Abstract
The NELF complex is a metazoan-specific factor essential for establishing transcription pausing. Although NELF has been implicated in cell fate regulation, the cellular regulation of NELF and its intrinsic role in specific lineage differentiation remains largely unknown. Using mammalian hematopoietic differentiation as a model system, here we identified a dynamic change of NELF-mediated transcription pausing as a novel mechanism regulating hematopoietic differentiation. We found a sharp decrease of NELF protein abundance upon granulocytic differentiation and a subsequent genome-wide reduction of transcription pausing. This loss of pausing coincides with activation of granulocyte-affiliated genes and diminished expression of progenitor markers. Functional studies revealed that sustained expression of NELF inhibits granulocytic differentiation, whereas NELF depletion in progenitor cells leads to premature differentiation towards the granulocytic lineage. Our results thus uncover a previously unrecognized regulation of transcription pausing by modulating NELF protein abundance to control cellular differentiation.
INTRODUCTION
Hematopoietic cell differentiation is initiated by hematopoietic stem cells (HSCs), a rare cell population with the capacity to self-renew and differentiate through hierarchically organized progenitor stages to generate all mature blood lineages including erythrocytes, platelet, granulocytes, monocyte and lymphocytes. This well characterized differentiation process has provided a powerful model to study transcriptional mechanisms in cellular differentiation and lineage fate selection.
Transcription mechanisms controlling hematopoietic differentiation can regulate different steps of transcription, including the pausing of RNA polymerase II (Pol II) during early transcription elongation. Unveiled by biochemical and genomic studies, transcriptionally engaged Pol II frequently pauses after initiation and accumulates at 20–60 nucleotide downstream of the promoter region, stabilized by pausing factors1, 2. The most extensively studied pausing factors are DSIF (the DRB sensitivity-inducing factor) and NELF (the negative elongation factor complex)3–5. Release of paused Pol II into productive elongation is triggered by the recruitment of P-TEFb (the positive transcription elongation factor b), which phosphorylates NELF, DSIF and the Pol II C-terminal domain, leading to the conversion of DSIF into an elongation-stimulating factor and the dissociation of NELF6–8. Unlike DSIF that has function in both pausing and elongation9, NELF is proposed mainly as a pausing factor to hold engaged Pol II within the promoter-proximal region10. Composed of four subunits (A, B, C/D and E), NELF executes its role in Pol II pausing beyond simply inhibiting transcription. Studies of transcription profiles in NELF-depleted cells revealed more down-regulated genes than up-regulated genes, suggesting a positive role of NELF in maintaining gene expression11. This was further supported by studies showing that NELF depletion causes either nucleosome reassembly on promoters or less recruitment of transcription initiation factors11–14, suggesting a role of paused Pol II in maintaining gene expression by facilitating initiation and generating a permissive chromatin state around the promoter region.
Despite the well-established role of NELF in regulating transcription pausing, the cellular regulation and function of NELF-mediated pausing in lineage differentiation remains to be explored. Using hematopoietic differentiation systems, here we identified a downregulation of NELF protein abundance upon induction of granulocytic differentiation from human and mouse hematopoietic progenitor cells. Genomic analyses further revealed a genome-wide change of transcription pausing correlated with NELF abundance. Manipulation of NELF expression demonstrated an inhibitory role of NELF-mediated Pol II pausing in granulocytic differentiation. Our studies unveil a novel regulation of NELF that is tightly linked to the biological function of pausing in cellular differentiation.
METHODS
Cell culture, treatment and transfection
Human hematopoietic CD34+ progenitor cells isolated from peripheral blood of GCSF-mobilized healthy volunteers were obtained from the Fred Hutchinson Cancer Research Center. Before differentiation, cells were expanded in StemSpan SFEM II medium (StemCell Technologies Inc.) with 1X CC100 cytokine mix (StemCell Technologies Inc.) and 2% penicillin-streptomycin (P/S) for 5–6 days. At the end of the expansion period, cells were reseeded in differentiation medium for up to 14 days with media change every other day. Erythroid differentiation medium contains SFEM II plus 2% P/S, 20ng/ml SCF, 1U/ml Epo, 5ng/ml IL-3, 2uM dexamethasone, and 1uM β-estradiol. Myeloid differentiation was done by culturing CD34+ cells in IMDM supplemented with 2% P/S, 10% FBS, 100ng/ml IL-3 and 100ng/ml SCF for two days, followed by addition of 10ng/ml G-CSF on day 3. Flavopiridol (Sigma) treatment was done by adding 50nM flavopiridol into differentiation medium.
Mouse 32Dcl3 cells were maintained in IMDM supplemented with 2% P/S, 10% FBS and 5ng/ml IL-3. Differentiation was induced by culturing cells in IMDM supplemented with 2% P/S, 15% FBS and 100ng/ml G-CSF.
Control and NELF siRNAs were purchased from Thermo Fisher (cat. 4390844, s24748, s15489). siRNAs were introduced into expanded CD34+ cells by the Neon transfection system (Fisher) following the manufacture’s protocol.
