Abstract
Background
Community-acquired pneumonia (CAP) is a leading cause of pediatric hospitalization. Pathogen identification fails in approximately 20% of children but is critical for optimal treatment and prevention of hospital-acquired infections. We used two broad-spectrum detection strategies to identify pathogens in test-negative children with CAP and asymptomatic controls.
Methods
Nasopharyngeal/oropharyngeal (NP/OP) swabs from 70 children <5 years with CAP of unknown etiology and 90 asymptomatic controls were tested by next-generation sequencing (RNA-seq) and pan viral group (PVG) PCR for 19 viral families. Association of viruses with CAP was assessed by adjusted odds ratios (aOR) and 95% confidence intervals controlling for season and age group.
Results
RNA-seq/PVG PCR detected previously missed, putative pathogens in 34% of patients. Putative viral pathogens included human parainfluenza virus 4 (aOR 9.3, P = .12), human bocavirus (aOR 9.1, P < .01), Coxsackieviruses (aOR 5.1, P = .09), rhinovirus A (aOR 3.5, P = .34), and rhinovirus C (aOR 2.9, P = .57). RNA-seq was more sensitive for RNA viruses whereas PVG PCR detected more DNA viruses.
Conclusions
RNA-seq and PVG PCR identified additional viruses, some known to be pathogenic, in NP/OP specimens from one-third of children hospitalized with CAP without a previously identified etiology. Both broad-range methods could be useful tools in future epidemiologic and diagnostic studies.
Keywords: RNA sequencing (RNA-seq), metagenomics, pan-viral group polymerase chain reaction (PVG PCR), pneumonia
Pneumonia is a leading cause of childhood death globally; approximately 1 million children die of pneumonia every year [1]. In the United States, up to 50% of children aged ≤5 years with community-acquired pneumonia (CAP) require hospitalization, accounting for 110 000 admissions annually [2]. Pathogens vary by age [3–5], but viruses are the most common cause of CAP in children aged ≤5 years, especially in the absence of lobar pneumonia and pleural effusion [3]. However, a pathogen cannot be identified in 14%–23% of children with CAP, even with extensive testing [4–11]. More effective pathogen identification will improve our understanding of pneumonia and guide treatment and site-of-care decisions.
Inability to detect etiologic agents may be due to incomplete test panels, genetic pathogen variants escaping molecular detection, unrecognized bacterial infections due to insensitive diagnostics, novel and emerging pathogens, or inadequate specimens. Most of these limitations could be overcome by unbiased pathogen detection [12, 13]. Shotgun metagenomic sequencing of DNA or RNA (RNA sequencing [RNA-seq]) and broad-range polymerase chain reaction (PCR) amplification of conserved pathogen genomic regions are two such methods. RNA sequencing enables sequence-independent detection of any pathogen with sufficient sequence homology to known viruses, bacteria, fungi, or parasites to allow their classification [13–15]. Panviral group (PVG) PCR uses broad-range PCR primers to detect known and novel members of relevant viral genera and families [16].
The aim of this study was to identify potential pathogens in children enrolled in a multicenter study of children hospitalized with CAP but without identifiable diagnosis after extensive testing [4].
METHODS
Study Population
Participants were children enrolled in the US Centers for Disease Control and Prevention Etiology of Pneumonia in the Community (EPIC) study. The EPIC study population, enrollment criteria, specimen collection, and etiologic testing have been described in detail [4]. Patients were included if they lived in the catchment area and were hospitalized for CAP, defined as acute infection, acute respiratory illness, and radio-graphically confirmed pneumonia. Exclusion criteria included recent hospitalization and severe immunosuppression.
For this analysis (University of Utah IRB 35409, CDC IRB 5827), we included children (n = 70) with CAP and asymptomatic pediatric control subjects aged <5 years enrolled at Primary Children’s Hospital in Salt Lake City, Utah (Table 1). Patients were enrolled between 1 January 2010 and 30 June 2012. Control subjects (n = 90) were same-day elective surgery patients and were enrolled between 1 February 2011 and 30 June 2012; if a control subject had respiratory symptoms or fever within 14 days of enrollment, they were excluded. Patients and control subjects were included if no pathogen was detected per EPIC study protocol [4].
Table 1.
Demographic and Clinical Information of Children With Pneumonia With No Identifiable Etiology and Control Subjects
| Patients (n = 70) |
Control subjects (n = 90) |
P value (χ2) | |
|---|---|---|---|
| Age group, no. (%) | .05 | ||
| <1 y | 14 (20) | 23 (26) | |
| 1 y | 26 (37) | 18 (20) | |
| 2–4 y | 30 (43) | 49 (54) | |
|
| |||
| Month of enrollment, no. (%) | .04 | ||
| January–March | 23 (33) | 13 (14) | |
| April–June | 25 (36) | 37 (41) | |
| July–September | 16 (23) | 25 (28) | |
| October–December | 6 (9) | 15 (17) | |
|
| |||
| Symptom, no. (%) | NA | ||
| Fever | 67 (96) | NA | |
| Cough | 58 (83) | NA | |
| Anorexia | 53 (76) | NA | |
| Dyspnea | 33 (47) | NA | |
|
| |||
| Underlying condition, no. (%) | ns | ||
| Asthma or reactive airway disease | 6 (9) | 3 (3) | |
| Preterm birth among children aged <2 y | 7 (10) | 7 (8) | |
|
| |||
| Radiographic findings, no. (%) | NA | ||
| Consolidation | 32 (45) | NA | |
| Alveolar or interstitial infiltrate | 23 (33) | NA | |
| Pleural effusion | 20 (29) | NA | |
|
| |||
| Hospitalization | NA | ||
| Length of stay, median (IQR) | 3 (2–4) | NA | |
| ICU admission, no. (%) | 19 (27) | NA | |
| Death in the hospital, no. (%) | 0 (0) | NA | |
Abbreviations: ICU, intensive care unit; IQR, interquartile range; NA, not applicable; NS, not significant.
