Abstract
The most prevalent pathological features of many neurodegenerative diseases are the aggregation of misfolded proteins and the loss of certain neuronal populations. Autophgy, as major intracellular machinery for degrading aggregated proteins and damaged organelles, has been reported to be involved in the occurance of pathological changes in many neurodegenerative disorders, including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease and amyotrophic lateral sclerosis. In this review, we summarized most recent research progress in this topic and provide a new perspective regarding autophagy regulation on the pathogenesis of neurodegenerative diseases. Finally, we further discussed the signaling molecules in autophagy-related pathways as therapeutic targets for the treatment of these diseases.
Keywords: autophagy, neurodegenerative diseases, protein aggregation
Introduction
Cellular aggregations of misfolded proteins are the most common pathological hallmark of many neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD) and amyotrophic lateral sclerosis (ALS) [3]. The pathological abnormalities of various neurodegenerative diseases are often associated with corresponding protein aggregations that reside in different cellular environment and subcellular compartments. Some of them are resulted from specific genetic mutations that cause autosomal recessive or dominant familial type of neurodegenerative diseases, while diverse mechanisms leading to impaired proteostasis contribute to the protein aggregations in sporadic type of neurodegenerative diseases.
Autophagy is one of the major intracellular machinery to eliminate misfolded proteins and maintain proteostasis. Dysregulated autophagy is increasingly considered to play key roles in most neurodegenerative diseases, and the regulation of autophagy is therefore proposed as a potential therapeutic avenue for these diseases [73,74,108]. Macroautophagy (referred as autophagy) literally means “self-eating” in Greek that is responsible for removal of long-lived proteins and damaged organelles which are too large for proteasome to degrade. Autophagy not only plays a vital role in development, cell differentiation, apoptosis, pathogen infection and starvation, but also contributes to cancer, immune diseases and neurodegenerative diseases [17,47,78]. Many studies have shown that autophagy is closely linked with neurodegenerative diseases. For example, the amount of autophagic vacuole, an intermediate vesicular compartment in the process of autophagy, are much more in the brains of neurodegenerative diseases than in health controls, suggesting impaired maturation of autophagosome to autolysosome [35]. Without any other causing factors, the depletion of key autophagy-related genes (such as Atg5, Atg7) can lead to neurodegeneration in mouse central nervous system [54,76].
Autophagy exerts a key role in degrading aggregate-prone proteins, which have been implicated in the pathogenesis of various neurodegenerative diseases, such as mutant α-synuclein in PD, mutant huntingtin in HD, and mutant TDP-43 in ALS. Once autophagy is inhibited, the clearance of these substrates is impeded. On the contrary, activation of autophagy may lead to enhanced clearance of those toxic proteins.
Lysosomal dysfunction in neurons is closely tied to neurodegeneration and cell death mechanisms [88]. Growing genetic and biochemical evidence implicates the dysfunction of endosomal-lysosomal and autophagic lysosomal pathways during the pathogenesis of many neurodegenerative disorders, including AD, PD, and ALS [31, 128]. The therapeutic efficacy of autophagy/lysosome modulators in animal models of these disorders [88,128] further underscores the significance of lysosomal impairments to the pathogenesis of neurodegenertive diseases.
In this review, we summarized recent research findings, which showing that the dysregulated autophagy contributes to protein aggregation, organelle impairment and neuronal loss, eventually leads to neurodegenerative diseases. Autophagy modulation can prevent the occuence and progression of diseases [132]. Even though various factors underlie the pathology of these diseases, we aim at providing an interaction between autophagy and the cause/progression of neurodegenerative diseases. We also reviewed autophagy-inducing agents, both mTOR-dependent and -independent, and evaluate their effectiveness in disease models of either in vitro or in vivo.
Autophagy mechanism
Autophagy includes three subtypes: macroautophagy, microautophagy and chaperon mediated autophagy. Although subtypes of autophagy differ from cargo recognition, mechanism of molecular chaperon, they share lysosome as the unique place for cargo digestion and products recycling. An intact autophagy process is depicted as autophagic flux including autophagosome formation, fusion of autophagosome and lysosome, and cargo degradation in lysosome [15,77]. Firstly, misfolded proteins and damaged organelles are enwrapped by newly formed membrane termed as phagophore that is potentially derived from plasma membrane, Golgi, mitochondria or endoplasmic reticulum (ER) [99,106,134]. Phagophore gradually sequesters cargoes through elongation till forming a closed autophagosome. By means of cytoskeletal microtubule systems, autophagosome traffics to lysosome and fuses with lysosome to form autolysosome. In autolysosome, cargoes are digested by lysosomal enzymes and recycled for reuse [2,53,55,64].
Autophagy is a multi-stage process containing numerous proteins, including several autophagy-related proteins identified in mammals [79,118]. Autophagy is initiated by two major complexes UN51-like Ser/ Thr kinases (ULK) complex and the class III phosphatidylinositol-3-kinase (PI3K), which are recruited to the phagophore assembly site (PAS) [2,91]. The ULK complex contains ULK1/2 family, FAK family kinase interacting protein of 200kDa (FIP200), and ATG13 [121]. The other complex PI3K, also named Beclin1 complex, consists of vacuolar protein sorting 34 (Vps34), p15 (VPS15), Beclin1 (ATG6), and Barkor (ATG14) [27]. Notably, Beclin1 which localizes on ER membrane is regulated by anti-apoptotic dimer BCL-2 and BCL-XL. When autophagy is activated, Beclin1 will be dissociated from BCL-2 complex to coordinate with Vps34 [36,69,95]. Subsequently, bulk phosphatidylinositol 3-phosphate [PI (3) P] will concentrate on the surface of phagophore [89,99].
The extension and closure of autophagosome are exerted by two ubiquitin like complexes. At the first, with the interaction of Atg7, Atg5 links with Atg12 covalently [114]. Then the covalent complex links with Atg16 to form Atg5-Atg12-Atg16 complex, responsible for elongating phagophore. Atg9 which binds Atg2 and Atg18 is essential for trafficking between the Trans-Golgi-network, endosomes and newly-formed autophagosomes. In another ubiquitin like complex, microtubule-associated protein 1 light chain 3 (LC3) is cleaved by Atg4B to generate LC3-I [122]. The Atg5-Atg12-Atg16 complex assists the transformation of LC3-I to phosphatidylethanolamine (PE)-conjugated LC3-II. Since LC3-II mainly resides on autophagosome, it is viewed as the significant marker for autophagosome [44].
