Abstract
Moderate to severe fatigue occurs in 14% to 96% of oncology patients undergoing active treatment. Current interventions for fatigue are not efficacious. A major impediment to the development of effective treatments is a lack of understanding of the fundamental mechanisms underlying fatigue. In the current study, differences in phenotypic characteristics and gene expression profiles were evaluated in a sample of breast cancer patients undergoing chemotherapy (CTX) who reported low (n=19) and high (n=25) levels of evening fatigue. Compared to the low group, patients in the high evening fatigue group reported lower functional status scores, higher comorbidity scores, and fewer prior cancer treatments. One gene was identified as up-regulated and eleven genes were identified to be down-regulated in the high evening fatigue group. Gene set analysis found 24 down-regulated and 94 simultaneously up and down perturbed pathways between the two fatigue groups. Transcript Origin Analysis found that differential expression originated primarily from monocytes and dendritic cell types. Query of public data sources found 18 gene expression experiments with similar differential expression profiles. Our analyses revealed that inflammation, neurotransmitter regulation, and energy metabolism are likely mechanisms associated with evening fatigue severity; that CTX may contribute to fatigue seen in oncology patients; and that the patterns of gene expression may be shared with other models of fatigue (e.g., physical exercise, pathogen-induced sickness behavior). These results suggest that the mechanisms that underlie fatigue in oncology patients are multi-factorial.
Keywords: gene expression, inflammation, cancer, fatigue, chemotherapy, diurnal variations in fatigue, breast cancer
INTRODUCTION
In women undergoing chemotherapy (CTX) for breast cancer, prevalence rates for fatigue range from 30% to 80% (Alcantara-Silva, Freitas-Junior, Freitas, & Machado, 2013). The severity of fatigue varies over the course of a day and displays marked inter-individual variability (Dhruva et al., 2010; B. A. Fletcher et al., 2009; Miaskowski et al., 2008; Molassiotis & Chan, 2004). Some of this inter-individual variability is explained by the fact that morning and evening fatigue are distinct but related symptoms (Dhruva et al., 2013; Dhruva et al., 2010; Miaskowski et al., 2008). Of note, morning and evening fatigue are distinguished by different phenotypic and genotypic characteristics. For example, a higher number of comorbid conditions was associated with more severe morning fatigue, whereas caring for children at home was associated with more severe evening fatigue (Dhruva et al., 2010). Variations in interleukin (IL) 8 and tumor necrosis factor alpha (TNFA) were associated with the severity of morning fatigue, whereas variations in interleukin 1 receptor 2 (IL1R2), IL4, IL6, and TNFA are associated with the severity of evening fatigue (Aouizerat et al., 2009; Dhruva et al., 2014; Miaskowski et al., 2010). A better understanding of the unique phenotypic and molecular characteristics associated with morning and evening fatigue would provide the means to identify high risk patients and to develop and test interventions for these devastating symptoms.
Difficulties in the diagnosis and treatment of fatigue are related to our lack of understanding of the fundamental mechanisms that underlie this debilitating symptom. Work by our group (Aouizerat et al., 2009; Miaskowski et al., 2010) and others (Bower et al., 2013; Bower et al., 2009; Collado-Hidalgo, Bower, Ganz, Irwin, & Cole, 2008; Reinertsen et al., 2011) suggest that inflammation plays a role in the development of fatigue. However, while it is safe to say that the mechanisms for fatigue are multi-factorial (Ryan et al., 2007), the causative pathways remain to be identified. One approach to identify additional mechanism(s) for fatigue is to evaluate for changes in gene expression associated with this symptom.
To date, only six studies, in four independent samples, have evaluated for differences in gene expression associated with fatigue in oncology patients (Bower, Ganz, Irwin, Arevalo, & Cole, 2011; Hsiao, Araneta, Wang, & Saligan, 2013; Hsiao, Wang, Kaushal, & Saligan, 2013; Landmark-Hoyvik et al., 2009; Light et al., 2013; Saligan et al., 2013). One study evaluated changes in gene expression in pathways solely involved in mitochondrial function (Hsiao, Wang, et al., 2013). In another study (Light et al., 2013), gene expression was evaluated for a number of specific pathways (e.g., adrenergic, monoamine and peptides). Of the four studies that collected whole-transcriptome measurements, three focused on select genes and pathways related to mitochondrial function (Hsiao, Araneta, et al., 2013) and/or inflammation and immune function (Bower et al., 2011; Hsiao, Araneta, et al., 2013; Saligan et al., 2013). Only one study reported findings on the whole transcriptome (Landmark-Hoyvik et al., 2009). In all six studies, a general measure of fatigue was used and diurnal variability in fatigue was not evaluated. Based on previous work on diurnal variations in fatigue (Aouizerat et al., 2009; Dhruva et al., 2013; Miaskowski et al., 2010), an evaluation of one dimension of fatigue (i.e., evening fatigue) may improve our ability to detect differences in gene expression between patients who do and do not experience fatigue. Additional studies are needed that apply hypothesis-generating approaches utilizing the entire transcriptome in order to identify novel pathways and processes associated with fatigue in oncology patients.
In this study, we evaluated for differences in gene expression in peripheral leukocytes of patients with low and high levels of evening fatigue. The role of central versus peripheral mechanisms in the development and maintenance of fatigue continues to be debated (Yavuzsen et al., 2009). However, peripheral changes in the expression of pro-inflammatory cytokine genes can influence neural and endocrine activity (Dantzer, O’Connor, Freund, Johnson, & Kelley, 2008) and contribute to a reciprocal regulation between the neural and innate immune systems termed the “neuro-immune circuit” (Irwin & Cole, 2011). This neuro-immune circuit originates with the innate immune system (Cole, Hawkley, Arevalo, & Cacioppo, 2011; Powell, Mays, Bailey, Hanke, & Sheridan, 2011). Pro-inflammatory cytokines can cross the blood brain barrier (Quan & Banks, 2007). In addition, increased synthesis of cytokines in the brain can occur in response to peripheral input (Quan & Banks, 2007). Therefore, studies of gene expression from peripheral leukocytes will provide valuable information on fatigue.
Evaluation of the parallel expression measures of a genome (e.g., the transcriptome) will increase our understanding of the functions of various genes as well as their contributions to the biology of an organism (Butte, 2002). Moreover, novel statistical approaches permit the identification of differential gene expression patterns at the level of a gene, a biological pathway, and the entire transcriptome. The aim of this study, in a sample of patients undergoing CTX for breast cancer (n=44), was to use high throughput methods to determine if there are changes in gene expression in peripheral leukocytes associated with high and low levels of evening fatigue. The analytic methods employed in this study included: the use of microarray data to identify genes and pathways associated with evening fatigue; the use of bioinformatic analyses to infer the cellular origin for the differences in gene expression detected in peripheral leukocytes; and an interrogation of publically available transcriptome gene expression experiments that share a similar pattern with the gene expression differences identified in our sample.
METHODS
A detailed description of the methods is found in Supplemental File 1.
Patients and Settings
This longitudinal study enrolled patients who were ≥18 years of age; had a diagnosis of breast, gastrointestinal, gynecological, or lung cancer; had received CTX within the preceding four weeks; were scheduled to receive at least two additional cycles of CTX; were able to read, write, and understand English; and gave written informed consent. Patients were recruited from two Comprehensive Cancer Centers, one Veteran’s Affairs hospital, one public hospital, and four community-based oncology practices. The first 44 eligible patients with breast cancer were included in this study.
Instruments
A demographic questionnaire obtained information on age, gender, ethnicity, marital status, living arrangements, education, employment status, and income. The Karnofsky Performance Status (KPS) scale was used to evaluate patients’ functional status (Karnofsky, Abelmann, Craver, & Burchenal, 1948). The Self-administered Comorbidity Questionnaire (SCQ) evaluated the occurrence, treatment, and functional impact of common comorbid conditions (e.g., diabetes, arthritis) (Sangha, Stucki, Liang, Fossel, & Katz, 2003).
The Lee Fatigue Scale (LFS) consists of 13 items designed to assess physical fatigue (K. A. Lee, Hicks, & Nino-Murcia, 1991). A total mean fatigue score was calculated with higher scores indicating greater fatigue severity. Patients were asked to rate each item based on how they felt prior to going to bed each night over the previous week (i.e., evening fatigue). The LFS has well-established validity and reliability. A score of >5.6 indicates a clinically meaningful level of evening fatigue (B. S. Fletcher et al., 2008). From the first 50 patients with breast cancer enrolled in the parent study, data from 44 patients were selected to enrich for low (LFS score <5.6, n=19) and high (LFS score ≥5.6, n=25) levels of evening fatigue.
Study Procedures
The study was approved by the Committee on Human Research at the University of California, San Francisco (UCSF) and by each of the study sites. A research staff member approached patients who had received at least one cycle of CTX in the infusion unit to discuss participation in the study. All patients signed written informed consent. Depending on the length of their CTX cycles, patients completed study questionnaires in their homes, a total of six times over two cycles of CTX (i.e., prior to the next CTX administration (enrollment), approximately 1 week after CTX administration, approximately 2 weeks after CTX administration). For this paper, mean evening fatigue scores at the time of enrollment were used in the analyses. Medical records were reviewed for disease and treatment information.
Gene Expression Measurements
Sample processing
Total ribonucleic acid (RNA) was extracted from whole blood collected into PAXgene RNA Stabilization tubes and processed using a standard protocol (Qiagen, USA). The blood specimen was collected prior to administration of CTX. RNA concentration was measured by NanoDrop UV spectrophotometry (ThermoScientific, USA). RNA integrity was evaluated using the RNA 6000 Nano Assay (Agilent, USA). All RNA samples were determined to be of good quality (i.e., RNA Integrity Number (RIN) ≥ 8) and were retained for gene expression profiling.
Microarray hybridization
For each sample, approximately 100 nanograms of total RNA was labeled using the Illumina Total Prep RNA Amplification Kit (Ambion, USA) and then hybridized to the HumanHT-12 v4.0 Expression BeadChip (47,214 transcripts) (Illumina, USA). The BeadChips were scanned using iScan system (Illumina, USA) at the UCSF Genomics Core Facility. Each HumanHT-12 BeadChip contains 12 sample BeadArrays. Forty-seven samples were measured (i.e., 44 patient specimens, 3 technical replicates). To assess for between-BeadChip variation, one sample was repeated on each of the four BeadChips at a different physical BeadArray position.
Microarray preprocessing and normalization
Summary level data from the uncorrected, non-normalized, and non-transformed summary intensities at the probe level were calculated. Data preparation and analyses were conducted using two well-established protocols (Gentleman et al., 2004; Luo, Friedman, Shedden, Hankenson, & Woolf, 2009). The quality control procedures and associated results are described in detail in Supplemental File 1 and summarized in Supplemental File 2. None of the samples displayed unusual distance between arrays or array signal intensity distributions. Background correction, quantile normalization, and log2 transformation were performed using limma (Smyth, 2005). Probes with insufficient expression measurements were excluded, leaving 34,267 assays spanning 16,980 genes for analysis (Supplemental File 2, Supplemental Figure 1, panel B). The reliability of the expression measurements were supported by a high level of correlation between quadruplicate arrays across all filtered assays (mean pairwise Pearson’s ρ = 0.95). These values were significantly higher than those observed between all samples (mean pairwise Pearson’s ρ = 0.92, Welch two sample t-test p<1.31×10−12). Finally, potential clustering of samples was evaluated by principal components analysis. No obvious clustering by fatigue group was observed (data not shown) and no adjustment for batch effects was warranted.
