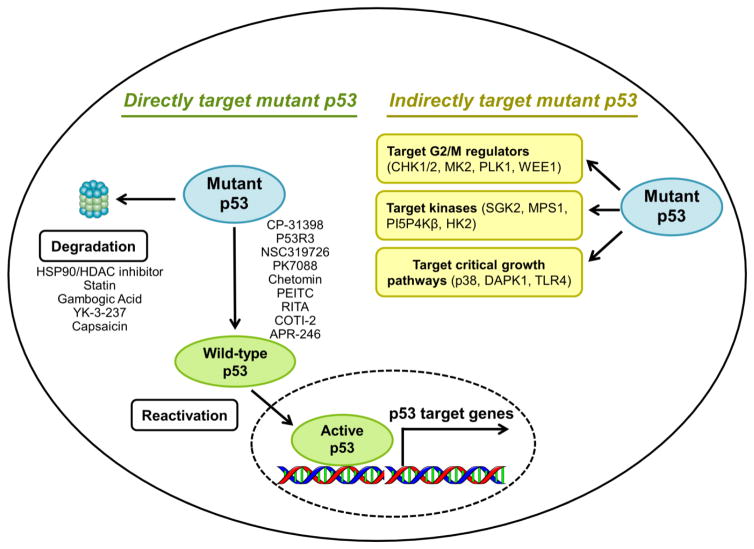
Given that p53 mutations are more frequent in cancers than normal cells, targeting mutant p53 through restoration of its wild-type tumor suppressive function (Table 1) or induction of its degradation (Table 2) has the potential to selectively kill cancer cells. Therefore, strategies have been taken to identify and develop small molecular compounds that could directly and specifically target p53-mutant p53 for effective anticancer therapy (Figure 1) (1).
TABLE 1.
Compounds that restore wile-type function of p53
| Compounds | Target Mutants | Mechanisms of Action | Reference |
|---|---|---|---|
| CP-31398 | V173A, S241F, R249S, R273H | Stabilize the DNA-binding core domain and induce conformational change | (27, 29) |
| P53R3 | R175H, R273H | Restore sequence-specific DNA binding and p53 transcriptional activities | (30) |
| NSC319726 | R175H, R172H | Restore wild-type p53 conformation and activity with MDM2-dependent degradation | (34) |
| PK7088 | Y220C | Bind to a p53Y220C-specific surface cavity and stabilize p53Y220C with restored wild-type p53 conformation | (36) |
| Chetomin | R175H | Increase Hsp40 (DNAJB1) levels and Hsp40-p53R175H binding, restoring wild-type p53 conformation, activity, and MDM2-dependent degradation | (37, 38) |
| PEITC | R175H | Sensitize the p53(R175H) mutant to proteasome-mediated degradation and further restore p53 WT conformation and transactivation | (39, 40) |
| RITA | R175H, R248Q, R273H, R280K | Reactivate p53 in mutant-p53 cancers by inhibiting the p53-HDM2 interaction | (41–43) |
| COTI-2 | R175H, R273H | Restore wildtype p53 activity by targeting and binding to misfolded p53 mutant | (46, 47) |
| PRIMA-1 and PRIMA-1Met | R175H, R273H | Bind to thiol groups in the core domain and restore wild-type conformation | (49, 50, 52, 54) |
TABLE 2.
Compounds that induce degradation of mutant p53
| Compounds | Target Mutants | Mechanisms of Action | Reference |
|---|---|---|---|
| Hsp90 inhibitors | R175H, L194F, R248Q, R273H, R280K | Reverse the Hsp90’s function to inactivate MDM2 and CHIP | (56, 57) |
| Statins | V157F, R172H, R175H, Y220C, R248W, R273H, R280K | Induce CHIP-dependent degradation of p53 with conformational or misfolded changes | (61) |
| HDAC inhibitors | R175H, R280K | Inhibit HDAC6 and disrupt the HDAC6/Hsp90/mutant p53 complex | (56–59) |
| Gambogic Acid | R175H, G266E, R273H, R280K | Inhibit the mutant p53-Hsp90 complex and induce CHIP-dependent degradation or induce autophagy | (64, 65) |
| YK-3-237 | V157F, M237I, R249S, R273H, R280K | Decrease mutant p53 levels through deacetylation at lysine 382 by activating SIRT1 | (66) |
| Capsaicin | R175H, R273H | Unknown | (67) |
Figure 1. Strategies to target mutant p53 in cancers.
Two approaches have been used to target mutant p53 for cancer treatment. The first approach is to using small molecular compounds to directly target mutant p53 by induction of its degradation or reactivation of its tumor-suppressive transcriptional activity. Several well-studied small molecular compounds are listed in the left part of this figure. The alternative approach is to targeting pathways that are critical for the survival and growth of p53 mutant cancers. Specific molecular targets, including G2/M regulators, kinases and are listed in the right part of this figure.
Compounds that restore wild-type p53 function
CP-31398
CP-31398 is a styrylquinazoline identified from a high throughput screen for therapeutic agents that restore the wild-type epitope on the DNA-binding domain of the p53 protein (27, 28). Studies found that CP-31398 not only restores p53 function in mutant p53-expressing cells but also significantly increases the protein expression of wild-type p53 through the stabilization of exogenous p53 in p53-mutant human cell lines. CP-31398 treatment increases the expression of p21 and cell cycle arrest. In addition, CP-31398 induces mitochondrial translocation of p53, leading to changes in mitochondrial membrane permeability pore transition and subsequent cytochrome c release and apoptosis in cancer cells (29). Treatment of CP-31398 dramatically inhibited tumor growth of both melanoma and colon xenografts with naturally mutated p53 (28). The compound appears to be safe at the dose of 200 mg/kg/day, as no mortality was observed for mice treated with CP-31398 for fourteen consecutive days. Tang et al. demonstrated that CP-31398 restored the tumor suppressive activity of UVB-induced mutant p53. Administration of CP-31398 not only inhibited the growth, but also prevented the development of UVB-induced skin cancers (29). These findings suggest that CP-31398 has potential applications for the treatment as well as chemoprevention for mutant p53 cancers.
P53R3
Another novel p53 “restorer”, named P53R3, was identified from a small library of compounds using an in vitro DNA binding assay (30). It restores sequence-specific DNA binding of endogenously expressed p53R175H and p53R273H mutants in glioma cell lines. P53R3 treatment inhibits cell proliferation by inducing the expression of p53 target genes, including MDM2, p21, PUMA and BAX. The P53R3-mediated increase of p53 target genes seems to be relatively more specific to mutant p53 cells, as little effects were observed in wildtyp-p53 cells. Furthermore, P53R3 strongly enhances expression of death receptor 5 (DR5) at the cell surface and sensitizes the cell to Apo2L/TRAIL induced cell death (30). This new p53 rescue compound opens up novel opportunities for the treatment of p53-mutant cancers.
NSC319726
It is known that p53 binds a single zinc ion near its DNA binding interface, which is critical for proper folding, site-specific DNA binding, and transcriptional activation (31). Insufficient zinc causes misfolding and functional loss of p53 (32). Treating tumor-bearing mice with zinc has been shown to restore DNA-binding activity of mutant p53, leading to tumor inhibition (33). Using the National Cancer Institute’s anticancer drug screen data, Yu et al. identified a compound named NSC319726 from the thiosemicarbazone family that exhibited selective growth inhibitory activity against mutant p53R175H, but not wild-type p53 cells (34). NSC319726 treatment restores wild-type structure and function of the p53R175H mutant and upregulates p53 target genes such as p21, MDM2 and PUMA (34). NSC319726 may also increase the degradation of p53R175H. Although high dose (10mg/kg/day) of NSC319726 shows strong toxicity to both p53WT and p53R175H mice, a lower dose (5mg/kg/day) induces xenograft inhibition with extensive apoptosis only in p53R172H, but not in p53WT mice (34). Thus, NSC319725 functions as an effective activator of p53R175H mutant and could be used for the treatment of p53R175H expressing cancers.
PK7088
The p53-mutant Y220C has a unique surface crevice that can be targeted by small-molecular stabilizers (35). PK7088 was identified from a compound library screen, and was found to specifically bind and stabilize the p53Y220C mutant. It restores wild-type p53 conformation and increases the expression of p21 and the proapoptotic protein NOXA (36). Consequently, treatment of PK7088 induces p53Y220C-dependent G2/M cell-cycle arrest, apoptosis and growth inhibition in cancer cells (36). In addition, PK7088 works synergistically with Nutlin-3 to further upregulate expression of p21 and NOXA (36).
Chetomin
Chetomin was identified as a specific mutant p53R175H activator from cell-based luciferase-reporter screen (37). It restores wild-type p53 transactivation and upregulates MDM2, p21 and PUMA expression. In mouse xenograft models, chetomin selectively inhibits the growth of tumor cells harboring p53R175H, but not p53R273H (37). Chetomin binds and increases the interaction of Hsp40 with p53R175H, leading to a conformational change of p53R175H and restoration of wild-type p53 function (37). However, further studies found that chetomin also suppresses tumor growth of colon cancer expressing wild-type p53, suggesting that chetomin may exert anti-cancer effects independently of mutant p53 (38).
PEITC
The natural compound PEITC (phenethyl isothiocyanate), derived from cruciferous vegetables, was recently demonstrated to reactivate the wild-type function of p53-mutant in cancer cells (39). Aggarwal et al. discovered that PEITC exhibits growth-inhibitory activity in cancer cells expressing p53R175H (40). Mechanistically, PEITC restores the wild-type conformation and transactivation of the p53R175H mutant. It also sensitizes the p53R175H mutant to proteasome-mediated degradation (40). Accordingly, PEITC treatment in p53R175H mutant cells induces apoptosis and a delay in S and G2/M phase, through the activation of canonical wild-type p53 targets. Further, dietary supplementation of PEITC in xenograft mouse model significantly inhibited tumor growth in vivo (40). No difference in body weights was observed between control and PEITC-treated groups, suggesting the safety of this natural compound. These findings provide the first example of mutant p53 reactivation by a dietary compound, and have important implications for the treatment of p53R175H mutant cancers.