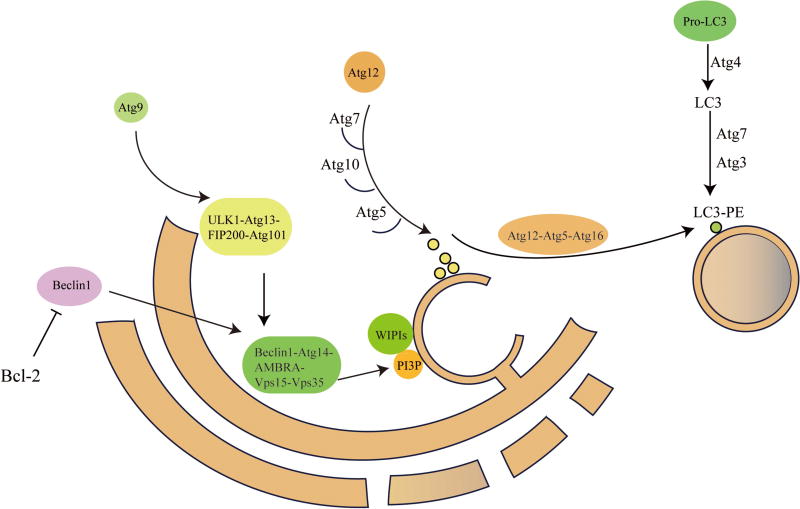
Subsequently, maturated autophagosome needs kinesin and motor proteins to move along microtubules [101]. Meanwhile, autophagosome fuses with lysosome in which multiple membrane proteins complexes such as the soluble NSF attachment protein receptor (SNAREs) are recruited [40]. After the formation of autolysosome is completed, cargoes carried by autophagosomes are degraded by proteolysis. A detailed illustration of autophagy process is shown in Figure 1.
Figure 1.
Autophagy induction and autophagosome formation. This diagram shows the process of autophagy induction and roles of Atg related proteins in autophagosome formation.
Autophagy plays an essential role in protein degradation and recycling. Though both of ubiquitin proteasome system and autophagy could clear ubiquitinated substrates, compared with proteasome, autophagy is the only one way to degrade large protein aggregates or impaired organelles which are too large to go into the narrow entrance of proteasome chamber.
Brains are the most vulnerable organ in most lysosome disorders, indicating that neurons might rely on autophagy even more heavily than other cells to maintain protein homeostasis. Unusual structures such as large dendritic and axonal cytoplasm in neurons cause difficulty for them to remove impaired organelles and other waste in time [5]. Two key components of autophagy locate on distinct places that autophagic vacuoles generated in axons should travel long distances to lysosomes mainly locating near the cell body. In addition, unlike other mitotic cells, neurons are not able to divide to disperse harmful substances [62]. Aging will worsen the situation that neurons are gradually losing the ability to efficiently clear wastes, eventually resulting in abnormal accumulated autophagic substrates. Altogether, neurons are prone to suffer from autophagic proteolytic damage.
Notably, numerous stress responses inducing autophagy promote misfolded protein aggregation at ER. ER stress is associated with unfolded protein response (UPR) [38], which is initiated by Inositol-requiring enzyme 1 α (IRE1α) [19]. IRE1 is an ER transmembrane sensor that activates the UPR to maintain the ER and cellular function and downstream target c-Jun N-terminal kinase (JNK) [50,90,93,130]. Notably, ER stress can regulate Beclin1 in autophagosome formation [14]. Protein misfolded diseases are usually accompanied with ER stress. UPR stands for a cell survival pathway to modulate autophagy to reduce protein aggregation and remain proteostasis [38,93]. In brief, various regulation mechanisms emerge to maintain proteostasis within cells making the modulation of autophagy as a promising strategy for therapeutic purposes.
Autophagy and Alzheimer’s disease
Alzheimer’s disease is the most common neurodegenerative disease that is characterized by extracellular amyloid-β (Aβ) plaques which are cleaved products of amyloid precursor proteins (APPs) and intracellular neurofibrillary tangles which are composed of aggregated hyperphosphorylated tau protein [135].
Under normal circumstance, autophagosome vesicles are rare in brains. Strikingly, detailed ultrastructural analyses have shown that dystrophic neurites in AD brains contain autophagosome vesicles [35,87]. Further study demonstrates that more autophagy vacuoles are found in Presenilin 1 (PS1)-rich locations [35]. Accumulation of autophagy vacuoles is likely arising from impaired clearance rather than the induction of autophagy itself, suggesting the modulation of late steps of autophagy as a possible therapeutic strategy for AD. Accordingly, treatment with autophagy enhancer rapamycin significantly increased autophagosome fusion with lysosome in vitro [35].
PS1 is a ubiquitous transmembrane protein, whose cleaved form is the catalytic subunit of γ-secretase complex, which induces the intra-membranous cleavage of APP [16]. Generally, APP is firstly cleaved by β-secretase to produce β-C-terminal fragment (βCTF). And then, βCTF is cleaved by Presenilin 1 (PS1) to produce Aβ. Mutant PS1 is considered to contribute to AD pathogenesis by interfering cleavage of APP. Recent investigations have shown that PS1 can also decrease Aβ levels by directing β-C-terminal fragment (βCTF) degradation through autophagy [8]. Morever, PS1 is invovled in the fusion of autophagsome and lysosome. Lack of phosphorylation on PS1 1 Ser367 impeded the fusion of autophagosome and lysosome in mouse brain. And then, this inhibition of autophagy reduced βCTF degradation leading to the accumulation of Aβ in the brain[7]. These observations imply that Presenilin 1 could be a promising target for the treatment of Alzheimer’s disease through autophagy.
However, PS1 is a vital mediator in lysosomal turnover of autophagic substrates. PS1 is an ER chaperone to facilitate maturation and targeting of the v-ATPase V0a1 subunit to lysosomes, which is a key component in acidification and substrate degradation [60]. Further investigation demonstrates that PS1 also maintains Ca2+ homeostasis by regulating acidification of lysosome [59]. Loss of acidification leads to dysfunction of lysosome that impedes fusion of autophagosome to lysosome, thereby accumulation of autophagosomes. In addition, lysosome dysfunction causes cargo-specific deficits of axonal transport leading to AD-like neuritic dystrophy [61]. Based on these observations, it is reasonable to suspect that restoring the proteolytic function of lysosome may enhance the removal of protein aggregations. In line with this notion, deletion of cystatin B, an inhibitor of lysosome cysteine protease in AD mice model promotes the clearance of abnormal protein aggregations in lysosomal compartments [133].
Genome-wide association studies (GWAS) have identified additional proteins involved in autophagy that are also closely linked with AD, such as the phosphatidylinositol binding clathrin assembly protein (PICALM/CALM). CALM is involved in endocytic trafficking to regulate endocytosis of SNAREs that enhance autophagy to clear tau aggregations [81].
Beclin1, a key factor in autophagosome formation has been shown to be transcriptionally suppressed in AD brains [96]. Under pathogenic conditions, Caspase 3, a key component in apoptosis pathway, may cleave Beclin1 protein and lead to autophagy disruption. The cleaved form of Beclin1 is therefore regarded as a common in vitro marker for apoptosis in AD pathogenesis [104]. Another potential marker for the pathology of AD is nuclear factor erythroid derived 2 like 2 (Nrf2). In response to oxidative stress, Nrf2 could induce autophagy receptor NDP52 [51] to stimulate autophagy and remove aggregated tau proteins [43]. Meanwhile, Nrf2 as a vital transcription factor can also regulate the transcription of autophagy related proteins [92].