Data Analyses
Demographic and clinical data
Demographic and clinical data were analyzed using SPSS version 22 (IBM, Armonk, NY) and Stata version 13.0 (StataCorp, College Station, TX). Descriptive statistics and frequency distributions were calculated for demographic and clinical characteristics as well as for fatigue severity. The two evening fatigue groups were defined as individuals with LFS scores below (<5.6, n=19) or above (≥5.6, n=25) the clinically meaningful cut-off score for evening fatigue (B. S. Fletcher et al., 2008). Independent sample t-tests, Mann-Whitney U tests, Chi square tests, and Fisher’s Exact tests were used to evaluate for differences in demographic and clinical characteristics between the two evening fatigue groups. Effect size was calculated using Cohen’s d statistic (Cohen, 1988).
Differential gene expression
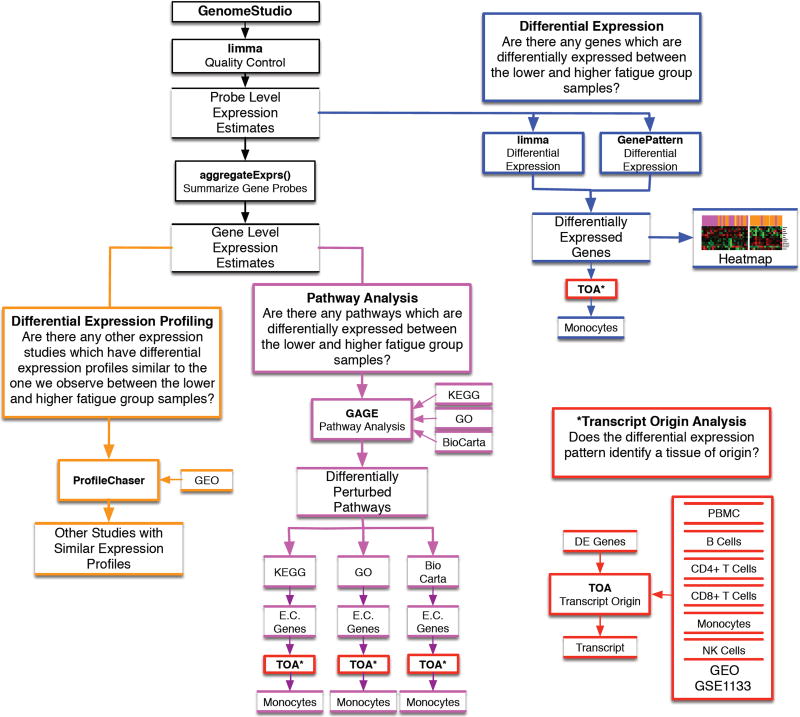
Differential expression (DE) of genes can offer insights into the biological processes that influence inter-individual variability in evening fatigue. Although numerous approaches are used to identify between group differences in DE (Jeanmougin et al., 2010), we selected two well-known methods, namely the t-test using GenePattern and an estimation of gene-by-gene variance with ‘limma’ (Figure 1, blue outline).
Figure 1. Experimental approach and analysis plan.
The diagram illustrates the analysis plan for the study, which includes differential expression (blue outline), differential expression profiling (orange outline), pathway analysis (purple outline), transcript origin analysis (red outline), and ProfileChaser (orange outline). Closed boxes denote analysis or software, and open boxes denote data or results.
Abbreviations: aggregateExprs() = a function contained in the PGSEA R statistical software package; DE = differentially expressed; E.C. Genes = essential contributing genes; GAGE = generally applicable gene set enrichment; GEO = Gene expression omnibus; GO = Gene Ontogeny; GSE = GEO series; KEGG = Kyoto Encyclopedia of Genes; NK = natural killer; PBMC = peripheral blood mononuclear cell (dendritic); TOA = Transcript Origin analysis.
Unsupervised clustering was used to evaluate the resolution of the two evening fatigue groups by the DE genes identified using limma and GenePattern. Evening fatigue group membership was then mapped onto the samples to visualize the degree to which the DE genes distinguished between patients with high versus low levels of evening fatigue (Figure 1, blue outline).
Differential pathway perturbation
Since pathway analysis is performed at the level of the gene, a summary signal estimate of expression was calculated from all valid probes spanning each gene (Reimers, 2010). Summary signal intensities were obtained for 16,980 genes. Differential pathway perturbation was performed using competitive analysis with the R package ‘GAGE’ (Generally Applicable Gene Set Enrichment) (Luo et al., 2009). By excluding genes with insufficient background expression levels and utilizing a whole-genome gene expression microarray, the impact of known limitations and spurious results associated with competitive approaches was minimized (Figure 1, purple outline) (Tripathi, Glazko, & Emmert-Streib, 2013).
Pathways and gene sets were defined using the 177 Kyoto Encyclopedia of Genes (KEGG) (Aoki-Kinoshita & Kanehisa, 2007), 259 BioCarta (Nishimura, 2001) and 17,202 Gene Ontology (GO) (Harris et al., 2004) annotated sets provided by the ‘gageData’ R package. Pathways model the complex interactions between genes in a biological setting and are not expected to be solely simultaneously all up- (or down-) regulated. Rather, perturbations are more likely to consist of a mixture of up- and down-regulation. As such, we tested for differential perturbations under three models: up-regulation, down-regulation, and both (simultaneous up/down or “2d”). While all of the genes in each pathway were included in this analysis, only a subset of these genes had discernible expression changes above background (termed “essential contributing (EC) genes”).
Transcript Origin Analysis (TOA)
Peripheral blood contains a heterogeneous population of nucleated immune cells (e.g., B-cells, CD4+ T-cells, CD8+ T-cells, dendritic cells, monocytes, natural killer (NK) cells). Each cell type is involved in unique biological processes and expresses different subsets of genes (e.g., genes involved in different biological pathways). In order to identify the cell type(s) of origin for genes and/or pathways that were significantly DE between the high and low evening fatigue groups, the TOA test was used (Figure 1, red outline) (Cole et al., 2011). We implemented the TOA methodology described by Cole et al. (Cole et al., 2011) using Python. Our implementation was diagnostic for B-cells, CD4+ T-cells, dendritic cells, monocytes, and NK cells (Supplemental File 3). We were not able to sufficiently validate for diagnosticity of CD8+ T cells and consider any results that identified this class of cells as unreliable.
Objective query of publically available transcriptome experiments
To better categorize and understand the biological significance of the “molecular signatures” associated with evening fatigue, a data driven approach was employed to leverage the collection of over 1800 data sets available in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) and to identify curated datasets with similar DE patterns (Figure 1, orange outline). Specifically, we employed ProfileChaser (Engreitz et al., 2010) (http://profilechaser.stanford.edu/) to identify DE profiles that existed in GEO similar to the ones that we identified between the low and high evening fatigue groups.
Two independent reviewers curated the abstracts for each published study from the candidate profiles with similar DE patterns for conceptual relevance to evening fatigue. If either reviewer selected an abstract for consideration, it was included for further evaluation. Then, the factor comparison identified by ProfileChaser was evaluated in order to exclude un-interpretable comparisons. While a large number of significant hits were expected, false positives needed to be identified and culled (Engreitz et al., 2010). As hypothesized, many of the significantly similar profiles were not appropriate for our study due to the content-agnostic fashion (i.e., no pairwise comparisons are excluded) in which the universe of target profiles were generated (e.g., a “sample ID” factor which splits samples based on the individual ID of samples), or easily interpretable in the context of our current study (e.g., a factor splitting samples based on expression at different time points of a yeast colony’s development). Manuscripts from profile matches for all studies of interest to either reviewer (KMK, BEA) were retained. From this list, the full manuscript and supplemental materials were collected and reviewed.
RESULTS
Patient Characteristics
The total sample consisted of 44 women undergoing CTX for breast cancer. As shown in Table 1, the majority of the patients were college graduates and Caucasian with a mean age of 56.1 (± 9.5) years. Patients had a mean KPS score of 80.9 (± 12.9) and a mean SCQ score of 5.8 (± 3.5). The mean fatigue score for the total sample was 5.6 (± 2.2). The scores for the low (n=19) versus the high (n=25) evening fatigue groups were 3.7 (± 1.7) and 7.1 (± 1.2), respectively (p<0.001).
Table 1.
Differences in Demographic and Clinical Characteristics Between the Low and High Evening Fatigue Groups
| Characteristic | Total Sample (n=44) Mean (SD) |
Low Evening Fatigue (n=19; 43.2%) Mean (SD) |
High Evening Fatigue (n=25; 56.8%) Mean (SD) |
Statistics |
|---|---|---|---|---|
|
| ||||
| Age (years) | 56.1 (9.5) | 57.3 (7.8) | 55.2 (10.7) | t=0.72, p=.474 |
|
| ||||
| Education (years) | 16.4 (2.4) | 15.9 (2.1) | 16.8 (2.5) | t=−1.27, p=.211 |
|
| ||||
| Body mass index (kg/m2) | 27.1 (6.4) | 25.4 (3.9) | 28.4 (7.6) | t=−1.64, p=.111 |
|
| ||||
| Karnofsky Performance Status score | 80.9 (12.9) | 86.3 (11.6) | 76.8 (12.5) | t=2.58, p=.014 |
|
| ||||
| Self-administered Comorbidity Questionnaire score | 5.8 (3.5) | 4.5 (2.8) | 6.8 (3.8) | t=−2.23, p=.031 |
|
| ||||
| Time since diagnosis (years) | 4.6 (7.7) | 5.5 (6.1) | 3.9 (8.8) | U, p=.142 |
|
| ||||
| Time since diagnosis (median, years) | 0.45 | 3.75 | 0.37 | |
|
| ||||
| Number of prior cancer treatments | 2.1 (1.8) | 2.8 (1.9) | 1.6 (1.5) | t=2.32, p=.025 |
|
| ||||
| Number of metastatic sites including lymph node involvement A | 1.6 (1.7) | 2.1 (1.9) | 1.3 (1.4) | t=1.56, p=.128 |
|
| ||||
| Number of metastatic sites excluding lymph node involvement | 1.1 (1.4) | 1.4 (1.5) | 0.8 (1.2) | t=1.26, p=.215 |
|
| ||||
| LFS evening fatigue score | 5.6 (2.2) | 3.7 (1.7) | 7.1 (1.2) | t=−7.66, p<.001 |
|
| ||||
| % (n) | % (n) | % (n) | ||
|
| ||||
| Self-reported ethnicity | χ2=4.96, p=.292 | |||
| White | 77.3 (35) | 68.4 (13) | 80.0 (20) | |
| Asian/Pacific Islander | 11.4 (5) | 15.8 (3) | 8.0 (2) | |
| Black Non-Hispanic | 4.5 (2) | 10.5 (2) | 4.0 (1) | |
| Hispanic/Mixed/Other | 6.8 (3) | 5.3 (1) | 8.0 (2) | |
|
| ||||
| Married or partnered (% yes) | 72.7 (32) | 84.2 (16) | 64.0 (16) | FE, p=.181 |
|
| ||||
| Lives alone (% yes) | 15.9 (7) | 10.5 (2) | 20.0 (5) | FE, p=.680 |
|
| ||||
| Currently employed (% yes) | 29.5 (13) | 31.6 (6) | 28.0 (7) | FE, p=1.00 |
|
| ||||
| Annual household income | χ2=2.83, p=.419 | |||
| Less than $30,000 | 2.8 (1) | 0.0 (0) | 4.3 (1) | |
| $30,000 to $70,000 | 16.7 (6) | 23.1 (3) | 13.0 (3) | |
| $70,000 to $100,000 | 8.3 (3) | 0.0 (0) | 13.0 (3) | |
| Greater than $100,000 | 72.2 (26) | 76.9 (10) | 69.6 (16) | |
|
| ||||
| Exercise on a regular basis (% yes) | 77.3 (34) | 78.9 (15) | 76.0 (19) | FE, p=1.00 |
|
| ||||
| Child care responsibilities (% yes) | 28.9 (11) | 17.6 (3) | 38.1 (8) | FE, p=.282 |
|
| ||||
| Elder care responsibilities (% yes) | 13.9 (5) | 12.5 (2) | 15.0 (3) | FE, p=1.00 |
|
| ||||
| AJCC Status | χ2=1.36, p=.715 | |||
| Stage 0 | 59.1 (26) | 63.2 (12) | 56.0 (14) | |
| Stage I | 4.5 (2) | 5.2 (1) | 4.0 (1) | |
| Stage IIA, IIB | 18.2 (8) | 10.5 (2) | 25.0 (6) | |
| Stage IIA,IIIB,IIIC,IV | 18.2 (8) | 21.1 (4) | 16.7 (4) | |
|
| ||||
| Prior cancer treatmentB | χ2=8.58, p=.035 | |||
| No prior treatment | 14.3 (6) | 11.8 (2) | 16.0 (4) | |
| Only surgery, CTX, or RT | 42.9 (18) | 23.5 (4) | 56.0 (14) | |
| Surgery and CTX, or surgery and RT, or CTX and RT | 9.5 (4) | 5.9 (1) | 12.0 (3) | |
| Surgery and CTX and RT | 33.3 (14) | 58.8 (10) | 16.0 (4) | |
|
| ||||
| Metastatic sites | χ2=6.99, p=.073 | |||
| No metastasis | 36.4 (16) | 31.6 (6) | 40.0 (10) | |
| Only lymph node metastasis | 20.5 (9) | 15.8 (3) | 24.0 (6) | |
| Only metastatic disease in other sites | 9.1 (4) | 0.0 (0) | 16.0 (4) | |
| Metastatic disease in lymph nodes and other sites | 34.1 (15) | 52.6 (10) | 20.0 (5) | |
Total number of metastatic sites evaluated was 9.
