Abstract
Introduction
Experiences of racial discrimination have been associated with poor health outcomes. Little is known, however, about how perceived racial discrimination influences DNA methylation (DNAm) among African Americans. Here, we examine the association of experiences of discrimination and DNAm among African American women in the Intergenerational Impact of Genetic and Psychological Factors on Blood Pressure Study (InterGEN).
Methods
The InterGEN study examines the effects of genetic and psychological factors on blood pressure among African American women and their children. Measures include the Major Life Discrimination (MLD) and the Race-Related Events (RES) scales. The 850K EPIC Illumina BeadChip was used for evaluating DNAm in the Epigenome-Wide Association Study (EWAS). In this analysis, we examined discrimination and DNAm at baseline in the InterGEN study.
Results
A total of 152 women contributed data for the RES-EWAS analysis and 147 for the MLD-EWAS analysis. Most were between 30-39 years old, non-smokers, had some college education, and had incomes <$15,000/year. After controlling for age, smoking and cell composition, MLD was significantly associated with DNAm at nine CpG sites (FDR corrected p<.05). For the RES-EWAS analysis,no DNAm sites passed the epigenome-wide significance level after genomic control. Suggestive associations were observed between DNAm and RES at CpG sites after genomic control (raw p<10-5).
Conclusion
We observed significant epigenetic associations between disease-associated genes (e.g. schizophrenia, bipolar disorder and asthma) and perceived discrimination as measured by the MLD scale. Future health disparities research should include epigenetics in high-risk populations to elucidate functional consequences induced by the psychosocial environment.
Keywords: DNA methylation, Epigenomics, African Americans, Women, Racism
Perceived experiences of discrimination have been associated with stress and poor physical and mental health outcomes for adults consistently in nursing and medical literature (Paradies et al., 2015; Williams & Mohammed, 2009), including increased risk of chronic illness such as hypertension (Orom, Sharma, Homish, Underwood, & Homish, 2016) and obesity (Cunningham et al., 2013), as well as preterm birth (Alhusen, Bower, Epstein, & Sharps, 2016) and depression (Molina & James, 2016). These experiences contribute to health disparities between racial/ethnic groups and adversely affect population health (Bailey et al., 2017). Experiences of perceived discrimination or racism have been defined as unfair treatment based on one's race or ethnicity (Clark, Anderson, Clark, & Williams, 1999). This social classification is based on phenotype (Jones, 2001). Studies have increasingly focused on how experiences of interpersonal racism affect biomarkers for disease, such as allostatic load and hormonal dysregulation (Paradies et al., 2015; Williams & Mohammed, 2009). The mechanisms by which these psychosocial experiences affect physical and mental health, however, are still unclear (Brondolo et al., 2011).
One potential mechanism for how negative social exposures (such as perceived discrimination) based on phenotype affect health is through DNAm across the epigenome. DNA methylation (DNAm), or the addition of methyl groups to promoter regions of genes, may effectively silence or activate gene expression. DNAm has been implicated in many disease processes, most notably in cancer (Liang & Weisenberger, 2017), but also in autoimmune diseases (Teruel & Sawalha, 2017), diabetes (Bansal & Pinney, 2017), schizophrenia (Shorter & Miller, 2015), and cardiovascular diseases (Muka et al., 2016). To our knowledge, there are no published studies thus far that have examined how perceived racism and discrimination as an environmental exposure is associated with DNAm across the epigenome among African American women. However, other studies have reported significant interactions between single nucleotide polymorphisms (SNPs) and experiences of discrimination associated with blood pressure among African Americans (Taylor et al., under review; Taylor et al., 2012).
African Americans commonly experience racism or discrimination (Stepanikova & Oates, 2017). A recent national survey by the Pew Research Center found that 71% of African Americans reported being treated unfairly because of their race, and 11% said this discrimination was a regular occurrence (Pew Research Center, 2016). One large, longitudinal study of multiethnic women reported that African Americans had the highest rates of perceived discrimination (35%), compared to Chinese (20%), Hispanic (12%) and Caucasian (3%) women (Jacobs et al., 2014). Considering that African Americans carry a disproportionate burden of incidence, morbidity and mortality from chronic diseases such as hypertension and obesity, (Centers for Disease Control and Prevention (CDC), 2005), it is worthwhile to examine how perceived racism and discrimination affect the epigenome. The purpose of the present study was to examine the influence of perceived racism and discrimination on DNAm in a sample of African American mothers in the Intergenerational Impact of Genetic and Psychological Factors on Blood Pressure Study (InterGEN).
Materials and Methods
The InterGEN study is an ongoing, longitudinal cohort study in Connecticut that examines the effects of genetic, epigenetic, and psychological factors on blood pressure. Recruitment began in April 2015, and 159 mother/child dyads have been enrolled as of April 2017. African American women were recruited from early child-care and education (ECE) centers, primary care clinics, and community events (i.e., health fairs). Eligibility criteria are that women: (a) are ≥ 21 years old; (b) identify as African American or Black (via self-report); (c) speak English; (d) not have a psychiatric or cognitive disorder which may limit accuracy of reporting of study data; and (e) have a biological child three to five years old. Full study procedures have been described elsewhere (Crusto, Barcelona de Mendoza, Connell, Sun, & Taylor, 2016; Taylor, Wright, Crusto, & Sun, 2016).
