Abstract
The dramatic increase in the prevalence and clinical impact of infections caused by bacteria producing carbapenemases is a global health concern. These carbapenemase-producing organisms (CPO) are especially problematic when encountered in members of the family Enterobacteriaceae. Due to their ability to readily spread and colonize patients in health care environments, preventing the transmission of these organisms is a major public health initiative and coordinated international efforts are needed to contain the risk of infection. Central to the treatment and control of CPO are phenotypic- (growth-/biochemical-dependent) and nucleic acid-based carbapenemase detection tests that identify carbapenemase activity directly or their associated molecular determinants. Importantly, bacterial isolates harboring carbapenemases are often resistant to multiple antibiotic classes resulting in limited therapy options. Emerging agents, novel antibiotic combinations and treatment regimens offer promise for management of these infections. This review highlights our current understanding of CPO with emphasis on their epidemiology, detection, treatment, and control.
Keywords: Carbapenem-producing organisms, Carbapenem-resistant Enterobacteriaceae, Carbapenemase, Metallo-beta-lactamase, Carbapenemase detection tests, Whole-genome sequencing, Antimicrobial therapy
Introduction
One of the most concerning forms of antimicrobial resistance (AMR) is resistance to the carbapenems, especially when observed in members of the family Enterobacteriaceae. A primary mechanism of carbapenem resistance in Gram-negative bacteria is acquired carbapenemases, enzymes that hydrolyze these antibiotics. In this review, the epidemiology, laboratory detection, approaches to combat widespread dissemination, and treatment strategies for carbapenemase-producing organisms (CPO), especially carbapenemase-producing carbapenem-resistant Enterobacteriaceae (CP-CRE), will be discussed.
The Biology and Epidemiology of CPO
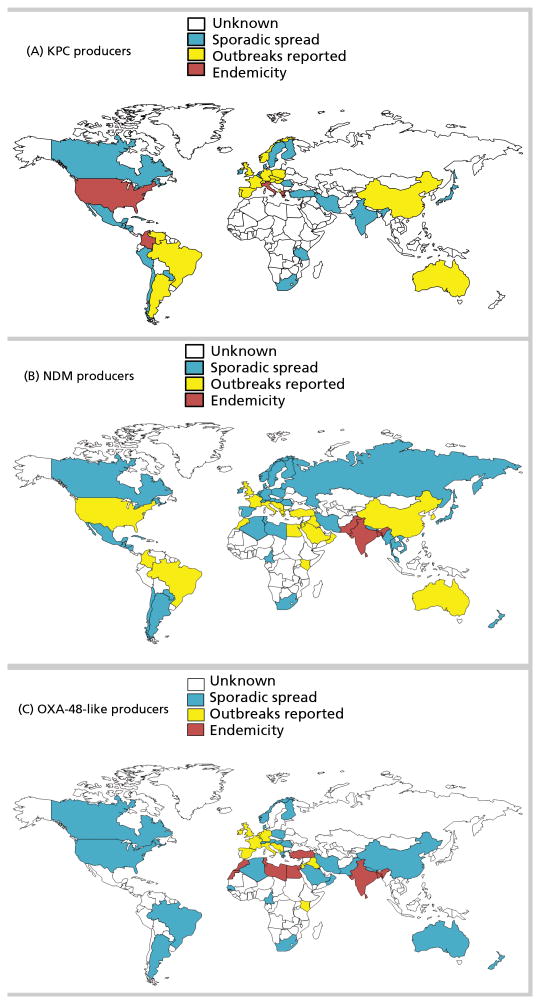
Phenotypic resistance to carbapenems in Gram-negative bacteria commonly results from acquisition of carbapenemases, or production of cephalosporinases combined with mutations that decrease permeability of the bacterial cell wall to entry of carbapenems. CPO may exhibit significant variation in carbapenem minimum inhibitory concentration (MIC) values depending on their permeability status, the rate of carbapenem hydrolysis by the associated enzyme, and the level of gene expression [1]. Carbapenemases belong to Ambler classes A, B, or D, with class A and D enzymes possessing a serine-based hydrolytic mechanism, and class B enzymes requiring one or two zinc ions for their catalytic activity [1]. There is a rare instance of class C beta-lactamase that can hydrolyze imipenem (CMY-10) [2]. Globally distributed in many genera of bacteria, certain carbapenemases are typically associated with specific regions or countries (Figure 1). However, in an era of widespread international travel and exposure to medical care, the association between a specific resistance mechanism and a given region or country may change, creating an urgent need for routine local and national surveillance.
Figure 1.
Worldwide distribution of carbapenemases. A) KPC producers in Enterobacteriaceae and P. aeruginosa. B) NDM producers in Enterobacteriaceae and P. aeruginosa. C) OXA-48 producers in Enterobacteriaceae.