RITA
RITA (Reactivation of p53 and Induction of Tumor cell Apoptosis) is another compound that can reactivate p53 function (41). It was originally identified as a molecule that inhibited growth of p53 wild type HCT 116 cells, but not HCT 116 p53−/− cells, by inhibiting the p53-HDM2 interaction and inducing p53-target genes, such as p21 and PUMA (41). Subsequent studies have demonstrated that RITA can also reactivate p53 in mutant-p53 cancers, but not p53-null cell lines, by inducing apoptotic genes such as NOXA, p21, and GADD45, in addition to suppressing oncogenic genes such as N-Myc and Bcl-2 (42, 43). However, another study in ovarian cancer showed that RITA-induced cell death occurred independently of p53, as RITA treatment induces caspase-dependent apoptosis in p53-null cells, as well as cells harboring wildtype p53 and mutant p53 (44). RITA enhances cisplatin-induced cytotoxicity in p53 wildtype head and neck cancers (45), and additional studies showed that reactivation of p53 by RITA synergistically enhances gemcitabine-induced apoptosis in p53 mutant pancreatic cancer (45).
COTI-2
COTI-2 was discovered by a pharmaceutical company named Critical Outcome Technologies as an orally available third generation thiosemicarbazone with ability to restore wildtype p53 from misfolded p53 mutants (R175H, R273H) (http://criticaloutcome.com/coti-2-and-pipeline/coti-2/). COTI-2 also acts as a negative regulator of the PI3K/AKT/mTOR pathway (46). Salim et al. recently demonstrated that COTI-2 treatment reduces cell proliferation and inhibits xenograft growth of multiple human cancer cell lines expressing either mutant p53 or wildtype p53 (47). This suggests that COTI-2 may affect mutant p53 in human tumors, but likely has additional effects on other targets in the PI3K/AKT/mTOR pathway in wildtype p53 tumors. Despite its potent anti-tumor efficacy, COTI-2 was well-tolerated and did not affect mice weights in vivo (47). Although the anti-tumor mechanisms of COTI-2 are still under investigation, it is currently undergoing a Phase I clinical trial at MD Anderson Cancer Center for the treatment of advanced or recurrent gynecologic malignancies (www.clinicaltrials.gov, NCT02433626).
PRIMA-1 and PRIMA-1Met (APR-246)
PRIMA-1 was identified as a small molecule with the ability to suppress growth of p53R175H and p53R273H cells through the reactivation of p53 target genes and induction of apoptosis by binding to the core DNA binding domain (34, 48, 49). This occurs by forming the compound methylene quinuclidinone, which covalently binds to thiol groups in mutant p53, refolding p53 into a wild-type conformation, thus restoring its anti-tumor transcriptional activity. Further testing of PRIMA-1 demonstrated that its methylated analogue, PRIMA-1Met (APR-246), is more potent and less toxic than the parental PRIMA-1 (50). In SCLC (small cell lung cancer) lines expressing mutant p53, APR-246 treatment induces apoptosis through activation of caspase-3 and upregulation of pro-apoptotic proteins such as Bax and Noxa. Moreover, APR-246 significantly inhibits in vivo tumor growth of p53-mutant SCLC models without apparent toxicity (51). Since many chemotherapy drugs depend on wild-type p53 for induction of tumor cell apoptosis, the restoration of wild-type p53 by APR-246 has the potential to sensitize p53-mutant cancers to these drugs. Indeed, APR-246 has showed strong synergy with traditional chemotherapy drugs such as cisplatin, 5-fluorouracil and doxorubicin in multiple p53-mutant expressing ovarian cancers (52). Furthermore, mutant p53 targeting with APR-246 effectively eliminates resistance to the proteasome inhibitor carfilzomib (53). In a triple negative breast cancer xenograft model, APR-246 combined with carfilzomib not only synergistically reduced primary tumor growth, but also efficiently eradicated lymph node and lung metastasis (53).
APR-246 is another p53-reactivating drug that has undergone clinical trials. The phase I/IIa clinical trial of APR-246, conducted in individuals with hematological malignancies and prostate cancer, showed both effectiveness and safety in patients with mutation of p53 (54). Combined APR-246 with carboplatin is currently being tested in phase Ib/II clinical trials for patients with recurrent high-grade serous ovarian cancer, 95% of which carry p53 mutations (www.clinicaltrials.gov, NCT02098343) (55).
Compounds that induce mutant p53 degradation
HSP90 and HDAC Inhibitors
In human cancers, mutant p53 is more stable than wild-type p53. This is mainly due to the interaction of mutant p53 with the HDAC6/Hsp90 chaperone complex (56). This complex stabilizes mutant p53 through preventing its degradation mediated by chaperone-associated E3 ubiquitin ligase. Therefore, disruption of HDAC6/HSP90 complex by either HDAC or Hsp90 inhibitors will induce the degradation of mutant p53 (57) (Figure 1). Heat shock protein 90 inhibitors (17-AAG and the more potent ganetespib) and HDAC inhibitors (such as SAHA or vorinostat) have been shown to promote proteasome-dependent degradation of mutant p53 (56, 58). These drugs are currently being tested in clinical trials for treatment of p53-mutant cancers. The Hsp90 inhibitor ganetespib entered phase III trials for the treatment of non-small cell lung cancer, but the trial was terminated following the first interim analysis because of futility (59). However, ganetespib is still in phase I and II trials for other cancers with mutant p53, including acute myeloid leukemia, ovarian, and breast cancers (59).
Statins
Statins are a class of cholesterol-lowering compounds used to treat and prevent cardiovascular disease (60). By conducting a chemical library screen to identify compounds that can degrade mutant p53, Parrales and colleagues recently discovered statins (lovastatin, atorvastatin and mevastatin) as biologically active compounds that preferentially induce degradation of p53 with conformational or misfolded mutation changes (V157F, R172H, R175H, Y220C, R248W, R273H, and R280K) (61). Mechanistic studies revealed that lovastatin treatment inhibits the mevalonate-5-phosphate pathway and subsequently induces CHIP (carboxyl terminus of Hsp70-interacting protein) ubiquitin ligase-mediated nuclear export, ubiquitylation, and degradation of mutant p53 by inhibiting the interaction of mutant p53 with DNAJA1 (DnaJ Heat Shock Protein Family (Hsp40) Member A1) (61). The effects of statins-mediated HMG-CoA inhibition and p53 degradation were highly specific to cancer cells expressing mutant p53 as minimal effects were observed in cancer cells expressing wild-type p53. Accordingly, lovastatin treatment reduces in vitro and in vivo tumor growth only in p53 mutant, but not in p53 wild type cancers (61). Furthermore, statins synergistically enhanced the anti-tumor effects of chemotherapeutic drugs such as doxorubicin exclusively in p53 mutant cancers (62). Thus, inhibition of the mevalonate pathway with statins may represent a novel and effective strategy to treat p53 mutant cancers.
Gambogic Acid
A natural product derived from the Garcinia hanburyi tree, gambogic acid, upregulates protein expression of p53 and induces apoptosis of wild-type p53 cells (63). It also down-regulates mutant p53 at post-transcription level by targeting the nuclear export of various p53 mutants for ubiquitination and subsequent protein degradation, mediated by the CHIP ubiquitin ligase (64). Furthermore, gambogic acid induces mutant p53 degradation through autophagy in cancer cells expressing the p53R280K and the p53S241F proteins, as the inhibition of autophagy with bafilomycin A1 or chloroquine counteracted mutant p53 degradation by gambogic acid (65).
YK-3-237
The small molecule compound YK-3-237 was discovered to induce degradation of mutant p53 in triple-negative breast cancers (TNBCs) (66). It reduces the acetylation of various p53 mutants (V157F, M237I R249S, R273H, and R280K) through activation of protein deacetylase SIRT1 (NAD-dependent deacetylase sirtuin-1) (66). Deacetylation of mutant p53 destabilizes its protein level and upregulates the expression of p53-target genes including PUMA and NOXA (66). YK-3-237 preferentially inhibits cell proliferation through induction of cell cycle arrest and PARP-dependent apoptotic cell death in TNBC harboring mutant p53. However, it remains to be determined whether YK-3-237 inhibits xenograft tumor growth of p53 mutant TNBCs.
Capsaicin
To explore natural compounds that induce degradation of mutant p53, Garufi et al. recently discovered that capsaicin, the constituent of peppers responsible for their pungency, could induce the protein degradation of mutant p53 (R175H, R273H) in both glioblastoma and breast cancer cell lines (67). Abrogation of mutant p53 by capsaicin treatment restored wild-type p53 activities, such as upregulation of PUMA and Bax, and induction of cancer cell death. Interestingly, capsaicin also decreased the expression of MDR1 (multidrug resistance gene) and therefore sensitized tumors to chemotherapy drugs such as cisplatin (67). However, the mechanism by which capsaicin induces degradation of mutant p53 remains unknown.
TARGETING PATHWAYS THAT ARE CRITICAL FOR SURVIVAL OF P53 MUTANT CANCERS
The concept of synthetic lethality originates from studies in drosophila model systems, in which a combination of mutations in two or more separate genes leads to cell death (68). In cancers, synthetic lethality can also occur by inhibiting a gene product in combination with another single-gene mutation (69). The relevance of synthetic lethal screens is supported by multiple observations that oncogenic mutations, or tumor suppressor defects, may lead to the development of secondary dependencies in cancer cells (70). Therefore, investigators are working to identify drugs targeting these critical survival pathways that specifically induce the death of cancer cells harboring oncogenic mutations (71, 72). In light of its high mutational rate across different tumors, p53 provides an attractive target for identifying candidate genes or pathways that are synthetically lethal with p53 mutations (73). The compounds that target these genes and pathways may selectively kill cancer cells expressing mutant p53 without affecting normal cells expressing wild-type p53 (Figure 1).
Multiple approaches, including RNA interference and chemical screens, have been used in search for synthetic lethality to p53 mutations (11, 74). Several different genes have shown synthetic lethality with p53 mutations, and these genes function through regulating cell cycle and critical kinase pathways. Targeted inhibition of these genes, or a block in relevant pathways, provides a unique therapeutic window to treat aggressive p53-mutant tumors.