Specimen Collection and Pathogen Detection in the EPIC Study
For children hospitalized with CAP, combined nasopharyngeal (NP) and oropharyngeal (OP) swabs were collected within 72 hours of hospital admission. Specimens were transferred into 3-mL universal transport media, refrigerated, and stored at −80°C within 24 hours. Bacteria (Haemophilus influenzae or other Gram-negative bacteria, Staphylococcus aureus, Streptococcus anginosus, Streptococcus mitis, Streptococcus pneumoniae, or Streptococcus pyogenes) were detected by culture (blood, endotracheal aspirate, bronchoalveolar-lavage specimen, pleural fluid) or PCR (whole blood, pleural fluid); Chlamydophila pneumoniae and Mycoplasma pneumoniae were detected by PCR from NP/OP swabs. Viruses (adenovirus [ADV], coronavirus, human metapneumovirus, human rhinovirus [HRV], human influenza, parainfluenza viruses 1–3 [HPIV], and respiratory syncytial virus) were detected by PCR of NP/OP swabs or serology of acute-and convalescent-phase serum (except for human rhinovirus and coronaviruses) [4]. Nasopharyngeal and OP swabs were also obtained from asymptomatic control subjects before elective surgery while in the operating room and tested for viral pathogens, C. pneumoniae, and M. pneumoniae [4].
RNA Sequencing
Nucleic Acid Extraction
Nasopharyngeal and OP swabs (75–200 µL) were extracted using the QIAamp Viral RNA extraction kit per manufacturer’s instructions with the addition of on-column DNase treatment after AW1 wash (10 µL of RNase-free DNase I plus 70 µL of Buffer RDD, Qiagen) at room temperature for 15 minutes and an additional wash step.
Library Generation
Indexed cDNA libraries were prepared with the TruSeq RNA Sample Prep Kit following manufacturer’s instructions (Illumina). Libraries were quantified with the Illumina Universal Library Quantification Kit (Kapa Biosystems) and the Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosciences). Library size and quality was assessed with a High Sensitivity DNA Analysis Kit on an Agilent 2100 Bioanalyzer (Agilent Technologies). Libraries from 24 samples were combined in equimolar ratios for a final concentration of 9.6 nM and sequenced in batches of 24 samples per lane on a HiSeq 2500 instrument (Illumina), generating an average of 1.7 × 107 sequencing reads (2 × 100 base pairs) per sample.
Analysis of Metagenomics Data
Matching paired-end reads were concatenated, adding an “N” between read 1 and read 2. The resulting sequences were analyzed by Taxonomer [14], an alignment-free, rapid, interactive metagenomic sequence analysis tool for microbial identification and results visualized through http://taxonomer.iobio.io. Taxa with only 1 read assigned to them were ignored. Viral taxa (other than phages) were confirmed manually by mapping the sequencing files against the relevant reference sequences in Geneious (version 8.1, Biomatters) and by comparing viral sequences to the National Center for Biotechnology Information nucleotide database [17, 18]. Viral taxa identified based on <100 reads were only considered if reads were not an identical match to any other sample within the same batch by manual analysis. These protocols had previously been shown to produce results comparable with a US Food and Drug Administration–cleared PCR panel [15].
Pan-Viral Group Family/Genus Polymerase Chain Reaction Panel
Nucleic Acid Extraction
Combined NP/OP swab samples (200 µL) in universal transport media were extracted using either a manual method by the QIAamp Viral RNA extraction kit (Qiagen) or an automated method by the BioSprint 96 One For All kit (Qiagen) on a Kingfisher 96 platform (Thermo) according to the manufacturer’s instructions.
Pan-Viral Goup Polymerase Chain Reaction
Pan-viral group PCR assays were designed to amplify known and potentially novel members of the viral families/genera listed below. They were designed using the consensus-degenerate hybrid oligonucleotide primer (CODEHOP) principle to conserved genes and regions [19, 20] and had analytical sensitivities of 10–500 copies (RNA viruses) and 10–1000 copies (DNA viruses) per reaction. Samples (5 µl of total nucleic acid) were tested with broadly reactive PVG PCR for the following viral families/genera: Adenoviridae, Anelloviridae, Arenaviridae, Astroviridae, Bunyaviridae, Caliciviridae, Circoviridae, Coronaviridae, Flaviviridae (Flavivirus), Herpesviridae, Orthomyxoviridae (influenza viruses A, B, and C), Paramyxoviridae, Parvoviridae, Picornaviridae (enterovirus and parechovirus; primers do not target all human rhinoviruses), Polyomaviridae, Reoviridae (aquareovirus, orthoreovirus, orbivirus, rotavirus, and seadornavirus), Rhabdoviridae, and Togaviridae (alphavirus) [21–28]. The Picornaviridae PCR targets only a subset of human rhinoviruses. First-round reverse-transcription PCR for RNA viruses was performed with Superscript III/Platinum Taq One Step kits (Invitrogen), and second-round PCR was performed with Titanium Taq (Clontech) kits. First- and second-round PCR for DNA viruses was performed with Hot Start Ex Taq kits (Takara). Positive and negative PCR controls containing mutation-engineered synthetic RNA transcript or DNA amplicon and nuclease-free water, respectively, were included in each run. Polymerase chain reaction products were visualized on 2% agarose gels.
Sequence Confirmation
Positive bands of the expected size that had strong signal and without additional bands were cleaned up using Exonuclease I (New England Biolabs) and Shrimp Alkaline Phosphatase (Roche). Samples were incubated at 37°C for 15 minutes followed by 80°C for 15 minutes to inactivate the Exonuclease and Shrimp Alkaline Phosphatase. Positive bands of the expected size with additional bands present in the PCR products were purified using QIAquick Gel Extraction kits (Qiagen). Purified PCR amplicons were sequenced with the PCR primers in both directions on an ABI Prism 3130 Automated Capillary Sequencer (Applied Biosystems) using Big Dye 3.1 cycle sequencing kits (Life Technologies).
Validation
We first assessed the ability of RNA-seq and PVG PCR to detect known respiratory pathogens using specimens from children with CAP (n = 63) and asymptomatic control (n = 52) subjects in whom viral or atypical bacterial pathogens had been detected using the EPIC study protocol. Nasopharyngeal and OP specimens were analyzed by RNA-seq and PVG PCR as described.