The class A Klebsiella pneumoniae carbapenemase (KPC) has been extensively reported in K. pneumoniae and other Enterobacteriaceae, but has also been identified in other Gram-negative pathogens including Pseudomonas aeruginosa [3]. KPC-producing K. pneumoniae are widespread in the United States, but are also endemic in some European countries such as Greece and Italy (Figure 1) [4].
Class B beta-lactamases, or metallo-beta-lactamases (MBL), are commonly identified in Enterobacteriaceae and in P. aeruginosa [5]. Among the MBLs, New Delhi Metallo-beta-lactamase (NDM)-, Verona Integron-encoded Metallo-beta-lactamase (VIM)-, and Imipenemase Metallo-beta-lactamase (IMP) enzymes, are the most frequently identified worldwide (Figure 1) [5]. IMP-producers are mainly detected in China, Japan, and Australia, mostly in Acinetobacter baumannii. VIM-producers are most often found in Italy and Greece (Enterobacteriaceae), and in Russia (P. aeruginosa) [6,7].
Acquired class D carbapenem-hydrolyzing beta-lactamases are commonly reported in A. baumannii (mainly OXA-23-, OXA-40-, and OXA-58-like enzymes), but not in P. aeruginosa. OXA-48 and derivatives (e.g., OXA-181 and OXA-232), have been detected in Enterobacteriaceae, hydrolyze narrow-spectrum beta-lactams and weakly hydrolyze carbapenems, but spare broad-spectrum cephalosporins [8]. OXA-48-producing Enterobacteriaceae have been endemic in Turkey since 2004, and are now also frequently discovered in several European countries (e.g., France and Belgium), and across North Africa (Figure 1) [9]. Ten variants of OXA-48 beta-lactamases are acknowledged and are increasingly reported worldwide [9], notably among nosocomial K. pneumoniae and community Escherichia coli isolates [10].
Carbapenemase genes are often located on mobile genetic elements further enhancing their spread. For example, the widespread dissemination of the blaOXA-48 gene was shown to be related to a successful and epidemic plasmid that conjugates at high rates within Enterobacteriaceae [11].
Other less common carbapenemases belonging to a variety of molecular classes (e.g., class A FRI-1 and IMI-like beta-lactamases, class B SPM-1 and GIM-1, and class D OXA-198) are reported sporadically and are found in specific species, likely because the corresponding genes are located on narrow host-range plasmids or chromosomes, which makes wide diffusion unlikely [10,12].
Laboratory Detection of CPO
Detection of carbapenemase-mediated carbapenem resistance is essential for patient management, infection control, and public health surveillance. The diversity of these enzymes and the range of associated susceptibility phenotypes makes detection challenging. Selection of a carbapenemase detection test (CDT) is contingent on several factors: epidemiology, diagnostic performance, labor intensity, complexity, and cost. The relative importance of turnaround time depends on whether the assay will be employed for therapeutic decision making and/or infection control or surveillance studies.
CDTs are broadly differentiated into two groups: phenotypic- (growth-/biochemical-dependent) and nucleic acid-based. Phenotypic assays monitor carbapenemase activity through a variety of methods: growth of a susceptible reporter strain following drug inactivation by a carbapenemase-producing test strain, observation of a pH change after beta-lactam ring hydrolysis, detection of carbapenem hydrolysis products, or via inhibition with small molecules. In contrast, nucleic acid assays detect genetic determinants associated with carbapenemases.
The modified Hodge test (MHT) is probably the most extensively described CDT used in Enterobacteriaceae. This assay demonstrates acceptable sensitivity for most carbapenemases, especially KPC enzymes, but low sensitivity for NDM-producing strains [13,14]. Additionally, it has poor specificity; isolates encoding cephalosporinases in conjunction with porin mutations often produce false-positive results [13,15]. While the MHT is inexpensive and uncomplicated to perform, it is often difficult to interpret and requires an additional 24-hour growth step after AST results are obtained.
Conceptually akin to the MHT, the carbapenem inactivation method (CIM) assesses growth of a susceptible reporter strain around a carbapenem disk previously incubated with a suspension of a suspected carbapenemase-producing test strain. If the test strain produces a carbapenemase, drug in the disk will be inactivated thus allowing growth of the reporter strain up to the edge of the disk, whereas a zone of growth inhibition indicates the antibiotic in the disk remains active, thus the test strain lacks carbapenemase activity. CIM sensitivity is reported to be between 98 and 100% [16,17], but again this technique requires an additional 24-hour culture step. A modified version of the CIM (mCIM) was evaluated in a multi-center study, demonstrating 97% sensitivity and 99% specificity for detection of carbapenemase production in Enterobacteriaceae [18]. Based on those data, the mCIM was added to the CLSI M100 document as a reliable method for detection of carbapenemase production in Enterobacteriaceae [19].