Targeting the G2/M checkpoint
When p53 is mutated, the G1 arrest function of p53 is lost, causing the cell to rely solely on the G2/M checkpoint for DNA repair (4, 8). Therefore, inhibition of cell cycle regulating genes or pathways that control the G2/M checkpoint can potentially induce synthetic lethality in p53-mutant cancers (75, 76). Several G2/M checkpoint regulators, including CHK1/2, MK2, PLK1 and WEE1, have been identified as druggable targets in p53-mutant cancers (Figure 1).
CHK1/2
The ATR/CHK1 signaling pathway is activated to regulate cell cycle arrest in response to explicative and genomic stress, such as DNA damage (77, 78). ATR/CHK1 activation prevents collapse of DNA-single strand breaks and stalled replication forks, and inhibits cell-cycle progression through the G2/M checkpoint (79, 80). Suppression of the G2/M checkpoint by inhibition of ATR/CHK1 in p53-mutant cells has been shown to induce loss of G1 and G2/M cell cycle checkpoints, resulting in cell death (81, 82). UCN-01 was the first CHK1 inhibitor that exhibited preclinical synergistic effects in combination with several DNA-damaging agents including irinotecan, a topoisomerase I inhibitor (83–85). Other potent and more selective CHK1 inhibitors, such as PF477736, A-690002, and SCH900776, have since been developed. These drugs potentiate the cytotoxicity of topoisomerase inhibitors and ionizing radiation in p53 mutant, but not p53 wild-type cancer cells (86, 87). In a recent study, Ma et al. employed elegant human-in-mouse models of triple-negative breast cancer and showed that combination treatment of AZD7762, a highly selective CHK1 inhibitor, with irinotecan, significantly inhibited tumor growth and prolonged survival in mice bearing p53-mutant or -deficient tumors (88). Furthermore, this selective CHK1/2 inhibitor can overcome the cisplatin resistance of head and neck p53-mutant cancer cells, reconfirming the feasibility of treating p53-mutant cancers with CHK inhibitors (89).
MK2
MAPKAP kinase 2 (MK2) is another important regulator of the G2/M checkpoint. In normal cells with wild-type p53, MK2 is a critical regulator of the cell cycle by sustaining the G2/M checkpoint in response to genotoxic stress or UV irradiation (90). Synthetic lethality between MK2 and p53-deficiency following genotoxic stress was identified using RNA interference (RNAi)-mediated depletion of MK2 in p53-proficient and p53-deficient settings (91). Knockdown of MK2 dramatically sensitized p53-deficient murine embryonic fibroblasts (MEFs), and H-RasV12-driven p53-deficient allografts, to the cytotoxic effects of cisplatin and the topoisomerase II inhibitor doxorubicin. In contrast, loss of MK2 in p53-proficient cells did not increase cell death or sensitivity to chemotherapy. Further mechanistic investigation revealed that MK2-depletion dramatically reduced phosphorylation of Cdc25A and B (91). Subsequent studies by Morandell et al. were conducted with isogenic MK2-proficient and deficient non-small-cell lung cancer tumors, which were oncogenically driven by K-Ras and a lack of p53. Using this model, they demonstrated that MK2−/− tumors are more sensitive to cisplatin treatment than MK2+/+ tumors, suggesting that MK2 is responsible for resistance of p53-deficient tumors to cisplatin (92). Therefore, targeting inhibition of MK2 in p53-deficient or mutant cancers may sensitize chemotherapy agents such as cisplatin.
PLK1
Polo-like kinase 1 (PLK1) is an enzyme that controls G2/M checkpoint, and its inhibition has been shown to induce synthetic lethality in cells with p53 mutations (93, 94). Transcriptome analysis revealed a consistent upregulation of PLK1 in P53-mutant (p53R248W and p53S241F), but not wild-type p53 colorectal cancer lines. Inhibition of PLK1 by small molecular inhibitor BI-2536 significantly enhanced cytotoxic effects of ionizing radiation in p53 mutant, but not p53 wild type cancers cells. Furthermore, BI-2536 treatment dramatically reduced the side effects of Nutlin-3 (MDM2 inhibitor) by protecting neutrophil depletion in nude mice bearing HCT116 p53−/− xenografts. Although clinical trials using PLK1 inhibitor as monotherapy have been terminated, the second-generation of PLK1 inhibitor, GSK461364, has shown greater sensitivity in p53-mutated cancer compared with that of p53-wild type cancer cells (95, 96). Anti-mitotic agents such as PLK1 inhibitors may synergistically enhance the therapeutic efficacy of chemotherapy agents in p53-mutant tumors.
WEE1
WEE1 is another checkpoint kinase that mediates G2/M cell cycle arrest through phosphorylation of CHK1, which inhibits mitotic transition (97, 98). Functional analysis identified WEE1 as an important survival factor in p53-mutant head and neck squamous cell carcinomas (99, 100). The synthetic lethality between WEE1 and mutant p53 was established by studies showing that MK-1775, a selective WEE1 inhibitor, sensitizes p53-mutant cancer cells to DNA-damaging agents such as cisplatin (100). Clinical trials utilizing a combination of MK-1775, carboplatin, and paclitaxel have been used in patients with p53-mutant ovarian cancer (101).
Targeting kinase pathways
SGK2/PAK3
SGK2 and PAK3 were identified as two novel kinases that when inhibited cause synthetic lethality with p53 dysfunction in cervical cancer cell lines in which p53 was inactivated by HPV infection (102). While loss of p53, SGK2 or PAK3 alone did not significantly affect cell viability, loss of p53 in combination with depletion of either SGK2 or PAK2 led to cell death in primary human epithelial cells derived from tissues of cervical carcinoma. Two different mechanisms of synthetic lethality in p53-deficient cervical carcinoma were proposed: SGK2 depletion induces autophagy, and PAK3 depletion increases apoptosis. However, it remains to be determined whether kinase activity of SGK2 or PAK3 is required for their synthetic interactions with p53 loss.
MPS1
Monopolar Spindle 1 (MPS1) kinase is a dual-specificity protein kinase that represents an essential component of the spindle assembly checkpoint (103, 104). It functions in a number of mitosis steps, including regulation of activities at the kinetochore in both chromosome attachment and spindle checkpoint (105). Using video microscopy and fluorescent TP53+/+ and TP53−/− human colon carcinoma cells, Jemaa et al. discovered that SP600125, a serine-threonine kinase inhibitor which acts on MPS1, kills more p53-deficient cells but not p53-profient cells (106). This preferential cytotoxicity of MSP1 inhibition is dependent on p53. TP53−/− cells treated with SP600125 failed to undergo cell cycle arrest and became polyploid upon mitotic abortion, resulting in apoptosis. The gene that encodes MPS1 is also significantly correlated with p53 mutation in several breast cancer datasets (107). Similarly, the inhibition of MSP1 by SP600125 reduced cell viability and increased cell death selectively in p53-mutant breast cancers (107). In addition, MSP1 inhibition sensitized breast cancers to conventional chemotherapy treatments (107). Therefore, MPS1 is a potential therapeutic target for TP53 mutated colon and breast cancers.
PI5P4Kβ
It has been shown that p53 supports cell survival by maintaining metabolic homeostasis by regulating mitochondrial respiration and limiting reactive oxygen species (ROS) (12, 108). Thus, p53-mutant cells lack the ability to cope with metabolic changes in conditions of low ROS (109). PI5P4Kβ (type 2 phosphatidylinositol-5-phosphate 4-kinase beta) was recently identified as a novel gene that, when inhibited, reduces the growth of p53-mutant breast cancers by inhibiting glucose metabolism and increasing levels of ROS (110, 111). PI5P4Kβ is found frequently amplified in breast cancers, often in co-occurrence with a p53 mutation. Knockdown of PI5P4Kβ specifically impaired tumor growth in p53-deficient breast cancers. These novel findings suggest that inhibitors of PI5P4Kβ could be effective in treating cancers with p53 mutations.
HK2
Wan et al. recently identified an interaction between p53 and hexokinase-2 (HK2), a metabolic-related kinase highly expressed in many cancer cells (112). HK2 catalyzes the phosphorylation of glucose in glycolysis, and increases glucose metabolism that is required for tumorigenesis (113, 114). The expression of HK2 was selectively upregulated by the combined loss of PTEN and p53 in prostate cancer. Genetic deletion of HK2 demonstrates that HK2-mediated aerobic glycolysis, known as the Warburg effect, is required for PTEN- and p53-deficienct tumor growth in xenograft mouse models of prostate cancer (112). Thus, targeting inhibition of HK2 might be effective for the treatment of prostate tumors with PTEN loss and a p53 mutation.
TARGETING PATHWAYS THAT ARE CRITICAL FOR GROWTH OF P53-MUTANT CANCERS
P38
p38 kinases are members of the mitogen-activated protein kinase (MAPK) family, which transduce signals from environmental stresses, growth factors, inflammatory cytokines to regulate cell growth, differentiation and apoptosis (115). During the onset of malignant transformation, p38 has been shown to exert tumor suppressive activity by phosphorylation and activation of p53, leading to cell cycle inhibition and apoptosis (116, 117). However, increased p38 expression and activation has been correlated with poor survival in patients with breast cancer or liver cancer, suggesting the potential oncogenic role of p38 (118, 119). Indeed, inhibition of p38 preferentially suppressed tumor growth of breast cancers expressing mutant, but not wild-type p53 (120). Another study showed that p38 inhibition also sensitizes breast cancer cells to cisplatin-induced apoptosis (121). Thus, targeting p38 by small molecular inhibitors may be clinically effective for the treatment of highly aggressive triple-negative breast cancers that harbor p53 mutations.
DAPK1
Death associated protein kinase 1 (DAPK1) is a calcium/calmodulin (CaM)-regulated protein kinase that activates death signaling in response to interferon-gamma (INF-γ), tumor necrosis factor-alpha (TNF-α), and transforming growth factor-beta (TGF-β) (122–124). It has been shown to be highly expressed in ER negative breast cancers when compared to ER positive breast cancers (125). Recent studies suggest that DAPK1 is essential for growth of p53-mutant cancers, which account for over 80% of TNBCs. DAPK1 expression is elevated in p53-mutant cancers compared to p53-wildtype cells. The depletion or inhibition of DAPK1 suppressed growth of p53-mutant, but not p53-wildtype breast cancer cells, suggesting that targeting DAPK1 may possess a therapeutic strategy for p53-mutant cancers.