Autophagy and Parkinson’s disease
PD is the second most common neurodegenerative disease that is characterized by selective loss of dopamine neurons in substantia nigra pars compacta, and intracellular inclusions of Lewy body and Lewy neurites composed of α-synuclein and polyubiquitinated proteins [20]. In the post-mortem brain samples of PD patients, dysfunctional lysosomes and accumulation of autophagosomes were observed in neurons [22], indicating a pathogenic role of autophagy in PD. The main component of Lewy bodies is misfolded and aggregated α-synuclein [20,45,123]. When lysosome is inhibited, the level of α-synuclein is increased, suggesting a close link between α-synuclein degradation and autophagy. Previous studies have shown that basically all forms of α-synuclein can be degraded by autophagy [22,37,58], while monomeric α-synuclein is also degraded by the proteasome [126]. Transcription factor EB (TFEB), a key modulator for autophagy [113], has been widely demonstrated to relieve pathology of neurodegenerative diseases. Over-expression of TFEB could decrease the damage of lysosome by inducing its biogenesis, thus ameliorating the α-synuclein pathology [21,49]. Taken together, these results suggest an essential role of autophagy in the prevention and treatment of synucleinopathy in PD.
Mutations in leucine rich repeat kinase 2 (LRRK2) represent the most common cause of autosomal dominant form of PD [123]. Over-expression of LRRK2 G2019S mutation in differentiated SH-SY5Y cells results in shortening dentric and autophagosomes aggregation [98]. In vivo experiments have demonstrated that the up-regulation of LRRK2 G2019S impairs autophagic flux with aging [107]. The VPS35 D620N mutation that causes autosomal-dominant PD destabilizes WASH complex leading to defect of autophagosome formation and compromises trafficking of autophagy protein ATG9 [137].
Besides, mutations in parkin RBR E3 ubiquitin protein ligase (PARKIN) and PTEN induced putative kinase 1 (PINK1) are the main causing factors for autosomal recessive forms of PD, accounting for 50% of familial cases in Europe [48]. These two proteins coordinate mitophagy to selectively degrade mitochondria by autophagy. Damaged mitochondria are delivered and sequestered within double membrane autophagosome, ultimately cleared by autolysosome. In this process, the proteasome-mediated degradation of PINK1 is stalled in depolarized mitochondria leading to accumulated PINK1 on the mitochondrial outer membrane where it phosphorylates ubiquitin and recruits parkin. In turn, the activated parkin can ubiquitinate outer membrane proteins, which are subsequently phosphorylated by PINK1. The outcome of these linkages greatly actives parkin and elicits a positive feedback involving more ubiquitinated proteins of mitochondria [46,56,70,85,86].
GWAS has identified a few lysosome related genes associated with PD. The protein ATP13A2 which is involved with lysosomal ATPase, is found mutated in autosomal recessive forms of early-onset Parkinsonism [24,100]. Down-regulation of ATP13A2 resulted in decreased lysosomal degradation in dopaminergic neurons and accumulation of α-synuclein protein [120]. Subsequently, depletion of ATP13A2 leads to ubiquitination and degradation of SYT11 that induces lysosome dysfunction and increases accumulation of mutant α-synuclein [4].
Autosomal recessive mutations in the gene GBA which encodes lysosomal hydrolase cause defects in autophagosome-lysosome pathway and aggregation of α-synuclein [1]. Depletion of ATP6AP2, which is essential for lysosomal acidification and function, has been associated with Parkinsonism [1]. Moreover, loss of VPS13C function causes mitochondrial dysfunction and lysosome dysfunction and is associated with autosomal recessive Parkinsonism [1,63].
Autophagy and Huntington’s disease
HD, the most common polyglutamine disease, is a devastating autosomal dominant neurodegenerative disease. HD is characterized by CAG repeat tri nucleotide in the first exon of the huntingtin (HTT) gene which leads to polyglutamine (polyQ) expansions and pathogenic aggregation [39,42].
Aggregated autophagosomes could be observed in HD models [68], although autophagosome formation is not affected by HD pathology. Huntingtin plays a key role in autophagosome transport. In HD models, depletion of huntingtin results in abnormal accumulation of autophagosomes with engulfed mitochondria which is indicative of impaired cargo degradation [140].
In addition, there are aberrant interactions between autophagy and onset of HD. One polymorphism in the Atg7 is associated with an earlier onset form of HD [75]. Beclin1 could reduce HTT mRNA level with aging [115]. Dysfunction of loading into autophagosomes has been observed in cellular and animal models of HD, causing a impaired autophagic protein degradation despite of increased autophagic vesicles contents. The autophagy selective substrate p62/SQSTM1 is commonly treated as a crucial marker for autophagic flux especially in the cargo-recognition machinery which transports substrates to autophagosomes. Deficiency in such machinery is prevalent in HD models [68]. Moreover, up-regulation of casein kinase 2 (CK2) which phosphorylates p62/SQSRM1 reduces large inclusion formation of mutant huntingtin [71].
Compared with mutant HTT, non-mutant HTT seems to coordinate with autophagy in a different manner. For example, wild-type HTT can bind p62 to enhance its role in autophagy and interact with ULK1 to evoke autophagy. In addition, Atg11 shares resemble structure with HTT to play a role in autophagosome formation. On the contrary, knock-out of dynein reveals increased levels of autophagosomes and impaired autolysosomes, accompanied with increased aggregation of mutant huntingtin [101].
Autophagy and ALS
Amyotrophic lateral sclerosis (ALS) is a paralytic and fatal disease characterized by selective loss of motor neurons in brain and spinal cord giving rise to muscle weakness and atrophy. Most cases in ALS are sporadic, while the familial ones account for approximately 10%. Mutations in superoxide dismutase 1 (SOD1), TAR DNA-binding protein (TDP43) and fused in sarcoma/translated in lip sarcoma (FUS/TLS) are common causes for familial type of ALS [94].
Several reports have demonstrated that autophagy is associated with ALS. Immunostaining experiments in transgenic mice with mutant SOD1 G93A have shown that autophagy is activated [82]. The aggregated autophagosomes in cytoplasm indicate taht autophagy is activated in degenerated motor neurons in ALS cases [82]. Notably, excess autophagosomes and autolysosomes are closely associated with p62/SQSTM1 positive inclusions, suggesting an impaired cargo digestion in lysosome [112]. Other studies have shown that increased autophagosomes are tightly related with the decreased phosphorylation of mTOR in numerous genetic ALS models [82].
