Abstract
To investigate cellular, molecular and behavioral mechanisms of noxious cold detection, we developed cold plate behavioral assays and quantitative means for evaluating the predominant noxious cold-evoked contraction behavior. To characterize neural activity in response to noxious cold, we implemented a GCaMP6-based calcium imaging assay enabling in vivo studies of intracellular calcium dynamics in intact Drosophila larvae. We identified Drosophila class III multidendritic (md) sensory neurons as multimodal sensors of innocuous mechanical and noxious cold stimuli and to dissect the mechanistic bases of multimodal sensory processing we developed two independent functional assays. First, we developed an optogenetic dose response assay to assess whether levels of neural activation contributes to the multimodal aspects of cold sensitive sensory neurons. Second, we utilized CaMPARI, a photo-switchable calcium integrator that stably converts fluorescence from green to red in presence of high intracellular calcium and photo-converting light, to assess in vivo functional differences in neural activation levels between innocuous mechanical and noxious cold stimuli. These novel assays enable investigations of behavioral and functional roles of peripheral sensory neurons and multimodal sensory processing in Drosophila larvae.
Keywords: Nociception, Noxious cold, Multimodal sensory processing, Calcium imaging, Optogenetics, Drosophila
Background
The capacity to sense and respond appropriately to environmental cues is one of the most fundamental aspects shared among the metazoans. Sensing potentially harmful stimuli, such as noxious temperature, chemical or mechanical insults, and responding appropriately is crucial for avoiding incipient damage that can lead to injury or death. Typically, upon sensing nociceptive stimuli an animal produces a set of avoidance behaviors that either mitigate or allow the animal to escape the noxious stimulus. Elucidating molecular, cellular, and behavioral level mechanisms in processing nociceptive stimuli is of great interest as there is potential for the identification and development of novel therapeutic interventions for aberrant sensory processing, which can lead to neuropathic pain. Sensory and behavioral responses to noxious chemical, mechanical and heat stimuli have been elucidated in Drosophila melanogaster larvae and adults, however, noxious cold detection has only recently been discovered in larvae (Im and Galko, 2012; Gorczyca et al., 2014; Guo et al., 2014; Mauthner et al., 2014; Turner et al., 2016). Drosophila larvae exhibit a distinct set of aversive behaviors in responses to noxious cold stimuli with the predominant cold-evoked response displaying as a bilateral anterior-posterior full body contraction (CT) (Turner et al., 2016). This behavioral response is mediated by class III md sensory neurons (Turner et al., 2016), which intriguingly have also been implicated in gentle touch mechanosensation revealing multimodality in these neurons (Tsubouchi et al., 2012; Yan et al., 2013). The Transient Receptor Potential (TRP) channels Pkd2, NompC, and Trpm are required for mediating noxious cold-evoked behavior and behavioral selection in response to innocuous mechanical vs. noxious cold stimuli is dependent upon class III neural activation levels providing insight into the mechanisms underlying cold nociception and multimodal sensory processing (Turner et al., 2016).
Materials and Reagents
Kimwipe (KCWW, Kimberly-Clark, catalog number: 34155)
25 x 75 mm microscope slide (Globe Scientific, catalog number: 1301)
22 x 22 mm No.1 thickness coverslip (Globe Scientific, catalog number: 1401-10)
24 x 50 mm No. 1 thickness coverslip (Genesee Scientific, catalog number: 29-118)
-
Pyrex 9 well glass spot plates (Fisher Scientific, catalog number: 13-748B)
Manufacturer: Corning, PYREX®, catalog number: 7220-85.
Amber glass dropper bottles (Fisher Scientific, Fisherbrand™, catalog number: 02-983B)
-
Bel-Art™ SP Scienceware™ wide mouth color-code safety labeled wash bottles (Fisher Scientific, catalog number: 22-288654)
Manufacturer: SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F11646-3739.
-
Bel-Art™ SP Scienceware Trigger Sprayers with 53 mm adapters (Fisher Scientific, catalog number: 01-189-100)
Manufacturer: SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F11620-0050.