The mouse Nelf-E cDNA was cloned in-frame into the pEF-FLAG-Biotag vector15 to generate an amino-terminal FLAG fusion molecule. 32D cells were transfected by the Neon transfection system with empty vector or pEF-FLAG-NelfE and cultured in medium with 1ug/ml puromycin to select stably transfected cells.
Protein extraction and Western blotting
Differentiating granulocytes were first incubated in 5.4mM diisopropyl fluorophosphate (DFP) for 15 minutes on ice to prevent protein degradation caused by neutrophil-derived protease16. Washed cells were then lysed directly in 1x SDS loading buffer followed by immediate boiling in the presence of 100mM DTT for 10 minutes before loading on SDS-PAGE for Western blotting. Antibodies used are: Anti-GAPDH (Bethyl, A300-641A), Anti-NELFA (Bethyl, A301-910A), Anti-NELFB (Bethyl, A301-912A), Anti-NELFD (Santa Cruz, sc393972), Anti-NELFE (Santa Cruz, sc377052), anti-Flag (Sigma, F1804).
RNA extraction and q-RT-PCR
RNA was isolated using the Trizol reagent (Thermo Fisher), and subsequently treated with the DNA-free kit (Thermo Fisher). cDNA was synthesized with the SuperScript III Kit (Thermo Fisher). Nascent RNA extraction was performed using the Click-iT Nascent RNA capture kit (Thermo Fisher) following the manufacture’s protocol. Quantitative PCR was performed on Roche LightCycler 480 real-time PCR machine using the iQ SYBR Green Mastermix (BioRad). Gene expression was analyzed relative to β-actin using the ΔΔCt method. Primer sequences are available upon request.
GRO-seq
Nuclei isolation, nuclear run-on, and library preparation were performed as previously described17. The GRO-seq data were analyzed using the groHMM software package described previously18. Further details about GRO-seq, including sample preparation and data analyses, can be found in the supplemental information.
Accession numbers
GRO-seq data are available on GEO under accession number GSE98555.
RESULTS
NELF protein levels are downregulated during granulocytic differentiation
To study the function of NELF in hematopoietic differentiation, we first examined NELF expression in hematopoietic progenitors and differentiated cells. Using human CD34+ hematopoietic stem and progenitor cells (HSPCs) derived from mobilized adult peripheral blood, we followed published protocols19, 20 to differentiate HSPCs into either erythroid or granulocytic lineage induced by erythropoietin (EPO) or granulocyte-colony stimulating factor (G-CSF), respectively. After confirming efficient differentiation of both lineages by May-Grünwald Giemsa (MGG) staining (Fig. S1A), cells at different stages of differentiation were examined for NELF protein by western blot analysis. Surprisingly, while NELF protein levels remained unchanged upon erythroid differentiation (Fig. S1B), granulocytic differentiation induced an evident decrease of NELF protein starting from early stages of differentiation (Fig. 1A). Consistent with previous studies that NELF subunits are interdependent for their protein stability21, all NELF subunits showed decreased protein levels with NELF-E protein showing the largest reduction (Fig. 1A and S1C). Q-RT-PCR analysis revealed no obvious change at the transcription level of NELF genes during early differentiation (Fig. 1B), suggesting that NELF expression in differentiating granulocytes is regulated by a post-transcriptional mechanism. To test if NELF is regulated by proteasome-mediated protein degradation, differentiating granulocytes were treated with the 26S proteasome inhibitor MG132. This treatment markedly enhanced NELF protein levels, especially the level of NELF-E (Fig. 1C, S1D). Because NELF-E is reduced faster than other subunits (Fig. 1A and S1C), these results suggest that NELF-E might be the primary target for protein turnover triggered by granulocytic differentiation.
Figure 1. Downregulation of NELF protein during granulocytic differentiation.
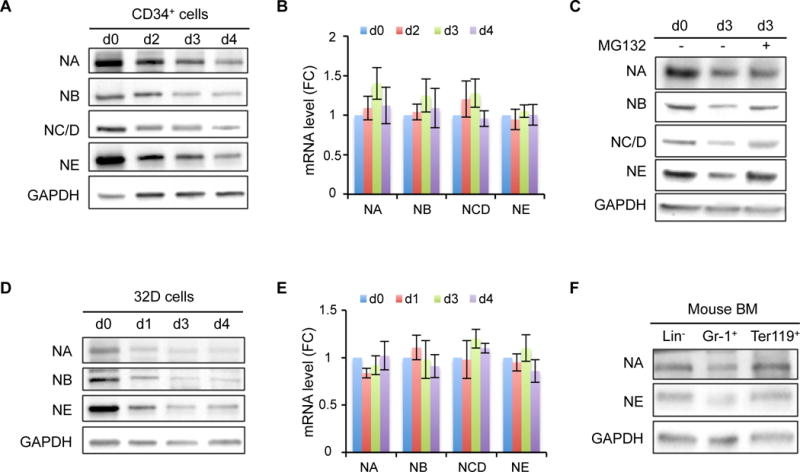
(A) Western blot of NELF subunits from different days during granulocytic differentiation of human CD34+ HSPCs. GAPDH serves as a loading control. NA: NELF-A, NB: NELF-B, NC/D: NELF-C/D, NE: NELF-E. Quantification results are in Figure S1C.