Growing evidence has shown that mutations in autophagy-related proteins are closely associated with the onset of ALS. Earlier studies have indicated that depletion in subunits of endosomal sorting complexes required for transport (ESCRT) causes abnormal multi vesicular bodies (MVBs) with autophagosomes and is considered to be associated with ALS [29]. In addition, mutations in ESCRT subunit charged multi vesicular body protein-2B (CHMP2B) are found in patients with ALS which impair ESCRT function leading to accumulation of ubiquitinated proteins and p62 [29]. Autophagy receptor p62/SQSTM1 which binds both LC3 and ubiquitin to target ubiquitinated substrates to autophagosomes has been involved in ALS cases. Clearance of mutant SOD1 via ubiquitin proteasome system or autophagy is coordinated by p62/SQSTM1. Similarly, over-expression of p62/SQSTM1 could reduce TDP-43 aggregation via autophagy or proteasome in vitro [6].
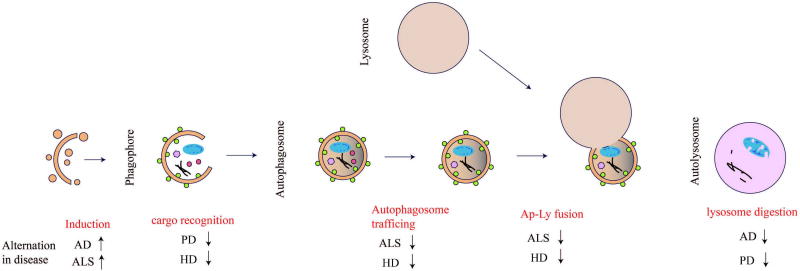
Moreover, multi groups have identified a link between serine/threonine kinase TANK-binding kinase 1(TBK1) and ALS [52]. One recent study has shown that TBK1 is the upstream regulator of autophagy receptor optineurin (OPTN) [90]. Both TBK1 and OPTN play key roles in mitophagy [129]. Since mitochondria are the place for not only the generation of energy but also the execution of cellular apoptosis, clearance of damaged mitochondria is essential for cellular homeostasis. These investigations suggest mitophagy as a new etiology of ALS. Ubiquilin2 (UBQLN2), a proteasome shuttle factor, plays a key role in formation of autophagosome. Mutations in UBQLN2 lead to cognitive deficits, shortened lifespan and neuron loss in mouse models [23,57]. A detailed illustration of alternations in neurodegenerative diseases in autophagic flux is shown in Figure 2.
Figure 2.
Overview of autophagic flux and impaired states in neurodegenerative diseases. This schematic diagram shows the procedures through the autophagic flux from formation of the autophagosome to fusion with the lysosome. Red text highlights refer to the dysfunctional steps in autophagy, along with related neurodegenerative diseases. Arrows’ directions stand for activation or inhibition.
Autophagy as a therapeutic target for neurodegenerative diseases
Links between autophagy and neurodegenerative diseases promote an intriguing question: whether the modulation of autophagy could slow down disease progression. Emerging evidence has shown that autophagy enhancement could efficiently ameliorate neuropathology and neurodegeneration via either an mTOR-dependent or -independent pathway. Thus various reagents targeting for autophagy have been investigated [102,116,127].
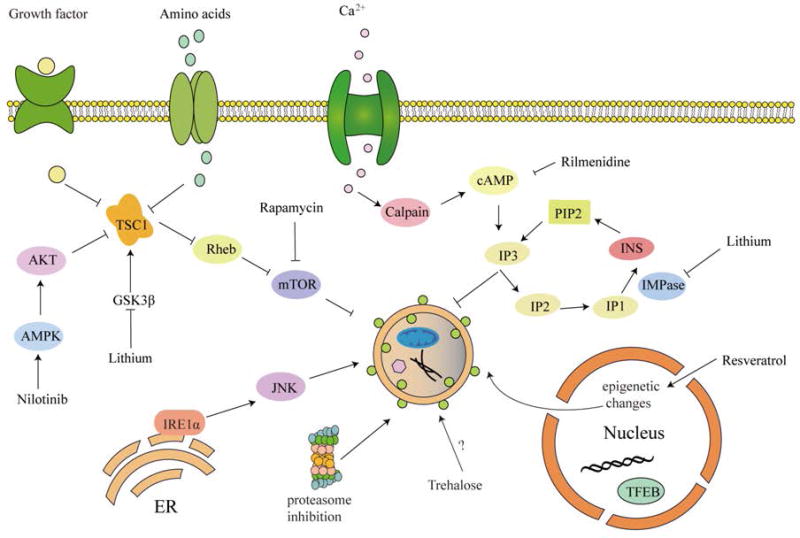
Autophagy is activated by diverse signaling classified as mammalian target of rapamycin (mTOR) dependent and mTOR independent pathway (Figure 3).
Figure 3.
Molecular targets of autophagy up-regulating agents. This schematic diagram shows representative molecular agents involved in autophagy activation through regulating autophagy-related pathways. Either the mTOR-dependent or mTOR-independent pathway could play a negative role in autophagy. In addition, suppression of these pathways will result in activation of autophagy.
mTOR dependent pathway
The serine/threonine protein kinase mTOR is a core component of two distinct complexes, mTOR complex 1 (mTORC1) and mTORC2. mTORC1 negatively regulates autophagy, while mTORC2 does it in an opposite way [32]. Under normal conditions, autophagy is suppressed by mTOR. Since mTORC1 phosphorylates and inhibits core autophagy complex composed of ULK1, Atg13 and FIP200. Rapamycin interacts with immunophilin FK506-binding protein (FKBP12) to form a complex which inhibits the kinase activity of mTORC1, thus inducing autophagy [11,66]. Additionally, some drugs indirectly target mTOR such as Nilotinib, which could stimulate AMPK pathway in mTOR dependent manner to induce autophagy [136].
mTOR independent pathway
Besides autophagy, mTOR-dependent pathway plays a wide role among several other biology reactions. To avoid potential adverse effects induced by mTOR multiple function, studies on mTOR-independent pathway are increasingly emphasized. For example, more and more pharmacological drugs have been screened for regulating autophagy and have been shown to influence diverse signaling pathways including calcium flux, inositol phosphophates and epigenetics.
Lithium decreases the level of Inositol 3-phosphates (IP3), which is a second messenger binding to its receptor on the ER leading to Ca2+ release into cytoplasm. Rilmenidine reduces level of cAMP [110] which could bind IP3 to regulate intracytosolic Ca2+ levels [10]. Resveratrol could active autophagy via epigenetic mechanisms [124]. Nilotinib can upregulate AMPK pathway to active autophagy [37, 65]. Although great achievements have been made in the discovering of novel mTOR-independent autophagy mordulators, till now, the detailed mechanisms underlying authopahgy regulating effects of these molecules remains exclusive and efforts are still needed for their clinical application.