Polypropylene vials (Genesee Scientific, catalog number: 32-120)
Droso-Plugs, Narrow vials (Genesee Scientific, catalog number: 59-200)
-
Drosophila stocks:
ChETA: y1 w*; wgSp-1/CyO, P{Wee-P.ph0}BaccWee-P20; P{20XUAS-CHETA.YFP}attP2/™6C, Sb1 Tb1 (Bloomington Drosophila Stock Center, catalog number: 36495)
CaMPARI: w*; P{UAS-CaMPARI}attP40 (Bloomington Drosophila Stock Center, catalog number: 58761)
GCaMP6 (medium variant): w1118; PBac{20XUAS-IVS-GCaMP6m}VK00005 (Bloomington Drosophila Stock Center, catalog number: 42750)
Class III md neuron driver: GAL419-12 and GAL4nompC (Bloomington Drosophila Stock Center, catalog numbers: 36369 and 36361) (Turner et al., 2016)
Class IV md neuron driver: GAL4pp1.9 and GAL4477 (Turner et al., 2016)
Control strain: w1118 (Bloomington Drosophila Stock Center, catalog number: 3605)
All trans-Retinal (ATR) (Sigma-Aldrich, catalog number: R2500)
Halocarbon oil #700 (LabScientific, catalog number: FLY-7000)
Ethyl ether anhydrous (Fisher Scientific, catalog number: E138-500)
NutriSoy, Soy Flour (Genesee Scientific, catalog number: 62-115)
Yellow cornmeal (Genesee Scientific, catalog number: 62-101)
Drosophila agar type II (Genesee Scientific, catalog number: 66-104)
Inactive dry yeast (Genesee Scientific, catalog number: 62-107)
Dry molasses (Genesee Scientific, catalog number: 62-119)
O-phosphoric acid (Fisher Scientific, catalog number: A242-212)
Propionic acid (Fisher Scientific, catalog number: A258-500)
Drosophila media (see Recipes)
Equipment
A. Cold plate assay
Brush (Craft Smart® round brush set golden taklon) (Michaels Stores, model: Size 3, catalog number: 10408282)
Nikon body plus lens combination (Nikon, model: D5300) and AF-S Nikkor 18–55 mm DX VRII (Nikon, model: AF-S DX)
Tripod for mounting DSLR
Cold plate cooler (TE Technology, model: CP-031)
Cold plate temperature controller (TE Technology, model: TC-720)
Cold plate power supply (TE Technology, model: PS-12-8.4A)
Infrared thermometer (Fluke, model: Fluke 62 MAX)
-
Aluminum plate–Laminated aluminum shim (Global Equipment, catalog number: WBB512969)
Notes:
Cut the shim to 7.5 by 11.5 mm at 0.22 mm thickness.
Paint the plate with black spray paint for high contrast.
Black spray paint–12 oz. black flat general purpose spray paint (Rust-Oleum)
B. In vivo GCaMP assay
PE120 Peltier stage (Linkam Scientific Instruments, Linkam Scientific, model: PE120)
T95 system controller (including T95 linkpad and PE95) from (Linkam Scientific Instruments, Linkam Scientific, model: T95)
Laser confocal microscope capable of imaging GFP (Carl Zeiss, model: LSM 780)
C. Optogenetic dose response assay
D5300 DSLR (Nikon, model: D5300)
Adapters for mounting Nikon DSLR onto Zeiss microscopes: T2-adapter for Nikon F (Carl Zeiss, model: T2-adapter, catalog number: 416009-0000-000) and Adapter 60N–T2 1.0x (Carl Zeiss, model: Adapter 60N, catalog number: 426103-0000-000)
Glass plate: 10 x 15 x 0.1 cm
D. CaMPARI Ca2+ integrator assay
-
Photo-conversion filter cube
612/69 BrightLine bandpass filter, 25 mm (IDEX Health & Science, Semrock, catalog number: FF01-612/69-25)
440 nm BrightLine SWP edge filter, 25 mm (IDEX Health & Science, Semrock, catalog number: FF01-440/SP-25)
562 BrightLine dichroic beamsplitter, 25.2 x 35.6 mm (IDEX Health & Science, Semrock, catalog number: FF562-Di03-25x36)
Axio Zoom.V16 (ZEISS, model: Axio Zoom.V16) with Illuminator HXP200c lamp (Carl Zeiss, model: HXP 200C)
Light touch stimulus: Nickel plated pin holder (Fine Science Tools, catalog number: 26018-17) with mounted single fine paint brush bristle
Noxious cold stimulus: TE technology cold plate cooler described in cold plate assay
E. Drosophila media preparation
Adventurer Pro II Analytical/Precision Balance (Ohaus, model: AX2202)
FastPette V2 Pipette Controller (Labnet International, catalog number: P2000)
10ml Serological Pipets (Genesee Scientific, catalog number: 12-104)
Droso-Filler, Narrow (Genesee Scientific, catalog number: 59-168)
Avantco Induction Range (Avantco Equipment, model: IC3500)
Stainless Stock Pot with lid (Thunder Group, catalog number: SLSPS020)
Narrow Fly Vial Reload Tray (Genesee Scientific, catalog number: 59-207)
Software
ImageJ (https://imagej.nih.gov/ij/)
Video to video converter (http://www.videotovideo.org/)
Zeiss Zen Blue Lite (https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html)
Procedure
A. Cold plate assay
Note: The following procedure describes how noxious cold stimulus is delivered to the ventral surface of D. melanogaster larvae. Larvae are placed on a black metal plate, to enable high contrast imaging of larvae, allowed to resume locomotion and then exposed to noxious thermal stimulus by placing the black metal plate on a chilled Peltier plate. The benefit of using this assay to assess noxious cold evoked behavioral responses is that this assay exposes the entire ventral surface of the animal to noxious cold stimulus and the behavior can be quantitatively assessed (see Data analysis section).