The Carba NP test (RAPIDEC® CARBA NP, bioMérieux, Durham, NC), its derivatives, and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), monitor the hydrolysis of carbapenems using bacterial extracts [20,21] and produce same-day results. In the Carba NP test, carbapenemase-dependent hydrolysis of imipenem causes a decrease in pH, registered by a pH indicator as a color change. The test exhibits excellent sensitivity [20], although the recognition of OXA-48-producing isolates may be challenging [17,22]. To aid in early identification, the Carba NP test has been successfully extended to detect the presence of CPO in positive blood cultures even before isolation of organism on solid media, providing value for antibiotic stewardship [23].
MALDI-TOF MS can identify carbapenem degradation products following incubation of a bacterial protein extract with a carbapenem substrate. Overall, the sensitivity of MALDI-TOF MS for this purpose is high, and sensitivity for OXA-48-producing isolates is enhanced by inclusion of bicarbonate in the reaction buffer [22]. Notwithstanding the promise of mass spectrometry-based assays, because they are complex to perform and interpret, widespread implementation in clinical microbiology laboratories may be unfeasible.
Conventional AST methods such as broth microdilution, disk diffusion, and gradient diffusion can be modified to detect different classes of carbapenemases by performing them in the absence and presence of small molecule inhibitors including phenylboronic acid, which inhibits serine active site enzymes, and ethylenediaminetetraacetic acid, an inhibitor of MBL activity. These assays have reportedly high sensitivities and specificities [24,25,26,27,28], are inexpensive, and generally easy to implement and interpret but require overnight incubation.
Nucleic acid-based CDT include commercially available and laboratory-developed PCR and microarray platforms to detect carbapenemase genes in bacterial isolates or directly from clinical specimens. They exhibit clinically relevant sensitivities and specificities and have same-day turnaround times [29,30,31,32,33], but are typically associated with high costs. In the setting of changing epidemiology or emergence of novel enzymes, the specificity of targeted PCR- or microarray-based platforms could be a shortcoming.
Whole-genome sequencing (WGS) platforms potentially represent the ultimate molecular CDT by interrogating the entire genomic content, chromosomal and extrachromosomal, of a bacterium to identify carbapenem resistance determinants [34,35,36]. Furthermore, WGS data provide an opportunity to query for extra information, including strain relatedness, plasmid types encoding the carbapenemase, other factors influencing carbapenem resistance (e.g., porin mutations), presence of additional resistance factors, and data can be analyzed in near real-time or archived for future inquiry. Notwithstanding the power and promise of WGS, these assays are still the purview of advanced clinical microbiology and public health laboratories, and require considerable expertise to perform and interpret. As algorithms improve, costs decrease, and commercialized options are brought to market, the clinical workforce is likely to become increasingly proficient at performing and interpreting these data allowing WGS to gain wider acceptance.
WGS for Investigation of the Epidemiology and Diversity of CPO
Recent studies indicate that WGS, combined with hospital epidemiology, may facilitate the tracking of transmissions within healthcare facilities with the level of precision necessary to guide the modification of infection control procedures and limit the spread of healthcare-associated infections [36,37,38,39]. One example is the National Institutes of Health Clinical Center outbreak in which a single patient colonized on admission with KPC-producing K. pneumoniae was eventually linked to CPO colonization in 18 additional patients. The epidemiologic data could not discriminate between undetected transmission from the index patient or introduction of a second strain. The extensive genetic similarity among KPC-producing K. pneumoniae in the United States prevented a definitive match to the index patient using standard outbreak investigation tools such as pulsed-field gel electrophoresis or repetitive element PCR. WGS revealed direct linkage of the index patient, with transmission originating from three different anatomic sites [34], indicating silent colonization, even in immunocompromised patients. In another healthcare-related outbreak, WGS was instrumental in identifying limited healthcare-associated transmission of CRE against a background of sporadic introduction of multiple other strains [36]. In other studies, WGS was key in determining the phylogeny of carbapenem resistant Enterobacter species and how gene regulation by insertion sequence elements impacted carbapenem and multidrug resistance in A. baumannii [40,41]. WGS has also been used to create a reference set capturing the diversity of plasmids and mobile elements that carry the KPC gene [36,42].
Novel Treatment Strategies for CPO
Treatment of CPO, especially CP-CRE, remains problematic. Patients with CP-CRE infection suffer unacceptably high mortality, emphasizing the need for novel diagnostics and therapies. Studies performed to date demonstrate a bias to report trials of successful combination chemotherapy, informed largely by results from in vitro studies. In most trials targeting CP-CRE, combination therapies have included the use of i) colistin (polymyxin E) and a carbapenem, ii) colistin and tigecycline, or colistin and fosfomycin, or iii) double carbapenem therapy. Interestingly, it was also shown in vitro that dual carbapenem combinations might work against carbapenemase-producing strains, with significant synergies observed when using imipenem and another carbapenem [43].