Statistics
Proportions of putative pathogens detected by each method individually or in combination were determined. Descriptive statistics were calculated, and proportions were compared by χ2 or Fisher exact test, as appropriate. To assess the association of virus detection with disease, we compared children with CAP with asymptomatic control subjects and calculated adjusted odds ratios (aORs) and 95% confidence intervals (CIs) using approximate exact logistic regression controlling for season and age group. All statistical analyses were performed with a 2-tailed α of .05 using R 3.2.2 (R Foundation for Statistical Computing). Approximate exact logistic regression models were developed using the elrm package for R with 1 000 000 Markov chain iterations and 20 000 burn-in iterations that were discarded when conducting the inference.
RESULTS
Validation of Respiratory Pathogen Detection by RNA Sequencing and Pan-Viral Group Polymerase Chain Reaction
We validated RNA-seq and PVG PCR methods to detect known respiratory pathogens by testing specimens using both methods from children with CAP (n = 63) and asymptomatic control subjects (n = 52) in whom viral or atypical bacterial pathogens had been detected using the EPIC study protocol. In children with pneumonia, RNA-seq detected 90% of pathogens detected by the EPIC study tests, and PVG PCR detected 57% (78% when excluding HRV and M. pneumoniae) (Table 2). In control subjects, the proportion detected was 64% by RNA-seq and 22% by PVG PCR (38% when excluding HRV) ( Table 2). Combining results of both methods, 93% of known pathogens in patients and 73% of known pathogens in control subjects were detected. Table 2 shows sensitivity and specificity for detection of each of the known pathogens. Calculations were based only on the tested samples. Results could be different when calculated for the entire EPIC study. A trend toward lower sensitivity in control subjects may be related to lower viral loads in asymptomatic children [29–34]. Of note, sensitivity was lowest for adenovirus for both methods; RNA-seq did not detect any of the 8 ADV-positive samples (n = 3 patients; n = 5 control subjects), whereas PVG PCR detected ADV in 1 of 3 patients and 3 of 5 control subjects. RNA sequencing detected HRV in 46 (92%) of 50 PCR-positive specimens, whereas PVG PCR detected 7 (14%) of 50. All M. pneumoniae infections were detected by RNA-seq but not targeted by PVG PCR.
Table 2.
Performance of RNA Sequencing and Pan-Viral Group Polymerase Chain Reaction Compared with Pathogen-Specific Real-Time Polymerase Chain Reaction Performed Per Etiology of Pneumonia in the Community Protocol
| Positive in this study
|
RNA-seq
|
PVG PCR
|
RNA-seq or PVG PCR |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EPIC positive | RNA-seq | PVG PCR | RNA-seq or PVG PCR |
Sens | Spec | Sens | Spec | Sens | Spec | |
| Patients (n = 63) | ||||||||||
| IAV | 0 | 0 | 0 | 0 | NA | 100% | NA | 100% | NA | NA |
| IBV | 1 | 1 | 0 | 1 | 100% | 100% | 0% | 100% | 100% | 100% |
| HMPV | 8 | 7 | 7 | 8 | 88% | 100% | 88% | 100% | 100% | 100% |
| HRVa | 12 | 12 | 2a | 12 | 100% | 100% | 17% | 100% | 100% | 100% |
| RSV | 24 | 23 | 21 | 23 | 96% | 100% | 88% | 100% | 96% | 100% |
| HPIV | 3 | 2 | 1 | 2 | 67% | 98% | 33% | 98% | 67% | 98% |
| ADV | 3 | 0 | 1 | 1 | 0% | 100% | 33% | 100% | 33% | 100% |
| HCoV | 2 | 2 | 2 | 2 | 100% | 100% | 100% | 100% | 100% | 100% |
| M. pneumoniae | 7 | 7 | ND | 7 | 100% | 98% | ND | ND | 100% | 98% |
|
| ||||||||||
| Control subjects (n = 52) | ||||||||||
| IAV | 1 | 0 | 0 | 0 | 0% | 100% | 0% | 100% | 0% | 100% |
| IBV | 0 | 0 | 0 | 0 | NA | 100% | NA | 100% | NA | 100% |
| HMPV | 4 | 0 | 0 | 0 | 0% | 100% | 0% | 100% | 0% | 100% |
| HRVa | 38 | 34 | 5a | 34 | 89% | 96% | 13% | 100% | 89% | 96% |
| RSV | 5 | 0 | 1 | 1 | 0% | 100% | 20% | 100% | 20% | 100% |
| HPIV | 3 | 3 | 2 | 3 | 100% | 98% | 67% | 100% | 100% | 98% |
| ADV | 5 | 0 | 3 | 3 | 0% | 100% | 60% | 100% | 60% | 100% |
| HCoV | 3 | 1 | 2 | 2 | 33% | 100% | 67% | 100% | 67% | 100% |
| M. pneumoniae | 0 | 0 | ND | 0 | NA | 100% | ND | ND | NA | 100% |
Abbreviations: ADV, adenovirus; EPIC, Etiology of Pneumonia in the Community; HCoV, human coronavirus; HMPV, human metapneumovirus; HPIV, human parainfluenza viruses; HRV, human rhinovirus; IAV, influenza A virus; IBV, influenza B virus; M. pneumoniae, Mycoplasma pneumoniae; ND, not done; PVG PCR, pan-viral group polymerase chain reaction; RSV, respiratory syncytial virus; Sens, sensitivity; Spec, specificity.
The Picornaviridae polymerase chain reaction targets only a subset of human rhinoviruses.