-
Cold plate assay
Prepare genetic cross of interest with approximately 30–40 virgins and 20–25 males. Place the vial in 29 °C. Allow adult flies to mate for two days and then transfer to a new vial to perform a timed egg collection for 4–6 h at 29 °C. After 4–6 h remove adult flies and age the vial to 96–102 h after egg lay (AEL) corresponding to the third instar larval stage of development.
Behavioral assays should be performed in a diffusely lit room.
-
At 96–102 h AEL, turn on the Peltier cold plate and set to desired temperature (6 °C). Allow few minutes for the plate to reach desired temperature and critically, make sure that the plate is able to hold desired temperature as measured with an infrared thermometer.
Note: At 6 °C make sure to wipe off excess condensation.
Turn on recording equipment, Nikon DSLR and lens set to 55 mm placed roughly 18 cm above the focal plane of the plate.
Select 6–9 third instar larvae (Figure 1A) at a time with a brush for cold plate assay and rinse them gently with tap water in a 9-well glass spot plate.
Quickly, transfer them to a wet Kimwipe.
-
Apply a thin fine mist of water via a trigger sprayer on a black metal plate.
Note: Too much water on the plate will create noise during the ImageJ/Fiji quantification steps.
-
Using a brush gently pick up each larva and place them on the metal plate. After all the larvae have been placed on the plate, allow roughly 30 sec for the larvae to acclimate and resume peristaltic movement on the plate.
Note: Make sure there are no residual food particles on larvae, excess water has not accumulated around the larvae, and there is enough space between larvae (Figure 1B).
Place the metal plate on the Peltier plate very gently so that light touch stimuli do not confound noxious thermal assay. Record the animals for up to 30 sec at 30 frames per second (Figure 1B and Video 1).
Repeat until desired number of larvae are recorded.
Representative image stills (Figures 1C and 1D) and videos of baseline larval behavior at 25 °C (Video 2) and in response to noxious cold stimulation (6 °C) (Video 3). At baseline temperature (25 °C) larvae exhibit peristaltic locomotion and head turning behavior consistent with surveying their environment, whereas upon noxious cold stimulation (6 °C), larvae exhibit bilateral anterior-posterior contraction (CT) behavior.
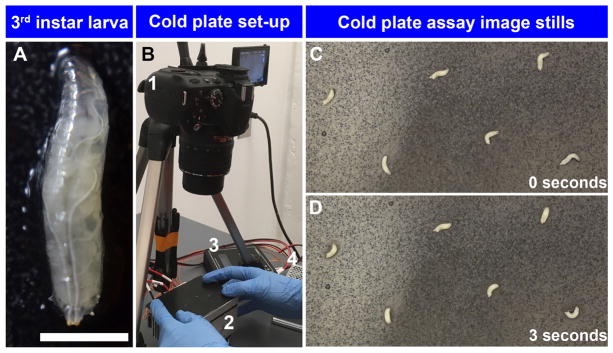
Figure 1. Set up of cold plate assay.