TLR
The Toll-like receptors (TLR) plays important role in activating early innate immunity in response to different pathogens and orchestrating late adaptive immune responses (126, 127). The functions of at least two TLRs including TLR4 and TLR3 have been linked to mutant p53 cancers. Toll-like receptor 4 (TLR4) is expressed on the cell surface of immune cells to activate innate immune response to gram-negative bacterial lipopolysaccharide (LPS) (126, 127). It is also expressed in breast epithelial tumors, and has oncogenic functions to promote tumor growth and drug resistance (128). Therefore, TLR4 appears to be a promising target for immune-based therapeutic options and is the focus of many drugs currently in development. However, a recent study demonstrated that TLR4 activation promotes cell growth in p53-mutant breast cancers, but inhibits cell growth in p53 wildtype breast cancers (129). The differential effects appear to be mediated by tumor cell cytokine secretions upon TLR4 activation. In p53 wild-type breast cancer cells, TLR4 activation increases type I IFN (IFN-γ), resulting in p21 expression and tumor inhibition. However, in p53-mutant breast cancer cells, TLR4 activation promotes tumor growth by inducing the secretion of CXCL1 and CD154. These findings suggest the need to determine p53 status for anti-TLR4 therapy, as the therapy may only be beneficial for p53-mutant tumors.
Recently studies suggest that wildtype-p53 can amplify immune response against cancer cells by regulating the expression of immune-related TLRs including TLR2, TLR3 and TLR5(130). Menendez and colleagues transfected several specific p53 mutants (R138V, P151H, R175H, H178Y) into HCT 116 p53−/− cells and found that those with p53 mutants decreased the expression of TLR3 and inhibited TLR3-mediated cytokine secretion and apoptotic response after stimulation by the cognate ligand ploy (131). Furthermore, treatment with the p53 reactivating agent RITA rescued TLR3 expression and enhanced DNA damage-induced apoptosis via TLR3 signaling in a p53 mutant lymphoma cell line, Raji, that harbors loss-of-function alleles (R213Q and Y234H)(131). Therefore, targeting mutant p53 to increase TLR3 expression may enhance anti-tumor immune responses in cancer cells harboring specific p53 mutations.
TARGETING P53 “GAIN-OF-FUNCTION” PATHWAYS
The fact that most tumors contain p53 missense mutations, rather than p53 deletions, raises the possibility that tumor cells harboring p53 mutations may acquire advantages over cells lacking p53 (22). Although mutations can occur throughout p53 protein, the most common mutations occur in the DNA-binding domain, of which the majority are substitution mutations in six codons which contain CpG dinucleotide sequences (5 missense: R175, G245, R248, R273, R282; 1 nonsense: R213*) (132). Accumulating evidence has shown that the mutations of p53 not only cause loss of transcriptional activity, but also allow the acquisition of novel oncogenic functions, which contribute to tumor development and progression. p53 mutants that localize in the cytoplasm have been shown to favor cancer cell survival by activating mTOR signaling. p53 mutants can also activate cell migratory pathways (e.g. PDGFRβ signaling, RhoA/ROCK, EGFR/integrin recycling, Myo10, Pla2g16) to promote cancer invasion and metastasis. Studies have also demonstrated that introducing p53 mutants into p53-null cancer cells resulted in an increase in tumor growth and enhanced tumor invasion through metabolic alterations, epigenetic modifications, and increased drug resistance, as detailed in Figure 2 (26). Therefore, targeted inhibition of the pathways that contribute to mutant p53’s GOF activities offers an alternative strategy for effectively treating p53-mutant cancers.
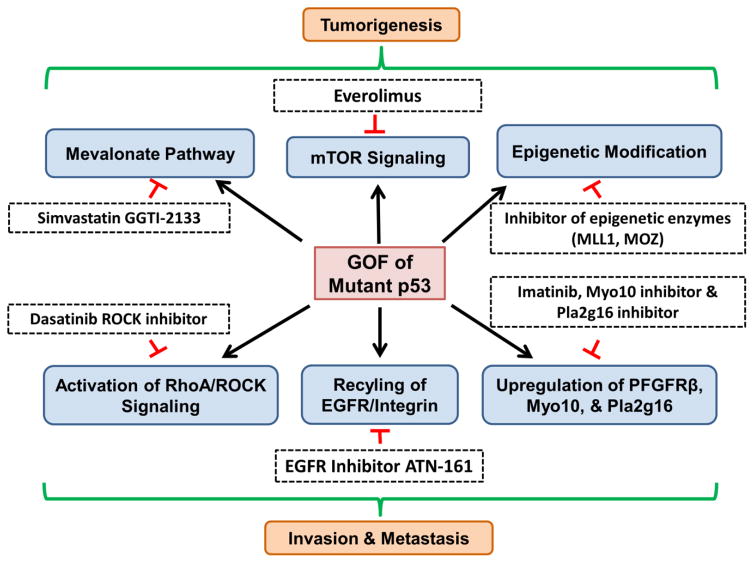
Figure 2. Strategies to target pathways induced by p53 GOF mutants.
Mutant p53 gains new functions to promote tumorigenesis by activation of the metabolic melanovate pathway or upregulation of epigenetic enzymes. Drugs that target the melanovate pathway (simvastatin, GGTI-2133) or epigenetic enzymes (MLL1, MOZ inhibitors) show promise for the treatment of p53 mutant cancers. Cytoplasmic mutant p53 also promote tumor growth through activation of mTOR signaling, providing an opportunity to using mTOR inhibitors to treat cancers with cytoplasmic mutant p53. In addition, mutant p53 often gains novel function(s) to promote tumor invasion and metastasis through activation of cell invasion pathways (PDGFRβ signaling, RhoA/ROCK, EGFR/integrin recycling, Myo10, Pla2g16). Small molecular inhibitors (ROCK inhibitor, EGFR inhibitor, Myo10 inhibitor and Pla2g16 inhibitor) have been used to target each pathway to inhibit the metastatic ability of p53 mutant cancers.
Targeting mTOR pathway induced by gain-of-function p53 mutants
Autophagy is an intracellular process by which damaged organelles and macromolecules are targeted by lysosomes for degradation via autophagic vesicles (133). Under normal conditions, autophagy functions to maintain cellular homeostasis by monitoring the integrity of long-lived proteins and organelles. Under cellular stress, such as nutrient starvation and oxidase stress, autophagy is crucial for maintaining primary biological activities to recycle intracellular contents as an alternative energy for cell survival (133). Accumulating data indicate that autophagy plays tumour-suppressive role during tumorigenesis (134). Previous studies showed that genomic stress can induce autophagy in a p53-dependent fashion and wild-type p53 can activate autophagy by upregulating autophagy-related gene DRAM1 (damage-regulated autophagy modulator 1) (135). However, several line of recent studies demonstrated that specific p53 mutants (R175H, R273H, R273L) that localize in the cytoplasmic can gain new function to inhibit autophagy either through blockade of AMPK signaling or activation of Akt/mTOR signaling (136, 137). In contrast, other p53 mutants (P151H, R282W) that localize in the nucleus failed to inhibit autophagy (138). Those cytoplasmic p53 mutants counteract the formation of autophagic vesicles and fusion with lysosomes through the repression of several key autophagy-related proteins and enzymes such as BECN1, DRAM1 and ATG12 (137). While AMPK activation triggers autophagy by inducing degradation of macromolecules, mTOR signaling inhibits autophagy by stimulating anabolic biosynthesis for cancer cell growth (139). Consequently, the inhibition of autophagy by cytoplasmic p53 mutants increases the proliferation and survival of cancer cells. In addition, the Akt/mTOR activation by GOF p53 mutants sensitizes cancer cells to the treatment with mTOR inhibitor everolimus (137). Therefore, targeting inhibition of mTOR signaling to induce autophagy has potential therapeutic applications to treat human cancers harboring cytoplasmic p53 mutants.
Targeting p53-activated invasion pathways
RhoA/ROCK pathway
It is known that aerobic glycolysis is primary utilized by tumor cells for energy production, a phenomenon known as the Warburg effect (140). A novel mutant p53 GOF was shown to be a driving of the Warburg effect and promotion of tumorigenesis in a murine knock-in p53R172H model (141). Tumor formation was promoted by the translocation of the glucose transporter GLUT1 to the plasma membrane, which was induced by increased RhoA/ROCK signaling in p53R172H knock-in mice. Similar studies using FLIM-FRET imaging to track RhoA activity in pancreatic cancer driven by the p53R172H mutant found that this mutant p53 increased RhoA activity in vivo (142). Dasatanib, a clinically used kinase inhibitor agent, inhibited the activity of RhoA and invasive ability of p53R172H cells in vivo (141). These studies therefore suggest that targeted inhibition of RhoA/ROCK signaling in p53-mutant cancers may inhibit tumorigenesis, and possibly metastasis.
EGFR/integrin recycling
Muller and coworkers showed that cells containing mutant p53 gain novel functions to promote cell migration and tumor metastasis (143). They demonstrated that p53R175H and p53R273H mutants promote recycling of EGFR and integrin alpha-5 beta-1 (α5β1) to the cell surface, thereby increasing invasive and metastatic potential (143). Recycling of integrin/EGFR by mutant p53 is linked to the transcriptional inhibition of p63. Consequently, the loss of p53 and p63 can phenocopy a p53-mutant cancer. Although the molecular details of how mutant p53 inhibits p63 remain to be determined, these findings suggest that inhibiting integrin α5β1 integrin or blocking EGFR signaling (Cetuximab) may have therapeutic benefits in cancers with GOF p53 mutations.
PDGFRβ
Platelet-derived growth factor receptor beta (PDGFRβ) is mainly expressed by stromal cells, where PFGFRβ signaling contributes to tumor-associated invasion and metastasis (144, 145). A recent study demonstrated that PDGFRβ signaling enhanced the expression of a GOF mutant p53 induced pancreatic cancer metastasis in PKC mice harboring one oncogenic allele of KrasG12D and one allele of p53R172H (146). Inhibition of PDGFRβ using imatinib effectively prevented cell invasion and metastasis of pancreatic cancer with p53 mutations (146). Given than over 90% of pancreatic cancers contain p53 mutations, these studies highlight the potential targeted therapy of blocking PDGFRβ signaling to treat p53-mutant pancreatic cancers.