Trained research assistants approached mothers for recruitment, conducted screening to verify eligibility, and obtained written, informed consent per established study protocols. InterGEN consists of four study visits, each approximately six months apart over the span of two years. During the baseline (Time 1) study visit, clinical measurements of blood pressure, height, weight, are taken, and saliva is collected for DNA analysis from both mother and child. Repeated clinical data and psychological measures are collected at the three follow-up visits. Demographic information, health history and psychological measures (including parenting, experiences of perceived racism and discrimination, and depression) are collected through mother's report using Audio Computer-Assisted Self-Interview (ACASI) software. Although an InterGEN research assistant is present for data collection, social desirability bias is minimized through use of computer-based data collection. For this analysis, we examined the association between discrimination (measured by Major Life Discrimination (MLD) and Race-Related Events (RES)) and DNA methylation (EWAS) at baseline (Time 1 in the InterGEN cohort study). Study procedures were reviewed and approved by Yale University's Institutional Review Board (approval #1311012986). Data are available via written request to the InterGEN study investigators.
Experiences of perceived racism and discrimination were measured using two scales. The 22-item Race-Related Events Scale (RES) assesses exposure to stressful and potentially traumatizing experiences of race-related stress in adults (Waelde et al., 2010). These events include being treated unfairly, harassed or hurt because of their race. Participants indicated if they had ever experienced each event (yes/no), and all 22 items were summed for a total RES score, ranging from 0-22. The RES has demonstrated good reliability (α = .78-.88) in a diverse sample of caregivers recruited from Head Start centers (Crusto, Dantzler, Roberts, & Hooper, 2015).
The 9-item Major Discrimination scale assesses experiences of unfair treatment in adults (Krieger, Smith, Naishadham, Hartman, & Barbeau, 2005; Williams, Yan, Jackson, & Anderson, 1997). Participants indicated if they had ever experienced each major discrimination event, such as being unfairly fired, denied a bank loan, or stopped and searched by the police (yes/no). Each event was attributed to one main reason from 11 options, including gender, race, age, skin color, religion, weight or height, and others. The 9 items were summed (0 = No, 1 = Yes) for a total Major Lifetime Discrimination score (MLD), ranging from 0-9.
Saliva samples for DNA were collected from mothers and children using the Oragene (OG)-500 format tubes (Bahlo et al., 2010), which requires participants to spit until the contents reach the fill line (2 mL). Detailed DNA collection and analysis procedures have been described elsewhere (Taylor et al., 2016). Samples were transported from the field to the research laboratory where they were refrigerated at 4°C until DNA extraction and analysis were completed. Standard protocol for DNA extraction and purification was conducted as indicated in the standard operating procedures guidelines using ReliaPrep kits; more details on DNA and methylation processing can be found in Taylor, Wright, Crusto & Sun, 2016. All tubes and plates that contain an individual's DNA are labeled with a barcode to ensure precise sample tracking, and recorded in the laboratory's computerized freezer inventory via barcodes upon arrival to the laboratory. All DNA pipetting is performed with robotic workstations that incorporate barcode scanning to track the transfer of the biological material from tube to tube, tube to plate, and plate to plate. These customized barcode scanning programs are integrated with the robotics deck configuration and are networked to the laboratory. This concurrent electronic sample tracking ensures that an individual's DNA in each well of every plate generated can be identified for correct merging to genotype calls. These quality control assurance procedures are monitored frequently and updated as needed for continued improvement of sample management. The Illumina Infinium Methylation EPIC (850K) BeadChip was used for epigenome wide DNAm measurement. Quantile-normalization of beta values for autosomal CpG sites was performed. All individual samples passed laboratory based quality control procedures (missing rate <10% and no sex mismatch). CpG sites were excluded if they had detection p-value greater than 0.01 (N=71), had a missing rate greater than 10% (N=514), overlapped with single nucleotide polymorphisms (N=14,184), or were listed in the recent Illumina quality notice (Illumina, 2017) (N=977). A total of 831,658 autosomal and 19,007 X-chromosomal CpG sites were included in the association analyses.
Participants self-reported age in years, whether they smoked cigarettes (yes/no) and other demographic data at the initial interview. We controlled for age and maternal smoking, which are accepted confounders in epigenetic studies (Klebaner et al., 2016). We also adjusted for batch effects, and potential heterogeneity in cell proportions from saliva using the reference-free EWAS (Epigenome-Wide Association Studies) method (Houseman, Molitor, & Marsit, 2014).
All variables were examined using univariate analyses and descriptive statistics, including mean and standard deviation for continuous and frequencies for categorical variables. The correlation coefficient was calculated between the two discrimination variables. The following linear regression model used to study the influence of perceived discrimination on DNAm:
where each discrimination variable (D) was modeled separately, controlling for maternal smoking and age confounding factors, to predict the dependent variable (Y), the beta value of DNAm. We applied the genomic control approach (Devlin & Roeder, 1999) to address residual unmeasured confounding and inflation of type I error. False discovery rate (FDR) was used to correct for multiple comparisons. All analyses were conducted in the R statistical computing environment (R Development Core Team, 2006), and a selection of packages from Bioconductor.
Results
Of the 159 InterGEN participants contributing data for this analysis, six had missing data for the RES discrimination variable, 11 were missing data for the MLD variable, and three were missing maternal smoking status. CpG sites with ≥ 50% missing rate were excluded, and all subjects with complete discrimination data were included in analyses, leaving N=152 for the RES-EWAS analysis and N=147 for the MLD-EWAS analysis. Most participants were between the ages of 30-39 years old, non-smokers, had achieved at least some college education or higher, and had an annual household income of less than $15,000/year. Other demographic information is presented in Table 1. The correlation coefficient was calculated between the two discrimination variables, and was 0.62, indicating moderate correlation between them.
