Abstract
Problem
The regulatory mechanisms governing differential expression of classical MHC class I (MHC-Ia) and non-classical MHC class I (MHC-Ib) genes are poorly understood.
Method of study
Quantitative RT-PCR was used to compare the abundance of MHC-I transcripts and related transcription factors in PBMC and placental trophoblast cells (PTC). Methylation of MHC-I CpG islands was detected by bisulfite treatment and next-generation sequencing. Demethylation of PBMC and PTC with 5′-aza-deoxycytidine was used to assess the role of methylation in gene regulation.
Results
MHC-I expression was higher in PBMC than PTC and was correlated with expression of IRF1, CIITA and STAT1. The MHC-Ia genes and BoLA-NC1 were devoid of CpG methylation in PBMC and PTC. In contrast, CpG sites in the gene body of BoLA-NC2, -NC3 and -NC4 were highly methylated in PBMC but largely unmethylated in normal PTC and moderately methylated in somatic cell nuclear transfer PTC. In PBMC demethylation resulted in upregulation of MHC-Ib by 2.8- to 6-fold, whereas MHC-Ia transcripts were elevated less than 2-fold.
Conclusion
DNA methylation regulates bovine MHC-Ib expression and is likely responsible for the different relative levels of MHC-Ib to MHC-Ia transcripts in PBMC and PTC.
Keywords: bovine, DNA methylation, non-classical MHC-I, transcription
Graphical Abstract
INTRODUCTION
Major histocompatibility complex class I (MHC-I) molecules are cell surface glycoproteins that play a critical role in triggering an immune response. MHC-I genes are classified into classical (MHC-Ia) and non-classical (MHC-Ib) subtypes according to their sequence polymorphism, expression patterns and gene structure.1,2 Classical MHC-I genes are highly polymorphic and are ubiquitously expressed in most somatic cells. MHC-Ia proteins activate cluster of differentiation 8 positive (CD8+) T lymphocytes by presenting peptide antigens derived from internal proteins. Activated T lymphocytes subsequently clear cells that display the same antigens from intracellular pathogens.3–5 In contrast, non-classical MHC-I genes exhibit limited polymorphism and are expressed in a more tissue-restricted manner, such as in the placenta during pregnancy.2,6 MHC-Ib proteins primarily function to inhibit immune responses.7 For example, the human MHC-Ib protein human leukocyte antigen (HLA)-E inhibits natural killer cells.8,9 During pregnancy, HLA-G inhibits both T cells and natural killer cells to provide an immunologically favorable environment at the maternal-fetal interface that protects the conceptus from the maternal immune system.6,10,11 HLA-G is also thought to contribute to immune evasion of tumor cells,9,12 and blocking HLA-G with a specific antibody may offer an innovative therapeutic strategy for cancer.13
In cattle, MHC-I genes are abundantly expressed in lymphocytes, but expression in placental trophoblast cells is very low, particularly during the first two trimesters of pregnancy.14 Abnormally high expression of MHC-I in placental trophoblast is linked to a higher rate of miscarriage in somatic cell nuclear transfer (SCNT) pregnancies.15 Moreover, MHC-Ia and -Ib genes are differentially expressed among various tissues in cattle. Microarray screening in bovine peripheral blood mononuclear cells (PBMC) showed that MHC-Ia accounted for more than 90% of total MHC-I transcripts, whereas in bovine placental trophoblast cells (PTC) MHC-Ia and -Ib accounted for 22% to 66% and 34% to 79% of total transcripts, respectively.2
Because both MHC-Ia and MHC-Ib genes play important roles in the regulation of immune responses, it is important to know how MHC-I gene expression is regulated. Previous studies investigating human MHC-I gene regulation focused on functional promoter elements.(reviewed in 16) Important cis-regulatory elements that regulate MHC-I expression include enhancer A,17 interferon (IFN)-stimulated regulatory element,18 and the SXY module,19,20 which are bound by nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), interferon regulatory factor 1 (IRF1) and MHC class II (MHC-II) enhancersome A, respectively.17–20 Regulation of MHC-I genes by cytokines and inflammatory factors, such as interferon gamma (IFN-γ),21 transforming growth factor beta (TGF-β),22 and tumor necrosis factor alpha (TNF-α),23 can partially explain differences in MHC-I expression patterns among tissues. However, the extremely different transcription patterns for MHC-Ia and -Ib genes in bovine PBMC and PTC suggests that other mechanisms, such as epigenetic regulation, may also be involved in controlling expression of MHC-I genes in cattle.
Methylation of cytosine residues at CpG dinucleotides is one of the best-studied epigenetic modifications in the mammalian genome and is known to have a prominent effect on gene expression.24 DNA hypermethylation silences the expression of HLA-A and HLA-B in certain cancers,25 and demethylation treatment induces the expression of HLA-G.26 Suarez-Alvarez et al. demonstrated that promoter methylation plays a very important role in regulating the expression of MHC-I in human embryonic stem cells.27
The objective of this study was to elucidate the regulatory mechanisms responsible for differential expression of MHC-Ia and MHC-Ib in bovine PBMC and PTC, with a focus on epigenetic mechanisms. Our working hypotheses were that: (1) bovine MHC-Ia and MHC-Ib genes are regulated by the same transcription factors, and (2) the relative abundance of bovine MHC-Ia and MHC-Ib transcripts in PBMC and PTC is controlled by DNA methylation.
MATERIALS AND METHODS
Animals
The Utah State University Institutional Animal Care and Use Committee approved all procedures for the handling and treatment of cattle used in this study (protocols #1171 and #1506). Cattle were maintained at the Utah State University Animal Science Farm or the Caine Dairy Center in Wellsville, Utah.
Isolation and culture of PBMC
Cattle blood was collected using vacutainer tubes containing acid citrate dextrose (BD, Franklin Lake, NJ). Whole blood was centrifuged at 200 g for 30 min. The buffy coat layer was transferred to a new conical tube and resuspended in 10 mL phosphate-buffered saline (PBS). This suspension was overlaid on Ficoll-Hypaque density gradient (Accurate Chemical & Scientific, Westbury, NY) and centrifuged at 1400 g for 30 min to obtain the PBMC fraction. The PBMC were then washed three times with PBS. For the gene expression experiments, RNA from three cows was prepared as described below. For the DNA methylation study, DNA from four cows was isolated as described below.
For the demethylation study, which required that PBMC be cultured for several days, PBMC were isolated from three healthy Angus cows and resuspended in RPMI 1640 medium supplemented with 1 mM L-glutamine (HyClone Laboratories, Logan, UT), 100 μg /mL penicillin (HyClone Laboratories), 100 μg/mL streptomycin (HyClone Laboratories), 0.125 μg/mL concanavalin A (GE Healthcare, Uppsala, Sweden) and 10% heat inactivated fetal bovine serum (FBS, HyClone Laboratories). Cells were cultured with or without 100 μM 5′-aza-2-deoxycytidine (Acros Organics, Geel, Belgium), which demethylates DNA, at 37°C in an atmosphere of 5% CO2. After 3 days in culture, RNA was isolated and converted to cDNA for measurement of MHC-I expression by quantitative reverse transcription polymerase chain reaction (qRT-PCR).