Alzheimer’s disease
Previous studies have reported that the application of rapamycin can reduce fibrillary tangles and amyloid plaques in brains and rescue cognitive deficits [5,9,67]. Rapamycin analogue temsirolimus also shows similar effects in AD mouse models [41]. Arctigenin, a natural product from Arctium lappa, can inhibit Aβ production and promote Aβ clearance by activating autophagy through inhibiting AKT/mTOR signaling [141]. Latrepirdine is a pro-neurogenic compound that reduces accumulation of Aβ42 by stimulating autophagy [117]. GTM-1, a novel small molecule, can attenuate Aβ oligomer-induced neurotoxicity via inducing autophagy in an mTOR-independent manner [18]. Nilotinib, a tyrosine kinase inhibitor, can enhance interaction of parkin and Beclin1 that lead to clearance of Aβ [65]. Notably, it also plays a clearance role in PD-related parkin mutant models [37]. Trehalose, a natural disaccharide, is beneficial for removing abnormal proteins. It has been demonstrated to reduce accumulation of Aβ [103]. Trehalose rescues the learning impairment by reducing Aβ deposits in APP/PS1 mice [25]. Since trehalose is free of toxic effects at high concentrations suggesting a promising prospect for clinical applications in human tauopathies.
Parkinson’s disease
Resveratrol induces autophagy via AMPK/SIRT1 pathway to protect neurons form rotenone induced toxicity in vitro [83]. Administration of Nilotinib contributes to clearing α-synuclein aggregation via autophagy, and rescues dopaminergic neuron loss [37]. Notably, in proteasome inhibition-induced mouse models, proteasome dysfunction lead to activation of autophagy that serves a compensatory mechanism to clear protein aggregation and decrease cell death [34]. Further enhancement of autophagy by pharmacological drugs or molecular inhibitors can attain similar effects. Trehalose contributes to reducing α-synuclein mutants in vitro [109]. In addition, trehalose increases the number of dopamine neurons and the dopaminergic activity in the midbrain in PD mouse models [103]. Lithium facilitates clearance of mutant α-synuclein in vitro [28].
Huntington’s disease
Rapamycin reduces huntingtin accumulation and cell death in cell models of HD [102,111]. Lithium could partially rescue cell death [12,110]. Trehalose could bind expanded polyglutamine to delay pathology in HD mouse models [109,119]. Rilmenidine could enhance autophagy to remove mutant huntingtin fragments in cell models via mTOR independent pathway [105]. Lithium could reduce mutant huntingtin protein aggregates and cell death [30].
Amyotrophic lateral sclerosis
Interestingly, rapamycin plays two opposite roles in ALS animal models. For example, rapamycin treatment in SOD1G93A mouse models further augments motor neuronal degeneration and lead to more death of ALS mice [139]. However, in mutant TDP-43 models, rapamycin treatment decreases pathology of ALS [125]. These contradictory findings may be due to different pathogenic proteins overexpressed and their different impact on autophagy in the two animal models of ALS. Further studies have demonstrated that rapamycin administration impairs autophagic flux, although it significantly increases the number of autophagosomes in mutant SOD1 models. Trehalose could induce autophagy via mTOR independent pathway and significantly decrease SOD1 aggregation, reduce ubiquitinated protein accumulation in the motor neurons of SOD1 mice [13,138].
In addition, developing novel chemicals to modulate autophagy reveals a promising prospect. For example, single-walled carbon nanotubes (SWNT) restore normal autophagy by reversing abnormal activation of mTOR signaling and deficits in lysosomal proteolysis, thereby facilitates elimination of autophagic substrates. These findings suggest that SWNT could serve as a novel neuroprotective approach to AD therapy [131].
Autophagy in clinical diagnosis of neurodegenerative diseases
Recently, emerging evidence in clinics implies that autophagy is in close association with neurodegenerative diseases. Biochemical analyses show the dramatic increase of the autophagosome marker LC3 in postmortem brains of AD patients and confirm its co-localization with hyperphosphorylated tau [97]. Besides, some proteins known to regulate autophagy have been newly implicated with the pathogenesis of AD. In postmortem brains of AD patients, increase of tetraspanin impedes the fusion of autophagosome with lysosome, which leads to the abnormal accumulation of APP [33]. Another study shows that decrease of immunophilin FKBP52(FK506-Binding Protein of MW~52 kDa) , a protein co-localized with lysosome, is accompanied with accumulation of neurofibrillary tangles [72]. In postmortem brains of PD patients, expression of toll-like receptor 2 (TLR2) is elevated in neurons and spatially correlated with the pathological α-synuclein aggregation and increase of autophagy receptor SQSTM1 [26]. Aberrant alternation of LAMP2, which is a significant marker in lysosome, is closely related with the early pathology of PD [84]. These findings suggest that autophagy is commonly abberant in neurodegenerative diseases. However, further studies are still required to explore specific autophagic pathways or signalings involved in different types or sub-types of neurodegenerative diseases.
Concluding remarks
Overall, increasing evidence indicates dysregulated autophagy plays a key role in the pathogenesis of neurodegenerative diseases, and implies potential therapeutic strategies to ameliorate neurodegenerative diseases through regulating autophagy. However, mechanisms of autophagy regulation on proteostasis and general metabolism remain to be further investigated, especially through linking the interplay between specific proteins involved in autophagy and progression of diseases. Furthermore, other resident cells in the brain such as microglia might also be involved in the process of autophagy. It remains largely unknown whether and how microglia cooperates with neurons to regulate autophagy. As to autophagy-inducing agents, treatment dose, period and design of the drugs should be carefully chosen and examined, as over-activation of autophagy could result in detrimental effects in accelerating the progression of neurodegenerative diseases.
Acknowledgments
This review is supported by the National Natural Science Foundation of China (81430021, 81370470), and is also support in part by the intramural program of National Institute on Aging, National Institutes of Health (HC: AG000928, 000959).