In an early study performed at a tertiary care center, Qureshi and colleagues reported that 28-day mortality was 13.3% in the combination therapy group (colistin and another agent) compared with 57.8% in the monotherapy group (P= 0.01), and that combination regimens were independently associated with better survival (P=0.02) [44]. Additionally, a multi-center retrospective cohort study conducted in three large Italian teaching hospitals examined death within 30 days of the first positive blood culture among 125 patients with blood stream infections caused by KPC-producing K. pneumoniae [45]. That investigation found 54.3% mortality in the monotherapy arm versus 34.1% mortality in the combination therapy group (P = 0.02); triple combination therapy (tigecycline, colistin, and meropenem) was associated with lowest mortality (P = 0.01). This study also revealed that patients infected by CP-CRE with imipenem MIC values of ≥4 μg/mL had worse outcomes than patients whose isolates had an MIC value of ≤2 μg/mL. The “dividing line” appears to be an MIC value between 2 and 4 μg/mL and predicted differences in mortality were notable (16.1% versus 76.9%; P <0.01); each imipenem MIC doubling dilution increased the probability of death two-fold.
In a subsequent review of 20 clinical studies involving 414 patients, Tzouvelekis and colleagues reported that a single active agent resulted in mortality rates not significantly different from those observed in patients administered no active therapy [46]. Consistent with the notions reported above, combination therapy with two or more agents active in vitro was superior to monotherapy, providing a clear survival benefit (mortality rate, 27.4% versus 38.7%; P <0.001). The lowest mortality rate (18.8%) was observed in patients treated with carbapenem-containing combinations.
In contrast, Falagas and partners in 2014 reported the largest meta-analysis performed to date [47], examining 20 studies involving 692 patients. Surprisingly, the authors reported 50% mortality in patients treated with tigecycline and gentamicin, 64% mortality for tigecycline and colistin, and 67% mortality for carbapenems and colistin. This comprehensive analysis called into question the conclusions drawn from the earlier retrospective, nonrandomized studies, and emphasized that unexplained molecular heterogeneity, and non-uniform microbiology testing might be confounding results. These differences suggest that studies concluding the superiority of combination therapy over monotherapy may not be sufficiently rigorous for us to accept their conclusions.
What about new drugs in development? Avibactam is a synthetic non-beta-lactam, bicyclic diazabicyclooctane beta-lactamase inhibitor (DBO), that inhibits the activities of Ambler class A and C beta-lactamases and some Ambler class D enzymes. Avibactam closely resembles portions of the cephem bicyclic ring system, and has been shown to bond covalently to beta-lactamases. Against carbapenemase-producing K. pneumoniae, the addition of avibactam significantly improves the activity of ceftazidime in vitro (∼four-fold MIC reduction). In surveillance studies, the combination of ceftazidime with avibactam restores in vitro susceptibility against all ESBLs and most KPCs tested. Studies comparing outcomes of infections with KPC-producing Gram-negative bacteria treated with ceftazidime-avibactam as monotherapy or in combination with colistin are ongoing. An important study comparing the outcomes of patients infected with CRE treated with colistin vs. ceftazidime/avibactam was recently performed [48]. Patients initially treated with either ceftazidime-avibactam or colistin for CRE infections were selected from the Consortium on Resistance Against Carbapenems in Klebsiella and other Enterobacteriaceae (CRACKLE), a prospective, multicenter, observational study. Thirty-eight patients were treated first with ceftazidime-avibactam and 99 with colistin either as monotherapy or combination therapy. Patients treated with ceftazidime-avibactam vs colistin (monotherapy or combination) had a higher probability of a better outcome as compared to patients treated with colistin. This study strengthens the notion that treatment with a highly active agent as monotherapy in the appropriate clinical setting may be better than therapy with a less desirable agent singly or in combination.
Relebactam, also a DBO, combined with imipenem/cilistatin, will soon be evaluated in clinical studies [49]. In vitro studies indicate that imipenem/cilistatin-relebactam is comparable to ceftazidime-avibactam. The role of the combination of imipenem versus ceftazidime with different DBOs remains to be defined.
The United States Food and Drug Administration (FDA) recently approved ceftazidime-avibactam based on data obtained in Phase II/III trials of complicated urinary tract infections and intra-abdominal infections (ceftazidime-avibactam combined with metronidazole). Despite encouraging results, the FDA cautioned that ceftazidime-avibactam should be reserved for situations when there are limited or no alternative drugs for treating an infection. The concern was that resistance to ceftazidime-avibactam would emerge in KPC-producing strains. Regrettably resistance is already being reported due to mutations occurring in the KPC enzyme and porin changes [50,51]
In summary, combination chemotherapies seem to be effective against KPC-producing bacteria (Table 1) [49], but we still need to design the right trial to answer the fundamental question as to why. We also need to carefully examine new drugs in the pipeline, and use clinical trials to define their best use. Other drugs in development are summarized in Table 2. The reader will note that there are some drugs specifically targeted for MBL producers (aztreonam/avibactam and cefidericol); these developments are awaited in earnest. Novel combinations (ceftazidime/avibactam paired with aztreonam) are also being explored [52]. In addition, optimizing pharmacokinetic and pharmacodynamic parameters are essential for ensuring efficacy in difficulty to treat infections. Activities such as testing in hollow fiber models, prolonged or continuous infusion are being aggressively evaluated to optimize drug dosing [53,54,55].