Post-hoc contrasts failed to reveal the subgroup(s) underlying the differences in prior cancer treatments observed in the high compared to the low evening fatigue groups.
Abbreviations: AJCC = American Joint Committee on Cancer; CTX = chemotherapy; FE = Fisher’s Exact; kg = kilograms; LFS = Lee Fatigue Scale; m2 = meters squared; SD = standard deviation; RT = radiation therapy; U = Mann-Whitney test; χ2 = Chi-square test.
Differences in Demographic and Clinical Characteristics Between the Two Fatigue Groups
Table 1 summarizes the differences in demographic and clinical characteristics between the low and high evening fatigue groups. Compared to the low group, patients in the high evening fatigue group reported a lower KPS score, had a higher comorbidity score, and had a lower number of prior cancer treatments.
Differences in Gene Expression Between the Two Fatigue Groups
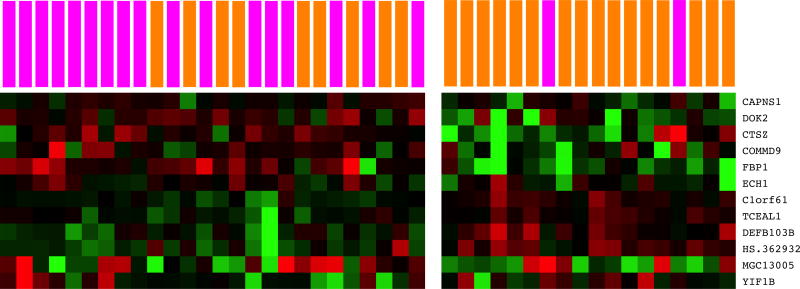
The transcriptomic analysis was done using the Human HT-12 Expression BeadChip. One DE gene was identified by limma and eleven DE genes were identified by GenePattern (Table 2). Among these twelve genes, one gene was up-regulated and eleven genes were down-regulated in the high evening fatigue group. A heatmap of the two class cluster analysis of these twelve genes revealed that the DE genes noticeably, but incompletely, distinguished between the low and high evening fatigue groups (Supplemental File 4 Figure S2).
Table 2.
Differentially Expressed Genes
| Probe | Symbol | Name | Fold Change |
ENTREZ Gene ID |
Direction | Padjusted | Primary Role |
|---|---|---|---|---|---|---|---|
| Limma | |||||||
| ILMN_1 684308 | DEFB103B | Defensin, beta 103B (DEFB103B) | 0.10 | 55894 | Down | 0.0489 | I |
| GenePattern | |||||||
| ILMN_1 873793 | Hs.650028 | cDNA FLJ25030 fis, clone CBL02631 | 0.18 | Up | <0.0001 | U | |
| ILMN_1655418 | CAPNS1 | Calpain, small subunit 1 | 0.58 | 826 | Down | <0.0001 | I |
| ILMN_1705605 | MGC13005 | PREDICTED: Homo sapiens hypothetical protein MGC13005 | 0.83 | Down | <0.0001 | U | |
| ILMN_1728799 | FBP1 | Fructose-1,6-bisphosphatase 1 | 0.75 | 2203 | Down | <0.0001 | M |
| ILMN_1808821 | COMMD9 | COMM domain containing 9 | 0.59 | 29099 | Down | <0.0001 | I |
| ILMN_2398408 | TCEAL1 | Transcription elongation factor A (SII)-like 1, transcript variant 2 | 0.18 | 9338 | Down | <0.0001 | I/M |
| ILMN_1653115 | ECH1 | Enoyl Coenzyme A hydratase 1, peroxisomal | 0.46 | 1891 | Down | <0.0001 | M |
| ILMN_1759652 | C1orf61 | chromosome 1 open reading frame 61 | 0.16 | 10485 | Down | <0.0001 | U |
| ILMN_1666269 | CTSZ | Cathepsin Z | 0.77 | 1522 | Down | <0.0001 | I |
| ILMN_2363668 | YIF1B | Yip1 interacting factor homolog B (S. cerevisiae), transcript variant 4 | 0.55 | 90522 | Down | <0.0001 | I/M/S |
| ILMN 1791211 | DOK2 | Docking protein 2, 56kDa | 0.75 | 9046 | Down | <0.0001 | I |
Abbreviations: Direction = Direction of gene expression in the high as compared to the low evening fatigue group; Padjusted = Benjamini-Hochberg adjusted p-value; Fold = fold change of the log2 transformed, background-corrected, quantile-normalized intensity gene expression values in the high as compared to low evening fatigue group.
The primary functional role(s) of a given gene is identified as follows: M = Metabolism, protein synthesis (n=4); I = Immune activation and immune cell replenishment (n=7); S = serotonergic activation (n=1); U = unknown (n=3).
Three of the genes identified (i.e., cDNA FLJ25030 fis, clone CBL02631 (Hs.650028); Homo sapiens hypothetical protein MGC13005; chromosome 1 open reading frame 61 (C1orf61)) do not have established functional roles. However, the remaining eight genes can be categorized into three groups based on their known function(s) (Summarized in Supplemental File 5 Table S2): immune activation (i.e., Calpain, small subunit 1 (CAPSN1), COMM domain containing 9 (COMMD9), Cathepsin Z (CTSZ), Defensin, beta 103B (DEF103B), Docking protein 2, 56kDa (DOK2), Transcription elongation factor A (SII)-like 1, transcript variant 2 (TCEAL1), Yip1 interacting factor homolog B (S. cerevisiae), transcript variant 4 (YIF1B)); energy metabolism and physical activity (i.e., TCEAL1, Enoyl Coenzyme A hydratase 1, peroxisomal (ECH1), YIF1B, Fructose-1,5-bisphosphatase 1 (FBP1)); and serotonergic activation (i.e., YIF1B).
Differentially Perturbed Pathways Between the Two Fatigue Groups
Gene set analysis was performed to discover differences between the low and high evening fatigue groups in perturbations of genes that operate together in pathways. KEGG pathways are the primary focus of this manuscript given their superior depth of annotation and rich usage in pathway analysis. No up-regulated, 24 down-regulated, and 94 2d perturbed KEGG pathways were identified that differentiated between the two evening fatigue groups (Table 3). Only those down-regulated pathways that were not identified as 2d perturbed are listed in Table 3 (i.e., 5 of 24). A listing of all 19 KEGG down-regulated, GO, and BioCarta analyses are provided in Supplemental File 6.
Table 3.
Differentially Perturbed Pathways
| KEGG ID | KEGG Pathway description | p-value | q-value | Fundamental Role A |
TOA B |
|---|---|---|---|---|---|
| Down-regulated | |||||
| hsa00280 | Valine, leucine and isoleucine degradation | 1.007E-03 | 1.612E-02 | M | M |
| hsa00410 | beta-Alanine metabolism | 1.946E-03 | 2.616E-02 | M | B |
| hsa00970 | Aminoacyl-tRNA biosynthesis | 2.610E-03 | 2.984E-02 | D | NK |
| hsa00640 | Propanoate metabolism | 4.953E-03 | 4.055E-02 | M | DC |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 5.068E-03 | 4.055E-02 | M | M |
| 2D perturbed | |||||
| hsa04145 | Phagosome | 1.626E-56 | 2.601E-54 | M | M |
| hsa04612 | Antigen processing and presentation | 8.693E-51 | 6.955E-49 | I | DC |
| hsa04640 | Hematopoietic cell lineage | 5.206E-47 | 2.777E-45 | I | DC |
| hsa04142 | Lysosome | 1.416E-36 | 5.663E-35 | M | M |
| hsa04380 | Osteoclast differentiation | 2.512E-35 | 8.038E-34 | I | M |
| hsa03010 | Ribosome | 5.333E-32 | 1.422E-30 | D | B |
| hsa00190 | Oxidative phosphorylation | 3.901E-24 | 8.916E-23 | M | M |
| hsa04141 | Protein processing in endoplasmic reticulum | 1.087E-22 | 2.174E-21 | M | DC |
| hsa04514 | Cell adhesion molecules (CAMs) | 3.541E-22 | 6.296E-21 | I | M |
| hsa03040 | Spliceosome | 5.169E-22 | 8.270E-21 | M | DC |
| hsa04650 | Natural killer cell mediated cytotoxicity | 3.430E-20 | 4.990E-19 | I | M |
| hsa04062 | Chemokine signaling pathway | 1.390E-18 | 1.853E-17 | I | id |
| hsa04660 | T cell receptor signaling pathway | 2.625E-18 | 3.230E-17 | I | NK |
| hsa04144 | Endocytosis | 9.210E-18 | 1.053E-16 | M | NK |
| hsa04666 | Fc gamma R-mediated phagocytosis | 5.354E-17 | 5.711E-16 | I | M |
| hsa04670 | Leukocyte transendothelial migration | 2.283E-16 | 2.283E-15 | I | id |
| hsa04662 | B cell receptor signaling pathway | 2.625E-16 | 2.471E-15 | I | M |
| hsa03050 | Proteasome | 2.087E-15 | 1.855E-14 | M | M |
| hsa04672 | Intestinal immune network for IgA production | 5.226E-14 | 4.401E-13 | I | DC |
| hsa04620 | Toll-like receptor signaling pathway | 8.205E-14 | 6.564E-13 | I | M |
| hsa04920 | Adipocytokine signaling pathway | 3.765E-13 | 2.869E-12 | I | M |
| hsa04621 | NOD-like receptor signaling pathway | 1.408E-12 | 1.024E-11 | I | DC |
| hsa04210 | Apoptosis | 2.992E-11 | 2.081E-10 | D | M |
| hsa00051 | Fructose and mannose metabolism | 3.914E-11 | 2.610E-10 | M | id |
| hsa04722 | Neurotrophin signaling pathway | 2.686E-10 | 1.719E-09 | I | M |
| hsa00240 | Pyrimidine metabolism | 1.290E-09 | 7.937E-09 | M | id |
| hsa03018 | RNA degradation | 2.241E-08 | 1.328E-07 | M | DC |
| hsa00030 | Pentose phosphate pathway | 3.228E-08 | 1.845E-07 | M | DC |
| hsa04910 | Insulin signaling pathway | 4.674E-08 | 2.579E-07 | M | NK |
| hsa00520 | Amino sugar and nucleotide sugar metabolism | 5.161E-08 | 2.752E-07 | M | id |
| hsa04330 | Notch signaling pathway | 6.687E-08 | 3.451E-07 | I | DC |
| hsa00010 | Glycolysis / Gluconeogenesis | 7.849E-08 | 3.925E-07 | M | DC |
| hsa00480 | Glutathione metabolism | 1.485E-07 | 7.201E-07 | M | M |
| hsa04630 | Jak-STAT signaling pathway | 1.625E-07 | 7.649E-07 | I | DC |
| hsa00910 | Nitrogen metabolism | 1.804E-07 | 8.248E-07 | M | M |
| hsa04120 | Ubiquitin mediated proteolysis | 4.660E-07 | 2.071E-06 | M | DC |
| hsa00052 | Galactose metabolism | 9.313E-07 | 4.027E-06 | M | DC |
| hsa00230 | Purine metabolism | 1.036E-06 | 4.364E-06 | M | DC |
| hsa04623 | Cytosolic DNA-sensing pathway | 4.788E-06 | 1.964E-05 | I | M |