AdPLA
Adipocyte phospholipase A2 (AdPLA), a transcript from the PLA2G16 gene, was recently identified in expression array studies comparing primary osteosarcomas from metastatic p53R172H/+ mice and non-metastatic p53+/− mice (147). AdPLA is a phospholipase that catalyzes phosphatidic acid into lysophosphatidic acid and free fatty acid, both of which are implicated in metastasis. Xiong et al. found that murine mutant p53R172H increased PLA2G16 expression in mouse osteosarcoma cells, suggesting that PLA2G16 is a p53-regulated gene (147). Indeed, ChIP analysis revealed that transcription factor EST2 recruited p53-mutant protein to bind the PLA2G16 promoter at the E26 transformation specific binding motif. Functionally, AdPLA knockdown inhibited migration and invasion in mutant p53-expressing cells. These studies identify PLA2G16 as a transcriptional target of mutant p53 and suggest that targeting AdPLA will inhibit invasion and metastasis of p53-mutant tumors.
Targeting cellular and nucleotide metabolism induced by gain-of-function p53 mutants
Several studies demonstrated that gain-of-function (GOF) p53 mutants regulate cellular and nucleotide metabolism (148–150). Using a three-dimensional (3D) culture model, Freed-Pastor et al. discovered that breast cancer cell lines expressing either p53R273H or p53R280K disrupted acinar morphology through upregulation of the mevalonate pathway, which is responsible for cholesterol synthesis (148). Treatment with simvastatin, a clinically approved statin that inhibits cholesterol synthesis, induced cell death in p53R273H cells and reduced the invasive morphology of the p53R280K cells. Geranylgeranyl transferase was identified to be the critical enzyme in this mevalonate pathway, as its inhibitor (GGTI-2133) significantly reduced the growth and 3D invasive morphology of p53R280K cells. Mechanistic studies revealed that SREBPs (sterol regulatory element-binding protein) recruited mutant p53 to gene promoters that encode mevalonate pathway enzymes, which increased protein geranylgeranylation and altered acinar morphogenesis and promoted tumorigenesis (148). These findings provide strong evidence for targeting the mevalonate pathway for the treatment of p53-mutant breast cancers.
A novel gain-of-function of mutant p53 in promoting cancer cell metabolism was reported in head and neck squamous cell carcinoma (23, 151). Under energy stress conditions, mutant p53, but not wildtype-p53, inhibits the activation of adenosine monophosphate-activated protein kinase (AMPK) by binding with its α subunit. Inhibition of AMPK by mutant p53 impaired metabolic checkpoint and increased aerobic tumor growth and progression (23). Given that wildtype p53 could activate AMPK activity through transcriptional activation of the gene encoding β subunit of AMPK (152), these results strongly support a transcription-independent mechanism by which mutant p53 promotes tumor progression by activating cancer cell metabolism. Regulation of nucleotide metabolism by mutant p53 was recently found to be functionally important for GOF activities (150). CHIP-seq analysis of mutant p53 knockdown breast cancer cells demonstrated that loss of mutant p53 reduced many nucleotide metabolism genes (NMGs) and substantially depleted nucleotide pools, which attenuated GTP-dependent protein activity and cell invasion (150). The reduction of invasion by mutant p53 knockdown was rescued by addition of exogenous GTP, suggesting that mutant p53 promotes invasion by increasing GTP. Mutant p53 appears to depend on deoxycytidine kinase, a nucleoside salvage pathway enzyme, to maintain a proper balance of the dNTP pools required for cell proliferation (150). These data reveal the mechanism underlying the lethal genetic interaction between mutant p53 and deoxycytidine kinase.
Targeting epigenetic changes induced by gain-of-function p53 mutants
A novel GOF mutant p53 activity was discovered by Zhu and colleagues, who demonstrated that chromatin regulation by mutant p53 can drive aggressive cancer growth. This group showed that mutant p53 regulates gene expression through induction of histone modifying proteins (153). Using genome-wide measurements of p53 chromatin occupancy in a panel of breast cancer cell lines, Zhu et al. demonstrated that mutant p53, but not wild-type p53, is recruited to chromatin via interaction with transcription factor ETS2. This interaction led to binding and upregulation of genes encoding epigenetic enzymes, including the methyltransferases KMT2A (MLL1) and KMT2D (MLL2), and acetyltransferase KAT6A (MOZ), resulting in genome-wide increases of histone methylation and acetylation (153). Chromatin regulatory genes, especially MLL1, appear to be responsible for cell proliferation driven by mutant p53, as genetic or pharmacological inhibition of MLL1 significantly attenuated cancer cell proliferation. In support of these observations, analysis of The Cancer Genome Atlas shows specific upregulation of epigenetic regulatory genes including MLL1, MLL2, and MOZ in GOF p53-mutant patient-derived tumors, but not in p53 wild-type or p53 null tumors (154). GOF p53 mutants were also recently shown to act with the SWI/SNF chromatin-remodeling complex to upregulate VEGFR2, which contributed to tumorigenesis (155). Taken together, the links between p53 mutation, chromatin regulation, and gene expression, could explain why so many genes are affected by the presence of mutant p53. In addition, these findings also highlight new therapeutic opportunities for designing combinatorial chromatin-based therapies to treat cancers harboring p53 mutations.
Targeting drug resistance induced by gain-of-function p53 mutants
An accumulating body of evidence suggests that GOF p53 mutants also mediate drug resistance through multiple mechanisms, including inhibiting apoptotic proteins and gene regulations (10, 156). Early studies have shown that p53 mutations correlated with resistance to chemotherapy drugs such as cisplatin, doxorubicin and paclitaxel (157, 158). The first line evidence comes from studies demonstrating that knockdown of mutant p53 in human squamous cell carcinoma A431 cells harboring p53R273H increases procaspase-3 and sensitizes cells to doxorubicin-induced apoptosis (159). Conversely, transfection of p53R273H into p53-null human osteosarcoma Saos-2 cells down-regulated procaspase-3 level and induced resistance to doxorubicin-induced apoptosis (159). Further studies found that expressing p53R248Q in p53-null liver cancer cells conferred cross-resistances to doxorubicin and paclitaxel through upregulation of P-glycoprotein, a known multidrug resistance protein that is responsible for decreased drug accumulation in multidrug-resistant cells (158). Therefore, combination of P-glycoprotein inhibitors with doxorubicin may overcome drug resistance in p53-mutant cancers. Subsequently studies investigating mutant p53 binding sites in Li-Fraumeni cell line MDAH087 harboring p53R248W revealed that mutant p53 regulates gene expression through binding to the promoters of ETS-binding motif and that ETS2 mediates the interaction with this motif (160). Do et al. identified TDP2 (tyrosyl-DNA phosphodiesterase 2), a DNA phosphodiesterase involved in the repair of DNA damage caused by chemotherapy drug etoposide, as a transcriptional target of mutant p53. Consequently, inhibition of TDP2 sensitizes cells to the treatment of etoposide, a chemotherapy drug for lung cancer. Since mutant p53 and TDP2 are frequently overexpressed in lung cancer, TDP2 may serve as a “druggable” target to increase chemotherapy sensitivity for p53-mutant lung cancer. In pancreatic cancers, Fiorini and colleagues recently demonstrated that the expression of mutant p53 confers chemoresistance to gemcitabine (161). Gemcitabine treatment induces phosphorylation and nuclear stabilization of mutant p53 which further upregulate Cdk1 and CCNB1, leading to increased cell proliferation. Restoration of wildtype-p53 function by p53-reactiving agent (RITA and CP-31398) induced apoptosis, resulting in synergistic anti-tumor effects with gemcitabine (161).
CONCLUDING REMARKS AND FUTURE DIRECTIONS
The fact that most cancers have mutation of p53 makes this molecule an attractive therapeutic target. Several approaches, including inactivation of mutant p53, degradation of mutant p53, and restoration of the wild type function of p53, have been studied. However, these strategies often work only for specific p53 mutations and not for others. An alternative strategy, targeting signaling pathways essential in cells p53 mutations, has been effective in many types of cancers. RNA interference and chemical genetic screens have enabled investigators to identify molecules critical for survival or growth of cells with p53 mutations that can be targeted for the selective treatment of p53-mutant cancers. Ongoing studies continue to identify such critical mutant p53-specific survival and growth regulatory pathways. Thus, novel drugs that target mutant p53 or the critical pathways activated by p53 mutation are highly promising for effective treatment of many cancers.
Acknowledgments
Financial Support: This work was funded by a Komen SAB grant (P.H.B.), a Breast Cancer Research Foundation grant (P.H.B.), a John Charles Cain Distinguished Chair Award (P.H.B.), a John F. & Julie Young Award (P.H.B.), and a Cancer Center Support Grant (5 P30 CA016672-38 P.H.B.).
We would like to thank Sam Short for her assistance with this submission.
Footnotes
Conflict of interest:
PH Brown is on the Scientific Advisory Board of Susan G. Komen for the Cure. All remaining authors declare no actual, potential, or perceived conflict of interest that would prejudice the impartiality of this article.