A. High magnification image of Drosophila third instar larva (scale bar represents 1.7 mm); B. 1. Nikon DSLR camera mounted on tripod, 2. TE Technology Peltier plate, 3. TE Technology temperature controller, and 4. TE Technology power supply. C. Cold plate assay of w1118 third instar larvae exposed 6 °C at 0 sec; D. Cold plate assay of w1118 third instar larvae exposed to 6 °C at 3 sec. Larvae exhibit contraction (CT) behavior in response to noxious cold stimulation.
B. In vivo calcium imaging via GCaMP
Note: We designed an experimental paradigm for imaging transient calcium responses in live intact D. melanogaster larvae using modern confocal imaging and fluorescence technology. These novel in vivo analyses are facilitated by investigations of larval md sensory neurons that are located just beneath a semi-transparent cuticle. We use larval md neuron class specific GAL4 drivers to direct the expression of GCaMP6, a transient fluorescent intracellular Ca2+ sensor. To assess in vivo GCaMP responses to noxious cold exposure we delivered stimuli via a Linkam Peltier system. Using this method one can assess GCaMP calcium responses in any PNS neuron in a live animal preparation.
-
Prepare before setting up genetic cross or stock for calcium imaging.
-
Prepare an amber glass dropper bottle containing halocarbon oil #700. This will be used for microscope slide preparation for in vivo imaging.
Note: Water will also work, but halocarbon oil #700 has greater viscosity that is ideal for mounting 22 x 22 mm coverslips not moving.
-
Optimize laser confocal microscope time-lapse imaging settings.
Notes:
Make sure that laser power and gain settings do not saturate the neuron throughout time-lapse acquisition period.
Set the highest possible image acquisition rate. At maximum image acquisition rate should be 1 frame per second for proper temporal resolution.
Image resolution should be at least 256 x 256 pixel resolution. There will ideally have to be a balance between image resolution and image acquisition rate.
-
Set up genetic cross or stock, as described above for the cold plate assay, prior to calcium imaging and place the vial in 29 °C.
Age embryo collection in food vial to 96–102 h AEL and select third instar larvae for imaging.
-
Set up water circulation system, Linkam PE 120 Peltier stage, mount Peltier stage onto the laser confocal microscope, and turn on confocal microscope (Figures 2A and 2B).
Notes:
Use double sided tape to mount the Peltier stage onto microscope stage.
Allow time for lasers to stabilize and maximal water circulation.
-
Program the pre-determined temperature profile using the T95 linkpad.
Ideally, 2–5 min of baseline at 25 °C, ramp down at 20 °C/min (maximal ramp speed) to desired temperature, hold at desired temperature for at least 10 sec and then ramp up at 20 °C/min to 25 °C and hold for up 1 min (Figure 2D).
Note: For multiple noxious thermal stimulations allow at least 1 min at 25 °C for GCaMP signal to return baseline.
-
Prepare microscope slide for calcium imaging
Obtain one microscope slide, two 22 x 22 mm coverslips, and one 24 x 50 mm long coverslip.
Place a small drop of halocarbon oil #700 on both ends of the slide.
Place two small coverslips on top of the small droplets. One at a time shimmy the coverslips, and stop after the coverslip is difficult to move.
Using a brush gently pick up one larva, expressing GCaMP in your desired cell type, and wash the larva in a 9-well glass spot plate. Place the larva on a wet Kimwipe.
-
Place the semi-wet larva on the middle of the slide.
Note: Depending on laser confocal microscope set up, the orientation of larva will differ either horizontal or vertical compared to the length of the slide.
Place a drop of water on top of both the small coverslips. If the larva is not straight on the slide, then use forceps to gently reorient the larva to the preferred direction.
Place a 24 x 50 mm coverslip on top of the larva and the two coverslips. Again, shimmy, this time gently, until the larva is completely flat, straight and the desired side of the animal facing up (Figure 2C).
Mount the slide containing the larva onto the Linkam PE 120 Peltier stage.
Focus onto the region of interest and acquire time-lapse images for duration of thermal cycle. Representative GCaMP response in class III md neuron at baseline (25 °C) and at noxious cold temperature (6 °C) (Figure 2E).