Isolation and culture of PTC
Day 35 PTC samples were collected from seven artificial insemination (AI) and five SCNT pregnancies. SCNT embryos were produced and transferred into recipient cows as previously described.28,29 Pregnant cows were euthanized on day 35 ± 1 of gestation at a USDA-inspected slaughter facility following standard protocols. Trophoblast cells were collected from the placenta by carefully peeling the distinct tan colored trophoblast layer away from the other layers of the placenta, snap frozen in liquid nitrogen, and maintained at −80°C until used for RNA and DNA isolation.
Trophoblast cells were also isolated from day 35 AI conceptuses for the demethylation experiment, which required culturing the cells for several days. To isolate PTC for cell culture, the trophoblast layer of the placenta was washed three times with PBS, cut into small pieces and treated with 20 mL 0.25% trypsin/DNase solution (HyClone Laboratories, Logan, UT) at 37°C on a plate shaker for 20 min. Samples were then filtered through four layers of cheesecloth and any undigested tissue was incubated in fresh trypsin/DNase solution for an additional 30 min. Next, dispersed cells were pooled and filtered through a 100 μm nylon cell strainer (BD science, Franklin Lake, NJ). The filtrate was overlaid on 40% Percoll (GE healthcare, Waukesha, WI) and centrifuged at 800 g for 10 min at room temperature. Cells at the Percoll interface were collected and washed three times with DMEM/F12 medium. Finally, cells were resuspended in DMEM/F12 medium with 10% FBS, 100 μg/mL penicillin and 100 μg/mL streptomycin and cultured with or without 100 μM 5′-aza-2-deoxycytidine (Acros Organics, Geel, Belgium) at 37°C in an atmosphere of 5% CO2. After 3 days in culture, RNA was isolated and converted to cDNA for measurement of MHC-I expression by qRT-PCR.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA from PBMC, and day 35 AI and SCNT PTC were isolated using an Ambion PureLink RNA mini isolation kit (Life Technologies, Carlsbad, CA). First-strand cDNA from each sample was generated from 1 μg total RNA using the SuperScript VILO cDNA synthesis kit (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. Two quantitative PCR platforms were used in this work. The primers that were used for MHC-I and transcription factor genes with both qRT-PCR platforms are listed in Table 1. To establish the relative abundance of bovine MHC-I subtypes and the change after demethylation treatment, real-time PCR was performed on an Eppendorf Realplex Mastercycler (Eppendorf, Hamburg, Germany). Each 20 μL reaction included 25 ng diluted cDNA, 200 nmol of each primer, and 10 μL SYBR-Green master mix (Life Technologies, Carlsbard, CA). The cycling parameters were 94°C for 2 min of hot-start, followed by 40 cycles of 94°C for 15 s, 60°C for 20 s and 72°C for 30 s. To study the correlation between expression of bovine MHC-I and related transcription factors in PTC and PBMC, and the level of expression of DNA methyltransferases, qRT-PCR was done using a Fluidigm 48.48 Dynamic Array chip with a Fluidigm BioMark high through-put, quantitative PCR system.30 Quantitative PCR data was analyzed using the formula for relative quantification described by Pfaffl.31 The relative mRNA levels were normalized to GAPDH mRNA levels.
Table 1.
Primers for qRT-PCR
| Gene name | Forward primer (5′-3′) | Reverse primer (5′-3′) | Efficiency |
|---|---|---|---|
| MHC-I | TTGTGGAGACCAGGCCTTCAG | GAGAACCAGGCCAACAATGATG | 0.99 |
| MHC-Ia | TTGTGGAGACCAGGCCTTCAG | AATGATGCCCATGGTGAGGAA | 0.99 |
| BoLA-NC1 | GGATCAAGAGACGCGGATACAA | CCGCAGCCGTGCATCCACT | 0.98 |
| BoLA-NC2 | GGGTGCGCTGATCCTCACT | CCACCCACCGCGCTGTA | 0.95 |
| BoLA-NC3 | CCAAGGAAAGTCAACAGGAATC | AATCTCTGCCGTCGTAGGC | 0.94 |
| BoLA-NC4 | AGCGATGACAAGAGATGCCAAGAA | CCGCACCGTCATAGGCGT | 0.96 |
| NFKB1 | CTGCTCACCACCCTCCTC | GCACTTTGTTAAGAGTTAGCAAG | 0.97 |
| RELA | CCTTTCAATGGACCCACCGA | GAGAGATGGCGTAAAGGAATAG | 0.95 |
| IRF1 | GATGCCTGTCTGTTTCGGA | TGGTGAGGGGTGGGAGCAT | 0.97 |
| CREB1 | CAGACCACTGATGGACAGCAA | TGGGGAAGACGCCATAACAACT | 0.99 |
| RFX5 | TGTATCTCTACCTTCAGCTCC | GGCAGGTGTCGGTATGCT | 0.97 |
| RFXAP | CTCAGGAAACGTCAAACTGGA | CACCACTTCTGGACTTCTTAGTAA | 0.94 |
| NFYC | TCCAAGTCCAGGGGCAGC | CTGGGCTTGACCTTGTGG | 0.97 |
| CIITA | CTGTGTCACCCGTTTCAGG | GAGATTGCCAAGGTCTTCCACA | 0.98 |
| STAT1 | TCATTTGTGGTGGAAAGACAGC | GTGCCCAGAATGTTGAACTTC | 0.97 |
| DNMT1 | TGCCTCAGTGCCTCCA | GCGTGGTTCGGAGGATCT | 0.95 |
| DNMT3A | ATGACGATGGCTATCAGTCCTA | TCTTCTTTGATGGCTGCCTG | 0.95 |
| DNMT3B | AGGACTGGAGTGTGCGTC | GAATCGCAGGGTATAACTTGG | 0.96 |
| GAPDH | GAGAAGGCTGGGGCTCACTTG | GCTGACAATCTTGAGGGTGTTG | 0.99 |
DNA sequencing of MHC-I CpG islands
Genomic DNA from PBMC and PTC was extracted using the Purelink Genomic DNA Mini Kit (Life Technologies, Carlsbad, CA). Primers for amplifying the CpG island regions within each MHC-I gene are listed in Table 2. PCR reactions using Phusion Hot-Start II High-Fidelity DNA Polymerase (New England BioLabs, Ipswich, MA) were performed at 98°C for 30s, followed by 30 cycles of 98°C for 15s, 62°C for 30s and 72°C for 30s, with a final extension at 72°C for 5 min. PCR products were cloned into the pCR-Blunt vector (Life Technologies, Carlsbad, CA) and transformed into E. coli DH5α. Plasmids were then isolated and sequenced on an ABI PRISM 3730 DNA Analyzer (ABI, Foster City, CA).
Table 2.