| hsa04110 | Cell cycle | 5.049E-06 | 2.020E-05 | D | NK |
| hsa04146 | Peroxisome | 1.758E-05 | 6.861E-05 | M | M |
| hsa00760 | Nicotinate and nicotinamide metabolism | 1.870E-05 | 7.125E-05 | M | M |
| hsa03013 | RNA transport | 2.209E-05 | 8.221E-05 | M | NK |
| hsa04070 | Phosphatidylinositol signaling system | 2.989E-05 | 1.087E-04 | I | NK |
| hsa00020 | Citrate cycle (TCA cycle) | 3.237E-05 | 1.151E-04 | M | M |
| hsa04010 | MAPK signaling pathway | 4.111E-05 | 1.430E-04 | I | M |
| hsa03015 | mRNA surveillance pathway | 5.529E-05 | 1.882E-04 | M | id |
| hsa03410 | Base excision repair | 6.074E-05 | 2.025E-04 | D | id |
| hsa00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate | 7.306E-05 | 2.386E-04 | M | id |
| hsa04966 | Collecting duct acid secretion | 7.703E-05 | 2.465E-04 | M | M |
| hsa04810 | Regulation of actin cytoskeleton | 9.502E-05 | 2.981E-04 | D | M |
| hsa04973 | Carbohydrate digestion and absorption | 9.893E-05 | 3.044E-04 | M | DC |
| hsa04130 | SNARE interactions in vesicular transport | 1.226E-04 | 3.703E-04 | M | M |
| hsa04710 | Circadian rhythm – mammal | 1.460E-04 | 4.325E-04 | C | NK |
| hsa04664 | Fc epsilon RI signaling pathway | 1.702E-04 | 4.950E-04 | I | DC |
| hsa00563 | Glycosylphosphatidylinositol(GPI) -anchor biosynthesis | 2.136E-04 | 6.102E-04 | M | DC |
| hsa00620 | Pyruvate metabolism | 2.266E-04 | 6.360E-04 | M | DC |
| hsa03450 | Non-homologous end-joining | 2.368E-04 | 6.533E-04 | D | id |
| hsa04520 | Adherens junction | 2.501E-04 | 6.781E-04 | I | NK |
| hsa00511 | Other glycan degradation | 3.379E-04 | 9.012E-04 | M | M |
| hsa00071 | Fatty acid metabolism | 3.661E-04 | 9.516E-04 | M | DC |
| hsa04964 | Proximal tubule bicarbonate reclamation | 3.687E-04 | 9.516E-04 | M | NK |
| hsa04012 | ErbB signaling pathway | 4.779E-04 | 1.214E-03 | I | id |
| hsa00604 | Glycosphingolipid biosynthesis - ganglio series | 5.013E-04 | 1.253E-03 | M | B |
| hsa00564 | Glycerophospholipid metabolism | 9.206E-04 | 2.266E-03 | M | id |
| hsa04370 | VEGF signaling pathway | 2.024E-03 | 4.907E-03 | I | id |
| hsa04960 | Aldosterone-regulated sodium reabsorption | 2.122E-03 | 5.061E-03 | M | id |
| hsa00270 | Cysteine and methionine metabolism | 2.151E-03 | 5.061E-03 | M | id |
| hsa03060 | Protein export | 2.188E-03 | 5.073E-03 | M | DC |
| hsa03320 | PPAR signaling pathway | 2.616E-03 | 5.979E-03 | M | M |
| hsa04622 | RIG-I-like receptor signaling pathway | 3.298E-03 | 7.433E-03 | I | id |
| hsa03030 | DNA replication | 3.354E-03 | 7.452E-03 | D | DC |
| hsa04971 | Gastric acid secretion | 5.221E-03 | 1.144E-02 | M | id |
| hsa04150 | mTOR signaling pathway | 5.602E-03 | 1.211E-02 | I | id |
| hsa04510 | Focal adhesion | 6.216E-03 | 1.326E-02 | I | M |
| hsa00561 | Glycerolipid metabolism | 7.320E-03 | 1.541E-02 | M | M |
| hsa03008 | Ribosome biogenesis in eukaryotes | 7.485E-03 | 1.555E-02 | M | DC |
| hsa00860 | Porphyrin and chlorophyll metabolism | 7.594E-03 | 1.558E-02 | M | NK |
| hsa04530 | Tight junction | 7.703E-03 | 1.560E-02 | I | B |
| hsa00900 | Terpenoid backbone biosynthesis | 8.277E-03 | 1.655E-02 | M | NK |
| hsa03420 | Nucleotide excision repair | 8.925E-03 | 1.763E-02 | D | NK |
| hsa00330 | Arginine and proline metabolism | 9.351E-03 | 1.825E-02 | M | DC |
| hsa04962 | Vasopressin-regulated water reabsorption | 1.022E-02 | 1.971E-02 | M | NK |
| hsa04310 | Wnt signaling pathway | 1.085E-02 | 2.048E-02 | I | id |
| hsa04720 | Long-term potentiation | 1.088E-02 | 2.048E-02 | L | id |
| hsa00510 | N-Glycan biosynthesis | 1.298E-02 | 2.414E-02 | M | id |
| hsa00770 | Pantothenate and CoA biosynthesis | 1.365E-02 | 2.511E-02 | M | M |
| hsa00380 | Tryptophan metabolism | 1.760E-02 | 3.199E-02 | M | M |
| hsa00531 | Glycosaminoglycan degradation | 1.889E-02 | 3.395E-02 | M | M |
| hsa00450 | Selenocompound metabolism | 2.839E-02 | 5.047E-02 | M | M |
| hsa04614 | Renin-angiotensin system | 2.943E-02 | 5.174E-02 | I | id |
| hsa00790 | Folate biosynthesis | 3.327E-02 | 5.787E-02 | M | id |
| hsa04260 | Cardiac muscle contraction | 4.953E-02 | 8.521E-02 | L | M |
| hsa03020 | RNA polymerase | 5.568E-02 | 9.477E-02 | D | id |
The primary functional role of a given pathway is identified as follows: M = Metabolism, protein synthesis (n=56); I = Immune activation and immune cell replenishment (n=30); D = DNA synthesis and repair, cell division (n=10); L = Long term potentiation (n=2); C = Circadian dysfunction (n=1).
Tissue of origin analysis (TOA) was employed in order to infer the cell type from which a differentially expressed pathway originates. The cell types identified by TOA are abbreviated as follows: M = monocytes (n=34); DC = dendritic cells (n=24); id = insufficient data to perform TOA (n=22); NK = natural killer cells (n=14); B = B cells (n=4).
Abbreviations: KEGG = Kyoto Encyclopedia of Genes and Genomes; id = insufficient data; TOA = Transcript Origin Analysis; 2D = Simultaneous two dimensional perturbation (up and down).
Differentially expressed pathways were broadly categorized into those associated with immune cell replenishment and activation (n=30); cellular metabolism and protein synthesis (n=56); DNA synthesis, repair and cell division (n=10); and neurological activity (n=3). TOA inferred that the DE pathways originated from B-cells, dendritic cells, monocytes, and NK cells. A dearth of DE pathways were identified as unambiguously originating from CD4+ T-cells.
When the two fatigue groups were compared, significantly differentially perturbed cytokine pathways were identified from KEGG (KEGG: hsa04920), GO (GO: 0019221, GO: 0071345, GO: 0034097, GO: 0019221, GO: 0071345, GO: 0034097), and BioCarta (gata3pathway), as well as inflammation and immune-response pathways from GO (GO: 0006954, GO:0002472, GO:0002252) and BioCarta (il1rpathway, il2pathway, il3pathway, il4pathway, il6pathway, il10pathway, il22pathway, nthipathway) (Table 3 and Supplemental File 6).
Prior to multiple hypothesis correction, several cytokine-related genes were DE between the fatigue groups. Among 57 (94 probes) of 70 measured genes that are involved in the adipocytokine signaling pathway (KEGG: hsa04920), five genes had probes identified by limma to be DE prior to statistical correction: retinoid X receptor, gamma (RXRG), v-akt murine thymoma viral oncogene homolog 3 (AKT3), acyl-CoA synthetase long-chain family member 4 (ACSL4), inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma (IKBKG), and signal transducer and activator of transcription 3 (acute-phase response factor) (STAT3) (all p< 0.03). Among 114 genes (160 probes) of 265 measured genes that are involved in the cytokine-cytokine receptor interaction pathway (KEGG: ksa04060), two genes had probes identified by limma to be DE prior to statistical correction: platelet factor 4 (PF4) and chemokine (C-X-C motif) receptor 5 (CXCR5) (all p<0.02). Gene expression studies of fatigue in a larger sample may result in the detection of cytokine gene expression differences that survive statistical correction for multiple testing.
Differentially perturbed pathways related to DNA synthesis, repair, and cell division included DNA replication (hsa03030), cell cycle (hsa04110), nucleotide excision repair (hsa03420), and base excision repair (hsa03410) (Table 3 and Supplemental File 6).
Cellular metabolism and protein synthesis pathways that were differentially perturbed include glycolysis/gluconeogenesis (hsa00010), oxidative phosphorylation (hsa000190), fructose and mannose metabolism (hsa00051), amino sugar and nucleotide sugar metabolism (hsa00520) and the citrate cycle (TCA cycle) (hsa00020) from KEGG, and mitochondrial matrix (GO:0005759), membrane (GO:0031966), inner membrane (GO:0005743) and envelope (GO:0005740), generation of precursor metabolites and energy (GO:0006091), oxidative phosphorylation (GO:0006119), and respiratory chain (GO:0070469) from GO (Table 3 and Supplemental File 6).
Finally, differentially perturbed pathways related to neurotransmission included: Long-term potentiation (hsa04720), soluble NSF [N-ethylmaleimide-sensitive factor] attachment protein receptors (SNARE) interactions in vesicular transport (hsa04130), Mitogen-activated protein kinase (MAPK) signaling (hsa04010) and epidermal growth factor receptor (avian erythroblastic leukemia viral (v-erb-b) oncogene homolog) (ErbB) signaling (hsa04012) pathways in KEGG and the gamma-aminobutyric acid (GABA) receptor lifecycle (gabapathway) and MAPKinase Signaling (mapkpathway) pathways in BioCarta (Table 3 and Supplemental File 6).