Table 1.
Clinical regimens used in observational studies for treating carbapenem-resistant Klebsiella pneumoniae where carbapenemase is identified [45].
| Beta-lactamases present | Regimen | Improved survival versus monotherapy |
|---|---|---|
| KPC- and MBL- producing K. pneumoniae | Carbapenem and tigecycline, plus aminoglycoside or colistin; Carbapenem and tigecycline; Carbapenem and aminoglycoside; Carbapenem and colistin |
Yes |
| KPC-producing K. pneumoniae | Colistin and aminoglycoside; Colistin and tigecycline; Colistin and quinolone; Colistin and carbapenem; Carbapenem and carbapenem |
Yes |
Table 2. Novel agents in development for treating carbapenem-resistant and CPO.
| Antibiotic | Drug class | Intended Indication/Activity/Comments |
|---|---|---|
| Cefiderocol | Siderophore-β-lactam (cephalosporin) | Complicated urinary tract infections (cUTIs), carbapenem-resistant Gram-negative bacterial infections Active against metallo-beta-lactamase producing strains |
| ceftaroline fosamil/avibactam | Cephalosporin and DBO BLI. | Currently undefined. |
| Eravacycline | Tetracycline | cIAI and cUTI Multi-drug resistant organisms (MDRO) |
| Imipenem/cilistatin/relebactam | Carbapenem and DBO beta-lactamase inhibitor (BLI). | cUTIs, intra-abdominal infections (cIAI), hospital acquired pneumonia (HAP) Active against ESBLs and KPCs |
| Meropenem-vaborbactam | Carbapenem and cyclic boronic acid beta-lactamase inhibitor | cUTI, catheter-related bloodstream infections, HAP/ventilator-associated bacterial pneumonia (VAP), cIAI due to CRE |
| Plazomycin | Aminoglycoside | cUTI, catheter-related bloodstream infections, HAP/ventilator-associated pneumonia, cIAI due to CPO and CRE |
Monitoring and Control of Carbapenemase-Producing Organisms
Approaches to addressing the rapid intercontinental spread of CPO and other multi-resistant organisms include surveillance and judicious use of infection prevention and control (IPC) practices. There is evidence that IPC efforts at the local and country-wide level are effective in reducing transmission of CPO [56], and the role of IPC in the overall control of CPO cannot be overemphasized. Regarding surveillance at a global level, the Global Antimicrobial Resistance Surveillance System (GLASS) program was launched in 2015 as part of the WHO Global Action Plan on AMR to support a standardized approach to collection, analysis, and sharing of AMR data to inform local and national decision-making, and provide the evidence base for action and advocacy. Another approach that has been suggested is the application of the International Health Regulations (IHR), which represents a legal framework for international efforts to reduce the risk from public health threats that may spread between countries [57]. IHR requires countries to report certain disease outbreaks, including smallpox, wild-type poliomyelitis, severe acute respiratory syndrome, new types of influenza, or any public health event of international concern (PHEIC) which may include “new or emerging antibiotic resistance” [57]. The rationale for declaring AMR, specifically CPO, as a PHEIC has been reported previously [58], and includes multi-drug resistance, propensity for rapid spread, absence of geographic/political boundaries, presence in E. coli (the most common cause of urinary tract infection globally), presence in microbes of high public health importance, namely Salmonella, Shigella, and Vibrio species, and carriage of resistance traits on very mobile broad-host range plasmids [59]. The emergence of plasmid-mediated colistin resistance in Enterobacteriaceae has created a potential scenario of pan-resistant CRE [60].
Although application of IHR to CPO may have potential benefits including increasing surveillance and response capacities to address the spread of AMR on a global basis [58], a counter reaction argues that it is difficult to appreciate how the global spread of AMR constitutes an “extraordinary event” and that it is neither pragmatic nor within the framework of the IHR to consider it a PHEIC [61]. The only PHEICs declared to date include H1N1 2009 global influenza pandemic, Ebola virus disease in 2014, and the recent clusters of microcephaly and neurological abnormalities associated with Zika virus. In addition to global efforts underway, country-specific guidelines, including The Combating Antibiotic Resistant Bacteria report and the President's Council of Advisors on Science and Technology strategic plans, provide practical recommendations to the United States government to facilitate addressing the problem of antimicrobial resistance. Canada and the European Union have made similar commitments.