Primers for MHC-I CpG island DNA sequencing and 454 methylation sequencing
| Category | Primer name | Sequence (5′-3′) |
|---|---|---|
| CpG island sequencing | MHC-IF | KATCRGGGCAAAGTCCCAG |
| MHC-IR | CGCAGCARCGTGTCCTTYCC | |
| Methylation sequencing round 1 | F1F | GACGGCTTGCGGCTACAGGAGGGATTAGGGTAAAGTTTTA |
| F1R | GGGCAGCGGACTGTTCTTTCCCTCCAAACCCCRCACTCACC | |
| F2F | GACGGCTTGCGGCTACANGTTATGRGGTYGYGAATTTTTT | |
| F2R | GGGCAGCGGACTGTTCTAACTACRTATCRTCCACRTAACC | |
| F3F | GACGGCTTGCGGCTACATAGGTTTTTATTTTWTGAGKTATTTT | |
| F3R | GGGCAGCGGACTGTTCTCRCRATAATTAAACYCAAACTA | |
| F4F | GACGGCTTGCGGCTACAATTAGAGYGAGGTYGGTGAGYG | |
| F4R | GGGCAGCGGACTGTTCTAAACCCCATTTTYCTCTCYTC | |
| Methylation sequencing round 2 | MID-PBMC-F | CGTATCGCCTCCCTCGCGCCATCAGACGAGTGCGTGACGGCTTGCGGCTACA |
| MID-PBMC-R | CTATGCGCCTTGCCAGCCCGCTCAGACGAGTGCGTGGGCAGCGGACTGTTCT | |
| MID-PTC-F | CGTATCGCCTCCCTCGCGCCATCAGAGACGCACTCGACGGCTTGCGGCTACA | |
| MID-PTC-R | CTATGCGCCTTGCCAGCCCGCTCAGAGACGCACTCGGGCAGCGGACTGTTCT | |
| MID-SCNT-F | CGTATCGCCTCCCTCGCGCCATCAGATCAGACACGGACGGCTTGCGGCTACA | |
| MID-SCNT-R | CTATGCGCCTTGCCAGCCCGCTCAGATCAGACACGGGGCAGCGGACTGTTCT |
Bisulfite-sequencing of MHC-I CpG islands
To determine the methylation status of the MHC-I CpG island regions in PBMC and PTC, 1 μg genomic DNA from each sample was bisulfite-treated using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA) according to the manufacturer’s recommended protocol. Because methylation-specific PCR is inefficient for reactions generating products longer than 500 bps, the MHC-I genetic region containing the CpG islands was split into 4 fragments. Specific primers for each genomic DNA fragment were designed using Methprimer32 and primer sequences are provided in Table 2. Roche 454 sequencing was performed as previously described,33 with minor modifications. Two PCR steps were performed to produce amplicons for 454 sequencing. First, primer pairs with MHC-I specific sequence and flanking adaptor sequence were used to amplify CpG island fragments from bisulfite-treated genomic DNA; the cycling parameters were 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 52–57°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 5 min. Sequencing adaptors and molecular ID tags (454 Life Sciences) were then added in a second round of PCR using primers that hybridized to the adaptors added in the first PCR. After the second PCR, equal amounts of each amplicon were mixed and then sequenced on a 454 GS FLX+ DNA sequencer (Roche, Indianapolis, IN). Sequencing reads were analyzed using DNASTAR Lasergene software (Madison, WI).
Statistical analysis
Quantitative PCR data was analyzed using the relative quantification method described by Pfaffl.31 For each sample the percentage of each MHC-I gene, or gene subset in the case of MHC-Ia, was based on the relative abundance compared to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), the internal control. The abundance was calculated using the equation: RMHC-X = EfficiencyMHC-X −CT(MHC-X) / EfficiencyGAPDH−CT(GAPDH). The percentage of MHC-X in each sample was obtained using the formula: MHC-X % = RMHC-x / (RMHC-Ia + RNC1 + RNC2 + RNC3 + RNC4).
All statistical analyses were performed using R software. One-way ANOVA with a Bonferroni post-hoc test for multiple group comparisons was performed to determine significance. A probability of p < 0.05 was considered statistically significant.
RESULTS
Expression of MHC-I is regulated by transcription factors
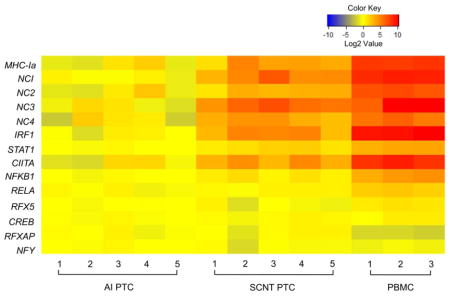
Expression of all MHC-I subtypes was significantly higher in PBMC than in PTC from AI or SCNT pregnancies (p < 0.001; Table 3). Expression of MHC-I subtypes was also significantly higher in SCNT PTC than in AI PTC. The transcription factor genes interferon regulatory factor 1 (IRF1) and signal transducer and activator of transcription 1 (STAT1), and the transcriptional coactivator gene class II MHC transactivator (CIITA) showed a similar expression pattern to MHC-I. Expression of nuclear factor kappa B subunit 1 (NFKB1; previously known as NFκB p50), RELA proto-oncogene (RELA; previously known as NFκB p65) and regulatory factor X5 (RFX5) were significantly higher in PBMC compared to PTC, while expression of these transcription factors did not differ in AI and SCNT PTC. No significant difference in the expression of transcription factor genes cyclic-AMP responsive element binding protein 1 (CREB1), regulatory factor X associated protein (RFXAP) and nuclear transcription factor Y subunit gamma (NFYC) was observed among the groups (Figure 1 and Table 3). Expression of MHC-I subtypes in PBMC and PTC from AI and SCNT pregnancies was positively correlated with elevated expression of IRF1, CIITA, STAT1, NFKB1 and RELA (Table 4).
Table 3.
Statistical analysis of gene expression
| Gene | Mean
|
One-way ANOVA
|
||||
|---|---|---|---|---|---|---|
| AI PTC | SCNT PTC | PBMC | AI PTC vs SCNT PTC | AI PTC vs PBMC | SCNT PTC vs PBMC | |
| MHC-Ia | 1.00 ± 1.05 | 9.52 ± 6.76 | 148.63 ± 14.8 | ** | *** | *** |
| BoLA-NC1 | 1.00 ± 0.50 | 18.43 ± 32.91 | 124.01 ± 58.68 | *** | *** | *** |
| BoLA-NC2 | 1.00 ± 1.21 | 4.60 ± 1.93 | 73.06 ± 8.41 | * | *** | ** |
| BoLA-NC3 | 1.00 ± 1.08 | 36.07 ± 17.44 | 65.21 ± 13.71 | *** | *** | * |
| BoLA-NC4 | 1.00 ± 1.62 | 9.98 ± 5.79 | 50.26 ± 20.11 | ** | *** | * |
| IRF1 | 1.00 ± 0.48 | 20.14 ± 13.28 | 729.61 ± 268.30 | *** | *** | *** |
| STAT1 | 1.00 ± 0.27 | 3.49 ± 1.45 | 10.71± 1.89 | * | *** | *** |
| CIITA | 1.00 ± 0.91 | 12.53 ± 7.61 | 228.62 ± 90.65 | * | *** | *** |
| NFKB1 | 1.00 ± 0.22 | 2.47 ± 0.66 | 15.86 ± 5.67 | ns | *** | *** |
| RELA | 1.00 ± 0.43 | 1.64 ± 0.64 | 3.97 ± 0.59 | ns | *** | *** |
| RFX5 | 1.00 ± 0.17 | 1.06 ± 0.66 | 2.12 ± 0.27 | ns | * | * |
| CREB1 | 1.00 ± 0.32 | 1.53 ± 0.67 | 1.53 ± 0.383 | ns | ns | ns |
| RFXAP | 1.00 ± 0.42 | 1.41 ± 0.81 | 0.25 ± 0.04 | ns | ns | ns |
| NFYC | 1.00 ± 0.27 | 0.95 ± 0.46 | 0.58 ± 0.08 | ns | ns | ns |
Mean values were calculated using GAPDH as an internal control. The fold change in gene expression for each PTC and PBMC sample was calculated with respect to the mean value for the AI PTC samples (AI PTC n = 5; SCNT PTC n = 5; PBMC n = 3). Statistical comparisons between groups were performed using one-way ANOVA with the Bonferroni post-hoc test. ns = not significant;
p < 0.05;
p < 0.01;
p < 0.001.