Detection of Additional, Previously Unrecognized Respiratory Pathogens in Validation Samples by RNA Sequencing and Pan-Viral Group Polymerase Chain Reaction
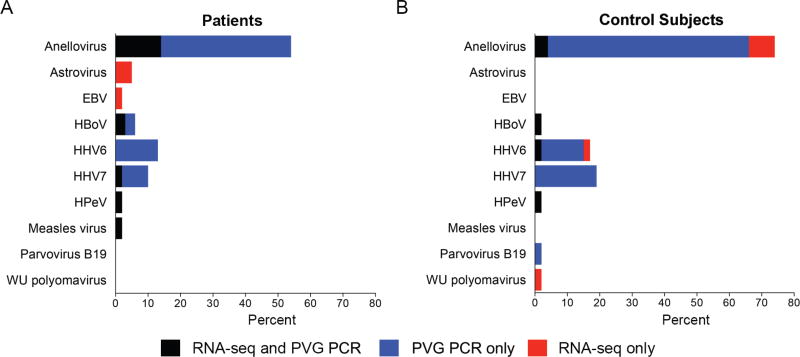
From these specimens in which pathogens were detected by PCR, RNA-seq and PVG PCR detected 58 previously unrecognized viruses in 63 children with CAP and 61 previously unrecognized viruses in 52 asymptomatic control subjects (Figure 1). Anelloviruses were the most commonly detected viruses (n = 34 patients; n = 38 control subjects), followed by human herpesvirus 6 (HHV6; n = 8 patients; n = 9 control subjects), and human herpesvirus 7 (HHV7; n = 6 patients; n = 10 control subjects). Other viruses of interest were human bocavirus (HBoV; n = 4 patients; n = 1 control subject), astrovirus (n = 3 patients; n = 0 control subjects), human parechovirus (n = 1 patient; n = 1 control subject), and measles virus (n = 1 patient).
Figure 1.
Detection of additional human viruses by RNA sequencing (RNA-seq) and pan-viral group polymerase chain reaction (PVG PCR) in children with community-acquired pneumonia (n = 63) and control subjects (n = 52) with positive pathogen-specific tests using the Etiology of Pneumonia in the Community (EPIC) study protocol. Human viruses detected by RNA-seq and PVG PCR that were not targeted in EPIC included human parechovirus, human bocavirus, Ebstein-Barr virus, human herpesvirus 6, and human herpesvirus 7. Abbreviatons: EBV, Epstein-Barr virus; HBoV, human bocavirus; HHV6, human herpesvirus 6; HHV7, human herpesvirus 7; HPeV, human parechovirus; PVG PCR, pan-viral group polymerase chain reaction; RNA-seq, RNA sequencing.
Pathogen Detection in Previously Test-Negative Specimens by RNA Sequencing and Pan-Viral Group Polymerase Chain Reaction
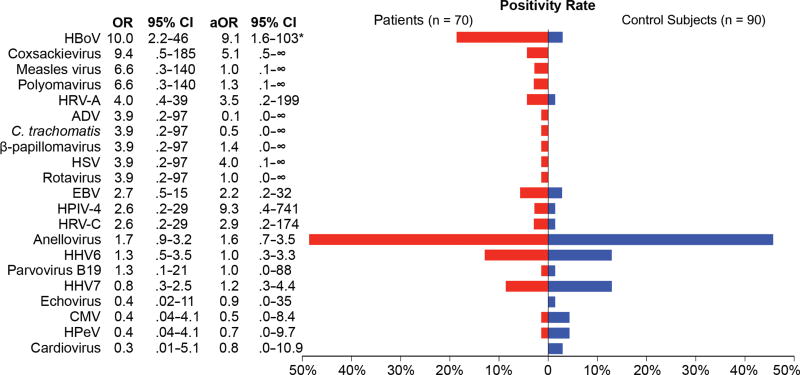
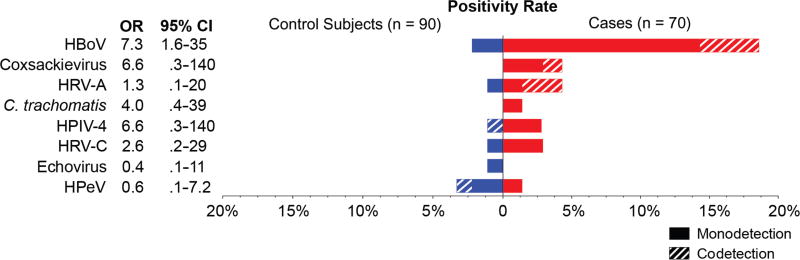
Human viruses were detected in 53 of 70 (76%) children with pneumonia and 55 of 90 (61%) control subjects (Figure 2, Table 3) who were test-negative using the EPIC study protocol. In patients compared with control subjects, the most commonly detected viruses were anelloviruses (49% vs 36%; aOR = 1.6; 95% CI = .7–3.5), HHV6 (13% vs 10%; aOR = 1.0; 95% CI = .3–3.3), and HHV7 (9% vs 10%; aOR = 1.2; 95% CI = .3–4.4). Human bocavirus was detected in a significantly greater proportion of patients (18.6%) than control subjects (2.2%; aOR = 9; 95% CI = 1.6–102.9; P < .01). Coxsackieviruses were detected in 3 patients (4.3%) and no control subjects, but this association was not statistically significant (aOR = 5.1; 95% CI = 0.5–∞; P = .09). Other potential pathogens were more commonly detected in patients than control subjects, but these differences were not statistically significant: HRV-A (aOR = 3.5; 95% CI = .2–199; P = .34), and HPIV-4 (aOR = 9.3; 95% CI = .4–741; P = .12). Additional viruses detected included Epstein-Barr virus (n = 4 patients; n = 2 control subjects), measles virus (n = 2 patients), polyomaviruses (n = 2 patients), HPV type 5 (n = 1 patient), herpes simplex virus (n = 1 patient), rotavirus (n = 1 patient), parvovirus B19 (n = 1 patient; n = 1 control subject), and echovirus (n = 1 control subject). Cytomegalovirus (CMV; n = 1 patient; n = 3 control subjects), human parechovirus (n = 1 patient; n = 3 control subjects), and cardioviruses (n = 2 control subjects) were more commonly detected in control subjects than in patients. Figure 3 shows the proportion of codetected putative pathogens; 24% of detections in patients and 22% of detections in control subjects were codetections. Monodetection of HBoV was significantly associated with CAP (OR = 7.3; 95% CI = 1.6–35).
Figure 2.