Table 1.
Baseline characteristics of study population, Intergenerational Blood Pressure Study, N=159.*
| n | % | |
|---|---|---|
| Primary covariates | ||
| Age | ||
| 20-29 | 64 | 40.2 |
| 30-39 | 77 | 48.4 |
| 40-49 | 18 | 11.3 |
| Maternal Smoker | ||
| No | 124 | 78.0 |
| Yes | 32 | 20.1 |
| Education | ||
| < High School | 8 | 5.1 |
| High School graduate | 57 | 36.3 |
| Some college/graduate | 92 | 58.6 |
| Annual household income | ||
| ≤$15,000 | 71 | 47.6 |
| >$15,000-$34,999 | 53 | 35.5 |
| >$35,000 | 25 | 16.7 |
| Health insurance type | ||
| Private | 19 | 12.1 |
| Medicaid | 96 | 61.5 |
| Government/ACA | 23 | 14.7 |
| None | 12 | 7.6 |
| Ever received diagnosis of high blood pressure | ||
| No | 131 | 83.9 |
| Yes | 25 | 16.0 |
| Current high blood pressure medication use | ||
| No | 148 | 94.2 |
| Yes | 9 | 5.7 |
| BMI | ||
| Underweight (<18.5) | 9 | 5.6 |
| Normal weight (18.5-24.9) | 40 | 25.1 |
| Overweight (25-29.9) | 44 | 27.6 |
| Obesity (≥30) | 66 | 41.5 |
Numbers may not sum to 159 due to rounding
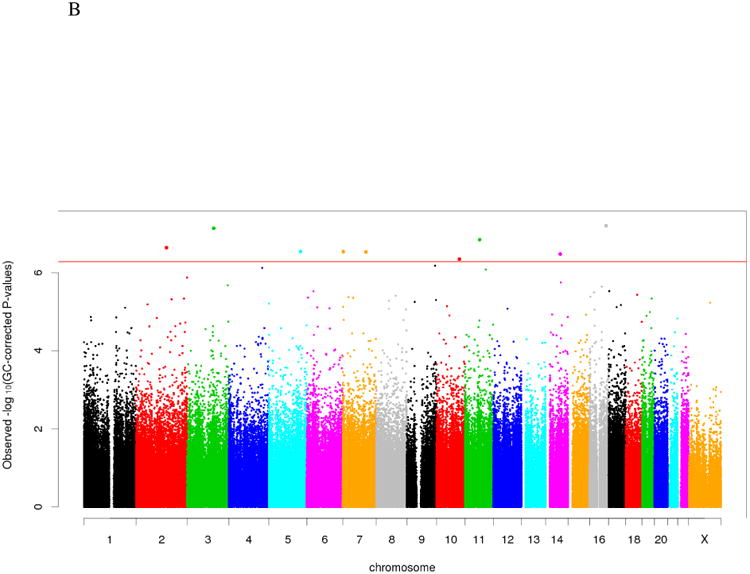
In the EWAS analyses, DNAm at nine CpG sites were found to be associated with MLD after genomic control (FDR corrected p<0.05; which indicates that these associations should not be false positive assuming a 0.05 false discovery rate for the whole epigenome (Figures 1a & 1b). Furthermore, reduced DNAm (hypomethylation) was observed with increasing MLD at most of the significantly associated CpG sites (seven out of nine, 77.8%) (Table 2). However, for the RES-EWAS analysis, no DNAm site passed the epigenome-wide significance level after genomic control. Marginal negative and positive associations were observed between DNAm and RES at six and five CpG sites after genomic control (p<10-5), respectively. No common DNA methylation site was identified in comparing the top 20 epigenetic associations of RES with the epigenome-wide significant associations of MLD.
Figure 1.
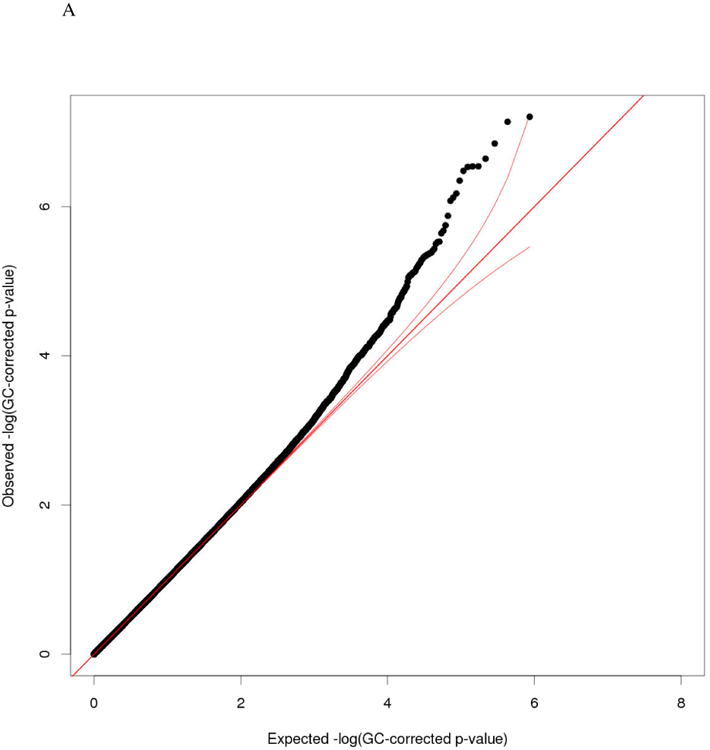
A. QQ plot - Epigenome-wide Association Analysis with MLD score among n=147 AA mothers. (Genomic control approach was applied on raw p-values. Inflation Factor = 1.00)
B. Manhattan plot - Epigenome-wide Association Analysis with MLD score among 147 AA mothers. (Genomic control approach was applied on raw p-values; Red line: FDR adjusted p-value = 0.05)
Table 2.