FIGURE 1.
qRT-PCR analysis of bovine MHC-I and transcription factor gene expression in PBMC and PTC. Data were normalized to GAPDH. The heatmap shows the fold change of gene expression relative to the average for the AI PTC samples.
Table 4.
Pearson correlation coefficients between MHC-I expression and transcription factor expression
| Gene | Transcription Factors
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IRF1 | STAT1 | CIITA | NFKB1 | RELA | CREB1 | RFX5 | RFXAP | NFYC | |
| MHC-Ia | 0.96** | 0.96** | 0.90** | 0.94** | 0.93** | 0.51 | 0.80** | 0.50 | 0.45 |
| BoLA-NC1 | 0.95** | 0.98** | 0.95** | 0.96** | 0.96** | 0.60* | 0.84** | 0.60* | 0.41 |
| BoLA-NC2 | 0.88** | 0.94** | 0.91** | 0.91** | 0.96** | 0.55 | 0.84** | 0.55 | 0.40 |
| BoLA-NC3 | 0.87** | 0.88** | 0.85** | 0.94** | 0.80** | 0.66* | 0.74** | 0.67* | 0.29 |
| BoLA-NC4 | 0.78** | 0.90** | 0.98** | 0.84** | 0.88** | 0.52 | 0.77** | 0.52 | 0.43 |
Correlation is significant at the 0.05 level (2-tailed).
Correlation is significant at the 0.01 level (2-tailed).
Expression of MHC-Ia and -Ib in bovine PBMC and PTC
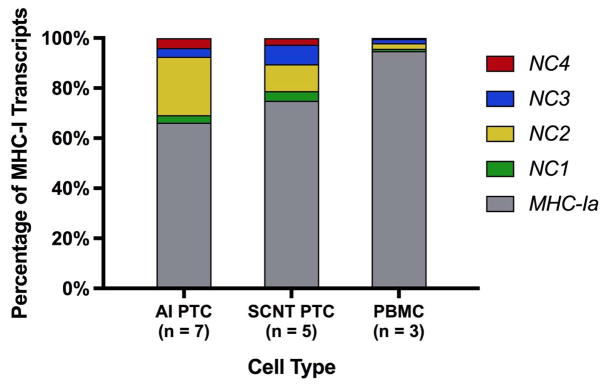
MHC-I gene expression was assessed using qRT-PCR. The majority of MHC-I transcripts in PBMC were from MHC-Ia loci (94.8%), whereas only 5.2% of MHC-I transcripts were from MHC-Ib (NC1-NC4) loci, confirming a previous report by Davies et al.2 MHC-Ib contributed a significantly larger proportion of the overall MHC-I transcript pool in PTC than PBMC, 33.8% and 25.1% in AI and SCNT PTC groups, respectively (Figure 2 and Table 5).
FIGURE 2.
MHC-Ia and MHC-Ib transcript percentages in PTC and PBMC. Expression of MHC-Ia and MHC-Ib (BoLA-NC1, -NC2, -NC3, -NC4) were detected by qRT-PCR. Average percentages are shown.
Table 5.
Analysis of MHC-I subtype percentage
| Gene | Percentage | One way ANOVA | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| AI PTC | SCNT PTC | PBMC | AI PTC vs SCNT PTC | AI PTC vs PBMC | SCNT PTC vs PBMC | |
| BoLA-NC1 | 3.02 ± 1.33% | 3.87 ± 1.09% | 0.96 ± 0.04% | ns | ns | * |
| BoLA-NC2 | 23.37 ± 8.72% | 10.72 ± 2.89% | 2.25 ± 0.86% | * | ** | * |
| BoLA-NC3 | 3.48 ± 3.03% | 7.85 ± 0.96% | 1.62 ± 0.79% | ns | * | ** |
| BoLA-NC4 | 3.95 ± 4.33% | 2.62 ± 0.74% | 0.38 ± 0.16% | ns | ** | ** |
| MHC-Ia | 66.14 ± 9.72% | 74.93 ± 4.49% | 94.78 ± 1.44% | ns | *** | ** |
For each sample the percentage of each MHC-I gene, or gene subset in the case of MHC-Ia, was calculated from the relative abundance compared to GAPDH, the internal control, as described in the methods section.31 The percentage of each subtype in each cell type is the mean value for all of the animals in the group (AI PTC n = 7; SCNT PTC n = 5; PBMC n = 3). Statistical comparisons between groups were performed using one-way ANOVA with the Bonferroni post-hoc test for multiple comparisons. ns = not significant;
p < 0.05;
p < 0.01;
p < 0.001.
Methylation of CpG islands in MHC-Ia and MHC-Ib genes
The DNA sequences of six MHC-I reference alleles were retrieved from GenBank (NW_001494163). In silico analysis (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) identified a CpG island of approximately 1500 bp spanning from approximately 300 bp upstream of the transcription start site to the third exon in each bovine MHC-I gene (Figure 3A). Methylation status of these CpG islands was determined by bisulfite conversion followed by sequencing on a 454 GS FLX+ sequencer. Information about the sequencing reads from four PBMC samples is provided in Table 6 and information about the sequencing reads from two AI PTC and two SCNT PCT samples is presented in Table 7. The methylation status of the CpG islands for two MHC-Ia and four MHC-Ib alleles per PBMC or PTC sample are summarized in Figure 3 and Figure 4, respectively. The results indicated that the MHC-Ia genes and the bovine leukocyte antigen non-classical 1 (BoLA-NC1) MHC-Ib gene were completely unmethylated in all three cell types. Exons 2 and 3 and the intervening intron of the BoLA-NC2, -NC3 and -NC4 MHC-Ib genes were methylated to varying degrees in the three types of samples. Methylation in this region was the highest in PBMC, the lowest in AI PTC, and intermediate in SCNT PTC.
FIGURE 3.
Methylation profile of CpG islands in MHC-Ia and MHC-Ib genes of PBMC. (A) Genomic organization of bovine MHC-I CpG islands, from −300 bp upstream of the transcription start site through the third exon, which is about 1,500 Kb. (B–E) Methylation status of the CpG sites of each MHC-I allele in 4 PBMC samples. Each horizontal line of circles represents the methylation status of an individual allele. Different colors of circles denote variation in the methylation level. The numbers above each line of circles stand for the position in the genomic sequence relative to the first base of the start codon.
Table 6.