References
- 1.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9(12):862–73. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 2.Mandinova A, Lee SW. The p53 pathway as a target in cancer therapeutics: obstacles and promise. Sci Transl Med. 2011;3(64):64rv1. doi: 10.1126/scitranslmed.3001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28(3):128–36. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pellegata NS, Antoniono RJ, Redpath JL, Stanbridge EJ. DNA damage and p53-mediated cell cycle arrest: a reevaluation. Proc Natl Acad Sci U S A. 1996;93(26):15209–14. doi: 10.1073/pnas.93.26.15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hainaut P, Wiman KG. 30 years and a long way into p53 research. Lancet Oncol. 2009;10(9):913–9. doi: 10.1016/S1470-2045(09)70198-6. [DOI] [PubMed] [Google Scholar]
- 6.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–87. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 8.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 9.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–17. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurpinar E, Vousden KH. Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol. 2015;25(8):486–95. doi: 10.1016/j.tcb.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 13.Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 2014;12(1):3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy MJ, Synnott NC, McGowan PM, Crown J, O’Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Cancer Treat Rev. 2014;40(10):1153–60. doi: 10.1016/j.ctrv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Corney DC, Flesken-Nikitin A, Choi J, Nikitin AY. Role of p53 and Rb in ovarian cancer. Adv Exp Med Biol. 2008;622:99–117. doi: 10.1007/978-0-387-68969-2_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–86. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26(15):2157–65. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 18.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60(24):6788–93. [PubMed] [Google Scholar]
- 20.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265(5170):346–55. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 21.Haupt S, Raghu D, Haupt Y. Mutant p53 Drives Cancer by Subverting Multiple Tumor Suppression Pathways. Front Oncol. 2016;6:12. doi: 10.3389/fonc.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, Patel AA, Ward AM, Sandulache VC, Jasser SA, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54(6):960–74. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Liu DP, Xu Y. The gain of function of p53 cancer mutant in promoting mammary tumorigenesis. Oncogene. 2013;32(23):2900–6. doi: 10.1038/onc.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturm I, Bosanquet AG, Hermann S, Guner D, Dorken B, Daniel PT. Mutation of p53 and consecutive selective drug resistance in B-CLL occurs as a consequence of prior DNA-damaging chemotherapy. Cell Death Differ. 2003;10(4):477–84. doi: 10.1038/sj.cdd.4401194. [DOI] [PubMed] [Google Scholar]
- 26.Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4(4):405–14. doi: 10.1158/2159-8290.CD-13-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wischhusen J, Naumann U, Ohgaki H, Rastinejad F, Weller M. CP-31398, a novel p53-stabilizing agent, induces p53-dependent and p53-independent glioma cell death. Oncogene. 2003;22(51):8233–45. doi: 10.1038/sj.onc.1207198. [DOI] [PubMed] [Google Scholar]
- 28.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286(5449):2507–10. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 29.Tang X, Zhu Y, Han L, Kim AL, Kopelovich L, Bickers DR, Athar M. CP-31398 restores mutant p53 tumor suppressor function and inhibits UVB-induced skin carcinogenesis in mice. J Clin Invest. 2007;117(12):3753–64. doi: 10.1172/JCI32481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinmann L, Wischhusen J, Demma MJ, Naumann U, Roth P, Dasmahapatra B, Weller M. A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 2008;15(4):718–29. doi: 10.1038/sj.cdd.4402301. [DOI] [PubMed] [Google Scholar]
- 31.Loh SN. The missing zinc: p53 misfolding and cancer. Metallomics. 2010;2(7):442–9. doi: 10.1039/c003915b. [DOI] [PubMed] [Google Scholar]
- 32.Hainaut P, Mann K. Zinc binding and redox control of p53 structure and function. Antioxid Redox Signal. 2001;3(4):611–23. doi: 10.1089/15230860152542961. [DOI] [PubMed] [Google Scholar]
- 33.Puca R, Nardinocchi L, Porru M, Simon AJ, Rechavi G, Leonetti C, Givol D, D’Orazi G. Restoring p53 active conformation by zinc increases the response of mutant p53 tumor cells to anticancer drugs. Cell Cycle. 2011;10(10):1679–89. doi: 10.4161/cc.10.10.15642. [DOI] [PubMed] [Google Scholar]
- 34.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21(5):614–25. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joerger AC, Fersht AR. The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol. 2010;2(6):a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Wilcken R, Joerger AC, Chuckowree IS, Amin J, Spencer J, Fersht AR. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 2013;41(12):6034–44. doi: 10.1093/nar/gkt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiraki M, Hwang SY, Cao S, Ramadhar TR, Byun S, Yoon KW, Lee JH, Chu K, Gurkar AU, Kolev V, et al. Small-Molecule Reactivation of Mutant p53 to Wild-Type-like p53 through the p53-Hsp40 Regulatory Axis. Chem Biol. 2015;22(9):1206–16. doi: 10.1016/j.chembiol.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kung AL, Zabludoff SD, France DS, Freedman SJ, Tanner EA, Vieira A, Cornell-Kennon S, Lee J, Wang B, Wang J, et al. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell. 2004;6(1):33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Gupta P, Srivastava SK. Antitumor activity of phenethyl isothiocyanate in HER2-positive breast cancer models. BMC Med. 2012;10:80. doi: 10.1186/1741-7015-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aggarwal M, Saxena R, Sinclair E, Fu Y, Jacobs A, Dyba M, Wang X, Cruz I, Berry D, Kallakury B, et al. Reactivation of mutant p53 by a dietary-related compound phenethyl isothiocyanate inhibits tumor growth. Cell Death Differ. 2016;23(10):1615–27. doi: 10.1038/cdd.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10(12):1321–8. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 42.Zhao CY, Grinkevich VV, Nikulenkov F, Bao W, Selivanova G. Rescue of the apoptotic-inducing function of mutant p53 by small molecule RITA. Cell Cycle. 2010;9(9):1847–55. doi: 10.4161/cc.9.9.11545. [DOI] [PubMed] [Google Scholar]
- 43.Burmakin M, Shi Y, Hedstrom E, Kogner P, Selivanova G. Dual targeting of wild-type and mutant p53 by small molecule RITA results in the inhibition of N-Myc and key survival oncogenes and kills neuroblastoma cells in vivo and in vitro. Clin Cancer Res. 2013;19(18):5092–103. doi: 10.1158/1078-0432.CCR-12-2211. [DOI] [PubMed] [Google Scholar]
- 44.Weilbacher A, Gutekunst M, Oren M, Aulitzky WE, van der Kuip H. RITA can induce cell death in p53-defective cells independently of p53 function via activation of JNK/SAPK and p38. Cell Death & Disease. 2014;5 doi: 10.1038/cddis.2014.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roh JL, Ko JH, Moon SJ, Ryu CH, Choi JY, Koch WM. The p53-reactivating small-molecule RITA enhances cisplatin-induced cytotoxicity and apoptosis in head and neck cancer. Cancer Lett. 2012;325(1):35–41. doi: 10.1016/j.canlet.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 46.Salim KY, Vareki SM, Danter WR, Koropatnick J. COTI-2, a new anticancer drug currently under clinical investigation, targets mutant p53 and negatively modulates the PI3K/AKT/mTOR pathway. Eur J Cancer. 2016;69:S19-S. [Google Scholar]
- 47.Salim KY, Vareki SM, Danter WR, Koropatnick J. COTI-2, a novel small molecule that is active against multiple human cancer cell lines in vitro and in vivo. Oncotarget. 2016;7(27):41363–79. doi: 10.18632/oncotarget.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8(3):282–8. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 49.Lambert JM, Moshfegh A, Hainaut P, Wiman KG, Bykov VJ. Mutant p53 reactivation by PRIMA-1MET induces multiple signaling pathways converging on apoptosis. Oncogene. 2010;29(9):1329–38. doi: 10.1038/onc.2009.425. [DOI] [PubMed] [Google Scholar]
- 50.Lambert JM, Gorzov P, Veprintsev DB, Soderqvist M, Segerback D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15(5):376–88. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 51.Zandi R, Selivanova G, Christensen CL, Gerds TA, Willumsen BM, Poulsen HS. PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin Cancer Res. 2011;17(9):2830–41. doi: 10.1158/1078-0432.CCR-10-3168. [DOI] [PubMed] [Google Scholar]
- 52.Fransson A, Glaessgen D, Alfredsson J, Wiman KG, Bajalica-Lagercrantz S, Mohell N. Strong synergy with APR-246 and DNA-damaging drugs in primary cancer cells from patients with TP53 mutant High-Grade Serous ovarian cancer. J Ovarian Res. 2016;9(1):27. doi: 10.1186/s13048-016-0239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walerych D, Lisek K, Sommaggio R, Piazza S, Ciani Y, Dalla E, Rajkowska K, Gaweda-Walerych K, Ingallina E, Tonelli C, et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol. 2016;18(8):897–909. doi: 10.1038/ncb3380. [DOI] [PubMed] [Google Scholar]
- 54.Lehmann S, Bykov VJ, Ali D, Andren O, Cherif H, Tidefelt U, Uggla B, Yachnin J, Juliusson G, Moshfegh A, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30(29):3633–9. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- 55.Singer G, Stohr R, Cope L, Dehari R, Hartmann A, Cao DF, Wang TL, Kurman RJ, Shih Ie M. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: a mutational analysis with immunohistochemical correlation. Am J Surg Pathol. 2005;29(2):218–24. doi: 10.1097/01.pas.0000146025.91953.8d. [DOI] [PubMed] [Google Scholar]
- 56.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18(12):1904–13. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, Moll UM. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9(5):577–88. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA, Lozano G, Dobbelstein M, Moll UM. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature. 2015;523(7560):352–6. doi: 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jhaveri K, Modi S. Ganetespib: research and clinical development. Onco Targets Ther. 2015;8:1849–58. doi: 10.2147/OTT.S65804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lippi G, Plebani M. Statins for Primary Prevention of Cardiovascular Disease. Trends Pharmacol Sci. 2017;38(2):111–2. doi: 10.1016/j.tips.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Parrales A, Ranjan A, Iyer SV, Padhye S, Weir SJ, Roy A, Iwakuma T. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nature Cell Biology. 2016;18(11):1233. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martirosyan A, Clendening JW, Goard CA, Penn LZ. Lovastatin induces apoptosis of ovarian cancer cells and synergizes with doxorubicin: potential therapeutic relevance. Bmc Cancer. 2010;10 doi: 10.1186/1471-2407-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu H, Wang X, Rao S, Wang J, Zhao J, Ren FL, Mu R, Yang Y, Qi Q, Liu W, et al. Gambogic acid mediates apoptosis as a p53 inducer through down-regulation of mdm2 in wild-type p53-expressing cancer cells. Mol Cancer Ther. 2008;7(10):3298–305. doi: 10.1158/1535-7163.MCT-08-0212. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Zhao Q, Qi Q, Gu HY, Rong JJ, Mu R, Zou MJ, Tao L, You QD, Guo QL. Gambogic acid-induced degradation of mutant p53 is mediated by proteasome and related to CHIP. J Cell Biochem. 2011;112(2):509–19. doi: 10.1002/jcb.22941. [DOI] [PubMed] [Google Scholar]