Abbreviations
- AD
Alzheimer’s disease
- PD
Parkinson’s disease
- HD
Huntington’s disease
- ALS
amyotrophic lateral sclerosis
- PAS
phagophore assembly site
- LC3
microtubule-associated protein 1 light chain 3
- PI3K
phosphatidylinositol-3-kinase
- ULK
UN51-like Ser/ Thr kinases
- FIP200
FAK family kinase interacting protein of 200kDa
- PI (3) P
phosphatidylinositol 3-phosphate
- SNARE
soluble NSF attachment protein receptor
- APPs
amyloid precursor proteins
- PS1
Presenilin 1
- GWAS
Genome-wide association studies
- PICALM/CALM
phosphatidylinositol binding clathrin assembly protein
- Nrf2
nuclear factor erythroid derived 2 like 2
- TFEB
transcription factor EB
- LRRK2
leucine rich repeat kinase 2
- PARKIN
parkin RBR E3 ubiquitin protein ligase
- INK1
PTEN induced putative kinase 1
- CK2
casein kinase 2
- ESCRT
endosomal sorting complexes. required. for transport
- CHMP2B
multi vesicular body protein-2B
- MVB
multi vesicular bodies
- FKBP12
FK506-binding protein
- UPR
unfolded protein response
- IRE1α
Inositol-requiring enzyme 1 α
- JNK
c-Jun N-terminal kinase
- IP3
Inositol 3-phosphates
References
- 1.Abeliovich A, Gitler AD. Defects in trafficking bridge Parkinson’s disease pathology and genetics. Nature. 2016;539:207–216. doi: 10.1038/nature20414. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal S, Tiwari SK, Seth B, et al. Activation of autophagic flux against xenoestrogen bisphenol-A-induced hippocampal neurodegeneration via AMP kinase (AMPK)/mammalian target of rapamycin (mTOR) pathways. J Biol Chem. 2015;290:21163–21184. doi: 10.1074/jbc.M115.648998. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Aguzzi A, O’Connor T. Protein aggregation diseases:pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 4.Bento CF, Ashkenazi A, Jimenez-Sanchez M, et al. The Parkinson/’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nat Commun. 2016;7:11803. doi: 10.1038/ncomms11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland B, Kumar A, Lee S, et al. Autophagy Induction and Autophagosome Clearance in Neurons:Relationship to Autophagic Pathology in Alzheimer’s Disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady OA, Meng P, Zheng Y, et al. Regulation of TDP-43 aggregation by phosphorylation andp62/SQSTM1. J Neurochem. 2011;116:248–259. doi: 10.1111/j.1471-4159.2010.07098.x. [DOI] [PubMed] [Google Scholar]
- 7.Bustos V, Pulina MV, Bispo A, et al. Phosphorylated Presenilin 1 decreases β-amyloid by facilitating autophagosome–lysosome fusion. Proc Natl Acad Sci. 2017:201705240. doi: 10.1073/pnas.1705240114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustos V, Pulina MV, Kelahmetoglu Y, et al. Bidirectional regulation of Aβ levels by Presenilin 1. Proc Natl Acad Sci. 2017:201705235. doi: 10.1073/pnas.1705235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caccamo A, Majumder S, Richardson A, et al. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-β, and Tau:Effects on cognitive impairments. J Biol Chem. 2010;285:13107–13120. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cárdenas C, Miller RA, Smith I, et al. Essential Regulation of Cell Bioenergetics By Constitutive InsP3 Receptor Ca2+ Transfer to Mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardenas ME, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmichael J, Sugars KL, Bao YP, et al. Glycogen synthase kinase-3β inhibitors prevent cellular polyglutamine toxicity caused by the Huntington’s disease mutation. J Biol Chem. 2002;277:33791–33798. doi: 10.1074/jbc.M204861200. [DOI] [PubMed] [Google Scholar]
- 13.Castillo K, Nassif M, Valenzuela V, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–1320. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 14.Castillo K, Rojas-Rivera D, Lisbona F, et al. BAX inhibitor-1 regulates autophagy by controlling the IRE1α branch of the unfolded protein response. EMBO J. 2011;30:4465–4478. doi: 10.1038/emboj.2011.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cecconi F, Levine B. The Role of Autophagy in Mammalian Development:Cell Makeover Rather than Cell Death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chávez-Gutiérrez L, Bammens L, Benilova I, et al. The mechanism of c -Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31(10):2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi AMK, Ryter SW. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 18.Chu C, Zhang X, Ma W, et al. Induction of Autophagy by a Novel Small Molecule Improves Aβ Pathology and Ameliorates Cognitive Deficits. PLoS One. 2013;8(6):e65367. doi: 10.1371/journal.pone.0065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Criollo A, Maiuri MC, Tasdemir E, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14:1029–39. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 20.Dauer W, Przedborski S. Parkinson’s disease:mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 21.Decressac M, Mattsson B, Weikop P, et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110:E1817–26. doi: 10.1073/pnas.1305623110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dehay B, Bové J, Rodríguez-Muela N, et al. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30:12535–12544. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult onset ALS and ALS/dementia. Nature. 2012;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djarmati A, Hagenah J, Reetz K, et al. ATP13A2 variants in early-onset Parkinson’s disease patients and controls. Mov Disord. 2009;24:2104–2111. doi: 10.1002/mds.22728. [DOI] [PubMed] [Google Scholar]
- 25.Du J, Liang Y, Xu F, et al. Trehalose rescues Alzheimer’s disease phenotypes in APP/PS1 transgenic mice. J Pharm Pharmacol. 2013;65:1753–1756. doi: 10.1111/jphp.12108. [DOI] [PubMed] [Google Scholar]
- 26.Dzamko N, Gysbers A, Perera G, et al. Toll-like receptor 2 is increased in neurons in Parkinson’s disease brain and may contribute to alpha-synuclein pathology. Acta Neuropathol. 2017;133:303–319. doi: 10.1007/s00401-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci U S A. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108\r1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng HL, Leng Y, Ma CH, et al. Combined Lithium and Valproate Treatment Delays Disease Onset , Reduces Neurological Deficits and prolongs survival in an amyotrophic lateral sclerosis mouse model. Neuroscience. 2008;155:567–572. doi: 10.1016/j.neuroscience.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filimonenko M, Stuffers S, Raiborg C, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fornai F, Longone P, Cafaro L, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2008;105:2052–7. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frakes AE, Ferraiuolo L, Haidet-phillips AM, et al. Microglia induce motor neuron death via the classical NF-κB pathway in amyotrophic lateral sclerosis. Neuron. 2015;81:1009–1023. doi: 10.1016/j.neuron.2014.01.013.Microglia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Granowitz BE, Vannier E, Poutsiaka DD, et al. Effect of Interleukin-1 (IL-1) Blockade on Cytokine Synthesis: II. IL-1 Receptor Antagonist Inhibits Lipopolysaccharide-Induced Cytokine Synthesis by Human Monocytes. Blood. 1992;79(9):2364–2369. [PubMed] [Google Scholar]
- 33.Guix FX, Sannerud R, Berditchevski F, et al. Tetraspanin 6:a pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol Neurodegener. 2017;12(1):25. doi: 10.1186/s13024-017-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo F, He XB, Li S, et al. A Central Role for Phosphorylated p38a in Linking Proteasome Inhibition-Induced Apoptosis and Autophagy. Mol Neurobiol. 2016:1–13. doi: 10.1007/s12035-016-0260-1. [DOI] [PubMed] [Google Scholar]
- 35.Haung Yu W, Cuervo AM, Kumar A, et al. Macroautophagy - A novel β-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C, Levine B. The Beclin-1 Interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hebron ML, Lonskaya I, Moussa CEH. Nilotinib reverses loss of dopamine neurons and improvesmotorbehavior via autophagic degradation of α-synuclein in parkinson’s disease models. Hum Mol Genet. 2013;22:3315–3328. doi: 10.1093/hmg/ddt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hetz C, Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci. 2014;15:233–249. doi: 10.1038/nrn3689. [DOI] [PubMed] [Google Scholar]