Viruses detected by RNA sequencing and/or pan-viral group polymerase chain reaction in children with pneumonia with no identifiable etiology (n = 70; red) and asymptomatic control subjects (n = 90; blue). A total of 20 different human viruses were detected in nasopharyngeal/oropharyngeal samples. In addition, Chlamydia trachomatis was detected in 1 newborn child with pneumonia. Fifteen viruses were more frequently detected in patients than control subjects (odds ratios >1), with human bocavirus (P < .001) having significant associations with community-acquired pneumonia. Abbreviations: ADV, adenovirus; aOR, adjusted odds ratio (adjusted for season and age group); C. trachomatis, Chlamydia trachomatis; CI, confidence interval; CMV, cytomegalovirus; HBoV, human bocavirus; HHV6, human herpesvirus 6; HHV7, human herpesvirus 7; HPeV, human parechovirus; HPIV-4, human parainfluenza virus type 4; HRV-A, human rhinovirus A; HRV-C, human rhinovirus C; HSV, herpes simplex virus; OR, odds ratio.
Table 3.
Putative Pathogens Detected in Children with Community-Acquired Pneumonia of Unknown Etiology by RNA Sequencing or Pan-Viral Group Polymerase Chain Reaction
| Patients
|
||
|---|---|---|
| Putative pathogen | No. | % |
| Chlamydia trachomatis | 1 | 1.4 |
|
| ||
| Coxsackievirus A6 | 1 | 1.4 |
|
| ||
| Coxsackievirus A6 and human bocavirus | 1 | 1.4 |
|
| ||
| Coxsackievirus B3 | 1 | 1.4 |
|
| ||
| Human bocavirus | 10 | 14.3 |
|
| ||
| Human parainfluenza virus 4 | 2 | 2.9 |
|
| ||
| Human parechovirus | 1 | 1.4 |
|
| ||
| Human rhinovirus A | 1 | 1.4 |
|
| ||
| Human rhinovirus A and human bocavirus | 2 | 2.9 |
|
| ||
| Human rhinovirus C | 2 | 2.9 |
|
| ||
| None | 48 | 65.7 |
|
| ||
| Total | 70 | 100 |
A putative pathogen was detected in 24 of 70 children (34.3%).
Figure 3.
Putative pathogens by RNA sequencing and/or pan-viral group polymerase chain reaction in children with pneumonia with no identifiable etiology (n = 70; red) and asymptomatic control subjects (n = 90; blue). In 31% of detections, other putative pathogens were codetected (hashed bars), whereas no other putative pathogen was detected in the remaining samples (monodetection). Odds ratios (ORs) and 95% confidence intervals (CIs) are shown. Only monodetection of human bocavirus was significantly associated with community-acquired pneumonia. Abbreviations: HBoV, human bocavirus; HPeV, human parechovirus HPIV-4, human parainfluenza virus type 4; HRV-A, human rhinovirus A; HRV-C, human rhinovirus C.
Comparison of Viral Detection by RNA Sequencing and Pan-Viral Group Polymerase Chain Reaction
In children with CAP with no identifiable etiology, 32% of all viruses were detected by both methods, 22% by RNA-seq only and 47% by PVG PCR only (Supplementary Figure 1). In control subjects, 19% of all viruses were detected by both methods, 20% by RNA-seq only, and 61% by PVG PCR only. The vast majority of viruses only detected by PVG PCR were anelloviruses, HHV6, and HHV7, which, combined, were detected in 35 of 41 (85%) patients and 37 of 39 (95%) control subjects (Supplementary Figure 1). In patients, the remaining viruses were most commonly detected by both methods (46%) or by RNA-seq only (38%; compared with 15% by PVG PCR only). In control subjects, the remaining viruses were more frequently detected by RNA-seq (64%) than by both methods (21%) or by PVG PCR only (14%). In 2 RNA-seq–positive/PVG PCR–negative samples, the entire viral genome could be determined. Two and 1 mismatches were identified in reverse primer binding sites of these Coxsackievirus and HRV-C genomes, respectively. Failure to detect these 2 viruses by PGV PCR was more likely due to reduced sensitivity of degenerate PCR primers than primer mismatches.
Bacteria in Children With Pneumonia With No Identifiable Etiology
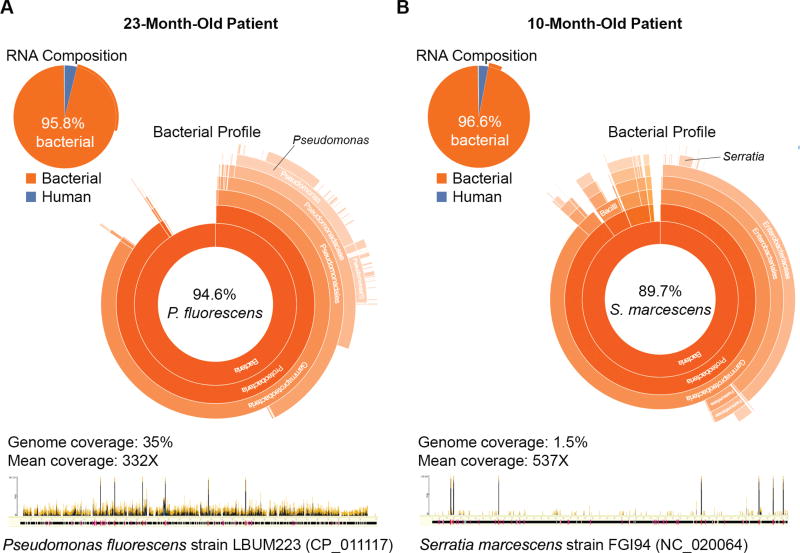
One infant with CAP and no identified pathogen by the EPIC study protocol had C. trachomatis detected by RNA-seq. In 2 patients with CAP, a potential bacterial pathogen was identified (Figure 4). Although a diverse set of bacteria consistent with upper respiratory tract flora was detected in most patients, the flora in these 2 patients was dominated by a single Gramnegative organism. In a 23-month-old patient with trisomy 21, Pseudomonas fluorescens dominated the bacterial sequences (approximately 95%, best matching strain LBUM223, accession number CP_011117). In a 10-month-old patient with spina bifida, chiari malformation, and history of aspiration pneumonia, approximately 90% of bacterial sequences originated from Serratia marcescens (best matching strain FGI94, accession number NC_020064).