(B) Quantitative RT-PCR of mRNA levels of NELF genes during granulocytic differentiation of CD34+ cells. Gene expression is normalized to β-actin and presented as fold change relative to d0 (n=3, mean ± SEM).
(C) Western blot of NELF subunits on differentiation day 3. MG132 treatment was done by adding 0.2uM MG132 for overnight before harvesting cells for protein extraction. Quantification results are in Figure S1D.
(D) Western blot of NELF subunits during neutrophil differentiation of mouse 32Dcl3 cell line. Quantification results are in Figure S1E.
(E) Quantitative RT-PCR of mRNA levels of Nelf genes during neutrophil differentiation of mouse 32Dcl3 cells. Gene expression is normalized to β-actin and presented as fold change relative to d0 (n=3, mean ± SEM).
(F) Western blot of NELF-A and -E subunits from cells isolated from mouse bone marrow.
To test whether differentiation-induced change of NELF is specific to human cells, we examine NELF expression in a mouse myeloblastic cell line 32Dcl3 that can be induced to neutrophils by G-CSF. Similar to human cells, differentiation induced a sharp decrease of NELF at the protein level but not the transcription level (Fig. 1D and 1E). Again, NELF-E is the most downregulated subunit (Fig. S1E). Finally, we tested whether NELF expression is differentially expressed in vivo during normal myeloid differentiation. Western blot analysis of cells isolated from mouse bone marrow revealed lower NELF protein levels in Gr-1+ granulocytes compared to lineage negative (Lin−) progenitor cells, whereas no such differences was detected in Ter119+ erythroid cells (Fig. 1F). Collectively these results suggest an evolutionarily conserved regulation of NELF protein abundance during granulocytic differentiation that may play a physiological function in hematopoiesis in vivo.
Genome-wide reduction of transcription pausing upon granulocytic differentiation
NELF is identified as a major transcription pausing factor. Down-regulation of NELF upon granulocytic differentiation prompted us to ask whether there is a reduction of transcription pausing in differentiating granulocytes. To test this, we performed global run-on sequencing (GRO-seq) in CD34+ HSPCs (day 0) and early-differentiated (day 3) granulocytes. GRO-seq measures nascent RNA transcription from transcriptionally engaged RNA polymerase in isolated nuclei, thus reflecting a high-resolution distribution map of transcriptionally competent Pol II22. We conducted GRO-seq in two biological replicates and confirmed the reproducibility by correlation analysis (Fig. S2A). Consistent with previous studies that Pol II pauses at majority of metazoan genes23–25, metagene analysis of GRO-seq data from progenitor cells (d0) revealed a typical genome-wide distribution pattern of transcriptionally engaged Pol II with high signals around the transcription start site (TSS) representing promoter-proximal paused Pol II, and low signals along the gene body representing elongating Pol II (Fig. 2A). In contrast, metagene analysis of early-differentiated granulocytes (d3) revealed a drastic reduction of Pol II occupancy around TSS, suggesting a genome-wide reduction of Pol II pausing upon granulocytic differentiation (Fig. 2A). To further verify metagene results, we calculated the pausing index (PI), a ratio of Pol II occupancy between the promoter (−300bp ~ +300bp centered at TSS) and the gene body (+300bp ~ up to 13kb). Higher PI indicates stronger pausing. Consistent with the metagene analysis, PI is significantly decreased in granulocytes compared to progenitor cells (Fig. 2B). Moreover, the cumulative distribution function analysis revealed that >85% genes in granulocytes have decreased PI (Fig. 2C). We concluded from these data that there is a genome-wide reduction of transcription pausing upon granulocytic differentiation, in line with the reduced abundance of NELF protein.
Figure 2. Genome-wide reduction of transcription pausing upon granulocytic differentiation.
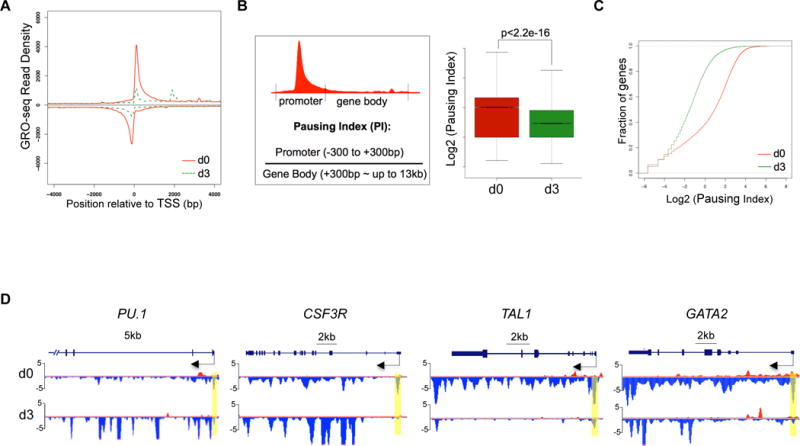
(A) Metagene analysis showing transcriptionally engaged Pol II occupancy measured by GRO-seq on both sense and anti-sense strands in undifferentiated human CD34+ HSPCs (d0) and 3-day differentiated cells (d3). TSS: transcription start site
(B) Left: An illustration for calculation of pausing index (PI) by the ratio of Pol II occupancy at promoter vs. gene body. The promoter is defined from 300bp upstream to 300bp downstream of TSS. The gene body is defined from 300bp downstream of TSS till the end of gene or up to 13kb for genes longer than 13kb. Right: boxplot analysis to compare PI between HSPCs (d0) and granulocytes (d3).