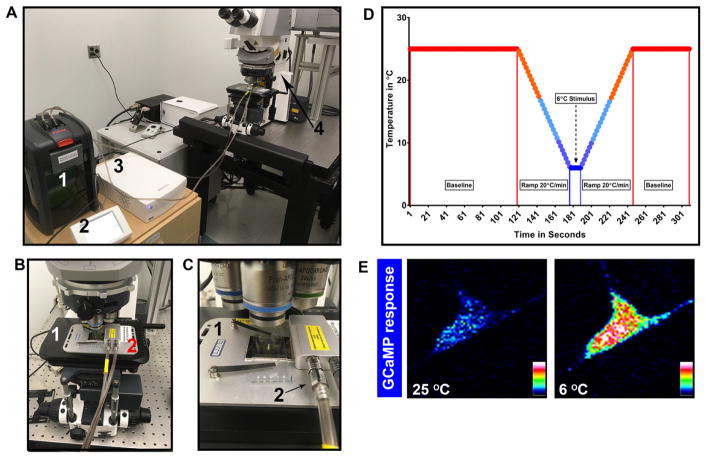
Figure 2. In vivo GCaMP set up, temperature profile and class III md neuron GCaMP response.
A. View of water circulation tank (1), temperature programmer (2), temperature controller (3), and Zeiss laser confocal (4). B. Close up of laser confocal microscope stage (1) with Peltier stage (2); C. Close up of how larva is mounted on to the Peltier stage (1) and water circulation pipes (2) connected to Peltier stage; D. Sample temperature profile for GCaMP assay; E. Class III md neuron GCaMP response to noxious cold. Fluorescence intensity is represented as lookup table. There is a large increase in GCaMP fluorescence in response to noxious cold temperature (6 °C) compared to baseline (25 °C).
C. Functional assays for multimodal sensory processing
Recent studies have revealed class III md neurons function in multimodal sensory processing (Turner et al., 2016). Class III neurons act as noxious cold sensing nociceptors, where the cold evokes bilateral head and tail contraction (CT), and innocuous mechanical sensors, where light touch primarily evokes only a head withdrawal (HW) response (Tsubouchi et al., 2012; Yan et al., 2013; Turner et al., 2016).
We developed an optogenetic dose response assay and implemented the use of the stable calcium integrator CaMPARI to investigate how a single class of md sensory neuron is able to mediate unique behaviors (HW and CT) in response to two distinct sensory stimuli. In the optogenetic dose response assay we titrate the amount of blue light intensity activating class III md sensory neurons, revealing that at high blue light intensities CT is the predominant behavior, whereas at lower blue light intensities HW is the predominant behavior and as expected at the lowest blue intensity, we failed to observe any CT behavior and only a few HW responders (Turner et al., 2016). Optogenetic dose response revealed that level of class III activation determines the behavioral output and suggested that class III neurons are high threshold cold nociceptors and low threshold mechanosensors.
To functionally assess how different levels of class III activation lead to distinct sensory behaviors, we examined intracellular calcium responses to gentle touch and noxious cold stimuli. Assessing intracellular calcium dynamics upon light touch, in an intact larvae, posed a technical challenge in the in vivo GCaMP assay.
Therefore, we implemented the use of class specific expression of CaMPARI, where the larva is freely behaving and either innocuous mechanical or noxious cold stimuli could be delivered. This technique allowed us to measure the amount of green-to-red fluorescence photo-conversion revealing that noxious cold evokes significantly greater photo-conversion than innocuous mechanical stimulus. Both of these techniques can be used to assess the multimodality of PNS neurons.
D. Optogenetic dose response assay
-
Optimize light delivery and recording systems.
-
Using a Zeiss AxioZoom.V16 microscope vary the light intensity delivered to the animal.
-
While keeping magnification and source of white light constant, change the aperture from 100-37% (Figure 3A).
Note: Preferred aperture settings are 100%, 90%, 80%, 70%, 50% 40% and 37%.
Use the dark field illumination setting on the microscope stage. Turn on the LED for bottom illumination, increase the brightness just enough to barely see the larva.
-
Mount recording equipment onto the microscope.
-
Prepare a genetic cross or stock as described above and age embryo collection to 96–102 h AEL prior to optogenetic dose response assay.
-
Prepare ATR supplemented food for optogenetic experiments
Final concentration of ATR needs to be 1,000 μM.
-
In a dimly lit room, add appropriate amount of ATR in liquefied food, mix thoroughly and allow the food to solidify.
Note: Maintain ATR and ATR supplemented food in the dark.
For the ATR condition: add the genetic cross/stock to ATR supplemented food containing vial.
For the no ATR control condition: add the genetic cross/stock to normal food containing vial.
Place both types of crosses in the dark.