Summary.
Infections caused by carbapenemase-producing bacteria have experienced unprecedented intercontinental spread and proliferation and continue to be a therapeutic challenge. The genetic features that facilitate widespread dissemination are becoming increasingly understood. Control requires efficient laboratory detection and treatment, and a coordinated international response.
Acknowledgments
The authors thank Jean B. Patel, Office of Antimicrobial Resistance, Centers for Disease Control and Prevention, Atlanta, Georgia, USA, for thoughtful review of the manuscript.
Funding: The authors received no specific financial support from agencies in the commercial, public, or not-for profit sectors for this work. RB, EB, JC, BL, JS, and LW have no affiliations or financial interests with organizations or entities in the subject matter discussed in this manuscript. LP is co-inventor of the Carba NP for which an international patent has been filled on behalf of INSERM (Paris, France).
Footnotes
Disclaimer: The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention, the National Institutes of Health, or the Department of Health and Human Services.
Contributor Information
Robert A. Bonomo, Medical Service, Louis Stokes Cleveland Department of Veterans Affairs Medical; Department of Veterans Affairs Medical Center, Departments of Medicine, Pharmacology, Molecular Biology, and Microbiology, Case Western Reserve University and Research Service, Cleveland, Ohio, USA
Eileen M. Burd, Department of Pathology and Laboratory Medicine and Department of Medicine, Division of Infectious Diseases, Emory University School of Medicine, Atlanta, GA, USA
John Conly, Departments of Medicine, Pathology and Laboratory Medicine, Microbiology, Immunology and Infectious Diseases, University of Calgary, Calgary, Alberta, Canada; Synder Institute for Chronic Diseases, University of Calgary, Calgary, Alberta, Canada; Cumming School of Medicine and Alberta Health Services, University of Calgary, Calgary, Alberta, Canada.
Brandi M. Limbago, Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention, Atlanta, Georgia, USA
Laurent Poirel, Medical and Molecular Microbiology Unit, Department of Medicine, Faculty of Science, University of Fribourg, Fribourg, Switzerland.
Julie A. Segre, Microbial Genomics Section, Translational and Functional Genomics Branch, National Human Genome Research Institute, Bethesda, MD, USA
Lars F. Westblade, Department of Pathology and Laboratory Medicine and Department of Medicine, Division of Infectious Diseases, Weill Cornell Medicine, New York, NY, USA
References
- 1.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim JY, Jung HI, An YJ, Lee JH, Kim SJ, Jeong SH, Lee KJ, Suh PG, Lee HS, Lee SH, Cha SS. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase. Molecular microbiology. 2006;60:907–916. doi: 10.1111/j.1365-2958.2006.05146.x. [DOI] [PubMed] [Google Scholar]
- 3.Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother. 2013;68:1757–1762. doi: 10.1093/jac/dkt094. [DOI] [PubMed] [Google Scholar]
- 4.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palzkill T. Metallo-β-lactamase structure and function. Ann N Y Acad Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-Lactamases: the Quiet before the Storm? Clin Microbiol Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelstein MV, Skleenova EN, Shevchenko OV, D'souza JW, Tapalski DV, Azizov IS, Sukhorukova MV, Pavlukov RA, Kozlov RS, Toleman MA, Walsh TR. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis. 2013;13:867–876. doi: 10.1016/S1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 9.Potron A, Poirel L, Dortet L, Nordmann P. Characterisation of OXA-244, a chromosomally-encoded OXA-48-like β-lactamase from Escherichia coli. Int J Antimicrob Agents. 2016;47:102–103. doi: 10.1016/j.ijantimicag.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 11.Potron A, Poirel L, Nordmann P. Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob Agents Chemother. 2014;58:467–471. doi: 10.1128/AAC.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Girlich D, Poirel L, Nordmann P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50:477–479. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasoo S, Cunningham SA, Kohner PC, Simner PJ, Mandrekar JN, Lolans K, Hayden MK, Patel R. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified hodge test for detection of carbapenemase-producing gram-negative bacilli. J Clin Microbiol. 2013;51:3097–3101. doi: 10.1128/JCM.00965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalhaes CG, Picão RC, Nicoletti AG, Xavier DE, Gales AC. Cloverleaf test (modified hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010;65:249–251. doi: 10.1093/jac/dkp431. [DOI] [PubMed] [Google Scholar]
- 16.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. The carbapenem inactivation method (CIM) a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative bacteria. PloS ONE. 2015;10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tijet N, Patel SN, Melano RG. Detection of carbapenemase activity in Enterobacteriaceae: comparison of the carbapenem inactivation method versus the Carba NP test. J Antimicrob Chemother. 2016;71:274–276. doi: 10.1093/jac/dkv283. [DOI] [PubMed] [Google Scholar]