(C) Cumulative distribution function analysis to compare PI distribution in HSPCs (d0) and granulocytes (d3).
(D) Genome browser captures of GRO-seq for granulocytic differentiation regulator genes (PU.1 and CSF3R) and progenitor genes (TAL1 and GATA2) in HSPCs (d0) and granulocytes (d3). Scale bar and gene diagram are depicted above the captures. Yellow shades highlight promoter peaks.
The elongating Pol II within gene bodies reflects the transcription strength of a gene. To identify genes that are differentially transcribed upon differentiation, we compared gene-body associated Pol II (+300bp~13kb) between progenitor cells and granulocytes. In total 1779 genes were identified with at least a 2-fold change, including 471 upregulated (up) genes and 1308 downregulated (down) genes (p<0.0001). Gene-Ontology (GO) analysis confirmed that upregulated genes contain several pathways involved in granulocyte function, such as cell migration, cell adhesion, proteolysis and chemotaxis (Fig. S2B). As expected, many genes involved in granulocytic differentiation, such as myeloid differentiation regulator genes PU.1 and CSF3R, and granulocytic primary granule genes (MPO, ELANE, AZU1 and PRTN3) appear to be upregulated with increased gene-body associated Pol II and a mild decrease of promoter-associated Pol II, suggesting enhanced Pol II release on these genes upon differentiation (Fig. 2D and S2C). In contrast, progenitor-related genes such as GATA2 and TAL1 have a dramatic reduction of Pol II occupancy from both promoters and gene bodies (Fig. 2D), consistent with the expected downregulation of progenitor genes during myeloid differentiation.
Forced expression of NELF inhibits granulocytic differentiation
The downregulation of NELF protein in differentiating granulocytes suggests an inhibitory role of NELF in granulocytic differentiation. To test this, we ectopically expressed NELF-E, which is the most downregulated subunit upon granulocytic differentiation (Fig. 1C and S1D), in the murine 32Dcl3 myeloblastic cells. A flag-tagged Nelf-E construct was stably transfected into cells and the expression of exogenous NELF-E was detected by anti-flag antibody (Fig. 3A). Transfected cells were induced to differentiation by adding G-CSF. Similar to the endogenous protein, differentiation caused a gradual reduction of the exogenous NELF-E (Fig. 3B), further supporting rapid turnover of NELF-E protein triggered by differentiation. Notably, the level of endogenous NELF-A is mildly increased in cells overexpressing NELF-E (Fig. 3B), supporting the view that NELF-E is the limiting subunit and increased NELF-E stabilizes other NELF components. Importantly, although the exogenous NELF-E level cannot be maintained at high level due to rapid protein turnover, neutrophil differentiation was still severely blocked (Fig. 3D), strongly suggesting that sustained NELF expression inhibits granulocytic differentiation.
Figure 3. Forced expression of NELF-E inhibits granulocytic differentiation.
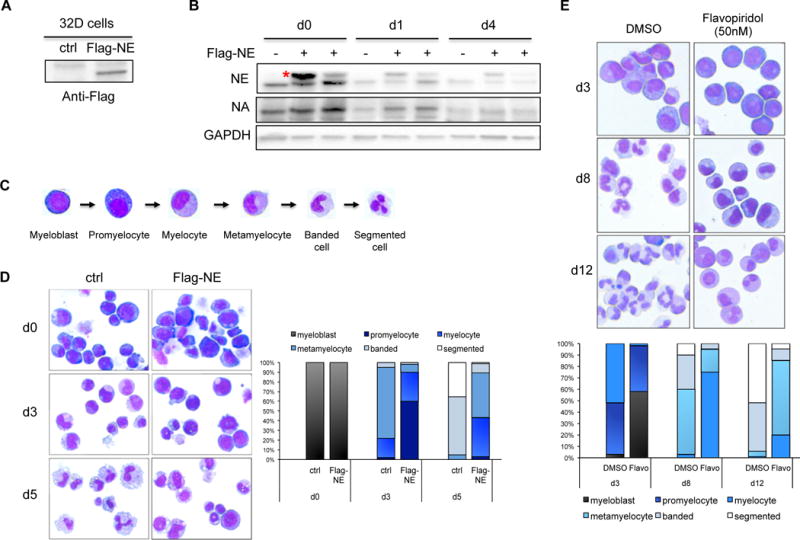
(A) Mouse 32Dcl3 cells were stably transfected with empty vector (ctrl) or flag-tagged NELF-E (Flag-NE) and blotted with anti-flag antibody.