Transcript Origin Analysis (TOA)
Peripheral blood contains a heterogeneous population of nucleated cells from which gene expression data are derived. TOA is used to identify the cell lineage with the highest degree of statistical significance for a group of DE genes (Cole et al., 2011). The cell types that can be distinguished included B cells, CD4+ T-cells, dendritic cells, monocytes, and NK cells. Diagnosticity scores were successfully mapped for: twelve out of the combined set of the thirteen genes in Table 2 that were found to be DE between fatigue groups; for 70 of 77 EC genes from all 22 down-regulated KEGG pathways; and for 907 of 980 EC genes from 94 2d perturbed KEGG pathways (Supplemental File 6). In total, TOA unambiguously inferred the origin of 77 of the 99 (78%) differentially perturbed KEGG pathways listed in Table 3. While the individual perturbed pathways originated primarily from monocytes, some originated from dendritic cells, NK cells, B-cells, and CD4+ T-cells (Table 4). The transcript origin for each differentially perturbed pathway is identified in Supplemental File 6 and is summarized in Supplemental File 7.
Table 4.
Summary of Transcript Origin Analysis results for differentially perturbed gene sets
| KEGG Down |
KEGG 2d |
GO Down |
GO 2d |
BioCarta Down |
BioCarta 2d |
|
|---|---|---|---|---|---|---|
| SPS | 22 | 94 | 249 | 2685 | 1 | 67 |
| SPS with E.C. Genes | 22 | 94 | 249 | 2685 | 1 | 67 |
| SPS with TOA signal | 22 | 72 | 230 | 2201 | 1 | 45 |
| Monocytes | 15 | 13 | 84 | 1081 | 0 | 16 |
| Dendritic cells | 14 | 23 | 80 | 617 | 0 | 13 |
| NK cells | 5 | 13 | 32 | 226 | 1 | 10 |
| B-cells | 2 | 3 | 31 | 213 | 0 | 4 |
| CD4+ T-cells | 0 | 0 | 3 | 33 | 0 | 0 |
Abbreviation: E.C. Genes = Essential contributing genes: Genes with substantial or above-background expression changes in the set; GO = Gene Ontogeny; KEGG = Kyoto Encyclopedia of Genes and Genomes; NK = natural killer; SPS = Significantly perturbed sets; TOA = Transcript origin analysis; 2d = Simultaneous two dimensional perturbation (up and down).
Identification of similar whole-transcriptome gene expression experiments
ProfileChaser was used to identify publically available gene expression studies and associated publications that shared a similar whole-transcriptome pattern of differential gene expression. The significant GEO DataSets (GDSs) identified across all five rounds of the split analyses (n=108) were retained (Supplemental Files 8 and 9). Abstracts for the original source publications obtained from the 108 GDSs (n=102), were reviewed by two independent reviewers (BEA, KMK) for their relevance to fatigue. A total of 44 abstracts were selected and the complete publication was evaluated. Of these 44 manuscripts, 20 were retained after full evaluation of the experimental profiles. These 20 publications, representing 18 unique GDSs, were reviewed and categorized in terms of providing “strong” (n=14), “moderate” (n=1), or “weak” (n=5) insights into the mechanism(s) that contribute to fatigue (Supplemental File 9).
DISCUSSION
Differences in Demographic and Clinical Characteristics
This study is the first to evaluate for differences in gene expressions and perturbed pathways in breast cancer patients who reported low and high levels of evening fatigue during CTX. The between group differences in fatigue severity scores represent not only statistically significant, but clinically meaningful differences (Cohen’s d =1.5) (Osoba, 1999). Consistent with previous reports, patients in the high fatigue group had a poorer functional status (Dhruva et al., 2013; Hofso, Miaskowski, Bjordal, Cooper, & Rustoen, 2012) and a more severe comorbidity profile (Berger, Gerber, & Mayer, 2012).
The more surprising and intriguing finding was the associations identified between a lower number of previous cancer treatments and membership in the high fatigue group. While this finding needs to be validated in an independent sample, a number of plausible explanations can be postulated. Patients who are in later stages of their disease trajectory may experience a “response shift” in their perception of fatigue. First used in oncology to describe changes over time in QOL (Schwartz & Sprangers, 1999), a “response shift” is an age-related psychological shift that represents a change in a person’s internal framework for the assessment of experiences (Costanzo, Ryff, & Singer, 2009). In this context, patients in the low fatigue group who had received prior cancer treatments may have changed their internal conceptualization of fatigue based on their previous experiences with the symptom. An alternative hypothesis is that with prolonged cancer treatment, patients may develop tolerance to the physiologic responses that contribute to the development of fatigue. Another alternative hypothesis is that there may be a “selection bias”, where more women who had previous cancer treatments (e.g., due to tolerance or increased survival) volunteered to participate in this study. Longitudinal studies, that assess both phenotypic and epigenetic trajectories associated with fatigue, are needed to confirm or refute these hypotheses.
Differences in Gene Expression
In this study, a data driven analysis was performed at the levels of the gene, pathway, and the entire transcriptome to evaluate for differences in gene expression between patients who reported high versus low levels of evening fatigue (Figure 1). Using TOA, differential gene expression and perturbed pathway expression originated from several cell types (Supplemental File 7), but the majority of the signals originated from monocytes, dendritic cells, B-cells, and NK cells (Table 4). Our profile is consistent with gene expression experiments that evaluated the effects of exhaustive physical exercise (Supplemental File 9). In addition, the expression profiles for the high evening fatigue group were consistent with a number of gene expression studies that evaluated sickness behavior (Calvano et al., 2005; Dennehy, 2007; Foteinou, Calvano, Lowry, & Androulakis, 2010; Rodriguez et al., 2007; Wurfel et al., 2005), circadian rhythm disruption (Bailey et al., 2009; Yang et al., 2007), and mechanisms of neuro-inflammation (Irwin & Cole, 2011). Of note, the DE genes (Table 2) and pathways (Table 3) identified using transcriptomic analyses in the current study are consistent in that these genes play roles in sickness behavior, inflammation, mitochondrial dysfunction, circadian rhythm disruption, and serotonin regulation. Taken together, our results and those of others described below provide a more complete picture of the mechanisms that underlie evening fatigue in oncology patients.
Twelve genes were identified that were DE between the low and high evening fatigue groups (Table 2). While the expression pattern of these twelve genes distinguished between patients in the two fatigue groups (Figure 2), this distinction was not perfect and suggests that additional genes remain to be identified. TOA of this set of twelve DE genes found that the expression patterns originated predominantly from monocytes. This finding is consistent with previous reports that noted that altered cytokine production in monocytes is associated with fatigue in oncology patients with breast cancer (Collado-Hidalgo, Bower, Ganz, Cole, & Irwin, 2006; Saligan & Kim, 2012).
Figure 2.
Inflammation and immune response
In general, cytoxic CTX kills rapidly proliferating cancer cells (Mitchison, 2012). While the targets are cancer cells, other rapidly dividing cells, including peripheral leukocytes, are depleted by CTX. The need for peripheral blood counts to recover is the primary reason that CTX regimens are administered in cycles. In addition, immune system effectors are impacted during and following CTX (Saligan & Kim, 2012). For example, increased CD4 T-cell counts, which may result in a prolonged pro-inflammatory state, are associated with increased fatigue in breast cancer survivors (Bower, Ganz, Aziz, Fahey, & Cole, 2003). Down-regulation of DEF103B in the high fatigue group may favor the production of pro-inflammatory cytokines, that are associated with increased fatigue severity (Aouizerat et al., 2009; Bower et al., 2011; Jager, Sleijfer, & van der Rijt, 2008; Miaskowski et al., 2010). Decreased expression of DOK2 in the high fatigue group may result in prolonged immune cell activation, which could lead to higher levels of evening fatigue. Decreased ECH1 gene expression in patients with higher evening fatigue may be associated with decreased energy production and delayed immune system recovery following CTX. While the function of YIF1B in peripheral leukocytes is unknown, reduction of YIF1B in the high fatigue group may result in decreases in HTR1A expression; decreases in intracellular cAMP; and decreases in immune activation (i.e., creates a pro-inflammatory state that results in fatigue). While it is not known whether lower YIF1B gene expression in the periphery is associated with lower gene expression in the central nervous system, increasing evidence suggests that peripheral gene expression reflects system wide biology (Liew, Ma, Tang, Zheng, & Dempsey, 2006). In addition to the identification of these DE genes with plausible inflammatory and immune mechanisms for evening fatigue, we identified a number of DE pathways associated with immune cell recovery following CTX.
Consistent with established associations between inflammatory cytokines and fatigue in oncology patients (Aouizerat et al., 2009; Bower et al., 2009; Miaskowski et al., 2010), two fatigue groups were compared, we identified significantly differentially perturbed cytokine pathways as well as inflammation-related pathways. In addition, the BioCarta pathway (nthipathway) that was significantly differentially perturbed between the two evening fatigue groups (Supplemental File 6) is consistent with the over-representation of the NF-κβ response elements reported by Bower and colleagues in breast cancer survivors (Bower et al., 2011). Common elements (i.e., genes) of the identified KEGG, GO, and Biocarta pathways that were identified as harboring polymorphisms associated with fatigue in oncology patients include: IL1B (Bower et al., 2013; Collado-Hidalgo et al., 2008; Reinertsen et al., 2011), IL4 (Doong et al., 2014), IL6 (Bower et al., 2013; Miaskowski et al., 2010; Reinertsen et al., 2011), and TNFA (Aouizerat et al., 2009; Bower et al., 2013; Dhruva et al., 2014). Genetic association studies with other members of the above-identified pathways should be evaluated in future studies.
Although DE genes and differentially perturbed pathways related to inflammation were observed, DE cytokines genes were not detected. This finding is consistent with previous research that failed to find an association between cytokine gene expression and levels of fatigue (Landmark-Hoyvik et al., 2009; Reinertsen et al., 2011). The lack of detectable differences in gene expression despite the repeated associations reported between cytokine genes and fatigue may be due to the timing of cytokine gene expression in relation to the experience of fatigue (i.e., may occur prior to the perception of fatigue). Alternatively, the DE genes and perturbed pathways detected in the current study represent upstream and/or downstream events in relation to cytokine gene expression. Finally, the conservative adjustment for multiple hypothesis testing may have resulted in the exclusion of gene expression signals that would be identified with larger samples.
Circadian rhythm
The co-occurrence of (Davidson, MacLean, Brundage, & Schulze, 2002) and common genetic risk factors for (Aouizerat et al., 2009; Miaskowski et al., 2010), fatigue and sleep disturbance suggest that their mechanisms may overlap. Circadian influences on immune function may be particularly relevant to explain the relationship between sleep and fatigue in oncology patients undergoing CTX. Altered expression of circadian clock genes in peripheral leukocytes was observed in healthy individuals who experienced sickness behavior when exposed to endotoxin (Haimovich et al., 2010). The majority of immune cells (including NK cells) demonstrate circadian rhythmicity in healthy individuals (Mazzoccoli et al., 2011). This rhythmicity may be perturbed during CTX. In support of this hypothesis, the KEGG circadian rhythm pathway (hs04710) was differentially perturbed in the high fatigue group. NK cells were the cell type of origin for this perturbation (Table 3). This observation is bolstered by the observation of malfunctioning NK cells in patients with chronic fatigue syndrome (Meeus, Mistiaen, Lambrecht, & Nijs, 2009) and warrants further study.