Number of 454 sequencing reads for PBMC
| Sample | Allele | Number of reads for each genomic region
|
||||||
|---|---|---|---|---|---|---|---|---|
| Promoter | Leading peptide | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | ||
| PBMC-1 | BoLA-3*00401 | 8 | 14 | 6 | 7 | 7 | 4 | 4 |
| BoLA-3*03301 | 5 | 10 | 5 | 2 | 2 | 3 | 3 | |
| BoLA-NC1*00301 | 2 | 12 | 10 | 4 | 4 | 7 | 7 | |
| BoLA-NC2*00101 | 2 | 9 | 7 | 11 | 11 | 17 | 17 | |
| BoLA-NC3*00101 | 7 | 10 | 3 | 11 | 11 | 5 | 5 | |
| BoLA-NC4*00301 | 6 | 9 | 3 | 22 | 22 | 16 | 16 | |
| PBMC-2 | BoLA-2*00801 | 3 | 5 | 2 | 4 | 4 | 5 | 5 |
| BoLA-1*01901 | 4 | 11 | 7 | 4 | 4 | 6 | 6 | |
| BoLA-NC1*00201 | 8 | 14 | 6 | 2 | 2 | 4 | 4 | |
| BoLA-NC2*00101 | 2 | 5 | 3 | 5 | 5 | 5 | 5 | |
| BoLA-NC3*00101 | 6 | 8 | 2 | 17 | 17 | 10 | 10 | |
| BoLA-NC4*00202 | 5 | 9 | 4 | 26 | 26 | 23 | 23 | |
| PBMC-3 | BoLA-3*00401 | 9 | 17 | 8 | 6 | 6 | 9 | 9 |
| BoLA-2*05401 | 6 | 11 | 5 | 3 | 3 | 10 | 10 | |
| BoLA-NC1*00201 | 3 | 8 | 3 | 3 | 3 | 8 | 8 | |
| BoLA-NC2*00101 | 3 | 5 | 2 | 3 | 3 | 12 | 12 | |
| BoLA-NC3*00101 | 12 | 18 | 6 | 4 | 4 | 2 | 2 | |
| BoLA-NC4*00201 | 4 | 7 | 3 | 26 | 26 | 18 | 18 | |
| PBMC-4 | BoLA-3*00402 | 4 | 10 | 6 | 5 | 5 | 8 | 8 |
| BoLA-3*05201 | 8 | 17 | 9 | 8 | 8 | 18 | 18 | |
| BoLA-NC1*00201 | 14 | 25 | 11 | 3 | 3 | 9 | 9 | |
| BoLA-NC2*00101 | 3 | 7 | 4 | 7 | 7 | 21 | 21 | |
| BoLA-NC3*00101 | 14 | 24 | 10 | 3 | 3 | 8 | 8 | |
| BoLA-NC4*00301 | 3 | 10 | 7 | 21 | 21 | 25 | 25 | |
Table 7.
Number of 454 sequencing reads for PTC
| Sample | Allele | Number of reads for each genomic region
|
||||||
|---|---|---|---|---|---|---|---|---|
| Promoter | Leading peptide | Intron 1 | Exon 2 | Intron 2 | Exon 3 | Intron 3 | ||
| AI PTC-1 | BoLA-3*00402 | 11 | 18 | 7 | 11 | 11 | 7 | 7 |
| BoLA-5*07201 | 6 | 8 | 2 | 3 | 3 | 2 | 2 | |
| BoLA-NC1*00501 | 2 | 19 | 17 | 9 | 9 | 12 | 12 | |
| BoLA-NC2*00103 | 2 | 4 | 2 | 23 | 23 | 13 | 13 | |
| BoLA-NC3*00101 | 10 | 12 | 2 | 7 | 7 | 5 | 5 | |
| BoLA-NC4*00301 | 3 | 6 | 3 | 23 | 23 | 6 | 6 | |
| AI PTC-2 | BoLA-2*05401 | 8 | 12 | 4 | 2 | 2 | 6 | 6 |
| BoLA-3*00801 | 9 | 17 | 8 | 3 | 3 | 10 | 10 | |
| BoLA-NC1*00201 | 2 | 6 | 4 | 2 | 2 | 3 | 3 | |
| BoLA-NC2*00101 | 3 | 5 | 2 | 3 | 3 | 4 | 4 | |
| BoLA-NC3*00101 | 6 | 9 | 3 | 4 | 4 | 3 | 3 | |
| BoLA-NC4*00301 | 3 | 7 | 4 | 8 | 8 | 10 | 10 | |
| SCNT PTC-1 | BoLA-2*00601 | 7 | 11 | 4 | 2 | 2 | 6 | 6 |
| BoLA-6*01402 | 5 | 10 | 5 | 2 | 2 | 11 | 11 | |
| BoLA-NC1*00201 | 4 | 22 | 18 | 2 | 2 | 13 | 13 | |
| BoLA-NC2*00101 | 2 | 5 | 3 | 7 | 7 | 7 | 7 | |
| BoLA-NC3*00101 | 15 | 30 | 30 | 6 | 6 | 4 | 4 | |
| BoLA-NC4*00201 | 4 | 13 | 9 | 8 | 8 | 10 | 10 | |
| SCNT PTC-2 | BoLA-2*00601 | 10 | 14 | 4 | 5 | 5 | 8 | 8 |
| BoLA-6*01402 | 6 | 8 | 2 | 5 | 5 | 5 | 5 | |
| BoLA-NC1*00201 | 9 | 14 | 5 | 5 | 5 | 5 | 5 | |
| BoLA-NC2*00101 | 2 | 4 | 2 | 5 | 5 | 4 | 4 | |
| BoLA-NC3*00101 | 4 | 9 | 5 | 7 | 7 | 8 | 8 | |
| BoLA-NC4*00201 | 3 | 8 | 5 | 8 | 8 | 7 | 7 | |
FIGURE 4.
Methylation profile of CpG islands in MHC-Ia and MHC-Ib genes of PTC. (A) Genomic organization of bovine MHC-I CpG islands. (B–C) Methylation status of the CpG sites of each MHC-I allele in 2 AI PTC samples and (D–E) in 2 SCNT PTC samples.
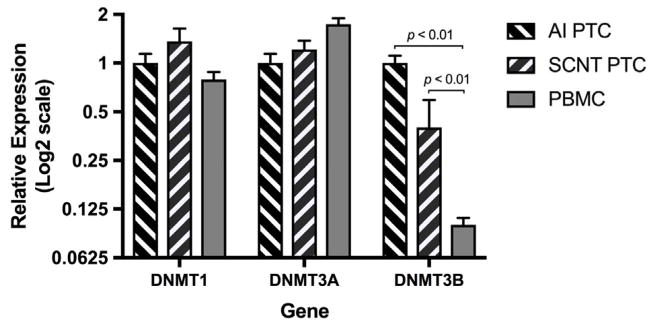
Expression of DNA methyltransferase (DNMT) isoforms
DNA methylation is catalyzed and maintained by three DNA methyltransferases: DNMT1, DNMT3A and DNMT3B.34 Expression of the three DNA methyltransferase genes in PBMC, AI and SCNT PTC were measured by qRT-PCR (Figure 5). Both DNMT1 and DNMT3A were expressed at a similar level among all groups. However, expression of DNMT3B was significantly lower in PBMC than that in either AI or SCNT PTC. Expression of DNMT3B in SCNT PTC was lower than in AI PTC but the difference was not statistically significant.