(B) Western blot of NELF-A and -E protein from 32D cells transfected with or without flag-tagged NELF-E (Flag-NE). Protein extraction was done in undifferentiated cells (d0), cells differentiated for 1 day (d1) and 4 days (d4). Two independent Flag-NE transfected cell lines were shown. Red asterisk indicates flag-NE that is slightly bigger than endogenous NELF-E. GAPDH is used as a loading control.
(C) Representative images of May-Grunwald-Giemsa (MGG) staining showing the morphology of human granulocytic differentiation from myeloblasts to mature segmented neutrophils.
(D) MGG staining to compare the morphology of murine 32D cells expressing flag-NE with control cells during neutrophil differentiation.
(E) MGG staining to compare morphology of differentiating cells from human CD34+ HSPCs treated with DMSO vs. flavopiridol.
On (D) and (E), the percentages of different cell types at various time points during differentiation are shown on the bar charts. Quantitative results represent average percentages by counting 100 cells per experiment from 3 independent experiments.
To further distinguish whether the inhibitory role of NELF is through regulating transcription pausing and subsequent elongation, or reflects a pausing-independent function of NELF, we asked whether transcription elongation is required for granulocytic differentiation. Indeed, we found that treating differentiating granulocytes with flavopiridol, an inhibitor of the essential transcription elongation factor P-TEFb, severely blocked granulocytic differentiation of human HSPCs (Fig. 3E). Together these results suggest that granulocytic differentiation requires proper Pol II elongation, which could be promoted by reducing NELF-mediated pausing.
NELF depletion leads to a genome-wide loss of transcription in progenitor cells
The above results suggest that downregulation of NELF-mediated pausing is necessary for proper granulocytic differentiation. We then asked how NELF depletion would affect progenitor cells. Using siRNAs targeting NELF-B or E subunit in human CD34+ HSPCs, more than 80% knockdown efficiency was achieved at the protein level (Fig. 4A, 4B). Moreover, knocking down NELF-B leads to a reduction of NELF-E, and vice versa (Fig. 4A), consistent with the interdependency of NELF subunits. GRO-seq was conducted in cells transfected with a scramble siRNA or either of the NELF-siRNAs. High correlations between biologic replicates were observed (Fig. S3A). Metagene analysis of GRO-seq data revealed a near complete abolishment of Pol II pausing in progenitor cells depleted with either NELF-B or -E (Fig. 4C and S3B). PI calculation and cumulative distribution plot assays further confirmed the genome-wide loss of Pol II pausing in NELF-depleted cells (Fig. 4D, 4E, S3C), in line with a view that NELF is a major pausing factor in HSPCs.
Figure 4. NELF depletion leads to a genome-wide loss of transcription in progenitor cells.
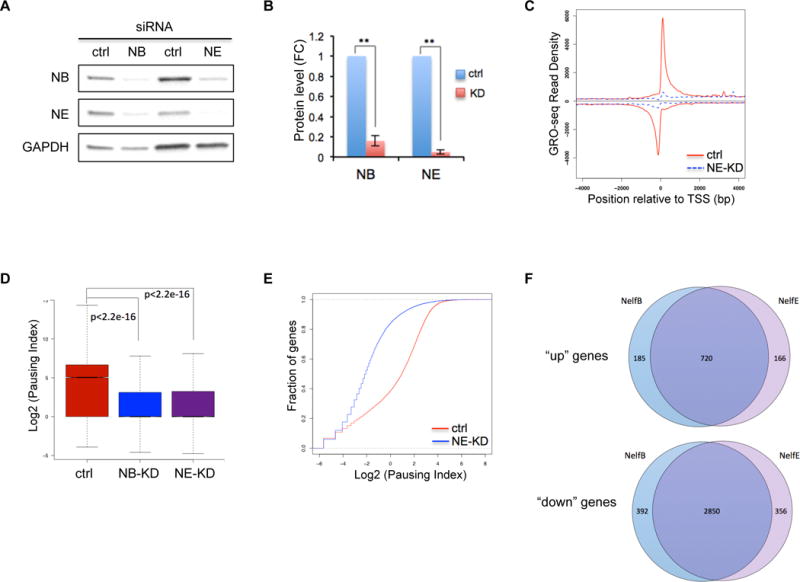
(A) Western blot of NELF-B and -E in human CD34+ HSPCs transfected with a scramble siRNA (ctrl), or siRNA targeting NELF-B or -E. Protein extraction was done on day 3 post-transfection. GAPDH is used as a loading control.
(B) Quantification of western blot in A by imageJ. Protein level is normalized to GAPDH and presented as fold change relative to control samples (n=3, mean ± SEM, **p<0.01).
(C) Metagene analysis showing Pol II occupancy measured by GRO-seq on both sense and anti-sense strands in human CD34+ HSPCs transfected with control-siRNA (ctrl) or siRNA targeting NELF-E (NE-KD). Cells were collected on day 3 post-transfection.
(D) Boxplot analysis to compare pausing index between cells transfected with a control-siRNA (ctrl), or siRNA targeting NELF-B (NB-KD) or –E (NE-KD).