-
On day of experimentation.
Turn on Zeiss Axiozoom.V16 microscope system.
Working in a dimly lit space, using a brush select third instar larvae and wash in a 9-well glass spot plate with tap water.
Place the larva on a wet Kimwipe.
Spray a thin fine mist of water via a trigger sprayer on a large piece of glass (10 x 15 x 0.1 cm).
-
Place the larva on the center of glass piece.
Note: Make sure there is not too much water around the animal, as it will make quantification difficult.
Allow animal to acclimate to the plate and resume peristaltic movement.
-
Use a timer for precise blue light delivery and expose the animal to the following blue light on/off cycles (Figure 3B).
10 sec off–5 sec on–10 sec off–5 sec on–10 sec off–5 sec on–10 sec off.
Executing multiple stimulations per animal will allow within animal comparisons of multiple stimulations.
Repeat the previous step until an N of 20 third instar larvae is achieved for each blue light dose.
Quantify changes in length as described in Data analysis section. (Figure 3C and Video 4)
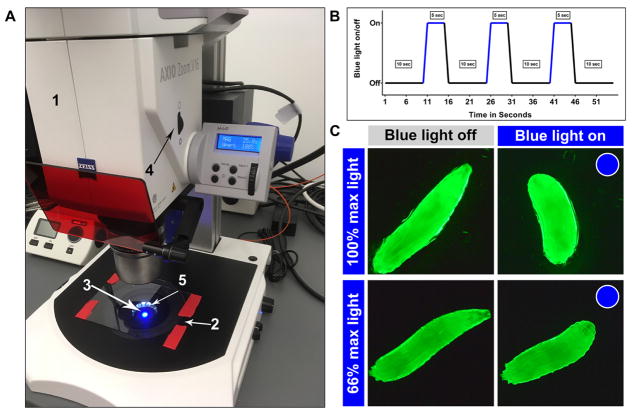
Figure 3. Zeiss AxioZoom.V16 set up for optogenetic dose response and blue light exposure diagram.
A. Optogenetic dose response set up in a brightly lit room for demonstration purpose, but conduct the assays in a dimly lit room. 1. Zeiss AxioZoom.V16, 2. Glass plate, 3. Blue light, 4. Aperture control and 5. Dark field illumination. B. Sample blue light on/off time course; C. Image stills of larvae expressing ChETA in class III md neurons being exposed 100% or 66% max blue light. 100% illumination elicits CT behavior (consistent with noxious cold evoked behavior), whereas 66% illumination elicits HW behavior (consistent with gentle touch evoked behavior).
E. CaMPARI Ca2+ integrator assay
As with GCaMP analysis, optimize red and green fluorescence for live confocal imaging.
-
Optimize photo-converting (PC) light intensity for consistent green to red photo-conversion.
PC light is delivered via Zeiss AxioZoom.V16 using previously described filter cube set.
Using Zeiss AxioZoom.V16 microscope and HXP200C lamp, deliver 84,000 lux for 20 sec of PC light, which reliably photo-converts CaMPARI from green to red in presence of high calcium.
Prepare 1:5 (v/v) ratio of ethyl ether:halocarbon oil #700 solution in an amber glass dropper bottle at least one day prior to imaging.
Prepare a genetic cross or stock as described above and age embryo collection to 96–102 h AEL prior to CaMPARI analysis.
-
Prepare microscope slide for calcium imaging.
Obtain one microscope slide, two 22 x 22 mm coverslips, and one 24 x 50 mm long coverslip.
Place a small drop of ethyl ether:halocarbon oil #700 solution on both ends of the slide.
Place two small coverslips on top of the small droplets. Shimmy the coverslips, one at a time, and stop after the coverslip is difficult to move.
Turn on your PC light delivery system.
-
Set up stimulus delivery equipment on the base of the microscope.
Using a brush gently pick up one larva, expressing CaMPARI in your desired cell type, and wash the larva in a 9-well glass spot plate. Place the larva on a wet Kimwipe.
-
Place the larva under Zeiss AxioZoom.V16. Simultaneously deliver stimulus and 20 sec of PC light.
For cold stimulus: place one larva on black metal plate. Gently place the metal plate on the Peltier plate (set to 6 °C) and turn on PC light.
For light touch stimulus: place one larva on black metal plate. Deliver light touch stimulus first to the head of the larva, followed by a stroke along the length of the animal, while the animal is exposed to PC light.