FIGURE 5.
qRT-PCR analysis of DNMT expression in PTC and PBMC. Data are presented as the fold change (mean ± SEM) of gene expression in other groups relative to that of the AI PTC group (AI PTC n = 5; SCNT PTC n = 5; PBMC n = 3). Statistical analyses were performed using one-way ANOVA.
Demethylation of MHC-Ib genes in PBMC
To test whether bovine MHC-I expression can be affected by demethylation, PBMC and AI PTC were cultured in medium containing 100 μM 5′-aza-2-deoxycytindine for 3 days to reduce overall methylation and these cells were compared to cells cultured without 5′-aza-2-deoxycytindine. Expression of each MHC-I gene was analyzed by qRT-PCR. The results indicated that MHC-Ib but not MHC-Ia expression was significantly upregulated in PBMC after demethylation treatment. Expression of the BoLA-NC1, -NC2, -NC3 and -NC4 genes in PBMC was increased 2.6-, 5.8-, 6.2- and 6.8-fold, respectively. However, in PTC MHC-Ia and MHC-Ib were only up-regulated 1.5- to 2.5-fold (Figure 6A). After demethylation, the percentage of MHC-Ib in the overall pool of MHC-I transcripts in PBMC increased to 41.3%, similar to PTC before (40.2%) and after (33.7%) treatment (Figure 6B).
FIGURE 6.
Change in MHC-I transcription following demethylation treatment of PBMC and PTC. Bovine PBMC and PTC were treated with 5′-aza-deoxycytidine for 3 days (n = 3 per group). (A) Fold change (mean ± SEM) of expression in the treated cells compared to cells cultured without 5′-aza-deoxycytidine. (B) Percentage of MHC-I subtypes in the overall pool of MHC-I transcripts.
DISCUSSION
In this study, we analyzed the expression of mRNA for transcription factors associated with MHC-I expression in bovine PTC and PBMC. Our results indicated that in comparison to PTC, PBMC express relatively high levels of mRNA for MHC-I transcription factors IRF1, CIITA, NFKB1 and RELA, and the cell signaling gene STAT1 (Table 3). In other studies, these transcription factors have been shown to induce MHC-I expression in humans and other mammals.16–20 The ability of these transcription factors to induce MHC-I expression in bovine cells was recently confirmed by transfection of plasmids encoding these transcription factors into bovine fibroblast cells (Shi and Davies, manuscript in preparation). The transcription factors IRF1 and CIITA appear to be particularly important for driving of MHC-I expression (Table 3 and Figure 1). The relatively low expression of these transcription factors in PTC is associated with a low level of MHC-I expression. In contrast, in PBMC these transcription factors and the MHC-I genes are expressed at a very high level. Our study also suggests that DNA methylation suppresses expression of three MHC-Ib genes in PBMC but not in PTC. We found that MHC-Ia and BoLA-NC1 were free of methylation at CpG sites in PBMC and PTC, whereas in PBMC the MHC-Ib genes BoLA-NC2, -NC3 and -NC4 were highly methylated in their second intron, and second and third exons (Figures 3 and 4). Accordingly, demethylation resulted in marked upregulation of MHC-Ib in bovine PBMC (Figure 6). Our study helps explain the abnormally high MHC-I expression in PTC from SCNT conceptuses and the differential expression of MHC-Ib in PTC and PBMC.2,15,28 These findings shed light on how the bovine conceptus regulates trophoblast MHC-I expression to protect itself from immunologically mediated rejection and how the immunological cross-talk between the placenta and the uterine immune system differs in normal and SCNT pregnancies.
The core promoters of the bovine MHC-Ia and MHC-Ib genes have a high level of homology, and include all of the conserved transcription factor binding sites found in humans and other mammals.35,36 In this study, we tested the expression of several transcription factor genes (IRF1, CIITA, NFKB1, RELA, CREB1, RFX5, RFXAP and NFYC) and the cell signaling gene STAT1 in three different types of cells. The expression patterns for all of the MHC-I subtypes were positively correlated with those of IRF1, STAT1, CIITA, NFKB1, RELA and RFX5 with the highest expression in PBMC and the lowest expression in AI PTC (Tables 3 and 4). IRF1 and CIITA are both IFN-γ stimulated genes37,38 and STAT1 is involved in the IFN-γ signaling pathway,38 suggesting that bovine MHC-I genes, like their human homologues, may be regulated by IFN-γ.
Although bovine MHC-Ia and MHC-Ib genes have a high level of homology in their promoter sequences, the relative levels of MHC-Ia and MHC-Ib in the total pool of MHC-I transcripts is distinct in different cell types. In the present study, the proportion of MHC-Ib in AI and SCNT PTC was 4.5- and 2.8-fold greater than that found in PBMC, respectively. This is reasonable, given the distinct functions of the two MHC-I subtypes and the different physiological roles of PBMC and PTC. In both humans and cattle, MHC-Ia proteins present peptide antigens to activate cytotoxic T cells that kill cells displaying antigens derived from intracellular pathogens, while the primary function of MHC-Ib molecules is regulation of the immune system. PBMC are important for immune surveillance and the high amounts of MHC-Ia expressed by these cells enable them to efficiently present peptide antigens to cytotoxic T lymphocytes. However, PTC are located at the maternal-fetal interface, which requires an immunologically favorable environment to foster fetal development. Thus, higher expression of MHC-Ib in PTC likely facilitates protection of the fetus from immune-mediated miscarriage.
The observation that the percentage of MHC-Ia and MHC-Ib transcripts differed between PBMC and PTC led us to determine the DNA methylation patterns for these genes. We found that the BoLA-NC2, -NC3 and -NC4 genes in PBMC were hypermethylated within the gene body, but that these genes were only slightly or moderately methylated in the same region in PTC. In contrast, the BoLA-NC1 and MHC-Ia genes were unmethylated in all of the samples that were examined. This may be explained by the genomic location of the BoLA-NC1 gene, which is located near the MHC-Ia genes and far away from the other MHC-Ib genes.1
DNA methylation in promoter regions is a repressive epigenetic mark that down-regulates gene expression.39,40 However, the BoLA-NC2, -NC3 and -NC4 genes were methylated in their gene body but not the promoter region. A review by Jones stated that gene body CpG methylation is not generally associated with transcription repression because transcription elongation is not sensitive to DNA methylation in mammals, but also points out that cytosine methylation does inhibit transcription elongation in Neurospora crassa.41 In contrast, Lorincz et al. reported that methylation in the intragenic region represses expression by inhibiting the elongation process of transcription in murine cells.42 Deaton et al. demonstrated that intragenic DNA methylation represses the GATA binding protein 3 (GATA3) gene in cluster of differentiation 4 positive (CD4+), IFN-γ+, interleukin 4 positive (IL-4+) T cells, and this repression is potentially responsible for the co-existence of Th1 and Th2 characteristics in this special subpopulation of CD4+ T cells.43 Huang et al. showed expression of the insulin like growth factor 2 (IGF2) gene was negatively regulated by intragenic DNA methylation in cattle.44 In this study, we observed that the methylated BoLA-NC2, -NC3 and -NC4 genes in PBMC contributed less to the overall MHC-I transcript abundance than their unmethylated counterparts did in PTC. Furthermore, demethylation treatment of PBMC with 5′-aza-deocycytidine resulted in an upregulation of the BoLA-NC2, -NC3 and -NC4 genes by 5- to 6-fold, but only slightly increased MHC-Ia and BoLA-NC1 expression. The increased expression level of these MHC-I genes is inversely correlated with their methylation status, which supports the concept that DNA methylation in the intragenic region of the BoLA-NC2, -NC3 and -NC4 genes alters the transcription of these genes. The argument that Jones made in his review that in mammals gene body methylation does not inhibit transcription was based on methylation changes in cancer cells where an increase in intragenic methylation of genes such as insulin-like growth factor 2 receptor (Igf2r), apolipoprotein E (APOE), and myogenic differentiation 1 (Myod1) was associated with increased expression.41 However, it is highly possible that there are a large number of genes repressed by intragenic DNA methylation, such as the GATA3 gene in humans and the IGF2 gene in cattle, as well as the BoLA-NC2, -NC3 and -NC4 genes characterized in this study.