- 39.Imarisio S, Carmichael J, Korolchuk V, et al. Huntington’s disease:from pathology and genetics to potential therapies. Biochem J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 40.Itakura E, Kishi-itakura C, Mizushima N. The Hairpin-type Tail-Anchored SNARE Syntaxin 17 Targets to Autophagosomes for Fusion with Endosomes / Lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Jiang T, Yu JT, Zhu XC, et al. Temsirolimus promotes autophagic clearance of amyloid-β and provides protective effects in cellular and animal models of Alzheimer’s disease. Pharmacol Res. 2014;81:54–63. doi: 10.1016/j.phrs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Jimenez-sanchez M, Licitra F, Underwood BR, et al. Huntington’s Disease:Mechanisms of Pathogenesis and Therapeutic Strategies. Cold Spring Harb Perspect Med. 2016 doi: 10.1101/cshperspect.a024240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jo C, Gundemir S, Pritchard S, et al. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat Commun. 2014;5:3496. doi: 10.1038/ncomms4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homolog of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalia LV, Kalia SK, McLean PJ, et al. α-synuclein oligomers and clinical implications for Parkinson disease. Ann Neurol. 2013;73:155–169. doi: 10.1002/ana.23746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kane LA, Lazarou M, Fogel AI, et al. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaushik S, Cuervo AM. Proteostasis and aging. Nat Med. 2015;21:1406–1415. doi: 10.1038/nm.4001. [DOI] [PubMed] [Google Scholar]
- 48.Kazlauskaite A, Muqit MMK. PINK1 and Parkin - Mitochondrial interplay between phosphorylation and ubiquitylation in Parkinson’s disease. FEBS J. 2015;282:215–223. doi: 10.1111/febs.13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kilpatrick K, Zeng Y, Hancock T, et al. Genetic and chemical activation of TFEB mediates clearance of aggregated α-synuclein. PLoS One. 2015;10:e0120819. doi: 10.1371/journal.pone.0120819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress:disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 51.Kim S, Lee D, Song JC, et al. NDP52 associates with phosphorylated tau in brains of an Alzheimer disease mouse model. Biochem Biophys Res Commun. 2014;454:196–201. doi: 10.1016/j.bbrc.2014.10.066. [DOI] [PubMed] [Google Scholar]
- 52.Kim YE, Oh KW, Noh MY, et al. Genetic and functional analysis of TBK1 variants in Korean patients with sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2017;50:170, e1–e6. doi: 10.1016/j.neurobiolaging.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 55.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 56.Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 57.Le NT, Chang L, Kovlyagina I, et al. Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS-FTD-linked UBQLN2 mutations. Proc Natl Acad Sci U S A. 2016;113(47):E7580–E7589. doi: 10.1073/pnas.1608432113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee HJ, Khoshaghideh F, Patel S. Clearance of α-Synuclein Oligomeric Intermediates via the Lysosomal Degradation Pathway. J Neurosci. 2004;24:1888–1896. doi: 10.1523/JNEUROSCI.3809-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JH, McBrayer MK, Wolfe DM, et al. Presenilin 1 Maintains Lysosomal Ca2+ Homeostasis via TRPML1 by Regulating vATPase-Mediated Lysosome Acidification. Cell Rep. 2015;12:1430–1444. doi: 10.1016/j.celrep.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JH, Yu WH, Kumar A, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S, Sato Y, Nixon RA. Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer-like neuritic dystrophy. Autophagy. 2011;7:1562–1563. doi: 10.4161/auto.7.12.17956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee S, Sato Y, Nixon RA. Lysosomal Proteolysis Inhibition Selectively Disrupts Axonal Transport of Degradative Organelles and Causes an Alzheimer’s-Like Axonal Dystrophy. J Neurosci. 2011;31:7817–7830. doi: 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lesage S, Drouet V, Majounie E, et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am J Hum Genet. 2016;98:500–513. doi: 10.1016/j.ajhg.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loos B, du Toit A, Hofmeyr J-HS. Defining and measuring autophagosome flux-concept and reality. Autophagy. 2014;8627:37–41. doi: 10.4161/15548627.2014.973338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lonskaya I, Hebron ML, Desforges NM, et al. Nilotinib-induced autophagic changes increase endogenous parkin level and ubiquitination, leading to amyloid clearance. J Mol Med. 2014;92:373–386. doi: 10.1007/s00109-013-1112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lorenz MC, Heitman J. TOR Mutations Confer Rapamycin Resistance by Preventing Interaction with FKBP12-Rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 67.Majumder S, Richardson A, Strong R, et al. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS One. 2011;6(9):e25416. doi: 10.1371/journal.pone.0025416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez-Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martyniszyn L, Szulc L, Boratyńska A, et al. Beclin 1 is involved in regulation of apoptosis and autophagy during replication of ectromelia virus in permissive L929 cells. Arch Immunol Ther Exp (Warsz) 2011;59:463–471. doi: 10.1007/s00005-011-0149-7. [DOI] [PubMed] [Google Scholar]
- 70.Matsuda N, Sato S, Shiba K, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsumoto G, Wada K, Okuno M, et al. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 72.Meduri G, Guillemeau K, Dounane O, et al. Caspase-cleaved Tau-D421 is colocalized with the immunophilin FKBP52 in the autophagy-endolysosomal system of Alzheimer’s disease neurons. Neurobiol Aging. 2016;46:124–137. doi: 10.1016/j.neurobiolaging.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 73.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 74.Menzies FM, Moreau K, Rubinsztein DC. Protein misfolding disorders and macroautophagy. Curr Opin Cell Biol. 2011;23:190–197. doi: 10.1016/j.ceb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Metzger S, Saukko M, Van Che H, et al. Age at onset in Huntington’s disease is modified by the autophagy pathway:Implication of the V471A polymorphism in Atg7. Hum Genet. 2010;128:453–459. doi: 10.1007/s00439-010-0873-9. [DOI] [PubMed] [Google Scholar]
- 76.Mizushima N, Hara T. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:302–304. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 77.Mizushima N, Komatsu M. Autophagy:Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 78.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 80.Moore AS, Holzbaur ELF. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A. 2016;113(24):E3349–58. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moreau K, Fleming A, Imarisio S, et al. PICALM modulates autophagy activity and tau accumulation. Nat Commun. 2014;5:4998. doi: 10.1038/ncomms5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morimoto N, Nagai M, Ohta Y, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 83.Morselli E, Mariño G, Bennetzen MV, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murphy KE, Gysbers AM, Abbott SK, et al. Lysosomal-associated membrane protein 2 isoforms are differentially affected in early Parkinson’s disease:Early loss of LAMP2A protein in PD. Mov Disord. 2015;30:1639–1647. doi: 10.1002/mds.26141. [DOI] [PubMed] [Google Scholar]
- 85.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8(1):e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nixon RA, Wegiel J, Kumar A, et al. Extensive Involvement of Autophagy in Alzheimer Disease:An Immuno-Electron Microscopy Study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 88.Nixon RA, Yang D-SS. Autophagy and Neuronal Cell Death in Neurological Disorders. Cold Spring Harb Perspect Biol. 2012;4(10):a008839. doi: 10.1101/cshperspect.a008839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Obara K, Ohsumi Y. Dynamics and function of PtdIns (3) P in autophagy. Autophagy. 2008;8627:952–954. doi: 10.4161/auto.6790. [DOI] [PubMed] [Google Scholar]