Figure 4.
An abundant bacterial flora (>95% of sequencing reads) dominated by a single potential pathogen was detected by RNA sequencing in nasopharyngeal/oropharyngeal (NP/OP) samples of 2 children with community-acquired pneumonia and no identified pathogen by the Etiology of Pneumonia in the Community study protocol. A, In a 23-month-old patient, 94.6% of sequencing reads generated from the NP/OP sample was identified as Pseudomonas fluorescens, covering 35% of the genome of strain LBUM223 (NCBI accession number CP_011117) at a mean of 332X (data analyzed as described in [14]). B, In a 10-month-old patient, 89.7% of sequencing reads were derived from Serratia marcescens, covering 1.5% of the genome sequence of strain FGI94 (NCBI accession number NC_020064) at a mean of 537X.
DISCUSSION
Using RNA-seq and PVG PCR, we identified additional viruses from upper respiratory tract specimens in >30% children hospitalized with clinical and radiographic pneumonia but in whom no pathogen was identified despite extensive testing by culture, molecular, and serologic methods. Human bocavirus, Coxsackieviruses, HPIV-4, HRV-C, and HRV-A were more commonly detected in children with CAP compared with control subjects, but only HBoV was statistically more common than in control subjects. This suggests that these pathogens may have played an etiologic role in CAP. Making an etiologic diagnosis in patients with pneumonia is important for understanding the epidemiology, providing appropriate therapy, and limiting unnecessary use of antimicrobials. However, extensive testing using standard approaches is unable to identify a pathogen in approximately 20% of children and approximately 60% of adults [4, 35]. Data from our proof-of-concept study of upper respiratory specimens suggest that RNA-seq and PVG PCR enable more comprehensive pathogen detection compared with virus-specific, real-time PCR-based tests. Although specimens from the upper respiratory tract can be collected without invasive procedure, they are most useful for identifying viral infections and have limited utility in testing for bacterial pneumonia.
The detected viruses in our study can be broadly categorized into 4 groups: (1) known respiratory pathogens, (2) viruses of unclear pathogenicity, (3) opportunistic viruses that are pathogenic in immunocompromised hosts, and (4) viruses not thought to play a pathogenic role in respiratory tract illness. Among known respiratory pathogens, we detected Coxsackievirus, HRV, ADV, HPIV, human parechovirus, and measles virus, which, combined, were detected in 33% of children with CAP. Coxsackievirus, HRV, ADV, HPIV, and measles virus were all more commonly detected in patients than control subjects (aOR > 2), but possibly due to the overall low frequency of these detections, these differences did not reach statistical significance. Human parechovirus and echovirus can cause respiratory tract infections but were detected infrequently in both patients and control subjects (aOR = 1.3 and 0.4, respectively). The 2 patients with measles virus detection did not show signs of measles but had been vaccinated 7 days (positive by RNA-seq only) and 9 days (positive by RNA-seq and PVG PCR) before sample collection, suggesting we detected vaccine strain. However, the low number of sequencing reads precluded demonstrating this by examining the complete viral genome. Cardioviruses, which are a possible cause of respiratory tract infections [36], were only detected in control subjects and not patients.
Human bocavirus was the most commonly detected virus among children with CAP and no identified pathogen (n = 13/70; 19%). Human bocavirus detection was strongly associated with CAP (aOR = 9.1; 95% CI = 1.6–103) (Figure 2). Three of these infections (23%) were codetections with other putative viral pathogens, and monodetection of HBoV was significantly associated with CAP (OR = 7.3; 95% CI = 1.6–35). Human bocavirus was not targeted as part of the EPIC study protocol due to uncertainty over its role as a human pathogen [37, 38]. Human bocavirus is a Parvovirus with a DNA genome and can be detected for weeks to months following acute infections, which makes it difficult to demonstrate its pathogenicity even in well-designed epidemiologic studies. Pan-viral group PCR detected HBoV DNA in 12 of 70 patients (17.1%) and 2 of 90 asymptomatic control subjects (2.2%). RNA sequencing identified HBoV mRNA in 10 of 70 patients (14.3%) and 0 of 90 asymptomatic control subjects (aOR = 31.4; 95% CI = 1.8– 546; P < .05). Sequencing reads spanning splice sites of the viral capsid mRNA [39] confirmed that mRNA rather than genomic DNA served as the sequencing template (data not shown). This strong association is in contrast with numerous PCR-based studies targeting viral genomic DNA [37, 40], suggesting that detection of HBoV mRNA may serve as a marker for acute (ie, clinically relevant) infections. Although these results will need to be confirmed in larger studies, our results suggest that HBoV may be associated with CAP and may be a true pathogen.
Human herpesviruses that can cause respiratory tract infections including pneumonia in immunocompromised hosts (eg, HSV, CMV, parvovirus B19, HHV6) were more frequently detected in patients than control subjects in our analysis. However, children with severely immune-compromising conditions were excluded from the EPIC study. These viruses were more frequently detected by PVG PCR (targeting viral genomic DNA) than by RNA-seq. In the absence of detectable RNA, active replication is unlikely, and their detection may be a result of reactivation or latent infection rather than acute infection. Lastly, we detected a number of viruses not known to cause respiratory tract infections, including EBV, anelloviruses, HHV7, polyomaviruses, and papillomavirus. Their detection in the nasopharynx and/or oropharynx of asymptomatic children as well as CAP patients (in validation and test-negative groups) is consistent with previous reports [41–47]. Their detection demonstrates both the power of comprehensive pathogen detection but also the importance of using appropriate controls. Interestingly, detection rates for these DNA viruses were much higher by DNA-based PVG PCR than by RNA-seq. It is possible that RNA-based testing may be more sensitive for DNA viruses during high-level replication when mRNA is abundant.