(E) Cumulative distribution function analysis to compare PI distribution in cells transfected with a control-siRNA (ctrl), or siRNA targeting NELF-E (NE-KD)
(F) Venn Diagram showing the overlap of upregulated (“up”) and downregulated genes (“down”) between NELF-B and NELF-E knockdown cells.
We clustered differentially transcribed genes with more than 2-fold change of gene-body associated Pol II level in NELF-depleted cells into “up” and “down” groups. In NELF-E knockdown cells, 886 genes were identified as “up” genes and 3206 genes were identified as “down” genes (p<0.0001), respectively. Similar number of genes was also found in NELF-B knockdown cells (905 for “up” genes and 3242 for “down” genes). Importantly, there is a tremendous overlap (80% for “up” genes and 88% for “down” genes) of dysregulated genes between NELF-B and -E knockdown cells (Fig. 4F), verifying the specificity of NELF knockdown.
Notably, metagene analysis revealed a relatively mild reduction of promoter-associated Pol II along with increased gene-body associated Pol II on upregulated genes (Fig. S3D), consistent with enhanced release of Pol II into elongation upon NELF knockdown. In contrast, the promoter-associated Pol II is almost completely abolished on downregulated genes along with reduced gene-body associated Pol II (Fig. S3D), indicating a complete loss of pausing on these genes upon NELF depletion. Moreover, “down” genes appear to be highly paused with significant higher pausing index (PI) than “up” genes in control cells (Fig. S3E). These results suggest that genes with a high level of pausing may require paused Pol II to maintain promoter accessibility thus transcription, therefore are prone to downregulation when pausing is lost. In contrast, genes with relatively low level of pausing may not rely on paused Pol II to maintain an open promoter, and are more likely to be upregulated through enhanced elongation in the absence of pausing.
NELF depletion in HSPCs promotes granulocytic differentiation
To test if loss of pausing by NELF depletion is sufficient to activate granulocyte gene transcription in progenitor cells, we examined Pol II levels on hematopoietic genes. We found that NELF-depletion in HSPCs causes similar transcription changes as those in early-differentiated granulocytes (Fig. 2D), with an increase of gene-body associated Pol II on granulocyte markers PU.1, CSF3R, ELANE, AZU1 and PRTN3, but an overall reduction of Pol II signal on progenitor genes (GATA2 and TAL1) (Fig. 5A, S4A). In addition, consistent with the activation of JAK2/STAT3 and MEK/ERK signaling pathways during myeloid differentiation, regulators of these pathways are also activated in both granulocytes and NELF-depleted HSPCs (Fig. 5B, S4B). The GRO-seq results were validated by q-RT-PCR on nascent transcripts (Fig. S4C). Moreover, comparison of differentially regulated gene lists revealed that many dysregulated genes are shared between early-differentiated granulocytes and NELF-depleted HSPCs (Fig. S4D). This is further supported by Gene Set Enrichment Analysis (GSEA), which revealed that “up” genes in NELF-depleted cells are markedly enriched for genes induced by granulocytic differentiation, whereas “down” genes in NELF-depleted cells are significantly enriched for genes inhibited by differentiation (Fig. 5C). Together these data strongly suggest a premature activation of granulocytic transcription program in progenitor cells when NELF is depleted.
Figure 5. NELF depletion in HSPCs promotes granulocytic differentiation.
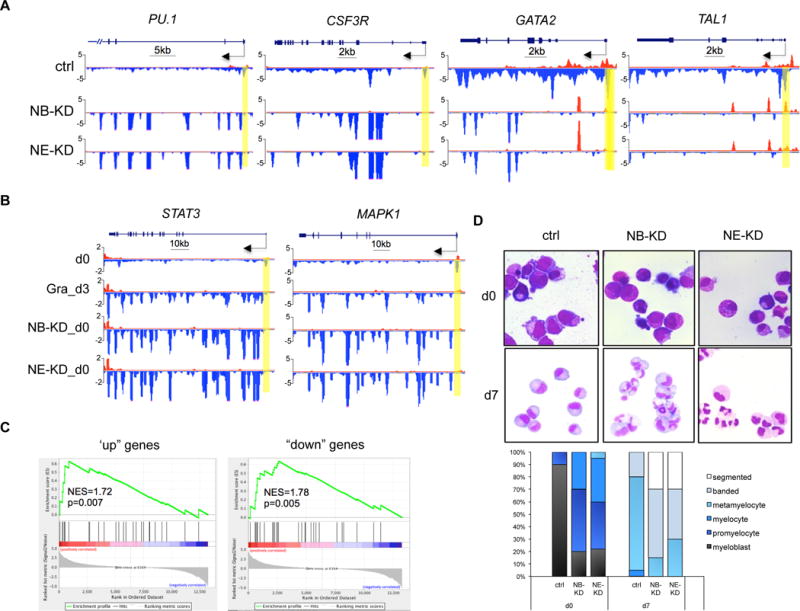
(A) Genome browser captures of GRO-seq for granulocytic differentiation regulator genes (PU.1 and CSF3R) and progenitor genes (TAL1 and GATA2) in NELF-depleted HSPCs. Scale bar and gene diagram are depicted above the captures. Yellow shades highlight promoter peaks.