Neurotransmission
An unexpected finding from the pathway analyses was the identification of pathways that participate in regulation of neurotransmission, including long-term potentiation of neurons (hsa04720, hsa04260), GABA receptor lifecyle (gabapathway), SNARE interactions in vesicular transport (hsa04130), and ErbB signaling pathway (hsa04012) (Table 3 and Supplemental File 6). It should be noted that the “long-term potentiation of neurons” pathway may be a misnomer (i.e, because it share genes with other pathways not related to neurons). In addition, the long-term potentiation of neurons has not been characterized specifically in PBMC. Centrally, long-term potentiation plays an important role in a number of physiologic processes (e.g., learning, memory, pain). While GABA has an effect in peripheral tissue (Erdo & Wolff, 1990), it is the main inhibitory neurotransmitter in the cortex (Petroff, 2002). Serotonin is a neurotransmitter that modulates GABA. SNARE proteins are involved in cell signaling in neurons (Gotte & von Mollard, 1998). ErbB receptor tyrosine kineases perform a complex array of functions including regulation of neurotransmitter receptors (Bublil & Yarden, 2007). However, many patients with breast cancer undergo treatment to block ErbB2 (Yarden & Sliwkowski, 2001), so this pattern may reflect differences in treatment rather than levels of fatigue. While these results, combined with the potential roles of the DE genes CAPNS1 and YIF1B in neurotransmission, are intriguing, it is unclear if these peripherally perturbed genes and pathways reflect changes in gene expression in the central nervous system (Cole, 2013; Liew et al., 2006) that are associated with fatigue. If replicated in an independent sample, these pathways would warrant further study.
Energy metabolism
Regulation and control of the expression of energy metabolism genes occur through a variety of processes that may be altered by the cancer or its treatment (Andrews, Morrow, Hickok, Roscoe, & Stone, 2004). Radiation treatment or CTX may result in a decrease in adenosine triphosphate (ATP) regeneration. This disruption of ATP metabolism may lead to a reduction in mechanical ability (Ryan et al., 2007). The differences in expression patterns in pathways related to mitochondrial or energy metabolism found in our study, parallel a report of associations between fatigue and mitochondrial function genes in patients receiving radiation therapy for prostate cancer (Hsiao, Wang, et al., 2013). The decreased expression of FBP1 in our high fatigue group may be associated with decreased energy production and an attempt to increase intracellular glucose in order to restore energy reserves. Importantly, mitochondrial dysfunction has pleiotropic effects (Chan, 2006) and is closely tied with other processes (e.g., inflammation (Liu et al., 2012)).
Whole-transcriptome differential expression similarities with other studies
An evaluation of the studies identified by ProfileChaser suggests that the mechanisms that underlie fatigue in oncology patients are multi-factorial. Cytokine-induced sickness behavior (B. N. Lee et al., 2004) is a long-standing mechanism associated with common symptoms experienced by oncology patients, including fatigue. Interrogation of publically available transcriptome data sets using ProfileChaser revealed a preponderance of similarities between our gene expression study and studies that featured an inflammatory component. Five of the studies identified by ProfileChaser and retained after in-depth review employed various experimental models (e.g., healthy adults, acute pediatric viral infection) that incorporated an endotoxin or viral challenge (Calvano et al., 2005; Foteinou et al., 2010; Rodriguez et al., 2007; Wang et al., 2007; Wurfel et al., 2005). Similar to pathogen-mediated induction of cytokines, four studies employed a more direct induction of cytokines (e.g., interferon, IL2) in a wide variety of experimental models (e.g., multiple sclerosis, rheumatoid arthritis) (Indraccolo et al., 2007; Zhang, Martino, & Faulon, 2007). Interestingly, in one murine model study that focused on radiation therapy (RT) (Goldrath, Luckey, Park, Benoist, & Mathis, 2004), a similar transcriptome profile to our study was found, which supports the extant literature that cancer treatments induce a pro-inflammatory response in peripheral leukocytes that is associated with increased fatigue.
One study described differences in the transcriptome of healthy volunteers who underwent exhaustive as compared to none or moderate physical exercise (Buttner, Mosig, Lechtermann, Funke, & Mooren, 2007). The gene expression differences between our low and high fatigue groups are similar to the differences found between the non-exhaustive and exhaustive exercise groups. These findings suggest that exhaustive exercise may result in gene expression differences that overlap with those that occur in patients with high evening fatigue.
Fatigue displays a diurnal variability (B. A. Fletcher et al., 2009; Miaskowski et al., 2008). In general, evening fatigue is more severe than morning fatigue and is associated with different risk factors (Dhruva et al., 2010). A novel study from ProfileChaser found diurnal variations in gene expression in the prefrontal cortex of rodents (Yang et al., 2007). The prefrontal cortex plays an important role in the regulation of sleep and fatigue. These findings corroborated the DE genes from our study known to be involved in diurnal processes (i.e., COMMD9 and CTSZ) (Yang et al., 2007). A complementary study identified diurnal variations in gene expression in the pineal gland of rodents (Bailey et al., 2009), that plays an important role in chronobiology and in diurnal variability of genes involved in immune/inflammatory responses.
To date, only two gene expression studies of fatigue in breast cancer survivors are published (Bower et al., 2011; Landmark-Hoyvik et al., 2009). While our findings and those of the previous studies identified differences in gene expression related to immune function and mitochondrial dysregulation, the specific genes and pathways differed. The lack of congruence among these studies may be due in part to different designs and study populations. The studies by Bower et al (Bower et al., 2011) and Landmark-Høyvik et al (Landmark-Hoyvik et al., 2009) evaluated breast cancer survivors with persistent fatigue and did not account for diurnal variations in fatigue severity. In the study by Bower et al. (Bower et al., 2011), only genes responsive to NF-κβ or down-regulation of glucocorticoid-responsiveness were evaluated and discussed. The analytic approaches employed in our study and the one by Landmark-Høyvik et al (Landmark-Hoyvik et al., 2009) were different. ProfileChaser did not identify either study because one (i.e., Bower et al) did not exist in the Gene Expression Omnibus and the other (i.e., Landmark-Høyvik et al) was submitted but not curated as a GEO dataset (which are used exclusively by ProfileChaser).
Limitations
Although the study findings do not provide direct support for the causal mechanisms for evening fatigue, they offer strong candidates for future functional, as well as intervention studies. Several limitations need to be acknowledged. While the sample size for this study is adequate or slightly larger than the typical gene expression study (Bower et al., 2011), a larger independent sample may identify additional DE genes and pathways that differentiate between patients with high and low levels of evening fatigue. While all of the gene expression studies of fatigue in oncology patients, including our own, identified differences in genes involved in inflammation, the specific genes and pathways identified differed, which may be due to differences in the patients studied (e.g., patient receiving CTX versus survivors). The study sample is heterogenous for type of treatment received, disease stage, and number of previous CTX cycles. Finally, because the preponderance of DE genes and pathways identified by ourselves and others are related to inflammation, one can hypothesize that this finding may be a byproduct of the tissues studied (i.e., peripheral leukocytes). However, the transciptome analyses (i.e., ProfileChaser) identified gene expression studies that were performed in non-immune cells, in numerous tissues, and under different experimental conditions. Until they are replicated, the current findings must be viewed as preliminary.
CONCLUSIONS
This study is novel in that differences between patients with high and low levels of evening fatigue were evaluated at the level of genes, pathways, and the entire transcriptome. Our analyses revealed that pathways involved in inflammation, neurotransmitter regulation, and energy metabolism are likely associated with evening fatigue severity; that CTX may contribute to the severity of evening fatigue; and that the patterns of gene expression may be shared with other models of fatigue (e.g., physical exercise, pathogen-induced sickness behavior). Importantly, our findings suggest that the molecular mechanisms associated with evening fatigue are multifactorial and that these mechanisms interplay among themselves (e.g., neurotransmitter regulation and inflammation; inflammation and mitochondrial dysfunction). Future research will need to evaluate this potential interplay among pathways to determine the mechanisms that underlie evening fatigue.
Supplementary Material
Additional File 2, Supplementary Figure S1. Quality control procedures and results.
The steps taken to filter out arrays that failed quality control criteria are illustrated in panel A. The counts of arrays from patients in each evening fatigue group are presented in parenthesis (i.e., low evening fatigue, high evening fatigue). The assay and gene level quality control filtering steps are shown in plate B.
Abbreviations: arrayQualityMetrics = an R statistical software package for microarray expression data quality metrics; BeadChip = Illumina HumanHT-12 Expression microarrays; MA = M (log ratios) and A (mean average) scale; QC = quality control; SNPs = single nucleotide polymorphisms; Da = Hoeffing's D-statistic.
Additional File 2, Supplementary Figure S2. K-means clustering heatmap of twelve differentially expressed genes in patients with high versus low levels of evening fatigue.
Illumina HumanHT-12 BeadChip microarrays identified eleven genes that were down-regulated and one gene that was up-regulated in patients undergoing chemotherapy for breast cancer who reported high (n=25) versus low (n=19) evening fatigue scores. Red indicates over-expression relative to the median and green indicates under-expression relative to the median. Magenta indicates patients with high evening fatigue and orange indicates patients with low evening fatigue. Arrays were k-means clustered (k=2) agnostic of fatigue group membership.
Acknowledgments
This study was funded by a grant from the National Cancer Institute (NCI, CA134900). Dr. Miaskowski is funded by a grant from the NCI (K05 CA168960) and is an American Cancer Society Clinical Research Professor.