Although both RNA-seq and PVG PCR provide broad-range detection of respiratory viruses, each method has potential advantages and disadvantages. RNA sequencing is highly unbiased, demonstrated by the detection of divergent enteroviruses not identified by PVG PCR, and enables identification of nonviral pathogens, as exemplified by detection of M. pneumoniae and C. trachomatis. In contrast, PVG PCR identified DNA viruses that were not detected by RNA-seq. This may have been due in part to shedding predominantly of viral particles (containing genomic DNA) with low levels of active replication (ie, production of mRNA) in the upper respiratory tract. Performing next-generation sequencing with both RNA-seq and DNA sequencing might increase the yield for DNA viruses and bacteria but at increased financial cost.
As hypothesized, broad-range pathogen detection enabled identification of viruses not part of comprehensive test panels (eg, HBoV, Coxsackievirus, HPIV-4, Echovirus, human parechovirus), genetically divergent strains escaping PCR-based detection (as can be seen with genetically diverse viruses; eg, HRV-A, HRV-C), and unrecognized bacterial infections (eg, C. trachomatis) [15]. In addition to the sequence data analysis described above, we also performed de novo assembly of RNA-seq results and searched resulting contiguous sequences for conserved protein profiles [48] on all data from children with CAP without identifying additional putative pathogens (data not shown). Despite these extensive efforts, a potential pathogen was still not detected in 46 children (65.7%) with CAP of unknown etiology. This could have been due to testing of NP/OP swabs and not lower respiratory tract samples, which are preferred for detection of bacterial and fungal pathogens; focus on viral pathogens; inadequate timing of sample collection; polymicrobial infections caused by bacterial or fungal pathogens; or noninfectious mimics. Use of broad-range methods may provide even greater benefits in the 60% of adults with CAP in whom no pathogen is detected using conventional approaches [35]. Our findings also highlight the limits of etiologic diagnosis of CAP with noninvasive samples. We cannot exclude that highly diverse viruses without homology to known human viral pathogens may have caused CAP in some of the children. Further advancing the diagnosis of CAP is likely to require additional sampling as well as host-based markers of infectious processes that may help confirm infectious etiologies even when a pathogen cannot be directly detected [49].
Supplementary Material
Acknowledgments
We thank all of the patients who enrolled in the EPIC study, their parents, and families.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the National Institutes of Health.
Financial support. This work was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health (award no. 1KL2TR001065 and UL1TR001067); the Primary Children’s Hospital Foundation; the ARUP Institute for Clinical and Experimental Pathology; and the Centers for Disease Control and Prevention (award no. U181P00030).
S. F., K. S., C. M., Y. Q., K. E., G. M., M. Y., and R. S. have a patent application pending for Taxonomer, which was licensed by IDbyDNA. M. Y. and R. S. own equity in and consult for IDbyDNA.
Footnotes
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Potential Conflicts Of Interest. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–40. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 2.Lee GE, Lorch SA, Sheffler-Collins S, Kronman MP, Shah SS. National hospital-ization trends for pediatric pneumonia and associated complications. Pediatrics. 2010;126:204–13. doi: 10.1542/peds.2009-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley JS, Byington CL, Shah SS, et al. Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis. 2011;53:e25–76. doi: 10.1093/cid/cir531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain S, Williams DJ, Arnold SR, et al. CDC EPIC Study Team. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372:835–45. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michelow IC, Olsen K, Lozano J, et al. Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics. 2004;113:701–7. doi: 10.1542/peds.113.4.701. [DOI] [PubMed] [Google Scholar]
- 6.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet. 2011;377:1264–75. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huguenin A, Moutte L, Renois F, et al. Broad respiratory virus detection in infants hospitalized for bronchiolitis by use of a multiplex RT-PCR DNA microarray system. J Med Virol. 2012;84:979–85. doi: 10.1002/jmv.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juvén T, Mertsola J, Waris M, et al. Etiology of community-acquired pneumonia in 254 hospitalized children. Pediatr Infect Dis J. 2000;19:293–8. doi: 10.1097/00006454-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Harris M, Clark J, Coote N, et al. British Thoracic Society guidelines for the management of community acquired pneumonia in children: update 2011. Thorax. 2011;66(suppl 2) doi: 10.1136/thoraxjnl-2011-200598. ii1-23. [DOI] [PubMed] [Google Scholar]
- 10.Wubbel L, Muniz L, Ahmed A, et al. Etiology and treatment of community-acquired pneumonia in ambulatory children. Pediatr Infect Dis J. 1999;18:98–104. doi: 10.1097/00006454-199902000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Pavia AT. What is the role of respiratory viruses in community-acquired pneumonia?: what is the best therapy for influenza and other viral causes of community-acquired pneumonia? Infect Dis Clin North Am. 2013;27:157–75. doi: 10.1016/j.idc.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipkin WI. The changing face of pathogen discovery and surveillance. Nat Rev Microbiol. 2013;11:133–41. doi: 10.1038/nrmicro2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu CY. Viral pathogen discovery. Curr Opin Microbiol. 2013;16:468–78. doi: 10.1016/j.mib.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flygare S, Simmon K, Miller C, et al. Taxonomer: an interactive metagenomics analysis portal for universal pathogen detection and host mRNA expression profiling. Genome Biol. 2016;17:111. doi: 10.1186/s13059-016-0969-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf EH, Simmon KE, Tardif KD, et al. Unbiased detection of respiratory viruses by use of RNA sequencing-based metagenomics: a systematic comparison to a commercial PCR panel. J Clin Microbiol. 2016;54:1000–7. doi: 10.1128/JCM.03060-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson LJ, Tong S. Identification and characterization of novel viruses. In: Novartis Foundation, editor. Novel and re-emerging respiratory viral disease. Vol. 290. Hoboken, NJ: Wiley; 2008. http://www.wiley.com/WileyCDA/WileyTitle/productCd-0470065389.html. [Google Scholar]
- 17.National Center for Biotechnology Information. Information NCfB. [Accessed 28 July 2015];Nucleotide Database. https://www.ncbi.nlm.nih.gov/nucleotide/