(B) Genome browser captures of GRO-seq for STAT3 and MAPK1 to compare transcription activity among HSPCs (d0), 3-day differentiated granulocytes (Gra_d3) and NELF-depleted HSPCs (NB-KD_d0 and NE-KD_d0).
(C) GSEA plots to compare differentially regulated genes between NELF-depleted HSPCs and 3-day differentiated granulocytes. Genes upregulated (left) or downregulated (right) in NELF-depleted HSPCs are indicated as black bars in the middle. A strong bias for these genes to also be upregulated (left) or downregulated (right) upon granulocytic differentiation is reflected in the top plots. NES: normalized enrichment score.
(F) MGG staining to compare cell morphology between control cells (ctrl) and NELF knockdown cells during granulocytic differentiation. The percentages of different cell types at various time points during differentiation are shown on the bar charts. Quantitative results represent average percentages by counting 100 cells per experiment from 3 independent experiments.
Consistent with the change of transcription profile, MGG staining revealed that NELF-depleted progenitor cells morphologically resemble early-stage granulocytes (Fig. 5D). Moreover, when induced to differentiation, NELF-depleted cells showed an accelerated maturation than control cells (Fig. 5D). In summary, these data indicate that removing Pol II pausing by depleting NELF promotes granulocytic differentiation.
DISCUSSION
Transcription pausing has emerged as a crucial control point downstream of transcription initiation in transcription regulation26, 27. As a major pausing factor, the NELF complex has been shown to play an important role in regulating the pattern of gene expression during development24, 28, however little is known of how this complex is manipulated to selectively affect gene transcription in different tissues. By combining high-resolution genomic analysis of transcriptionally competent Pol II with a functional dissection of NELF activity in mammalian hematopoietic progenitors, our current work has identified a dynamic change of transcription pausing via regulating NELF protein stability as a novel mechanism regulating hematopoietic differentiation. In both murine myeloblastic cell line and human primary HSPCs, induction of granulocytic differentiation triggers a rapid turnover of NELF protein mediated by the proteasome, leading to a genome-wide reduction of transcription pausing. Functional studies revealed that sustained expression of NELF inhibits granulocytic differentiation, whereas depleting NELF in HSPCs causes loss of Pol II pausing accompanied by activation of granulocytic transcription program and diminished expression of progenitor markers, resulting in accelerated granulocytic differentiation. Thus our studies not only reveal a cellular intrinsic requirement for altering NELF-mediated Pol II pausing during granulocytic differentiation, but also unveiled a cell-context specific mechanism regulating pausing via modulating NELF protein stability.
NELF subunits are interdependent for their protein stability21. Our western blot analysis revealed that NELF-E is the most prominently downregulated subunit in both human and mouse cells, suggesting it as the primary member among the four subunits targeted for active protein turnover upon granulocytic differentiation. Supporting this view, overexpression of NELF-E alone was sufficient to block granulocytic differentiation and partially restore the expression of other subunits. Regulation of protein stability has been found as a mechanism affecting other regulators of Pol II elongation, including the components in the super elongation complexes (SEC) such as AFF1, AFF4 and ELL2, which are regulated by Siah1-mediated polyubiquitination29. The fact that NELF downregulation is found in granulocytes but not erythrocytes suggests the existance of lineage specific regulators. There are multiple ubiquitin E3 ligases highly expressed in granulocyes. Testing these factors in future will help indentify the regulators of NELF protein.
NELF and transcription pausing have been proposed to play both positive and negative roles in transcription. On some genes, loss of pausing leads to gene activation by increasing release of Pol II into their gene body9. On the other hand, previous studies also found that paused Pol II is required for gene expression by antagonizing nucleosome assembly around the promoter to maintain permissive chromatin state11, 12. The identification of both upregulated and downregulated genes in our study is consistent with the dual role of pausing in transcription regulation. Moreover, our results extend previous studies by suggesting that whether pausing inhibits or promotes gene expression may depend on the level of pausing on the target genes. Genes with a high pausing level may rely more on paused Pol II to maintain transcription than genes with a low level of pausing, possibly because high-level paused Pol II competes better with nucleosomes to maintain promoter accessibility.
An important role of preloaded, paused Pol II is to keep genes in a poised state for rapid and synchronized activation upon receipt of developmental cues30–33. A major mechanism to achieve rapid release of paused Pol II is through the recruitment of P-TEFb27, 34–36, which leads to NELF phosphorylation and subsequent dissociation from chromatin. Our finding provides an alternative mechanism for rapid pause release, which is through modulating NELF abundance. To our knowledge, this is the first study showing such a regulation mechanism of NELF. Although this novel regulation of pausing was found in granulocytes but not in erythroid cells, it is known that erythroid differentiation also requires rapid pause release, which is mediated by P-TEFb recruitment as reported in our previous studies37. Thus both mechanisms (P-TEFb recruitment and NELF modulation) are used in hematopoiesis to regulate lineage differentiation. It will be interesting to investigate how distinct mechanisms are selected to regulate different blood lineages. Notably, regulation of Pol II pausing by modulating the abundance of pausing or elongation factors is certainly not limited to hematopoietic tissues. A recent study revealed differential expression of some SEC components between neural stem cells and their differentiated progeny38. Moreover, a variety of human cancer cells have altered expression of NELF subunits39–41. Our work is in line with these studies, suggesting that regulating the abundance of pausing/elongation factors may play an important role in normal and malignant cellular development. Understanding the mechanisms involved in these regulation processes is therefore of the upmost importance for understanding and treating human disease associated with dysregulated transcription elongation.