- 65.Foggetti G, Ottaggio L, Russo D, Monti P, Degan P, Fronza G, Menichini P. Gambogic acid counteracts mutant p53 stability by inducing autophagy. Biochim Biophys Acta. 2016;1864(2):382–92. doi: 10.1016/j.bbamcr.2016.11.023. [DOI] [PubMed] [Google Scholar]
- 66.Yi YW, Kang HJ, Kim HJ, Kong Y, Brown ML, Bae I. Targeting mutant p53 by a SIRT1 activator YK-3-237 inhibits the proliferation of triple-negative breast cancer cells. Oncotarget. 2013;4(7):984–94. doi: 10.18632/oncotarget.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garufi A, Pistritto G, Cirone M, D’Orazi G. Reactivation of mutant p53 by capsaicin, the major constituent of peppers. J Exp Clin Cancer Res. 2016;35(1):136. doi: 10.1186/s13046-016-0417-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobzhansky T. Genetics of Natural Populations. Xiii. Recombination and Variability in Populations of Drosophila Pseudoobscura. Genetics. 1946;31(3):269–90. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455–70. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
- 70.Fece de la Cruz F, Gapp BV, Nijman SM. Synthetic lethal vulnerabilities of cancer. Annu Rev Pharmacol Toxicol. 2015;55:513–31. doi: 10.1146/annurev-pharmtox-010814-124511. [DOI] [PubMed] [Google Scholar]
- 71.McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014;371(18):1725–35. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 72.Garber K. Synthetic lethality: killing cancer with cancer. J Natl Cancer Inst. 2002;94(22):1666–8. doi: 10.1093/jnci/94.22.1666. [DOI] [PubMed] [Google Scholar]
- 73.Weidle UH, Maisel D, Eick D. Synthetic lethality-based targets for discovery of new cancer therapeutics. Cancer Genomics Proteomics. 2011;8(4):159–71. [PubMed] [Google Scholar]
- 74.Reinhardt HC, Jiang H, Hemann MT, Yaffe MB. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 2009;8(19):3112–9. doi: 10.4161/cc.8.19.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawabe T. G2 checkpoint abrogators as anticancer drugs. Mol Cancer Ther. 2004;3(4):513–9. [PubMed] [Google Scholar]
- 76.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20(51):7453–63. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 77.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 78.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3(5):421–9. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 79.Furusawa Y, Iizumi T, Fujiwara Y, Zhao QL, Tabuchi Y, Nomura T, Kondo T. Inhibition of checkpoint kinase 1 abrogates G2/M checkpoint activation and promotes apoptosis under heat stress. Apoptosis. 2012;17(1):102–12. doi: 10.1007/s10495-011-0660-7. [DOI] [PubMed] [Google Scholar]
- 80.Yu H. Chk1: a double agent in cell cycle checkpoints. Dev Cell. 2007;12(2):167–8. doi: 10.1016/j.devcel.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 81.Origanti S, Cai SR, Munir AZ, White LS, Piwnica-Worms H. Synthetic lethality of Chk1 inhibition combined with p53 and/or p21 loss during a DNA damage response in normal and tumor cells. Oncogene. 2013;32(5):577–88. doi: 10.1038/onc.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18(6):721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17(2):88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma CX, Ellis MJ, Petroni GR, Guo Z, Cai SR, Ryan CE, Craig Lockhart A, Naughton MJ, Pluard TJ, Brenin CM, et al. A phase II study of UCN-01 in combination with irinotecan in patients with metastatic triple negative breast cancer. Breast Cancer Res Treat. 2013;137(2):483–92. doi: 10.1007/s10549-012-2378-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fracasso PM, Williams KJ, Chen RC, Picus J, Ma CX, Ellis MJ, Tan BR, Pluard TJ, Adkins DR, Naughton MJ, et al. A Phase 1 study of UCN-01 in combination with irinotecan in patients with resistant solid tumor malignancies. Cancer Chemother Pharmacol. 2011;67(6):1225–37. doi: 10.1007/s00280-010-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8(7):547–66. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 87.Chen Z, Xiao Z, Gu WZ, Xue J, Bui MH, Kovar P, Li G, Wang G, Tao ZF, Tong Y, et al. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int J Cancer. 2006;119(12):2784–94. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- 88.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, Lin L, Hoog J, Goiffon RJ, Prat A, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122(4):1541–52. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gadhikar MA, Sciuto MR, Alves MV, Pickering CR, Osman AA, Neskey DM, Zhao M, Fitzgerald AL, Myers JN, Frederick MJ. Chk1/2 inhibition overcomes the cisplatin resistance of head and neck cancer cells secondary to the loss of functional p53. Mol Cancer Ther. 2013;12(9):1860–73. doi: 10.1158/1535-7163.MCT-13-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AE, Yaffe MB. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell. 2005;17(1):37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 91.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11(2):175–89. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morandell S, Reinhardt HC, Cannell IG, Kim JS, Ruf DM, Mitra T, Couvillon AD, Jacks T, Yaffe MB. A reversible gene-targeting strategy identifies synthetic lethal interactions between MK2 and p53 in the DNA damage response in vivo. Cell Rep. 2013;5(4):868–77. doi: 10.1016/j.celrep.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zitouni S, Nabais C, Jana SC, Guerrero A, Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nat Rev Mol Cell Biol. 2014;15(7):433–52. doi: 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- 94.Sur S, Pagliarini R, Bunz F, Rago C, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci U S A. 2009;106(10):3964–9. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yim H. Current clinical trials with polo-like kinase 1 inhibitors in solid tumors. Anticancer Drugs. 2013;24(10):999–1006. doi: 10.1097/CAD.0000000000000007. [DOI] [PubMed] [Google Scholar]
- 96.Degenhardt Y, Greshock J, Laquerre S, Gilmartin AG, Jing J, Richter M, Zhang X, Bleam M, Halsey W, Hughes A, et al. Sensitivity of cancer cells to Plk1 inhibitor GSK461364A is associated with loss of p53 function and chromosome instability. Mol Cancer Ther. 2010;9(7):2079–89. doi: 10.1158/1535-7163.MCT-10-0095. [DOI] [PubMed] [Google Scholar]
- 97.O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16(3):545–54. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vera J, Raatz Y, Wolkenhauer O, Kottek T, Bhattacharya A, Simon JC, Kunz M. Chk1 and Wee1 control genotoxic-stress induced G2-M arrest in melanoma cells. Cell Signal. 2015;27(5):951–60. doi: 10.1016/j.cellsig.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 99.Moser R, Xu C, Kao M, Annis J, Lerma LA, Schaupp CM, Gurley KE, Jang IS, Biktasova A, Yarbrough WG, et al. Functional kinomics identifies candidate therapeutic targets in head and neck cancer. Clin Cancer Res. 2014;20(16):4274–88. doi: 10.1158/1078-0432.CCR-13-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Osman AA, Monroe MM, Ortega Alves MV, Patel AA, Katsonis P, Fitzgerald AL, Neskey DM, Frederick MJ, Woo SH, Caulin C, et al. Wee-1 kinase inhibition overcomes cisplatin resistance associated with high-risk TP53 mutations in head and neck cancer through mitotic arrest followed by senescence. Mol Cancer Ther. 2015;14(2):608–19. doi: 10.1158/1535-7163.MCT-14-0735-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brana I, Moore KN, Shapira-Frommer R, Welch S, Jou YM, Marinucci M, Freshwater T, Rose S, Oza AM. Targeting p53 mutant ovarian cancer: Phase I results of the WEE1 inhibitor MK-1775 with carboplatin plus paclitaxel in patients (pts) with platinum-sensitive, p53-mutant ovarian cancer (OC) J Clin Oncol. 2013;31(15) [Google Scholar]
- 102.Baldwin A, Grueneberg DA, Hellner K, Sawyer J, Grace M, Li WL, Harlow E, Munger K. Kinase requirements in human cells: V. Synthetic lethal interactions between p53 and the protein kinases SGK2 and PAK3. P Natl Acad Sci USA. 2010;107(28):12463–8. doi: 10.1073/pnas.1007462107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Winey M, Huneycutt BJ. Centrosomes and checkpoints: the MPS1 family of kinases. Oncogene. 2002;21(40):6161–9. doi: 10.1038/sj.onc.1205712. [DOI] [PubMed] [Google Scholar]
- 104.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Bio. 2013;14(1):25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.London N, Biggins S. Signalling dynamics in the spindle checkpoint response. Nat Rev Mol Cell Bio. 2014;15(11):735–47. doi: 10.1038/nrm3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jemaa M, Galluzzi L, Kepp O, Boileve A, Lissa D, Senovilla L, Harper F, Pierron G, Berardinelli F, Antoccia A, et al. Preferential killing of p53-deficient cancer cells by reversine. Cell Cycle. 2012;11(11):2149–58. doi: 10.4161/cc.20621. [DOI] [PubMed] [Google Scholar]
- 107.Gyorffy B, Bottai G, Lehmann-Che J, Keri G, Orfi L, Iwamoto T, Desmedt C, Bianchini G, Turner NC, de The H, et al. TP53 mutation-correlated genes predict the risk of tumor relapse and identify MPS1 as a potential therapeutic kinase in TP53-mutated breast cancers. Molecular Oncology. 2014;8(3):508–19. doi: 10.1016/j.molonc.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Olovnikov IA, Kravchenko JE, Chumakov PM. Homeostatic functions of the p53 tumor suppressor: regulation of energy metabolism and antioxidant defense. Semin Cancer Biol. 2009;19(1):32–41. doi: 10.1016/j.semcancer.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab. 2013;18(5):617–33. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, Bell EL, Shim HS, Lamia KA, Rameh LE, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013;155(4):844–57. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lokody I. Signalling: a new target for p53-null tumours. Nat Rev Cancer. 2014;14(1):8–9. doi: 10.1038/nrc3651. [DOI] [PubMed] [Google Scholar]