Epigenome-wide Significant Associations with MLD and RES score among AA mothers (n=147).
| CpG Sites | Chr | bp | Gene | Exposure | Beta | SE | T-stat | P-value after GCa | FDR adjusted p-valueb |
|---|---|---|---|---|---|---|---|---|---|
| cg09437479 | 16 | 78539954 | WWOX | RES | -1.73E-03 | 7.47E-04 | -2.32 | 3.56E-02 | 0.994 |
| MLD | -1.13E-02 | 1.88E-03 | -6.03 | 6.25E-08 | 0.031 | ||||
| cg16143945 | 3 | 126168382 | ZXDC | RES | 5.96E-04 | 1.36E-04 | 4.38 | 1.02E-04 | 0.766 |
| MLD | 2.27E-03 | 3.79E-04 | 6.00 | 7.30E-08 | 0.031 | ||||
| cg03317714 | 11 | 69912795 | LOC101928443 | RES | -1.85E-03 | 5.65E-04 | -3.28 | 3.16E-03 | 0.941 |
| MLD | -8.69E-03 | 1.49E-03 | -5.85 | 1.43E-07 | 0.035 | ||||
| cg19958721 | 2 | 144455439 | ARHGAP15 | RES | -1.45E-03 | 5.67E-04 | -2.56 | 2.07E-02 | 0.981 |
| MLD | -8.23E-03 | 1.43E-03 | -5.74 | 2.28E-07 | 0.035 | ||||
| cg05711042 | 5 | 150913871 | FAT2 | RES | -2.59E-03 | 6.59E-04 | -3.93 | 4.50E-04 | 0.823 |
| MLD | -1.14E-02 | 2.00E-03 | -5.69 | 2.88E-07 | 0.035 | ||||
| cg02578462 | 7 | 2028754 | MAD1L1 | RES | -1.59E-03 | 4.42E-04 | -3.60 | 1.24E-03 | 0.888 |
| MLD | -6.49E-03 | 1.14E-03 | -5.69 | 2.89E-07 | 0.035 | ||||
| cg07726178 | 7 | 110748868 | LRRN3; IMMP2L | RES | -1.62E-03 | 4.10E-04 | -3.95 | 4.27E-04 | 0.808 |
| MLD | -6.58E-03 | 1.16E-03 | -5.68 | 2.94E-07 | 0.035 | ||||
| cg15209133 | 14 | 70020151 | RES | 2.21E-03 | 7.21E-04 | 3.06 | 5.76E-03 | 0.957 | |
| MLD | 1.13E-02 | 1.99E-03 | 5.66 | 3.32E-07 | 0.035 | ||||
| cg04928761 | 10 | 108756503 | SORCS1 | RES | -3.16E-03 | 1.01E-03 | -3.13 | 4.87E-03 | 0.954 |
| MLD | -1.46E-02 | 2.62E-03 | -5.59 | 4.49E-07 | 0.042 |
GC: genomic control;
FDR adjustment applied after GC approach
Discussion
In this study, we found a significant inverse relationship between perceived racial discrimination measured by the MLD scale, and DNAm in a sample of African American women enrolled in the InterGEN study. Women who reported greater perceived discrimination as measured by the MLD scale had decreased DNAm on 9 genes/CpG sites. However, there were no significant associations found between the RES and DNAm.
The genes associated with increased reports of perceived racism and discrimination and hypomethylation were as follows: WWOX, LOC101928443 (uncharacterized in HapMap), ARHGAP15, FAT2, MAD1L1, LRRN3, and SORCS1. The WW domain-containing oxidoreductase (WWOX)is a protein-coding tumor suppressor gene that has been implicated in various cancers such as osteosarcoma (Wen et al., 2017), colorectal (Tian et al., 2017), and bone and breast cancer (Maroni, Matteucci, Bendinelli, & Desiderio, 2017), among others. Epigenetic mechanisms of WWOX have been implicated in influencing the phenotype of certain cancers (Maroni et al., 2017). The functional pathways for WWOX includes the transcriptional regulation by the AP-2 (TFAP2) family of transcription factors and gene expression (Gene Cards, 2017), and have also been associated with lung function (Burdett et al., 2017). ARHGAP15 is also a protein-coding gene that has been associated with cancers and cognitive function in animal models (Sun et al., 2017; Zamboni et al., 2016). The ARHGAP15 gene is associated with three pathways; GPCR downstream signaling, signaling by GPCR and signal transduction (Path Cards, 2017). FAT Atypical Cadherin 2 (FAT2) is another protein-coding tumor suppressor gene that has been linked with skin, spinal, and slower growth of other carcinomas (Cao et al., 2016). ARHGAP15 is also associated with asthma, and interestingly, with educational attainment (Burdett et al., 2017), suggesting education could be used as a proxy phenotype to study genetic influences on mental illness (Okbay et al., 2016). MAD1 Mitotic Arrest Deficient Like 1 (MAD1L1) is another tumor suppression protein-coding gene that has been associated with various cancers, but when hypomethylated, recurrence of tumors has followed (Cui et al., 2016). This locus is also associated with Schizophrenia and bipolar disease (Burdett et al., 2017). However, diverse DNAm profiles have been documented on MAD1L1 across differing multi-ethnic populations residing in the same city (Giuliani et al., 2016). This diversity suggests that the DNAm profile of individuals is influenced by genetic ancestry as well as environmental and demographic stressors (Giuliani et al., 2016).