- 18.Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, Bobenchik AM, Lockett ZC, Charnot-Katsikas A, Ferraro MJ, Thomson RB, Jr, Jenkins SG, Limbago BM, Das S. The Modified Carbapenem Inactivation Method (mCIM) for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J Clin Microbiol. 2017 Apr 5; doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. 27th. CLSI; Wayne, PA, USA: 2017. CLSI Supplement M100. [Google Scholar]
- 20.Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hrabák J, Študentová V, Walková R, Žemličková H, Jakubů V, Chudáčková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papagiannitsis CC, Študentová V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabák J. Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol. 2015;53:1731–1735. doi: 10.1128/JCM.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dortet L, Bréchard L, Poirel L, Nordmann P. Rapid detection of carbapenemase-producing Enterobacteriaceae from blood cultures. Clin Microbiol Infect. 2014;20:340–344. doi: 10.1111/1469-0691.12318. [DOI] [PubMed] [Google Scholar]
- 24.Migliavacca R, Docquier JD, Mugnaioli C, Amicosante G, Daturi R, Lee K, Rossolini GM, Pagani L. Simple microdilution test for detection of metallo-beta-lactamase production in Pseudomonas aeruginosa. J Clin Microbiol. 2002;40:4388–4390. doi: 10.1128/JCM.40.11.4388-4390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsakris A, Poulou A, Pournaras S, Voulgari E, Vrioni G, Themeli-Digalaki K, Petropoulou D, Sofianou D. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrob Chemother. 2010;65:1664–1671. doi: 10.1093/jac/dkq210. [DOI] [PubMed] [Google Scholar]
- 26.Miriagou V, Tzelepi E, Kotsakis SD, Daikos GL, Bou Casals J, Tzouvelekis LS. Combined disc methods for the detection of KPC- and/or VIM-positive Klebsiella pneumoniae: improving reliability for the double carbapenemase producers. Clin Microbiol Infect. 2013;19:E412–E415. doi: 10.1111/1469-0691.12238. [DOI] [PubMed] [Google Scholar]
- 27.Girlich D, Halimi D, Zambardi G, Nordmann P. Evaluation of Etest strips for detection of KPC and metallo-carbapenemases in Enterobacteriaceae. Diagn Microbiol Infect Dis. 2013;77:200–201. doi: 10.1016/j.diagmicrobio.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 28.van Dijk K, Voets GM, Scharringa J, Voskuil S, Fluit AC, Rottier WC, Leverstein-Van Hall MA, Cohen Stuart JW. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl boronic acid, dipicolinic acid and temocillin. Clin Microbiol Infect. 2014;20:345–349. doi: 10.1111/1469-0691.12322. [DOI] [PubMed] [Google Scholar]
- 29.Cuzon G, Naas T, Bogaerts P, Glupczynski Y, Nordmann P. Evaluation of a DNA microarray for the rapid detection of extended beta-lactamases (TEM, SHV and CTX-M), plasmid-mediated cephalosporinases (CMY-2-like, DHA, FOX, ACC-1, ACT/MIR and CMY-1-like/MOX) and carbapenemases (KPC, OXA-48, VIM, IMP and NDM) J Antimicrob Chemother. 2012;67:1865–1869. doi: 10.1093/jac/dks156. [DOI] [PubMed] [Google Scholar]
- 30.Kaase M, Szabados F, Wassill L, Gatermann SG. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J Clin Microbiol. 2012;50:3115–3118. doi: 10.1128/JCM.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodémont M, De Mendonça R, Nonhoff C, Roisin S, Denis O. Performance of the Verigene gram-negative blood culture assay for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2014;52:3085–3087. doi: 10.1128/JCM.01099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A, Kanack KJ. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol. 2016;54:687–698. doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tato M, Ruiz-Garbajosa P, Traczewski M, Dodgson A, McEwan A, Humphries R, Hindler J, Veltman J, Wang H, Cantón R. Multisite evaluation of Cepheid Xpert-Carba-R assay for the detection of carbapenemase-producing organisms in rectal swabs. J Clin Microbiol. 2016;54:1814–1819. doi: 10.1128/JCM.00341-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group. Henderson DK, Palmore TN, Segre JA. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012 Aug 22;4(148):148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathers AJ, Stoesser N, Sheppard AE, Pankhurst L, Giess A, Yeh AJ, Didelot X, Turner SD, Sebra R, Kasarskis A, Peto T, Crook D, Sifri CD. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother. 2015;59:1656–1663. doi: 10.1128/AAC.04292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pecora ND, Li N, Allard M, Li C, Albano E, Delaney M, Dubois A, Onderdonk AB, Bry L. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. mBio. 2015;6:e01030–15. doi: 10.1128/mBio.