- 112.Wang L, Xiong H, Wu FX, Zhang YJ, Wang J, Zhao LY, Guo XL, Chang LJ, Zhang Y, You MJ, et al. Hexokinase 2-Mediated Warburg Effect Is Required for PTEN- and p53-Deficiency-Driven Prostate Cancer Growth. Cell Reports. 2014;8(5):1461–74. doi: 10.1016/j.celrep.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tan VP, Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy. 2015;11(6):963–4. doi: 10.1080/15548627.2015.1042195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, Hawkins C, Guha A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. Journal of Experimental Medicine. 2011;208(2):313–26. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9(8):537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 116.Hui L, Bakiri L, Mairhorfer A, Schweifer N, Haslinger C, Kenner L, Komnenovic V, Scheuch H, Beug H, Wagner EF. p38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat Genet. 2007;39(6):741–9. doi: 10.1038/ng2033. [DOI] [PubMed] [Google Scholar]
- 117.Dolado I, Swat A, Ajenjo N, De Vita G, Cuadrado A, Nebreda AR. p38alpha MAP kinase as a sensor of reactive oxygen species in tumorigenesis. Cancer Cell. 2007;11(2):191–205. doi: 10.1016/j.ccr.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 118.Merlin JL, Harle A, Lion M, Ramacci C, Leroux A. Expression and activation of P38 MAP kinase in invasive ductal breast cancers: correlation with expression of the estrogen receptor, HER2 and downstream signaling phosphorylated proteins. Oncol Rep. 2013;30(4):1943–8. doi: 10.3892/or.2013.2645. [DOI] [PubMed] [Google Scholar]
- 119.Wang SN, Lee KT, Tsai CJ, Chen YJ, Yeh YT. Phosphorylated p38 and JNK MAPK proteins in hepatocellular carcinoma. Eur J Clin Invest. 2012;42(12):1295–301. doi: 10.1111/eci.12003. [DOI] [PubMed] [Google Scholar]
- 120.Chen L, Mayer JA, Krisko TI, Speers CW, Wang T, Hilsenbeck SG, Brown PH. Inhibition of the p38 kinase suppresses the proliferation of human ER-negative breast cancer cells. Cancer Res. 2009;69(23):8853–61. doi: 10.1158/0008-5472.CAN-09-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pereira L, Igea A, Canovas B, Dolado I, Nebreda AR. Inhibition of p38 MAPK sensitizes tumour cells to cisplatin-induced apoptosis mediated by reactive oxygen species and JNK. EMBO Mol Med. 2013;5(11):1759–74. doi: 10.1002/emmm.201302732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cohen O, Inbal B, Kissil JL, Raveh T, Berissi H, Spivak-Kroizaman T, Feinstein E, Kimchi A. DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J Cell Biol. 1999;146(1):141–8. doi: 10.1083/jcb.146.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jang CW, Chen CH, Chen CC, Chen JY, Su YH, Chen RH. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nature Cell Biology. 2002;4(1):51–8. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 124.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a Novel Serine Threonine Kinase and a Novel 15-Kd Protein as Potential Mediators of the Gamma-Interferon-Induced Cell-Death. Gene Dev. 1995;9(1):15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 125.Zhao J, Zhao DK, Poage GM, Mazumdar A, Zhang Y, Hill JL, Hartman ZC, Savage MI, Mills GB, Brown PH. Death-associated protein kinase 1 promotes growth of p53-mutant cancers. Journal of Clinical Investigation. 2015;125(7):2707–20. doi: 10.1172/JCI70805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nature Reviews Cancer. 2009;9(1):57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 127.Xu H, Yan J, Zhu ZQ, Hussain LR, Huang YM, Wen YJ, Ildstad ST. A Critical Role of Innate Immunity Via TLR4 Signaling Via the TRIF Pathway in Allogeneic Bone Marrow Cell Rejection. Am J Transplant. 2010;10:99-. [Google Scholar]
- 128.Rajput S, Volk-Draper LD, Ran S. TLR4 Is a Novel Determinant of the Response to Paclitaxel in Breast Cancer. Molecular Cancer Therapeutics. 2013;12(8):1676–87. doi: 10.1158/1535-7163.MCT-12-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Haricharan S, Brown P. TLR4 has a TP53-dependent dual role in regulating breast cancer cell growth. P Natl Acad Sci USA. 2015;112(25):E3216–E25. doi: 10.1073/pnas.1420811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shatz M, Menendez D, Resnick MA. The Human TLR Innate Immune Gene Family Is Differentially Influenced by DNA Stress and p53 Status in Cancer Cells. Cancer Research. 2012;72(16):3948–57. doi: 10.1158/0008-5472.CAN-11-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Menendez D, Lowe JM, Snipe J, Resnick MA. Ligand dependent restoration of human TLR3 signaling and death in p53 mutant cells. Oncotarget. 2016;7(38):61630–42. doi: 10.18632/oncotarget.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hainaut P, Pfeifer GP. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb Perspect Med. 2016;6(11) doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.White E. The role for autophagy in cancer. Journal of Clinical Investigation. 2015;125(1):42–6. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xie XL, Le L, Fan YX, Lv L, Zhang JJ. Autophagy is induced through the ROS-TP53-DRAM1 pathway in response to mitochondrial protein synthesis inhibition. Autophagy. 2012;8(7):1071–84. doi: 10.4161/auto.20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dando I, Cordani M, Donadelli M. Mutant p53 and mTOR/PKM2 regulation in cancer cells. Iubmb Life. 2016;68(9):722–6. doi: 10.1002/iub.1534. [DOI] [PubMed] [Google Scholar]
- 137.Cordani M, Oppici E, Dando I, Butturini E, Pozza ED, Nadal-Serrano M, Oliver J, Roca P, Mariotto S, Cellini B, et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Molecular Oncology. 2016;10(7):1008–29. doi: 10.1016/j.molonc.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, Vicencio JM, Soussi T, Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy (vol 7, pg 3056, 2008) Cell Cycle. 2014;13(14):2311. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 139.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology. 2011;13(2):132–U71. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Heiden MGV, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324(5930):1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang C, Liu J, Liang YJ, Wu R, Zhao YH, Hong XH, Lin MH, Yu HY, Liu LX, Levine AJ, et al. Tumour-associated mutant p53 drives the Warburg effect. Nature Communications. 2013;4 doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Timpson P, McGhee EJ, Morton JP, von Kriegsheim A, Schwarz JP, Karim SA, Doyle B, Quinn JA, Carragher NO, Edward M, et al. Spatial Regulation of RhoA Activity during Pancreatic Cancer Cell Invasion Driven by Mutant p53. Cancer Research. 2011;71(3):747–57. doi: 10.1158/0008-5472.CAN-10-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Muller PAJ, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. Mutant p53 Drives Invasion by Promoting Integrin Recycling. Cell. 2009;139(7):1327–41. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 144.Steller EJ, Raats DA, Koster J, Rutten B, Govaert KM, Emmink BL, Snoeren N, van Hooff SR, Holstege FC, Maas C, et al. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia. 2013;15(2):204–17. doi: 10.1593/neo.121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jechlinger M, Sommer A, Moriggl R, Seither P, Kraut N, Capodiecci P, Donovan M, Cordon-Cardo C, Beug H, Grunert S. Autocrine PDGFR signaling promotes mammary cancer metastasis. J Clin Invest. 2006;116(6):1561–70. doi: 10.1172/JCI24652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weissmueller S, Manchado E, Saborowski M, Morris JPt, Wagenblast E, Davis CA, Moon SH, Pfister NT, Tschaharganeh DF, Kitzing T, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157(2):382–94. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y, Jackson JG, Suh YA, Elizondo-Fraire AC, Yang P, et al. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc Natl Acad Sci U S A. 2014;111(30):11145–50. doi: 10.1073/pnas.1404139111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, Chicas A, Li WC, Polotskaia A, et al. Mutant p53 Disrupts Mammary Tissue Architecture via the Mevalonate Pathway. Cell. 2012;148(1–2):244–58. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alderton GK. MORPHOLOGY Oncogenic morphs of p53. Nature Reviews Cancer. 2012;12(3) doi: 10.1038/nrc3230. [DOI] [PubMed] [Google Scholar]
- 150.Kollareddy M, Dimitrova E, Vallabhaneni KC, Chan A, Le T, Chauhan KM, Carrero ZI, Ramakrishnan G, Watabe K, Haupt Y, et al. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nature Communications. 2015;6 doi: 10.1038/ncomms8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhou G, Myers JN. Mutant p53 exerts oncogenic functions by modulating cancer cell metabolism. Mol Cell Oncol. 2014;1(3):e963441. doi: 10.4161/23723548.2014.963441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Feng ZH, Hu WW, de Stanchina E, Teresky AK, Jin SK, Lowe S, Levine AJ. The regulation of AMPK beta 1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Research. 2007;67(7):3043–53. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 153.Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte-Lovejoy D, Al-awar R, Katona BW, Shilatifard A, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525(7568):206–11. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Prives C, Lowe SW. Cancer: Mutant p53 and chromatin regulation. Nature. 2015;525(7568):199–200. doi: 10.1038/nature15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pfister NT, Fomin V, Regunath K, Zhou JY, Zhou W, Silwal-Pandit L, Freed-Pastor WA, Laptenko O, Neo SP, Bargonetti J, et al. Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells. Genes Dev. 2015;29(12):1298–315. doi: 10.1101/gad.263202.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Parrales A, Iwakuma T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Front Oncol. 2015;5:288. doi: 10.3389/fonc.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Reles A, Wen WH, Schmider A, Gee C, Runnebaum IB, Kilian U, Jones LA, El-Naggar A, Minguillon C, Schonborn I, et al. Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer. Clin Cancer Res. 2001;7(10):2984–97. [PubMed] [Google Scholar]
- 158.Chan KT, Lung ML. Mutant p53 expression enhances drug resistance in a hepatocellular carcinoma cell line. Cancer Chemother Pharmacol. 2004;53(6):519–26. doi: 10.1007/s00280-004-0767-4. [DOI] [PubMed] [Google Scholar]
- 159.Wong RP, Tsang WP, Chau PY, Co NN, Tsang TY, Kwok TT. p53-R273H gains new function in induction of drug resistance through down-regulation of procaspase-3. Mol Cancer Ther. 2007;6(3):1054–61. doi: 10.1158/1535-7163.MCT-06-0336. [DOI] [PubMed] [Google Scholar]
- 160.Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V, Chauhan K, Daniels SR, Boccetta M, Garrett MR, et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26(8):830–45. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fiorini C, Cordani M, Padroni C, Blandino G, Di Agostino S, Donadelli M. Mutant p53 stimulates chemoresistance of pancreatic adenocarcinoma cells to gemcitabine. Bba-Mol Cell Res. 2015;1853(1):89–100. doi: 10.1016/j.bbamcr.2014.10.003. [DOI] [PubMed] [Google Scholar]