Leucine Rich Repeat Neuronal 3 (LRRN3) is a well-known protein-coding gene that has been significantly associated with inflammation, cigarette smoking and chronic obstructive pulmonary disease (Martin, Talikka, Hoeng, & Peitsch, 2015; Obeidat et al., 2016; Poussin et al., 2017). The LRRN3 gene has also recently been linked with differential gene expression by age after traumatic brain injury (TBI) (Cho et al., 2016). Lastly, the Sortilin Related VPS10 Domain Containing Receptor 1 (SORCS1) is a protein-coding gene that is also associated with a brain disorder, however unlike LRRN3 it is not TBI but narcolepsy and neurodegeneration in Alzheimer's (Knight et al., 2016; Printy, Verma, Cowperthwaite, Markey, & Alzheimer's Disease Neuroimaging Initiative, 2014; Scheinfeldt et al., 2015).
There were 2 genes in which the significant association between MLD and DNAm was not inversely correlated: one uncharacterized gene and the ZXD Family Zinc Finger C (ZXDC). The ZXDC gene is a protein-coding gene that has been linked with cardiovascular related diseases such as cerebral arterial disease (Shoemaker et al., 2015). Though the relationship between these genes and cardiovascular diseases requires further examination and study, these genes hold promise to improve understanding of the complex and multigenic response to environmental stimuli and how these responses may affect health.
Other studies have examined how perceived racism and discrimination relate to genetics and the development of chronic diseases on the population level, with much of the focus on hypertension and other common, chronic diseases. One such study investigated how SNPs on genes associated with hypertension interacted with experiences of discrimination to influence blood pressure among African Americans enrolled in the Jackson Heart Study (N=2937) (Taylor et al., under review). In that study, two SNPs were identified that interacted significantly with MLD on both systolic and diastolic blood pressure in African Americans (Taylor et al., under review). These SNPs are located near the sodium bicarbonate co-transporter gene (SLC4A5). Others have examined the relationship between genetic polymorphisms and self-reported skin color (as a proxy for racial discrimination) on high blood pressure in a sample of 137 African American women from Detroit (Taylor et al., 2012). They found that one SNP-skin color interaction was significant, also on the SLC4A5 gene (Taylor et al., 2012).
Researchers have also examined how family environment influences DNAm in African American populations. Brody and colleagues studied family environments (including parent-child conflict, parental emotional support and chaotic home environment) and epigenetic aging in adolescents from Georgia using two replication cohorts. They found that supportive family environments buffered epigenetic aging and DNAm among African American adolescents exposed to high levels of discrimination (Brody, Miller, Yu, Beach, & Chen, 2016). They also observed that adolescents in less supportive family environments had faster epigenetic aging in immune system cells in peripheral blood (Brody et al., 2016). Wright and colleagues also observed that higher parenting stress was associated with differential DNAm among a sample of African American women (n=74) also enrolled in the InterGEN study, especially in pathways associated with stress signaling (Wright et al., under review).
In the present study, discrimination was measured two ways, and interestingly, statistically significant results were only observed with the MLD variable. The MLD measures all experiences of discrimination over the lifespan, while the RES only captures those related to race. It may be that the cumulative effects of discrimination measured by the MLD resulted in longer and larger effects on the epigenome, which made it easier to detect epigenetic modifications. A large study of African American and Latino adults (N=617) from New York reported interaction between age and lifetime experiences of discrimination on ambulatory blood pressure, noting that effects of discrimination were not observed in younger participants (Beatty Moody et al., 2016). These findings support the idea that the accumulation of experiences of discrimination may be more important than the type of discrimination, when predicting long-term health effects. The measurement of discrimination can also include influences of structural racism as well as other, less studied indices, so that health disparities research can produce meaningful interventions (Riley, 2017). Further research on specific health outcomes accounting for perceived racism and discrimination and DNAm effects in this population are needed to truly understand and address health disparities.
This study had both strengths and limitations. Strengths included measures of discrimination as an environmental factor that has not traditionally been studied in the context of epigenetics, and in an all African American sample. The choice of saliva for EWAS analysis is also a strength as it is an accessible and acceptable tissue type, and a potential limitation, as its generalizability to other tissues may be limited due to tissue/cell type specificity. Some limitations are that our sample was relatively small, and this may have resulted in inadequate power to fully examine the effects of the RES on DNAm. It is encouraging that several significant CpG sites were identified after stringent controls for multiple testing, confounding, and genomic inflation for MLD. Future replication studies with similar samples are needed to further validate the findings.
In conclusion, this study adds to the sparse literature examining the influence of perceived racial discrimination on DNAm in African Americans. This research is important for nursing practice as a better understanding of social determinants of health and their interplay with epigenetics could lead to improved, individualized treatments for chronic illness, as well as a better delineation of disease risk. Future health disparities research should include how environmental factors contribute to epigenetic changes in this high-risk population. Nursing research can and should incorporate both genome wide and epigenome wide methodologies to explore –omic interactions with perceived racism and discrimination on health outcomes among African Americans.
Acknowledgments
Funding: This work was supported by the National Institutes of Health, National Institute of Nursing Research [R01NR013520].
Footnotes
Declaration of Conflicting Interests: The Authors declare that there are no conflict of interests.
Contributor Information
Veronica Barcelona de Mendoza, Email: veronica.barcelona@yale.edu.
Yunfeng Huang, Email: yunfeng.huang@emory.edu.