References
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nature Reviews Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Awabdh S, Miserey-Lenkei S, Bouceba T, Masson J, Kano F, Marinach-Patrice C, Darmon M. A new vesicular scaffolding complex mediates the G-protein-coupled 5-HT1A receptor targeting to neuronal dendrites. Journal of Neuroscience. 2012;32:14227–14241. doi: 10.1523/JNEUROSCI.6329-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara-Silva TR, Freitas-Junior R, Freitas NM, Machado GD. Fatigue related to radiotherapy for breast and/or gynaecological cancer: a systematic review. Journal of Clinical Nursing. 2013;22:2679–2686. doi: 10.1111/jocn.12236. [DOI] [PubMed] [Google Scholar]
- Andrews PLR, Morrow GR, Hickok JT, Roscoe J, Stone P. Mechanisms and models of fatigue associated with cancer and its treatment: Evidence from preclinical and clinical studies. In: Armes J, Krishnasamy M, Higginson I, editors. Fatigue in Cancer. Oxford: Oxford University Press; 2004. pp. 51–87. [Google Scholar]
- Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods in Molecular Biology. 2007;396:71–91. doi: 10.1007/978-1-59745-515-2_6. doi: 1-59745-515-6:71. [DOI] [PubMed] [Google Scholar]
- Aouizerat BE, Dodd M, Lee K, West C, Paul SM, Cooper BA, Miaskowski C. Preliminary evidence of a genetic association between tumor necrosis factor alpha and the severity of sleep disturbance and morning fatigue. Biological Research for Nursing. 2009;11:27–41. doi: 10.1177/1099800409333871. doi: 1099800409333871. [DOI] [PubMed] [Google Scholar]
- Aune TM, Golden HW, McGrath KM. Inhibitors of serotonin synthesis and antagonists of serotonin 1A receptors inhibit T lymphocyte function in vitro and cell-mediated immunity in vivo. Journal of Immunology. 1994;153:489–498. [PubMed] [Google Scholar]
- Bailey MJ, Coon SL, Carter DA, Humphries A, Kim JS, Shi Q, Klein DC. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. Journal of Biological Chemistry. 2009;284:7606–7622. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AM, Gerber LH, Mayer DK. Cancer-related fatigue: implications for breast cancer survivors. Cancer. 2012;118:2261–2269. doi: 10.1002/cncr.27475. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Aziz N, Fahey JL, Cole SW. T-cell homeostasis in breast cancer survivors with persistent fatigue. Journal of the National Cancer Institute. 2003;95:1165–1168. doi: 10.1093/jnci/djg0019. [DOI] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Irwin MR, Arevalo JM, Cole SW. Fatigue and gene expression in human leukocytes: increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain Behavior and Immunity. 2011;25:147–150. doi: 10.1016/j.bbi.2010.09.010. doi: S0889-1591(10)00468-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Irwin MR, Castellon S, Arevalo J, Cole SW. Cytokine genetic variations and fatigue among patients with breast cancer. Journal of Clinical Oncology. 2013;31:1656–1661. doi: 10.1200/JCO.2012.46.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JE, Ganz PA, Tao ML, Hu W, Belin TR, Sepah S, Aziz N. Inflammatory biomarkers and fatigue during radiation therapy for breast and prostate cancer. Clinical Cancer Research. 2009;15:5534–5540. doi: 10.1158/1078-0432.CCR-08-2584. 1078-0432.CCR-08-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Current Opinion in Cell Biology. 2007;19:124–134. doi: 10.1016/j.ceb.2007.02.008. doi: S0955-0674(07)00022-1. [DOI] [PubMed] [Google Scholar]
- Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, Duckett CS. COMMD proteins, a novel family of structural and functional homologs of MURR1. Journal of Biological Chemistry. 2005;280:22222–22232. doi: 10.1074/jbc.M501928200. doi: M501928200. [DOI] [PubMed] [Google Scholar]
- Butte A. The use and analysis of microarray data. Nature Reviews in Drug Discovery. 2002;1:951–960. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- Buttner P, Mosig S, Lechtermann A, Funke H, Mooren FC. Exercise affects the gene expression profiles of human white blood cells. Journal of Applied Physiology. 2007;102:26–36. doi: 10.1152/japplphysiol.00066.2006. doi: 00066.2006. [DOI] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ Host Response to Injury Large Scale Collaborative Research Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. doi: nature03985. [DOI] [PubMed] [Google Scholar]
- Camins A, Verdaguer E, Folch J, Pallas M. Involvement of calpain activation in neurodegenerative processes. CNS Drug Reviews. 2006;12:135–148. doi: 10.1111/j.1527-3458.2006.00135.x. doi: CNS135 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. doi: S0092-8674(06)00768-9. [DOI] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. 2. Hillsdale, NJ: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- Cole SW. Nervous system regulation of the cancer genome. Brain Behavior and Immunity. 2013;(30 Suppl):S10–18. doi: 10.1016/j.bbi.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proceedings of the National Academy of Science. 2011;108:3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Hidalgo A, Bower JE, Ganz PA, Cole SW, Irwin MR. Inflammatory biomarkers for persistent fatigue in breast cancer survivors. Clinical Cancer Research. 2006;12:2759–2766. doi: 10.1158/1078-0432.CCR-05-2398. doi: 12/9/2759. [DOI] [PubMed] [Google Scholar]
- Collado-Hidalgo A, Bower JE, Ganz PA, Irwin MR, Cole SW. Cytokine gene polymorphisms and fatigue in breast cancer survivors: early findings. Brain Behavior and Immunity. 2008;22:1197–1200. doi: 10.1016/j.bbi.2008.05.009. doi: S0889-1591(08)00270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo ES, Ryff CD, Singer BH. Psychosocial adjustment among cancer survivors: findings from a national survey of health and well-being. Health Psychology. 2009;28:147–156. doi: 10.1037/a0013221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotel F, Exley R, Cragg SJ, Perrier JF. Serotonin spillover onto the axon initial segment of motoneurons induces central fatigue by inhibiting action potential initiation. Proceedings of the National Academy of Science. 2013;110:4774–4779. doi: 10.1073/pnas.1216150110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nature Review Neuroscience. 2008;9:46–56. doi: 10.1038/nrn2297. doi: nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JR, MacLean AW, Brundage MD, Schulze K. Sleep disturbance in cancer patients. Social Science and Medicine. 2002;54:1309–1321. doi: 10.1016/s0277-9536(01)00043-0. [DOI] [PubMed] [Google Scholar]
- Dennehy PH. Rotavirus vaccines - success after failure. Med Health R I. 2007;90(10):321–324. [PubMed] [Google Scholar]
- Dhruva A, Aouizerat BE, Cooper B, Paul SM, Dodd M, West C, Miaskowski C. Differences in morning and evening fatigue in oncology patients and their family caregivers. European Journal of Oncology Nursing. 2013;17:841–848. doi: 10.1016/j.ejon.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhruva A, Aouizerat BE, Cooper B, Paul SM, Dodd M, West C, Miaskowski C. Cytokine gene associations with self-report ratings of morning and evening fatigue in oncology patients and their family caregivers. Biological Research Nursing. 2014 doi: 10.1177/1099800414534313. doi: 1099800414534313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhruva A, Dodd M, Paul SM, Cooper BA, Lee K, West C, Miaskowski C. Trajectories of fatigue in patients with breast cancer before, during, and after radiation therapy. Cancer Nursing. 2010;33(3):201–212. doi: 10.1097/NCC.0b013e3181c75f2a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong SH, Dhruva A, Dunn LB, West C, Paul SM, Cooper BA, Miaskowski C. Associations between cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression in patients prior to breast cancer surgery. Biol Res Nurs. 2014 doi: 10.1177/1099800414550394. doi: 1099800414550394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Morgan AA, Dudley JT, Chen R, Thathoo R, Altman RB, Butte AJ. Content-based microarray search using differential expression profiles. BMC Bioinformatics. 2010;11:603. doi: 10.1186/1471-2105-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdo SL, Wolff JR. gamma-Aminobutyric acid outside the mammalian brain. Journal of Neurochemistry. 1990;54(2):363–372. doi: 10.1111/j.1471-4159.1990.tb01882.x. [DOI] [PubMed] [Google Scholar]
- Fairfax BP, Vannberg FO, Radhakrishnan J, Hakonarson H, Keating BJ, Hill AV, Knight JC. An integrated expression phenotype mapping approach defines common variants in LEP, ALOX15 and CAPNS1 associated with induction of IL-6. Human Molecular Genetics. 2010;19(4):720–730. doi: 10.1093/hmg/ddp530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Bai L, Yan J, Li Y, Shen W, Wang Y, Liu J. Mitochondrial dynamic remodeling in strenuous exercise-induced muscle and mitochondrial dysfunction: regulatory effects of hydroxytyrosol. Free Radical Biology & Medicine. 2011;50:1437–1446. doi: 10.1016/j.freeradbiomed.2011.03.001. [DOI] [PubMed] [Google Scholar]
- FitzPatrick DR, Germain-Lee E, Valle D. Isolation and characterization of rat and human cDNAs encoding a novel putative peroxisomal enoyl-CoA hydratase. Genomics. 1995;27:457–466. doi: 10.1006/geno.1995.1077. doi: S0888-7543(85)71077-4. [DOI] [PubMed] [Google Scholar]
- Fletcher BA, Schumacher KL, Dodd M, Paul SM, Cooper BA, Lee K, Miaskowski C. Trajectories of fatigue in family caregivers of patients undergoing radiation therapy for prostate cancer. Research in Nursing and Health. 2009;32:125–139. doi: 10.1002/nur.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher BS, Paul SM, Dodd MJ, Schumacher K, West C, Cooper B, Miaskowski CA. Prevalence, severity, and impact of symptoms on female family caregivers of patients at the initiation of radiation therapy for prostate cancer. Journal of Clinical Oncology. 2008;26:599–605. doi: 10.1200/JCO.2007.12.2838. doi: 26/4/599 [pii] [DOI] [PubMed] [Google Scholar]
- Focht BC, Clinton SK, Devor ST, Garver MJ, Lucas AR, Thomas-Ahner JM, Grainger E. Resistance exercise interventions during and following cancer treatment: a systematic review. Journal of Supportive Oncology. 2013;11:45–60. [PubMed] [Google Scholar]
- Foteinou PT, Calvano SE, Lowry SF, Androulakis IP. Multiscale model for the assessment of autonomic dysfunction in human endotoxemia. Physiological Genomics. 2010;42:5–19. doi: 10.1152/physiolgenomics.00184.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. doi: gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldrath AW, Luckey CJ, Park R, Benoist C, Mathis D. The molecular program induced in T cells undergoing homeostatic proliferation. Proceedings of the National Academy of Science. 2004;101:16885–16890. doi: 10.1073/pnas.0407417101. doi: 0407417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotte M, von Mollard GF. A new beat for the SNARE drum. Trends in Cell Biology. 1998;8:215–218. doi: 10.1016/s0962-8924(98)01272-0. doi: S0962892498012720. [DOI] [PubMed] [Google Scholar]
- Haimovich B, Calvano J, Haimovich AD, Calvano SE, Coyle SM, Lowry SF. In vivo endotoxin synchronizes and suppresses clock gene expression in human peripheral blood leukocytes. Critical Care Medicne. 2010;38:751–758. doi: 10.1097/CCM.0b013e3181cd131c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research. 2004;32:D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofso K, Miaskowski C, Bjordal K, Cooper BA, Rustoen T. Previous chemotherapy influences the symptom experience and quality of life of women with breast cancer prior to radiation therapy. Cancer Nursing. 2012;35:167–177. doi: 10.1097/NCC.0b013e31821f5eb5. [DOI] [PubMed] [Google Scholar]
- Hopkins SJ. Central nervous system recognition of peripheral inflammation: a neural, hormonal collaboration. Acta Bio-medica. 2007;78:231–247. [PubMed] [Google Scholar]
- Hsiao CP, Araneta M, Wang XM, Saligan LN. The association of IFI27 expression and fatigue intensification during localized radiation therapy: implication of a para-inflammatory bystander response. International Journal of Molecular Science. 2013;14:16943–16957. doi: 10.3390/ijms140816943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CP, Wang D, Kaushal A, Saligan L. Mitochondria-related gene expression changes are associated with fatigue in patients with nonmetastatic prostate cancer receiving external beam radiation therapy. Cancer Nursing. 2013;36:189–197. doi: 10.1097/NCC.0b013e318263f514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indraccolo S, Pfeffer U, Minuzzo S, Esposito G, Roni V, Mandruzzato S, Amadori A. Identification of genes selectively regulated by IFNs in endothelial cells. Journal of Immunology. 2007;178(2):1122–1135. doi: 10.4049/jimmunol.178.2.1122. doi: 178/2/1122. [DOI] [PubMed] [Google Scholar]
- Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nature Reviews Immunology. 2011;11:625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager A, Sleijfer S, van der Rijt CC. The pathogenesis of cancer related fatigue: Could increased activity of pro-inflammatory cytokines be the common denominator? European Journal of Cancer. 2008;44:175–181. doi: 10.1016/j.ejca.2007.11.023. doi: S0959-8049(07)00968-9. [DOI] [PubMed] [Google Scholar]