Supplementary Material
KEY POINTS.
Granulocytic differentiation triggers a decrease of NELF protein abundance.
Downregulation of NELF-mediated transcription pausing is necessary and sufficient for granulocytic differentiation.
Acknowledgments
We thank Anusha Nagari and Tulip Nandu for advices on bioinformatics, and Drs. Murakami and Franco for technical consulting on GRO-seq experiments. This work was supported by NIH (R00DK088963 and R01DK105287 for X.B., R01HL089966 for L.J.H), the Cancer Prevention Research Institute of Texas (CPRIT R1115 for X.B.) and the Cecil H. and Ida Green Center Training Program in Reproductive Biology Sciences Research at UTSW.
Footnotes
Contributions: X.L. performed experiments, analyzed results and contributed to writing; A.A.G. and V.S.M. analyzed GRO-seq data; M.T., K.N., H.Y. and L.J.H. contributed to some experiments; X.B. designed and supervised all experiments and wrote the manuscript.
Conflict-of-interest disclosure: the authors declare no competing financial interests.
References
- 1.Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118(1):1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 2.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13(10):720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiba K, Yamamoto J, Yamaguchi Y, Handa H. Promoter-proximal pausing and its release: molecular mechanisms and physiological functions. Exp Cell Res. 2010;316(17):2723–2730. doi: 10.1016/j.yexcr.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 4.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282(30):21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim Biophys Acta. 2013;1829(1):98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17(24):7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26(16):6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi Y, Takagi T, Wada T, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97(1):41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 11.Gilchrist DA, Nechaev S, Lee C, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22(14):1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilchrist DA, Dos Santos G, Fargo DC, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143(4):540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilchrist DA, Fromm G, dos Santos G, et al. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26(9):933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan H, Qin K, Guo Z, et al. Negative elongation factor controls energy homeostasis in cardiomyocytes. Cell Rep. 2014;7(1):79–85. doi: 10.1016/j.celrep.2014.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo AJ, Moran TB, Schindler YL, et al. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol Cell Biol. 2008;28(8):2675–2689. doi: 10.1128/MCB.01945-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amrein PC, Stossel TP. Prevention of degradation of human polymorphonuclear leukocyte proteins by diisopropylfluorophosphate. Blood. 1980;56(3):442–447. [PubMed] [Google Scholar]
- 17.Franco HL, Nagari A, Kraus WL. TNFalpha signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell. 2015;58(1):21–34. doi: 10.1016/j.molcel.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chae M, Danko CG, Kraus WL. groHMM: a computational tool for identifying unannotated and cell type-specific transcription units from global run-on sequencing data. BMC Bioinformatics. 2015;16:222. doi: 10.1186/s12859-015-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 20.Gupta D, Shah HP, Malu K, Berliner N, Gaines P. Differentiation and characterization of myeloid cells. Curr Protoc Immunol. 2014;104 doi: 10.1002/0471142735.im22f05s104. Unit 22F 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narita T, Yamaguchi Y, Yano K, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23(6):1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muse GW, Gilchrist DA, Nechaev S, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39(12):1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeitlinger J, Stark A, Kellis M, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39(12):1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25(7):742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145(4):502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Kraus WL, Bai X. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem Sci. 2015;40(9):516–525. doi: 10.1016/j.tibs.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife. 2013;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Hsu J, Chan C, Li Z, Zhou Q. The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Mol Cell. 2012;46(3):325–334. doi: 10.1016/j.molcel.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaertner B, Johnston J, Chen K, et al. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep. 2012;2(6):1670–1683. doi: 10.1016/j.celrep.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagha M, Bothma JP, Esposito E, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153(5):976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danko CG, Hah N, Luo X, et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell. 2013;50(2):212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams LH, Fromm G, Gokey NG, et al. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell. 2015;58(2):311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14(7):792–803. [PMC free article] [PubMed] [Google Scholar]
- 35.Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol. 2009;29(5):1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10(10):721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 37.Bai X, Kim J, Yang Z, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142(1):133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu K, Shen D, Shen J, et al. The Super Elongation Complex Drives Neural Stem Cell Fate Commitment. Dev Cell. 2017;40(6):537–551 e536. doi: 10.1016/j.devcel.2017.02.022. [DOI] [PubMed] [Google Scholar]
- 39.Sun J, Watkins G, Blair AL, et al. Deregulation of cofactor of BRCA1 expression in breast cancer cells. J Cell Biochem. 2008;103(6):1798–1807. doi: 10.1002/jcb.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