Cindy A. Crusto, Email: cindy.crusto@yale.edu.
Yan V. Sun, Email: yan.v.sun@emory.edu.
References
- Alhusen JL, Bower KM, Epstein E, Sharps P. Racial discrimination and adverse birth outcomes: An integrative review. Journal of Midwifery & Women's Health. 2016;61(6):707–720. doi: 10.1111/jmwh.12490[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahlo M, Stankovich J, Danoy P, Hickey PF, Taylor BV, Browning SR, et al. Rubio JP. Saliva-derived DNA performs well in large-scale, high-density single-nucleotide polymorphism microarray studies. Cancer Epidemiology, Biomarkers & Prevention: A Publication of the American Association for Cancer Research, Cosponsored by the American Society of Preventive Oncology. 2010;19(3):794–798. doi: 10.1158/1055-9965.EPI-09-0812[doi]. [DOI] [PubMed] [Google Scholar]
- Bailey ZD, Krieger N, Agenor M, Graves J, Linos N, Bassett MT. Structural racism and health inequities in the USA: Evidence and interventions. Lancet (London, England) 2017;389(10077):1453–1463. doi: 10.1016/S0140-6736(17)30569-X. doi:S0140-6736(17)30569-X[pii] [DOI] [PubMed] [Google Scholar]
- Bansal A, Pinney SE. DNA methylation and its role in the pathogenesis of diabetes. Pediatric Diabetes. 2017;18(3):167–177. doi: 10.1111/pedi.12521[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty Moody DL, Waldstein SR, Tobin JN, Cassells A, Schwartz JC, Brondolo E. Lifetime racial/ethnic discrimination and ambulatory blood pressure: The moderating effect of age. Health Psychology: Official Journal of the Division of Health Psychology, American Psychological Association. 2016;35(4):333–342. doi: 10.1037/hea0000270[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody GH, Miller GE, Yu T, Beach SR, Chen E. Supportive family environments ameliorate the link between racial discrimination and epigenetic aging: A replication across two longitudinal cohorts. Psychological Science. 2016;27(4):530–541. doi: 10.1177/0956797615626703[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondolo E, Hausmann LR, Jhalani J, Pencille M, Atencio-Bacayon J, Kumar A, et al. Schwartz J. Dimensions of perceived racism and self-reported health: Examination of racial/ethnic differences and potential mediators. Annals of Behavioral Medicine: A Publication of the Society of Behavioral Medicine. 2011;42(1):14–28. doi: 10.1007/s12160-011-9265-1[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett T, Hall PN, Hastings E, Hindorff LA, Junkins HA, Klemm AK, et al. Welter D. The NHGRI-EBI catalog of published genome-wide association studies. 2017 doi: 10.1093/nar/gkw1133. Retrieved from https://www.ebi.ac.uk/gwas. [DOI] [PMC free article] [PubMed]
- Cao LL, Riascos-Bernal DF, Chinnasamy P, Dunaway CM, Hou R, Pujato MA, et al. Sibinga NE. Control of mitochondrial function and cell growth by the atypical cadherin Fat1. Nature. 2016;539(7630):575–578. doi: 10.1038/nature20170[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Health disparities experienced by Black or African Americans--United States. MMWR Morbidity and Mortality Weekly Report. 2005;54(1):1–3. doi:mm5401a1[pii] [PubMed] [Google Scholar]
- Cho YE, Latour LL, Kim H, Turtzo LC, Olivera A, Livingston WS, et al. Gill J. Older age results in differential gene expression after mild traumatic brain injury and is linked to imaging differences at acute follow-up. Frontiers in Aging Neuroscience. 2016;8:168. doi: 10.3389/fnagi.2016.00168[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R, Anderson NB, Clark VR, Williams DR. Racism as a stressor for African Americans. A biopsychosocial model. The American Psychologist. 1999;54(10):805–816. doi: 10.1037//0003-066x.54.10.805. [DOI] [PubMed] [Google Scholar]
- Crusto CA, Dantzler J, Roberts YH, Hooper LM. Psychometric evaluation of data from the Race-Related Events Scale. Measurement and Evaluation in Counseling and Development. 2015 Apr 6;:1–12. doi: 10.1177/0748175615578735. 2015. [DOI] [Google Scholar]
- Crusto CA, Barcelona de Mendoza V, Connell CM, Sun YV, Taylor JY. The intergenerational impact of genetic and psychological factors on blood pressure study (InterGEN): Design and methods for recruitment and psychological measures. Nursing Research. 2016;65(4):331–338. doi: 10.1097/NNR.0000000000000163[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Lu Z, Yang L, Gao Y, Liu W, Gu L, et al. Sun ZS. Genome-wide identification of differential methylation between primary and recurrent hepatocellular carcinomas. Molecular Carcinogenesis. 2016;55(7):1163–1174. doi: 10.1002/mc.22359[doi]. [DOI] [PubMed] [Google Scholar]
- Cunningham TJ, Berkman LF, Kawachi I, Jacobs DR, Jr, Seeman TE, Kiefe CI, Gortmaker SL. Changes in waist circumference and body mass index in the US CARDIA cohort: Fixed-effects associations with self-reported experiences of racial/ethnic discrimination. Journal of Biosocial Science. 2013;45(2):267–278. doi: 10.1017/S0021932012000429[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Gene Cards. WWOX gene. 2017 Retrieved from http://www.genecards.org/cgi-bin/carddisp.pl?gene=WWOX.