The pattern of expression of the BoLA-NC1 gene was similar to that of the other MHC-Ib subtypes, even though this gene was unmethylated in all tissues examined, suggesting that BoLA-NC1 may be regulated by other mechanisms. The co-activator CIITA acts as a platform for recruiting histone acetyltransferases to enhance MHC-I transcription45 or recruiting histone deacetylases to inhibit the transactivation of CIITA,46 it is possible that histone deacetylase-associated CIITA binds to the BoLA-NC1 promoter and interferes with its transcription in PBMC. This form of regulation may also influence expression of other MHC-I genes and needs to be investigated in future studies. It is also possible that BoLA-NC1 is regulated by microRNAs, possibly encoded by the pseudogenes that surround the locus.1 This is known to be the case for the human MHC-Ib gene HLA-G which has been reported to be down-regulated by microRNAs miR-148a, miR-152 and miR-133a binding to its 3′ untranslated region (UTR).47,48 Other regulatory mechanisms for BoLA-NC1 as well as other MHC-I subtypes warrant further investigation.
During embryogenesis DNA is almost completely demethylated at fertilization,49 and then gradually remethylated as development progresses.50 DNA methyltransferases are responsible for transferring methyl groups to DNA, with DNMT1 predominantly methylating hemimethylated CpG dinucleotides to maintain methylation status after DNA replication and DNMT3A/3B responsible for de novo methylation. DNMT3B was differently expressed among cells, with higher expression in AI and SCNT PTC than in PBMC. This result suggests that the PTC derived from day 34 embryos are undergoing a rigorous remethylation process. However, MHC-Ib expression in PTC does not decrease as pregnancy progresses, suggesting that the MHC-Ib genes in PTC remain unmethylated. We also observed a difference in MHC-Ib methylation between AI PTC and SCNT PTC, with the latter higher than the former. Dean et al. reported that DNA methylation in an in vitro fertilized bovine embryo is reduced up to the 8 cell stage, and then increased by de novo methylation beginning at the 16 cell stage.51 However, DNA demethylation occurred only at the one-cell stage in SCNT embryos.51 Since DNMT genes are expressed at a similar level in both types of PTC, the higher DNA methylation level of the BoLA-NC2, -NC3 and -NC4 genes in SCNT PTC is probably caused by insufficient DNA demethylation during embryogenesis.
In conclusion, our study suggests that bovine MHC-I expression is regulated by both genetic and epigenetic factors. Expression of the MHC-I genes was highly correlated with the expression of transcription factors IRF1, CIITA, NFKB1, RELA, RFX5 and STAT1, with IRF1 and CIITA exhibiting particularly high relative expression levels in both SCNT PTC and PBMC compared to AI PTC from day 35 conceptuses that are essentially negative for expression of MHC-I proteins.15,28 In addition, our data indicate that expression of bovine MHC-Ib genes (BoLA-NC2, -NC3 and -NC4) is regulated by intragenic DNA methylation. Although we cannot rule out other epigenetic regulatory mechanisms for MHC-I, DNA methylation appears to be an important mechanism for regulating tissue specific expression of MHC-Ib genes.
Acknowledgments
This work was supported by Agriculture and Food Research Initiative Competitive Grant no. 2011-67015-30008 from the USDA, National Institute of Food and Agriculture, and grant 1R01HD055502 from the NIH, National Institute of Child Health and Human Development. The authors thank Dr. Wei Li for assistance with the data analysis.
References
- 1.Birch J, Codner G, Guzman E, Ellis SA. Genomic location and characterisation of nonclassical MHC class I genes in cattle. Immunogenetics. 2008;60:267–273. doi: 10.1007/s00251-008-0294-2. [DOI] [PubMed] [Google Scholar]
- 2.Davies CJ, Eldridge JA, Fisher PJ, Schlafer DH. Evidence for expression of both classical and non-classical major histocompatibility complex class I genes in bovine trophoblast cells. Am J Reprod Immunol. 2006;55:188–200. doi: 10.1111/j.1600-0897.2005.00364.x. [DOI] [PubMed] [Google Scholar]
- 3.Wick MJ, Ljunggren HG. Processing of bacterial antigens for peptide presentation on MHC class I molecules. Immunol Rev. 1999;172:153–162. doi: 10.1111/j.1600-065x.1999.tb01363.x. [DOI] [PubMed] [Google Scholar]
- 4.Shastri N, Cardinaud S, Schwab SR, Serwold T, Kunisawa J. All the peptides that fit: the beginning, the middle, and the end of the MHC class I antigen-processing pathway. Immunol Rev. 2005;207:31–41. doi: 10.1111/j.0105-2896.2005.00321.x. [DOI] [PubMed] [Google Scholar]
- 5.Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 6.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. Faseb Journal. 2005;19:681–693. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 7.O’Callaghan CA, Bell JI. Structure and function of the human MHC class Ib molecules HLA-E, HLA-F and HLA-G. Immunol Rev. 1998;163:129–138. doi: 10.1111/j.1600-065x.1998.tb01192.x. [DOI] [PubMed] [Google Scholar]
- 8.Tomasec P, Braud VM, Rickards C, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031–1033. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 9.Braud VM, Allan DSJ, O’Callaghan CA, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 10.Gros F, Cabillic F, Toutirais O, Le Maux A, Sebti Y, Amiot L. Soluble HLA-G molecules impair natural killer/dendritic cell crosstalk via inhibition of dendritic cells. Eur J Immunol. 2008;38:742–749. doi: 10.1002/eji.200736918. [DOI] [PubMed] [Google Scholar]
- 11.Akhter A, Faridi RM, Das V, Pandey A, Naik S, Agrawal S. In vitro up-regulation of HLA-G using dexamethasone and hydrocortisone in first-trimester trophoblast cells of women experiencing recurrent miscarriage. Tissue Antigens. 2012;80:126–135. doi: 10.1111/j.1399-0039.2012.01884.x. [DOI] [PubMed] [Google Scholar]
- 12.Rouas-Freiss N, Moreau P, Ferrone S, Carosella ED. HLA-G proteins in cancer: Do they provide tumor cells with an escape mechanism? Cancer Res. 2005;65:10139–10144. doi: 10.1158/0008-5472.CAN-05-0097. [DOI] [PubMed] [Google Scholar]
- 13.Agaugue S, Carosella ED, Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood. 2011;117:7021–7031. doi: 10.1182/blood-2010-07-294389. [DOI] [PubMed] [Google Scholar]
- 14.Davies CJ. Why is the fetal allograft not rejected? J Anim Sci. 2007;85:E32–E35. doi: 10.2527/jas.2006-492. [DOI] [PubMed] [Google Scholar]