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. Whole-genome sequencing for analysis of an outbreak of methicillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Köser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8(8):e1002824. doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright MS, Jacobs MR, Bonomo RA, Adams MD. Transcriptome Remodeling of Acinetobacter baumannii during Infection and Treatment. MBio. 2017 Mar 7;8(2) doi: 10.1128/mBio.02193-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright MS, Iovleva A, Jacobs MR, Bonomo RA, Adams MD. Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med. 2016 Mar 3;8(1):26. doi: 10.1186/s13073-016-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program. Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med. 2014 Sep 17;6(254):254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poirel L, Kieffer N, Nordmann P. In vitro evaluation of dual carbapenem combinations against carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2016;71:156–161. doi: 10.1093/jac/dkv294. [DOI] [PubMed] [Google Scholar]
- 44.Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob Agents Chemother. 2012;56:2108–2113. doi: 10.1128/AAC.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, Spanu T, Ambretti S, Ginocchio F, Cristini F, Losito AR, Tedeschi S, Cauda R, Bassetti M. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae : importance of combination therapy. Clin Infect Dis. 2012;55:943–950. doi: 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 46.Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2014;20:862–872. doi: 10.1111/1469-0691.12697. [DOI] [PubMed] [Google Scholar]
- 47.Falagas ME, Lourida P, Poulikakos P, Rafailidis PI, Tansarli GS. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: systematic evaluation of the available evidence. Antimicrob Agents Chemother. 2014;58:654–663. doi: 10.1128/AAC.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Duin D, Lok J, Earley M, Cober E, Richter S, Perez F, Salata R, Kalayjian R, Watkins R, Doi Y, Kaye K, Fowler V, Paterson D, Bonomo R, Evans S. Colistin vs. ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clinical Infectious Diseases [in press] 2017 doi: 10.1093/cid/cix783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez F, El Chakhtoura NG, Papp-Wallace KM, Wilson BM, Bonomo RA. Treatment options for infections caused by carbapenem-resistant Enterobacteriaceae: can we apply “precision medicine” to antimicrobial chemotherapy? Expert Opin Pharmacother. 2016;17:761–781. doi: 10.1517/14656566.2016.1145658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis. 2016;63:1615–1618. doi: 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spellberg B, Bonomo RA. Editorial commentary: ceftazidime-avibactam and carbapenem-resistant Enterobacteriaceae: “We're gonna need a bigger boat”. Clin Infect Dis. 2016;63:1619–1621. doi: 10.1093/cid/ciw639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papp-Wallace KM, Bonomo RA. New β-Lactamase Inhibitors in the Clinic. Infectious Disease Clinics of North America. 2016;30:441–464. doi: 10.1016/j.idc.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Drusano GL. From lead optimization to NDA approval for a new antimicrobial: Use of pre-clinical effect models and pharmacokinetic/pharmacodynamic mathematical modeling. Bioorg Med Chem. 2016;24:6401–6408. doi: 10.1016/j.bmc.2016.08.034. [DOI] [PubMed] [Google Scholar]
- 54.Carpentier A, Metellus P, Ursu R, Zohar S, Lafitte F, Barrié M, Meng Y, Richard M, Parizot C, Laigle-Donadey F, Gorochov G, Psimaras D, Sanson M, Tibi A, Chinot O, Carpentier AF. Intracerebral administration of CpG oligonucleotide for patients with recurrent glioblastoma: a phase II study. Neuro Oncol. 2010;12:401–408. doi: 10.1093/neuonc/nop047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mattoes HM, Kuti JL, Drusano GL, Nicolau DP. Optimizing antimicrobial pharmacodynamics: dosage strategies for meropenem. Clin Ther. 2004;26:1187–1198. doi: 10.1016/s0149-2918(04)80001-8. [DOI] [PubMed] [Google Scholar]
- 56.Schwaber MJ, Lev B, Israeli A, Solter E, Smollan G, Rubinovitch B, Shalit I, Carmeli Y, Israel Carbapenem-Resistant Enterobacteriaceae Working Group Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally-implemented intervention. Clin Infect Dis. 2011;52:1–8. doi: 10.1093/cid/cir025. [DOI] [PubMed] [Google Scholar]
- 57.World Health Organization. International health regulations (2005) Geneva: World Health Organization; 2008. [Accessed December 2016]. Available at: http://www.who.int/ihr/9789241596664/en/ [Google Scholar]
- 58.Wernli D, Haustein T, Conly J, Carmeli Y, Kickbusch I, Harbarth S. A call for action: the application of The International Health Regulations to the global threat of antimicrobial resistance. PLoS Med. 2011;8(4):e1001022. doi: 10.1371/journal.pmed.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 60.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 61.Kamradt-Scott A. A public health emergency of international concern? Response to a proposal to apply the International Health Regulations to antimicrobial resistance. PLoS Med. 2011;8(4):e1001021. doi: 10.1371/journal.pmed.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]