- Giuliani C, Sazzini M, Bacalini MG, Pirazzini C, Marasco E, Fontanesi E, et al. Garagnani P. Epigenetic variability across human populations: A focus on DNA methylation profiles of the KRTCAP3, MAD1L1 and BRSK2 genes. Genome Biology and Evolution. 2016;8(9):2760–2773. doi: 10.1093/gbe/evw186[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics (Oxford, England) 2014;30(10):1431–1439. doi: 10.1093/bioinformatics/btu029[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina. April 2017 quality notice PQN0223. 2017 doi: https://mkt.illumina.com/rs/600-XEX-927/images/PQN0223_Methylation%20EPIC.pdf?mkt_tok=eyJpIjoiTURSa01ESm1aRGRtT0RnNCIsInQiOiJCcFBMc0pEUkFqUVwvc3g1OFJuOUdTYW1NWThIXC9EenVaemI4QkRzcXJDUGtcL0J0T1NkM05SaGxWcHNsZElEY0RyXC9jVFNNVnFGZ01seFVnZHlOSjFGMjdmVFwvVWxtR3.
- Jacobs EA, Rathouz PJ, Karavolos K, Everson-Rose SA, Janssen I, Kravitz HM, et al. Powell LH. Perceived discrimination is associated with reduced breast and cervical cancer screening: The Study of Women's Health Across the Nation (SWAN) Journal of Women's Health. 2014;23(2):138–145. doi: 10.1089/jwh.2013.4328[doi]. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP. Invited commentary: “Race,” racism, and the practice of epidemiology. American Journal of Epidemiology. 2001;154(4):299–304. doi: 10.1093/aje/154.4.299. discussion 305-6. [DOI] [PubMed] [Google Scholar]
- Klebaner D, Huang Y, Hui Q, Taylor JY, Goldberg J, Vaccarino V, Sun YV. X chromosome-wide analysis identifies DNA methylation sites influenced by cigarette smoking. Clinical Epigenetics. 2016;8 doi: 10.1186/s13148-016-0189-2[doi]. 20-016-0189-2. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight EM, Ruiz HH, Kim SH, Harte JC, Hsieh W, Glabe C, et al. Gandy S. Unexpected partial correction of metabolic and behavioral phenotypes of Alzheimer's APP/PSEN1 mice by gene targeting of diabetes/Alzheimer's-related Sorcs1. 16-016-0282-y. Acta Neuropathologica Communications. 2016;4 doi: 10.1186/s40478-016-0282-y[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger N, Smith K, Naishadham D, Hartman C, Barbeau EM. Experiences of discrimination: Validity and reliability of a self-report measure for population health research on racism and health. Social Science & Medicine (1982) 2005;61(7):1576–1596. doi: 10.1016/j.socscimed.2005.03.006. doi:S0277-9536(05)00097-3[pii] [DOI] [PubMed] [Google Scholar]
- Liang G, Weisenberger DJ. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics. 2017:1–17. doi: 10.1080/15592294.2017.1311434[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroni P, Matteucci E, Bendinelli P, Desiderio MA. Functions and epigenetic regulation of WWOX in bone metastasis from breast carcinoma: Comparison with primary tumors. International Journal of Molecular Sciences. 2017;18(1) doi: 10.3390/ijms18010075. 10.3390/ijms18010075. doi:E75[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Talikka M, Hoeng J, Peitsch MC. Identification of gene expression signature for cigarette smoke exposure response--from man to mouse. Human & Experimental Toxicology. 2015;34(12):1200–1211. doi: 10.1177/0960327115600364[doi]. [DOI] [PubMed] [Google Scholar]
- Molina KM, James D. Discrimination, internalized racism, and depression: A comparative study of African American and Afro-Caribbean adults in the US. Group Processes & Intergroup Relations: GPIR. 2016;19(4):439–461. doi: 10.1177/1368430216641304[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muka T, Koromani F, Portilla E, O'Connor A, Bramer WM, Troup J, et al. Franco OH. The role of epigenetic modifications in cardiovascular disease: A systematic review. International Journal of Cardiology. 2016;212:174–183. doi: 10.1016/j.ijcard.2016.03.062[doi]. [DOI] [PubMed] [Google Scholar]
- Obeidat M, Ding X, Fishbane N, Hollander Z, Ng RT, McManus B, et al. Sin DD. The effect of different case definitions of current smoking on the discovery of smoking-related blood gene expression signatures in chronic obstructive pulmonary disease. Nicotine & Tobacco Research: Official Journal of the Society for Research on Nicotine and Tobacco. 2016;18(9):1903–1909. doi: 10.1093/ntr/ntw129[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. Benjamin DJ. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533(7604):539–542. doi: 10.1038/nature17671[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom H, Sharma C, Homish GG, Underwood W, 3rd, Homish DL. Racial discrimination and stigma consciousness are associated with higher blood pressure and hypertension in minority men. Journal of Racial and Ethnic Health Disparities. 2016 doi: 10.1007/s40615-016-0284-2[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies Y, Ben J, Denson N, Elias A, Priest N, Pieterse A, et al. Gee G. Racism as a determinant of health: A systematic review and meta-analysis. PloS One. 2015;10(9):e0138511. doi: 10.1371/journal.pone.0138511[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Path Cards. Pathway network for signaling by GPCR. 2017 Retrieved from http://pathcards.genecards.org/card/signaling_by_gpcr.