- 15.Hill JR, Schlafer DH, Fisher PJ, Davies CJ. Abnormal expression of trophoblast major histocompatibility complex class I antigens in cloned bovine pregnancies is associated with a pronounced endometrial lymphocytic response. Biol Reprod. 2002;67:55–63. doi: 10.1095/biolreprod67.1.55. [DOI] [PubMed] [Google Scholar]
- 16.van den Elsen PJ, Holling TM, Kuipers HF, van der Stoep N. Transcriptional regulation of antigen presentation. Curr Opin Immunol. 2004;16:67–75. doi: 10.1016/j.coi.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Gobin SJP, Keijsers V, van Zutphen M, van den Elsen PJ. The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor kappa B. J Immunol. 1998;161:2276–2283. [PubMed] [Google Scholar]
- 18.Gobin SJP, van Zutphen M, Woltman AM, van den Elsen PJ. Transactivation of classical and nonclassical HLA class I genes through the IFN-stimulated response element. J Immunol. 1999;163:1428–1434. [PubMed] [Google Scholar]
- 19.Gobin SJP, Peijnenburg A, van Eggermond M, van Zutphen N, van den Berg R, van den Elsen PJ. The RFX complex is crucial for the constitutive and CIITA-mediated transactivation of MHC class I and beta(2)-microglobulin genes. Immunity. 1998;9:531–541. doi: 10.1016/s1074-7613(00)80636-6. [DOI] [PubMed] [Google Scholar]
- 20.Gobin SJP, van Zutphen M, Westerheide SD, Boss JM, van den Elsen PJ. The MHC-specific enhanceosome and its role in MHC class I and beta(2)-microglobulin gene transactivation. J Immunol. 2001;167:5175–5184. doi: 10.4049/jimmunol.167.9.5175. [DOI] [PubMed] [Google Scholar]
- 21.Baldeon ME, Neece DJ, Nandi D, Monaco JJ, Gaskins HR. Interferon-gamma independently activates the MHC class I antigen processing pathway and diminishes glucose responsiveness in pancreatic beta-cell lines. Diabetes. 1997;46:770–778. doi: 10.2337/diab.46.5.770. [DOI] [PubMed] [Google Scholar]
- 22.Geiser AG, Letterio JJ, Kulkarni AB, Karlsson S, Roberts AB, Sporn MB. Transforming growth factor beta 1 (TGF-beta 1) controls expression of major histocompatibility genes in the postnatal mouse: aberrant histocompatibility antigen expression in the pathogenesis of the TGF-beta 1 null mouse phenotype. Proc Natl Acad Sci U S A. 1993;90:9944–9948. doi: 10.1073/pnas.90.21.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Israel A, Lebail O, Hatat D, et al. TNF stimulates expression of mouse MHC class I genes by inducing an NF kappa B-like enhancer binding activity which displaces constitutive factors. EMBO J. 1989;8:3793–3800. doi: 10.1002/j.1460-2075.1989.tb08556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 25.Nie Y, Yang GY, Song YL, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615–1623. doi: 10.1093/carcin/22.10.1615. [DOI] [PubMed] [Google Scholar]
- 26.Moreau P, Mouillot G, Rousseau P, Marcou C, Dausset J, Carosella ED. HLA-G gene repression is reversed by demethylation. Proc Natl Acad Sci U S A. 2003;100:1191–1196. doi: 10.1073/pnas.0337539100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suarez-Alvarez B, Rodriguez RM, Calvanese V, et al. Epigenetic mechanisms regulate MHC and antigen processing molecules in human embryonic and induced pluripotent stem cells. Plos One. 2010;5:e10192. doi: 10.1371/journal.pone.0010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutigliano HM, Thomas AJ, Wilhelm A, et al. Trophoblast major histocompatibility complex class I expression is associated with immune-mediated rejection of bovine fetuses produced by cloning. Biol Reprod. 2016;95:39, 1–9. doi: 10.1095/biolreprod.115.136523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aston KI, Li GP, Hicks BA, et al. The developmental competence of bovine nuclear transfer embryos derived from cow versus heifer cytoplasts. Anim Reprod Sci. 2006;95:234–243. doi: 10.1016/j.anireprosci.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. Plos One. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 33.Taylor KH, Kramer RS, Davis JW, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 34.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 35.Barker N, Young JR, Morrison WI, Ellis SA. Sequence diversity present within the 5′ upstream regions of BoLA class I genes. Immunogenetics. 1997;46:352–354. doi: 10.1007/s002510050283. [DOI] [PubMed] [Google Scholar]
- 36.Harms JS, Li WM, Splitter GA. The cattle major histocompatibility complex (MHC) class-I possesses HLA-like promoters. Gene. 1995;160:249–252. doi: 10.1016/0378-1119(95)00051-7. [DOI] [PubMed] [Google Scholar]
- 37.Steimle V, Siegrist CA, Mottet A, Lisowskagrospierre B, Mach B. Regulation of MHC class-II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 38.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 39.Decock A, Ongenaert M, Hoebeeck J, et al. Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biology. 2012;13:R95. doi: 10.1186/gb-2012-13-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farthing CR, Ficz G, Ng RK, et al. Global Mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 42.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 43.Deaton AM, Cook PC, De Sousa D, Phythian-Adams AT, Bird A, MacDonald AS. A unique DNA methylation signature defines a population of IFN-/IL-4 double-positive T cells during helminth infection. Eur J Immunol. 2014;44:1835–1841. doi: 10.1002/eji.201344098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang YZ, Zhan ZY, Sun YJ, et al. Intragenic DNA methylation status down-regulates bovine IGF2 gene expression in different developmental stages. Gene. 2014;534:356–361. doi: 10.1016/j.gene.2013.09.111. [DOI] [PubMed] [Google Scholar]
- 45.Nekrep N, Fontes JD, Geyer M, Peterlin BM. When the lymphocyte loses its clothes. Immunity. 2003;18:453–457. doi: 10.1016/s1074-7613(03)00086-4. [DOI] [PubMed] [Google Scholar]
- 46.Zika E, Greer SF, Zhu XS, Ting JPY. Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation. Mol Cell Biol. 2003;23:3091–3102. doi: 10.1128/MCB.23.9.3091-3102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manaster I, Goldman-Wohl D, Greenfield C, et al. MiRNA-mediated control of HLA-G expression and function. Plos One. 2012;7:e33395. doi: 10.1371/journal.pone.0033395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang XH, Li B, Wang J, et al. Evidence that miR-133a causes recurrent spontaneous abortion by reducing HLA-G expression. Reprod Biomed Online. 2012;25:415–424. doi: 10.1016/j.rbmo.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 49.Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 50.Meehan RR. DNA methylation in animal development. Semin Cell Dev Biol. 2003;14:53–65. doi: 10.1016/s1084-9521(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 51.Dean W, Santos F, Stojkovic M, et al. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]