- Jeanmougin M, de Reynies A, Marisa L, Paccard C, Nuel G, Guedj M. Should we abandon the t-test in the analysis of gene expression microarray data: a comparison of variance modeling strategies. PLoS One. 2010;5:e12336. doi: 10.1371/journal.pone.0012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnofsky D, Abelmann WH, Craver LV, Burchenal JH. The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer. 1948;1:634–656. [Google Scholar]
- Katafuchi T, Kondo T, Take S, Yoshimura M. Enhanced expression of brain interferon-alpha and serotonin transporter in immunologically induced fatigue in rats. European Journal of Neuroscience. 2005;22:2817–2826. doi: 10.1111/j.1460-9568.2005.04478.x. doi: EJN4478. [DOI] [PubMed] [Google Scholar]
- Klotman ME, Chang TL. Defensins in innate antiviral immunity. Nature Reviews Immunology. 2006;6:447–456. doi: 10.1038/nri1860. doi: nri1860. [DOI] [PubMed] [Google Scholar]
- Landmark-Hoyvik H, Reinertsen KV, Loge JH, Fossa SD, Borresen-Dale AL, Dumeaux V. Alterations of gene expression in blood cells associated with chronic fatigue in breast cancer survivors. Pharmacogenomics Journal. 2009;9:333–340. doi: 10.1038/tpj.2009.27. doi: tpj200927. [DOI] [PubMed] [Google Scholar]
- Lee BN, Dantzer R, Langley KE, Bennett GJ, Dougherty PM, Dunn AJ, Cleeland CS. A cytokine-based neuroimmunologic mechanism of cancer-related symptoms. Neuroimmunomodulation. 2004;11:279–292. doi: 10.1159/000079408. [DOI] [PubMed] [Google Scholar]
- Lee KA, Hicks G, Nino-Murcia G. Validity and reliability of a scale to assess fatigue. Psychiatry Research. 1991;36:291–298. doi: 10.1016/0165-1781(91)90027-m. [DOI] [PubMed] [Google Scholar]
- Lemaigre FP, Rousseau GG. Transcriptional control of genes that regulate glycolysis and gluconeogenesis in adult liver. Biochemistry Journal. 1994;303:1–14. doi: 10.1042/bj3030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. Journal of Laboratory and Clinical Medicine. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. doi: S0022-2143(05)00383-5. [DOI] [PubMed] [Google Scholar]
- Light KC, Agarwal N, Iacob E, White AT, Kinney AY, VanHaitsma TA, Light AR. Differing leukocyte gene expression profiles associated with fatigue in patients with prostate cancer versus chronic fatigue syndrome. Psychoneuroendocrinology. 2013;38:2983–2995. doi: 10.1016/j.psyneuen.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TF, Brown CM, El Gazzar M, McPhail L, Millet P, Rao A, McCall CE. Fueling the flame: bioenergy couples metabolism and inflammation. Journal of Leukocyte Biology. 2012;92:499–507. doi: 10.1189/jlb.0212078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoccoli G, Sothern RB, De Cata A, Giuliani F, Fontana A, Copetti M, Tarquini R. A timetable of 24-hour patterns for human lymphocyte subpopulations. Journal of Biological Regulators and Homeostatic Agents. 2011;25:387–395. [PubMed] [Google Scholar]
- Meeus M, Mistiaen W, Lambrecht L, Nijs J. Immunological similarities between cancer and chronic fatigue syndrome: the common link to fatigue? Anticancer Research. 2009;29:4717–4726. doi: 29/11/4717. [PubMed] [Google Scholar]
- Miaskowski C, Dodd M, Lee K, West C, Paul SM, Cooper BA, Aouizerat BE. Preliminary evidence of an association between a functional interleukin-6 polymorphism and fatigue and sleep disturbance in oncology patients and their family caregivers. Journal of Pain and Symptom Management. 2010;40:531–544. doi: 10.1016/j.jpainsymman.2009.12.006. doi: S0885-3924(10)00251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaskowski C, Paul SM, Cooper BA, Lee K, Dodd M, West C, Wara W. Trajectories of fatigue in men with prostate cancer before, during, and after radiation therapy. Journal of Pain and Symptom Management. 2008;35:632–643. doi: 10.1016/j.jpainsymman.2007.07.007. doi: S0885-3924(08)00051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. The proliferation rate paradox in antimitotic chemotherapy. Molecular Biology of the Cell. 2012;23:1–6. doi: 10.1091/mbc.E10-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi C, Stojanovic A, Urbina M, Carreira I, Lima L. Differential cAMP levels and serotonin effects in blood peripheral mononuclear cells and lymphocytes from major depression patients. International Immunopharmacology. 2004;4:1125–1133. doi: 10.1016/j.intimp.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Molassiotis A, Chan CW. Fatigue patterns in Chinese patients receiving radiotherapy. European Journal of Oncology Nursing. 2004;8:334–340. doi: 10.1016/j.ejon.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Nishimura D. BioCarta. Biotech Software & Internet Report. 2001;2:117–120. [Google Scholar]
- Noguchi M, Sarin A, Aman MJ, Nakajima H, Shores EW, Henkart PA, Leonard WJ. Functional cleavage of the common cytokine receptor gamma chain (gammac) by calpain. Proceedings of the National Academy of Science. 1997;94:11534–11539. doi: 10.1073/pnas.94.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoba D. Interpreting the meaningfulness of changes in health-related quality of life scores: lessons from studies in adults. International Journal of Cancer. 1999;12:132–137. doi: 10.1002/(sici)1097-0215(1999)83:12+<132::aid-ijc23>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Petroff OA. GABA and glutamate in the human brain. Neuroscientist. 2002;8(6):562–573. doi: 10.1177/1073858402238515. [DOI] [PubMed] [Google Scholar]
- Pingel LC, Kohlgraf KG, Hansen CJ, Eastman CG, Dietrich DE, Burnell KK, Brogden KA. Human beta-defensin 3 binds to hemagglutinin B (rHagB), a non-fimbrial adhesin from Porphyromonas gingivalis, and attenuates a pro-inflammatory cytokine response. Immunology and Cell Biology. 2008;86:643–649. doi: 10.1038/icb.2008.56. [DOI] [PubMed] [Google Scholar]
- Powell ND, Mays JW, Bailey MT, Hanke ML, Sheridan JF. Immunogenic dendritic cells primed by social defeat enhance adaptive immunity to influenza A virus. Brain Behavior and Immunity. 2011;25:46–52. doi: 10.1016/j.bbi.2010.07.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan N, Banks WA. Brain-immune communication pathways. Brain Behavior and Immunity. 2007;21(6):727–735. doi: 10.1016/j.bbi.2007.05.005. doi: S0889-1591(07)00115-8. [DOI] [PubMed] [Google Scholar]
- Reimers M. Making informed choices about microarray data analysis. PLoS Computational Biology. 2010;6:e1000786. doi: 10.1371/journal.pcbi.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinertsen KV, Grenaker Alnaes GI, Landmark-Hoyvik H, Loge JH, Wist E, Kristensen VN, Edvardsen H. Fatigued breast cancer survivors and gene polymorphisms in the inflammatory pathway. Brain Behavior and Immunity. 2011;25:1376–1383. doi: 10.1016/j.bbi.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez N, Mages J, Dietrich H, Wantia N, Wagner H, Lang R, Miethke T. MyD88-dependent changes in the pulmonary transcriptome after infection with Chlamydia pneumoniae. Physiological Genomics. 2007;30:134–145. doi: 10.1152/physiolgenomics.00011.2007. doi: 00011.2007. [DOI] [PubMed] [Google Scholar]
- Ryan JL, Carroll JK, Ryan EP, Mustian KM, Fiscella K, Morrow GR. Mechanisms of cancer-related fatigue. Oncologist. 2007;12:22–34. doi: 10.1634/theoncologist.12-S1-22. doi: 12/suppl_1/22 [pii] [DOI] [PubMed] [Google Scholar]
- Saligan LN, Hsiao CP, Wang D, Wang XM, St John L, Kaushal A, Dionne RA. Upregulation of alpha-synuclein during localized radiation therapy signals the association of cancer-related fatigue with the activation of inflammatory and neuroprotective pathways. Brain Behavior and Immunity. 2013;27:63–70. doi: 10.1016/j.bbi.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saligan LN, Kim HS. A systematic review of the association between immunogenomic markers and cancer-related fatigue. Brain Behavior and Immunity. 2012;26:830–848. doi: 10.1016/j.bbi.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangha O, Stucki G, Liang MH, Fossel AH, Katz JN. The Self-Administered Comorbidity Questionnaire: a new method to assess comorbidity for clinical and health services research. Arthritis and Rheumatism. 2003;49:156–163. doi: 10.1002/art.10993. [DOI] [PubMed] [Google Scholar]
- Schwartz CE, Sprangers MA. Methodological approaches for assessing response shift in longitudinal health-related quality-of-life research. Social Science and Medicine. 1999;48:1531–1548. doi: 10.1016/s0277-9536(99)00047-7. doi: S0277953699000477. [DOI] [PubMed] [Google Scholar]
- Semple F, MacPherson H, Webb S, Cox SL, Mallin LJ, Tyrrell C, Dorin JR. Human beta-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. European Journal of Immunology. 2011;41:3291–3300. doi: 10.1002/eji.201141648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple F, Webb S, Li HN, Patel HB, Perretti M, Jackson IJ, Dorin JR. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. European Journal of Immunology. 2010;40:1073–1078. doi: 10.1002/eji.200940041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Inoue A, Toyama-Sorimachi N, Nagai Y, Yasuda T, Suzuki H, Yamanashi Y. Dok-1 and Dok-2 are negative regulators of lipopolysaccharide-induced signaling. Journal of Experimental Medicine. 2005;201:333–339. doi: 10.1084/jem.20041817. doi: jem.20041817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. Limma: Linear Models for Microarray Data. In: Gentleman RC, Carey VJ, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Tripathi S, Glazko GV, Emmert-Streib F. Ensuring the statistical soundness of competitive gene set approaches: gene filtering and genome-scale coverage are essential. Nucleic Acids Research. 2013;41:e82. doi: 10.1093/nar/gkt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc F, Vrbova G. Stabilisation of neuromuscular junctions by leupeptin increases motor unit size in partially denervated rat muscles. Brain Research. Developmental Brain Research. 1995;88:186–193. doi: 10.1016/0165-3806(95)00096-v. doi: 016538069500096V. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dennehy PH, Keyserling HL, Tang K, Gentsch JR, Glass RI, Jiang B. Rotavirus infection alters peripheral T-cell homeostasis in children with acute diarrhea. Journal of Virology. 2007;81:3904–3912. doi: 10.1128/JVI.01887-06. doi: JVI.01887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurfel MM, Park WY, Radella F, Ruzinski J, Sandstrom A, Strout J, Martin TR. Identification of high and low responders to lipopolysaccharide in normal subjects: an unbiased approach to identify modulators of innate immunity. Journal of Immunology. 2005;175:2570–2578. doi: 10.4049/jimmunol.175.4.2570. doi: 175/4/2570. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang K, Valladares O, Hannenhalli S, Bucan M. Genome-wide expression profiling and bioinformatics analysis of diurnally regulated genes in the mouse prefrontal cortex. Genome Biology. 2007;8:R247. doi: 10.1186/gb-2007-8-11-r247. doi: gb-2007-8-11-r247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature Reviews in Molecular and Cell Biology. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yavuzsen T, Davis MP, Ranganathan VK, Walsh D, Siemionow V, Kirkova J, Yue GH. Cancer-related fatigue: central or peripheral? Journal of Pain and Symptom Management. 2009;38:587–596. doi: 10.1016/j.jpainsymman.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Martino A, Faulon JL. Identification of expression patterns of IL-2-responsive genes in the murine T cell line CTLL-2. Journal of Interferon and Cytokine Research. 2007;27:991–995. doi: 10.1089/jir.2006.0169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional File 2, Supplementary Figure S1. Quality control procedures and results.
The steps taken to filter out arrays that failed quality control criteria are illustrated in panel A. The counts of arrays from patients in each evening fatigue group are presented in parenthesis (i.e., low evening fatigue, high evening fatigue). The assay and gene level quality control filtering steps are shown in plate B.
Abbreviations: arrayQualityMetrics = an R statistical software package for microarray expression data quality metrics; BeadChip = Illumina HumanHT-12 Expression microarrays; MA = M (log ratios) and A (mean average) scale; QC = quality control; SNPs = single nucleotide polymorphisms; Da = Hoeffing's D-statistic.
Additional File 2, Supplementary Figure S2. K-means clustering heatmap of twelve differentially expressed genes in patients with high versus low levels of evening fatigue.
Illumina HumanHT-12 BeadChip microarrays identified eleven genes that were down-regulated and one gene that was up-regulated in patients undergoing chemotherapy for breast cancer who reported high (n=25) versus low (n=19) evening fatigue scores. Red indicates over-expression relative to the median and green indicates under-expression relative to the median. Magenta indicates patients with high evening fatigue and orange indicates patients with low evening fatigue. Arrays were k-means clustered (k=2) agnostic of fatigue group membership.