-
Mount larva on to a microscope slide (Figure 4B).
-
Place the larva on the middle of the slide.
Note: Depending on laser confocal microscope set up orientation of larva will differ either horizontal or vertical compared to the length of the slide.
Place a drop of ethyl ether:halocarbon oil #700 solution on both small coverslips. If the larva is not straight on the slide, then use forceps to reorient the larva to the preferred direction. Generously add drops of ethyl ether:halocarbon oil #700 solution on to and around the larva.
Place a 24 x 50 mm coverslip on top of the larva and the two coverslips. Again, shimmy, this time gently, until the larva is completely flat, straight and desired side of the animal facing up.
-
-
Image z-stacks of neuronal cell bodies in the neural type of interest.
Imaging multiple segments and/or both hemisegments will allow more detailed analysis of spatial CaMPARI response.
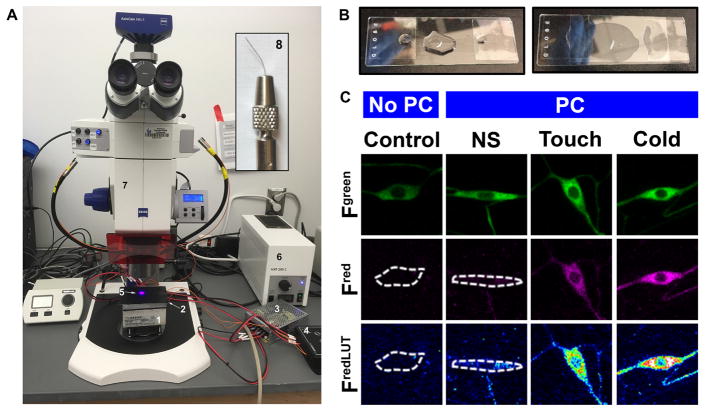
Figure 4. Microscope slide preparation.
A. A view of the Zeiss AxioZoom.V16 with Peltier plate placed on the microscope stage, where larvae can be stimulated via cold and delivered PC light. 1. TE tech cold plate, 2. Black metal plate, 3. TE tech. power supply, 4. TE tech. temperature controller, 5. PC light, 6. HXP200C white light source, 7. Zeiss AxioZoom.V16 microscope and 8. Inset shows image of fine brush bristle mounted to a nickel plated pin holder. B. For live confocal imaging, larval mounted on the microscope slide with dorsal side up and fully stretched. C. Representative images of class III neurons expressing CaMPARI. PC (photo-converting light) and NS (no stimulus).
Data analysis
-
Behavioral quantitation for cold plate and optogenetic dose response assays
Uncompress video files using Video to video converter (videotovideo.org).
Open uncompressed video file as gray scale in ImageJ.
Use threshold function to make the larva black and white (Figure 5).
Use ImageJ function Remove Outliers using the setting: radius = 2.0 pixels, threshold = 50, and which outliers: select dark.
Apply the following functions: make binary and skeletonize. Watch the entire video to make sure that larval skeletons do not have any aberrant branches or protrusions (Figure 5).
Using the analyze function, measure area, which will give the larval length for the duration of the video.
-
Perform percentage change from max length calculation: % Max length = (Lmax - Ln)/Lmax
Lmax is the maximum length of the larva.
Ln is the length of larva in one frame.
-
In vivo calcium imaging data analysis conducted using ImageJ/Fiji
Discard videos with excessive movement in either x, y, or z axis.
Upload the time-lapse data file onto ImageJ.
-
Run ImageJ plugin StackReg for lateral motion stabilization.
Note: Rigid body and translation are two of the best options to use during image stabilization.
Draw a region of interest around the cell body and using ImageJ function called Plot z-axis profile, which measures the mean fluorescence intensity normalized to area for each frame.
Smooth the raw data using a 2 sec moving average.
-
Calculate ΔF/F0 using the following equation: ΔF/F0 = (F − F0)/F0 x 100
F0 is the average fluorescence during baseline
F is the fluorescence at one frame
-
CaMPARI Ca2+ integrator assay data analysis conducted using Zeiss Zen Blue Lite (Figure 4C)
Draw region of interest around maximum intensity projections of neural cell bodies.
Quantify red and green fluorescence.
-
Calculate fluorescence change as previously described in Fosque et al. (2015).
CaMPARI photo-conversion = Fred/Fgreen
Figure 5.