- 90.Ogata M, Hino S, Saito A, et al. Autophagy Is Activated for Cell Survival after Endoplasmic Reticulum Stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 92.Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, et al. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park HS, Jun DY, Han CR, et al. Proteasome inhibitor MG132-induced apoptosis via ER stress-mediated apoptotic pathway and its potentiation by protein tyrosine kinase p56 lck in human Jurkat T cells. Biochem Pharmacol. 2011;82:1110–1125. doi: 10.1016/j.bcp.2011.07.085. [DOI] [PubMed] [Google Scholar]
- 94.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis:insights from genetics. Nat Rev Neurosci. 2016;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 95.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 96.Pickford F, Masliah E, Britschgi M, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid-β accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Piras A, Collin L, Grüninger F, et al. Autophagic and lysosomal defects in human tauopathies:analysis of post-mortem brain from patients with familial Alzheimer disease, corticobasal degeneration and progressive supranuclear palsy. Acta Neuropathol Commun. 2016;4:22. doi: 10.1186/s40478-016-0292-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Plowey ED, Cherra SJ, Liu YJ, et al. Role of autophagy in G2019S–LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Puri C, Renna M, Bento CF, et al. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramirez A, Heimbach A, Gründemann J, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 101.Ravikumar B, Acevedo-Arozena A, Imarisio S, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 102.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 103.Rodríguez-Navarro JA, Rodríguez L, Casarejos MJ, et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010;39:423–438. doi: 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 104.Rohn TT, Wirawan E, Brown RJ, et al. Depletion of Beclin-1 due to proteolytic cleavage by caspases in the Alzheimer’s disease brain. Neurobiol Dis. 2011;43(1):68–78. doi: 10.1016/j.nbd.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rose C, Menzies FM, Renna M, et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum Mol Genet. 2010;19:2144–2153. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 107.Saha S, Ash PE, Gowda V, et al. Mutations in LRRK2 potentiate age-related impairment of autophagic flux. Mol Neurodegener. 2015;10:26. doi: 10.1186/s13024-015-0022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways:autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans. 2013;41:1103–30. doi: 10.1042/BST20130134. [DOI] [PubMed] [Google Scholar]
- 109.Sarkar S, Davies JE, Huang Z, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and α-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 110.Sarkar S, Floto RA, Berger Z, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sarkar S, Ravikumar B, Floto R, et al. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 112.Sasaki S. Autophagy in Spinal Cord Motor Neurons in Sporadic Amyotrophic Lateral Sclerosis. J Neuropathol Exp Neurol. 2011;70(5):349–359. doi: 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- 113.Settembre C, Polito VA, Garcia M, et al. Manuscripts TFEB Links Autophagy to Lysosomal Biogenesis. Science. 2013;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shao Y, Gao Z, Feldman T, et al. Stimulation of ATG12-ATG5 Conjugation by Ribonucleic Acid. Autophagy. 2007;3(1):10–16. doi: 10.4161/auto.3270. [DOI] [PubMed] [Google Scholar]
- 115.Shibata M, Lu T, Furuya T, et al. Regulation of intracellular accumulation of mutant huntingtin by beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 116.Son JH, Shim JH, Kim KH, et al. Neuronal autophagy and neurodegenerative diseases. Exp Mol Med. 2012;44:89. doi: 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Steele JW, Lachenmayer ML, Ju S, et al. Latrepirdine improves cognition and arrests progression of neuropathology in an Alzheimer’s mouse model. Mol Psychiatry. 2013;18:889–897. doi: 10.1038/mp.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes to Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 119.Tanaka M, Machida Y, Niu S, et al. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 120.Usenovic M, Tresse E, Mazzulli JR, et al. Deficiency of ATP13A2 leads to lysosomal dysfunction, α-synuclein accumulation and neurotoxicity. J Neurosci. 2012;257:4240–4246. doi: 10.1523/JNEUROSCI.5575-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fujita N, Hayashi-Nishino M, Fukumoto H, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;(11):19. 4651–9. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vekrellis K, Xilouri M, Emmanouilidou E, et al. Pathological roles of α-synuclein in neurological disorders. Lancet Neurol. 2011;10:1015–1025. doi: 10.1016/S1474-4422(11)70213-7. [DOI] [PubMed] [Google Scholar]
- 124.Vingtdeux V, Giliberto L, Zhao H, et al. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-β peptide metabolism. J Biol Chem. 2010;285:9100–9113. doi: 10.1074/jbc.M109.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang IF, Guo BS, Liu YC, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A. 2012;109:15024–9. doi: 10.1073/pnas.1206362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Webb JL, Ravikumar B, Atkins J, et al. α-synuclein Is Degraded by Both Autophagy and the Proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 127.Williams AJ, Paulson HL. Polyglutamine neurodegeneration:protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wolfe DM, Lee J hyun, Kumar A, et al. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013;37:1949–1961. doi: 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wong YC, Holzbaur ELF. Temporal dynamics of PARK2/parkin and OPTN/optineurin recruitment during the mitophagy of damaged mitochondria. Autophagy. 2015;11:422–424. doi: 10.1080/15548627.2015.1009792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu C, Bailly-Maitre B, Reed J. Endoplasmic reticulum stress:cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xue X, Wang LR, Sato Y, et al. Single-walled carbon nanotubes alleviate autophagic/lysosomal defects in primary glia from a mouse model of alzheimers disease. Nano Lett. 2014;14:5110–5117. doi: 10.1021/nl501839q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins:A role for aggrephagy in neurodegeneration. Neurobiol Dis. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang DS, Stavrides P, Saito M, et al. Defective macroautophagic turnover of brain lipids in the TgCRND8 Alzheimer mouse model:Prevention by correcting lysosomal proteolytic deficits. Brain. 2014;137:3300–3318. doi: 10.1093/brain/awu278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yen WL, Shintani T, Nair U, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yoon SY, Kim DH. Alzheimer’s disease genes and autophagy. Brain Res. 2016;1649(Pt B):201–209. doi: 10.1016/j.brainres.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 136.Yu H, Lin C, Tai W, et al. Nilotinib Induces Autophagy in Hepatocellular Carcinoma through AMPK Activation. J Biol Chem. 2013;288:18249–18259. doi: 10.1074/jbc.M112.446385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zavodszky E, Seaman MN, Moreau K, et al. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat Commun. 2014;5:3828. doi: 10.1038/ncomms4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang X, Chen S, Song L, et al. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy. 2014;10:588–602. doi: 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang X, Li L, Chen S, et al. Rapamycin treatment augments motor neuron degeneration in SOD1 G93A mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 140.Zheng S, Clabough EBD, Sarkar S, et al. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhu Z, Yan J, Jiang W, et al. Arctigenin effectively ameliorates memory impairment in Alzheimer’s disease model mice targeting both beta-amyloid production and clearance. J Neurosci. 2013;33:13138–13149. doi: 10.1523/JNEUROSCI.4790-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]