- Pew Research Center. On views of race and inequality, whites and blacks are worlds apart. 2016 Retrieved from http://www.pewsocialtrends.org/2016/06/27/on-views-of-race-and-inequality-blacks-and-whites-are-worlds-apart/
- Poussin C, Belcastro V, Martin F, Boue S, Peitsch MC, Hoeng J. Crowd-sourced verification of computational methods and data in systems toxicology: A case study with a heat-not-burn candidate modified risk tobacco product. Chemical Research in Toxicology. 2017;30(4):934–945. doi: 10.1021/acs.chemrestox.6b00345[doi]. [DOI] [PubMed] [Google Scholar]
- Printy BP, Verma N, Cowperthwaite MC, Markey MK Alzheimer's Disease Neuroimaging Initiative. Effects of genetic variation on the dynamics of neurodegeneration in Alzheimer's disease. Conference Proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual Conference. 2014;2014:2464–2467. doi: 10.1109/EMBC.2014.6944121[doi]. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R foundation for statistical computing; Vienna, Austria: 2006. R: A language and environment for statistical computing. Retrieved from http://www.R-project.org. [Google Scholar]
- Riley AR. Neighborhood disadvantage, residential segregation, and beyond-lessons for studying structural racism and health. Journal of Racial and Ethnic Health Disparities. 2017 doi: 10.1007/s40615-017-0378-5[doi]. [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, Gharani N, Kasper RS, Schmidlen TJ, Gordon ES, Jarvis JP, et al. Christman MF. Using the coriell personalized medicine collaborative data to conduct a genome-wide association study of sleep duration. American Journal of Medical Genetics Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics. 2015;168(8):697–705. doi: 10.1002/ajmg.b.32362[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker LD, Clark MJ, Patwardhan A, Chandratillake G, Garcia S, Chen R, et al. Steinberg GK. Disease variant landscape of a large multiethnic population of moyamoya patients by exome sequencing. G3 (Bethesda, Md) 2015;6(1):41–49. doi: 10.1534/g3.115.020321[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter KR, Miller BH. Epigenetic mechanisms in schizophrenia. Progress in Biophysics and Molecular Biology. 2015;118(1-2):1–7. doi: 10.1016/j.pbiomolbio.2015.04.008[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanikova I, Oates GR. Perceived discrimination and privilege in health care: The role of socioeconomic status and race. American Journal of Preventive Medicine. 2017;52(1S1):S86–S94. doi: 10.1016/j.amepre.2016.09.024. doi:S0749-3797(16)30466-4[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Zhang B, Wang C, Fu T, Li L, Wu Q, et al. Wang J. Forkhead box P3 regulates ARHGAP15 expression and affects migration of glioma cells through the Rac1 signaling pathway. Cancer Science. 2017;108(1):61–72. doi: 10.1111/cas.13118[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JY, Sun YV, Barcelona de Mendoza V, Ifatunji M, Rafferty J, Fox E, et al. Jackson JS. The combined effects of genetic risk and perceived discrimination on blood pressure among African Americans in the Jackson Heart Study. doi: 10.1097/MD.0000000000008369. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JY, Wright ML, Crusto CA, Sun YV. The intergenerational impact of genetic and psychological factors on blood pressure (InterGEN) study: Design and methods for complex DNA analysis. Biological Research for Nursing. 2016 doi: 10.1177/1099800416645399. doi:1099800416645399[pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JY, Wu CY, Darling D, Sun YV, Kardia SL, Jackson JS. Gene-environment effects of SLC4A5 and skin color on blood pressure among African American women. Ethnicity & Disease. 2012;22(2):155–161. [PMC free article] [PubMed] [Google Scholar]
- Teruel M, Sawalha AH. Epigenetic variability in systemic lupus erythematosus: What we learned from genome-wide DNA methylation studies. Current Rheumatology Reports. 2017;19(6) doi: 10.1007/s11926-017-0657-5[doi]. 32-017-0657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Chen C, Yang F, Tang J, Pei J, Shi B, et al. Zhang J. Establishment of apoptotic regulatory network for genetic markers of colorectal cancer and optimal selection of traditional Chinese medicine target. Saudi Journal of Biological Sciences. 2017;24(3):634–643. doi: 10.1016/j.sjbs.2017.01.036[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waelde LC, Pennington D, Mahan C, Mahan R, Kabour M, Marquett R. Psychometric properties of the race-related events scale. Psychological Trauma: Theory, Research, Practice and Policy. 2010;2(1):4–11. [Google Scholar]
- Wen J, Xu Z, Li J, Zhang Y, Fan W, Wang Y, et al. Li J. Decreased WWOX expression promotes angiogenesis in osteosarcoma. Oncotarget. 2017 doi: 10.18632/oncotarget.17126[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Mohammed SA. Discrimination and racial disparities in health: Evidence and needed research. Journal of Behavioral Medicine. 2009;32(1):20–47. doi: 10.1007/s10865-008-9185-0;10.1007/s10865-008-9185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Yan Y, Jackson JS, Anderson NB. Racial differences in physical and mental health: Socio-economic status, stress and discrimination. Journal of Health Psychology. 1997;2(3):335–351. doi: 10.1177/135910539700200305[doi]. [DOI] [PubMed] [Google Scholar]
- Wright ML, Huang Y, Hui Q, Newhall K, Crusto C, Sun Y, Taylor JY. Parenting stress and DNA methylation among African Americans inthe InterGEN study. doi: 10.1017/cts.2018.3. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni V, Armentano M, Saro G, Ciraolo E, Ghigo A, Germena G, et al. Merlo GR. Disruption of ArhGAP15 results in hyperactive Rac1, affects the architecture and function of hippocampal inhibitory neurons and causes cognitive deficits. Scientific Reports. 2016;634877 doi: 10.1038/srep34877[doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]