ImageJ screenshots for quantitative behavioral analysis
Recipes
-
Drosophila media
-
Ingredients:
18.4 g soyfluor
132 g cornmeal
12 g Drosophila agar
36 g inactive dry yeast
194 g dry molasses
2.15 L water
9 ml propionic acid
-
1.56 ml O-phosphoric acid
Note: Makes ~2 L of Drosophila media, enough for 200 vials.
Mix the following ingredients in a large cooking vessel: soy, drosophila agar, cornmeal, inactive dry yeast, dry molasses, and water
Cook at medium to high heat until food comes to boil. Mix thoroughly every 5–10 min
Let the fly food low boil for 30 min and then turn off the stove
Wait until cooking vessel is warm to touch and add appropriate amount of acids (O-phosphoric acid and propionic acid)
Mix thoroughly and pour the fly food into Droso-Filler for making fly food vials
Pour the desired volume in a vial loaded tray
-
Supplementary Material
Video of a cold plate assay setup.
Video of w1118 larvae at 25 °C.
Video of w1118 larvae at 6 °C.
Video of larvae expressing ChETA in class III md neurons being exposed to either 100% or 66% max blue light.
Acknowledgments
We thank Kevin Armengol for initial designs of the cold plate assay, in vivo GCaMP assay and quantitative behavioral analysis described in Turner et al. (2016). The Cox laboratory is supported by NINDS R01 NS086082, NIMH R15 MH086928, Brains & Behavior Seed Grant awards and a 2CI Neurogenomics and Molecular Basis of Disease award (GSU) to D.N.C. A.A.P. is funded by 2CI Neurogenomics Fellowship (GSU).
References
- 1.Fosque BF, Sun Y, Dana H, Yang CT, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, Jayaraman V, Looger LL, Schreiter ER. Neural circuits. Labeling of active neural circuits in vivo with designed calcium integrators. Science. 2015;347(6223):755–760. doi: 10.1126/science.1260922. [DOI] [PubMed] [Google Scholar]
- 2.Gorczyca DA, Younger S, Meltzer S, Kim SE, Cheng L, Song W, Lee HY, Jan LY, Jan YN. Identification of Ppk26, a DEG/ENaC channel functioning with Ppk1 in a mutually dependent manner to guide locomotion behavior in Drosophila. Cell Rep. 2014;9(4):1446–1458. doi: 10.1016/j.celrep.2014.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y, Wang Y, Wang Q, Wang Z. The role of PPK26 in Drosophila larval mechanical nociception. Cell Rep. 2014;9(4):1183–1190. doi: 10.1016/j.celrep.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Im SH, Galko MJ. Pokes, sunburn, and hot sauce: Drosophila as an emerging model for the biology of nociception. Dev Dyn. 2012;241(1):16–26. doi: 10.1002/dvdy.22737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mauthner SE, Hwang RY, Lewis AH, Xiao Q, Tsubouchi A, Wang Y, Honjo K, Pate Skene JH, Grandl J, Tracey WD., Jr Balboa (PPK-26) binds to Pickpocket in vivo and is required for mechanical nociception in Drosophila larvae. Curr Biol. 2014;24(24):2920–2925. doi: 10.1016/j.cub.2014.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsubouchi A, Caldwell JC, Tracey WD. Dendritic filopodia, Ripped Pocket, NOMPC, and NMDARs contribute to the sense of touch in Drosophila larvae. Curr Biol. 2012;22(22):2124–2134. doi: 10.1016/j.cub.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner HN, Armengol K, Patel AA, Himmel NJ, Sullivan L, Iyer SC, Bhattacharya S, Iyer EP, Landry C, Galko MJ, Cox DN. The TRP channels Pkd2, NompC, and Trpm act in cold-sensing neurons to mediate unique aversive behaviors to noxious cold in Drosophila. Curr Biol. 2016;26(23):3116–3128. doi: 10.1016/j.cub.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Z, Zhang W, He Y, Gorczyca D, Xiang Y, Cheng LE, Meltzer S, Jan LY, Jan YN. Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature. 2013;493(7431):221–225. doi: 10.1038/nature11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video of a cold plate assay setup.
Video of w1118 larvae at 25 °C.
Video of w1118 larvae at 6 °C.
Video of larvae expressing ChETA in class III md neurons being exposed to either 100% or 66% max blue light.