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Rose TM. CODEHOP-mediated PCR—a powerful technique for the identification and characterization of viral genomes. Virol J. 2005;2:20. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate hybrid oligonucleotide primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–6. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conrardy C, Tao Y, Kuzmin IV, et al. Molecular detection of adenoviruses, rhabdoviruses, and paramyxoviruses in bats from Kenya. Am J Trop Med Hyg. 2014;91:258–66. doi: 10.4269/ajtmh.13-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkbeiner SR, Li Y, Ruone S, et al. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol. 2009;83:10836–9. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phaneuf CR, Oh K, Pak N, et al. Sensitive, microliter PCR with consensus degenerate primers for Epstein Barr virus amplification. Biomed Microdevices. 2013;15:221–31. doi: 10.1007/s10544-012-9720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schatzberg SJ, Li Q, Porter BF, et al. Broadly reactive pan-paramyxovirus reverse transcription polymerase chain reaction and sequence analysis for the detection of canine distemper virus in a case of canine meningoencephalitis of unknown etiology. J Vet Diagn Invest. 2009;21:844–9. doi: 10.1177/104063870902100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao Y, Shi M, Conrardy C, et al. Discovery of diverse polyomaviruses in bats and the evolutionary history of the Polyomaviridae. J Gen Virol. 2013;94:738–48. doi: 10.1099/vir.0.047928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–8. doi: 10.1128/JCM.00192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong S, Conrardy C, Ruone S, et al. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg Infect Dis. 2009;15:482–5. doi: 10.3201/eid1503.081013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong S, Singh J, Ruone S, et al. Identification of adenoviruses in fecal specimens from wild chimpanzees (Pan trogylodytes schweinfurthii) in western Tanzania. Am J Trop Med Hyg. 2010;82:967–70. doi: 10.4269/ajtmh.2010.09-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan XL, Li YN, Tang YJ, et al. Clinical characteristics and viral load of respiratory syncytial virus and human metapneumovirus in children hospitaled for acute lower respiratory tract infection. J Med Virol. 2017;89:589–97. doi: 10.1002/jmv.24687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee N, Chan MC, Lui GC, et al. High viral load and respiratory failure in adults hospitalized for respiratory syncytial virus infections. J Infect Dis. 2015;212:1237–40. doi: 10.1093/infdis/jiv248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esposito S, Daleno C, Scala A, et al. Impact of rhinovirus nasopharyngeal viral load and viremia on severity of respiratory infections in children. Eur J Clin Microbiol Infect Dis. 2014;33:41–8. doi: 10.1007/s10096-013-1926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roussy JF, Carbonneau J, Ouakki M, et al. Human metapneumovirus viral load is an important risk factor for disease severity in young children. J Clin Virol. 2014;60:133–40. doi: 10.1016/j.jcv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 33.DeVincenzo JP, Wilkinson T, Vaishnaw A, et al. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am J Respir Crit Care Med. 2010;182:1305–14. doi: 10.1164/rccm.201002-0221OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houben ML, Coenjaerts FE, Rossen JW, et al. Disease severity and viral load are correlated in infants with primary respiratory syncytial virus infection in the community. J Med Virol. 2010;82:1266–71. doi: 10.1002/jmv.21771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain S, Self WH, Wunderink RG, et al. CDC EPIC Study Team. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373:415–27. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin TL, Lin TH, Chiu SC, et al. Molecular epidemiological analysis of Saffold car-diovirus genotype 3 from upper respiratory infection patients in Taiwan. J Clin Virol. 2015;70:7–13. doi: 10.1016/j.jcv.2015.06.100. [DOI] [PubMed] [Google Scholar]
- 37.Williams JV. Déjà vu all over again: Koch’s postulates and virology in the 21st century. J Infect Dis. 2010;201:1611–4. doi: 10.1086/652406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schildgen O, Müller A, Allander T, et al. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin Microbiol Rev. 2008;21:291–304. doi: 10.1128/CMR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen AY, Cheng F, Lou S, et al. Characterization of the gene expression profile of human bocavirus. Virology. 2010;403:145–54. doi: 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellett PE. Indictment by association: once is not enough. J Infect Dis. 2015;212:509–12. doi: 10.1093/infdis/jiv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berger C, Hug M, Gysin C, et al. Distribution patterns of beta- and gamma-her-pesviruses within Waldeyer’s ring organs. J Med Virol. 2007;79:1147–52. doi: 10.1002/jmv.20899. [DOI] [PubMed] [Google Scholar]
- 42.Burián Z, Szabó H, Székely G, et al. Detection and follow-up of torque teno midi virus (“small anelloviruses”) in nasopharyngeal aspirates and three other human body fluids in children. Arch Virol. 2011;156:1537–41. doi: 10.1007/s00705-011-1021-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Zhu N, Li Y, et al. Metagenomic analysis of vvviral genetic diversity in respiratory samples from children with severe acute respiratory infection in China. Clin Microbiol Infect. 2016;22:458. doi: 10.1016/j.cmi.2016.01.006. e451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunes MC, Kuschner Z, Rabede Z, et al. Polyomaviruses-associated respiratory infections in HIV-infected and HIV-uninfected children. J Clin Virol. 2014;61:571–8. doi: 10.1016/j.jcv.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song X, Van Ghelue M, Ludvigsen M, Nordbø SA, Ehlers B, Moens U. Characterization of the non-coding control region of polyomavirus KI isolated from nasopharyngeal samples from patients with respiratory symptoms or infection and from blood from healthy blood donors in Norway. J Gen Virol. 2016;97:1647–57. doi: 10.1099/jgv.0.000473. [DOI] [PubMed] [Google Scholar]
- 46.Knör M, Tziridis K, Agaimy A, Zenk J, Wendler O. Human papillomavirus (HPV) prevalence in nasal and antrochoanal polyps and association with clinical data. PLoS One. 2015;10:e0141722. doi: 10.1371/journal.pone.0141722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue XC, Chen XP, Yao WH, Zhang Y, Sun GB, Tan XJ. Prevalence of human pap-illomavirus and Epstein-Barr virus DNA in Chinese children with tonsillar and/or adenoidal hypertrophy. J Med Virol. 2014;86:963–7. doi: 10.1002/jmv.23894. [DOI] [PubMed] [Google Scholar]
- 48.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsalik EL, Henao R, Nichols M, et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med. 2016;8:322ra311. doi: 10.1126/scitranslmed.aad6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.