Abstract
Exposure to industrial solvent and water pollutant trichloroethylene (TCE) can promote autoimmunity, and expand effector/memory (CD62L) CD4+ T cells. In order to better understand etiology reduced representation bisulfite sequencing was used to study how a 40-week exposure to TCE in drinking water altered methylation of ∼337 770 CpG sites across the entire genome of effector/memory CD4+ T cells from MRL+/+ mice. Regardless of TCE exposure, 62% of CpG sites in autosomal chromosomes were hypomethylated (0–15% methylation), and 25% were hypermethylated (85–100% methylation). In contrast, only 6% of the CpGs on the X chromosome were hypomethylated, and 51% had mid-range methylation levels. In terms of TCE impact, TCE altered (≥ 10%) the methylation of 233 CpG sites in effector/memory CD4+ T cells. Approximately 31.7% of these differentially methylated sites occurred in regions known to bind one or more Polycomb group (PcG) proteins, namely Ezh2, Suz12, Mtf2 or Jarid2. In comparison, only 23.3% of CpG sites not differentially methylated by TCE were found in PcG protein binding regions. Transcriptomics revealed that TCE altered the expression of ∼560 genes in the same effector/memory CD4+ T cells. At least 80% of the immune genes altered by TCE had binding sites for PcG proteins flanking their transcription start site, or were regulated by other transcription factors that were in turn ordered by PcG proteins at their own transcription start site. Thus, PcG proteins, and the differential methylation of their binding sites, may represent a new mechanism by which TCE could alter the function of effector/memory CD4+ T cells.
Keywords: trichloroethylene, immunotoxicity, autoimmunity, DNA methylation, polycomb proteins
Introduction
Approximately 24 million Americans have one or more autoimmune disease (e.g. Type I diabetes, systemic lupus erythematosus, autoimmune hepatitis). These chronic and incurable diseases disproportionately affect women, and are among the leading causes of death for young and middle-age women. In order to prevent these chronic incurable diseases we need to know more about the factors that trigger and maintain their pathology.
Effector/memory CD4+ T cells that secrete IFN-γ or IL-17 are critical mediators of both idiopathic and experimental autoimmune disease [1, 2]. These CD4+ T cells can persist for years in humans and animals without causing disease, while maintaining a memory phenotype, a stable cytokine response pattern, and the capacity for induced autoimmune attack [1]. Several adoptive transfer studies have shown that memory CD4+ T cells that have differentiated into Th1 or Th17 cells and reactivated can provide crucial help to cytotoxic CD8+ T cells, promote generation of pathogenic autoantibodies, and secrete tissue-damaging pro-inflammatory cytokines [3, 4].
Understanding how autoimmune disease triggers differentiation of naive CD4+ T cells into pro-inflammatory effector/memoryTh1 or Th17 cells is important for defining etiology. Twin concordance studies have shown that although genetics may increase susceptibility to autoimmunity, environmental triggers are required to initiate disease. Our work examining the link between the environment and CD4+ T cell differentiation has focused on the volatile organic compound trichloroethylene (TCE). Although the use of TCE as a solvent in the USA has declined as its toxicity became more apparent, over 31 million pounds of TCE-containing waste was released or disposed of in this country in the last decade alone (https://www.epa.gov/toxics-release-inventory-tri-program). Because of its improper disposal over the years, TCE has contaminated many of the water systems in the USA [5].
TCE is one of the first 10 chemicals selected for risk evaluation by the EPA under the newly revised TSCA (Toxic Substances Control Act) (https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/evaluating-risk-existing-chemicals-under-tsca#chemicalnames). This list was compiled based on the highest combined hazard, exposure, persistence and bio-accumulation characteristics. One of the most sensitive non-cancer outcomes of TCE exposure is immunotoxicity [6]. Specifically, chronic exposure to TCE (occupational or environmental) has been linked to a variety of autoimmune diseases and other hypersensitivity disorders [7–15].
CD4+ T cells are especially susceptible to the effects of TCE. Even if overt disease is not diagnosed, increased numbers of activated CD4+ T cells are often found in humans exposed to TCE [7, 16–19]. An expansion of peripheral blood CD4+ T cells is also a biomarker for patients with TCE-induced hypersensitivity [20]. Finally, as shown by ourselves and others, TCE exposure increased the percentage of effector/memory IFN-γ- and Th17-secreting CD4+ T cells in mice that went on to develop CD4+ T cell-mediated autoimmune hepatitis [21–23].
It has recently been reported that CD4+ T cell differentiation into different effector/memory CD4+ T cell subsets (Th1, Th2, Th17 and Treg) is at least partially regulated by gene-specific (i.e. Ifng, Il17A, Ctla, Tnfsf14, and Foxp3) increases or decreases in DNA methylation [24, 25]. This differentiation process can be disrupted during the development of autoimmunity, resulting in inappropriate DNA methylation and associated expression of genes that encode pro-inflammatory cytokines, chemokines, adhesion molecules, or suppressive mediators (e.g. LTA, CD11α, CD70, CD40L, FOXP3) [26–34]. The dysregulated methylome in autoimmune disease can enhance heterogeneity or plasticity in CD4+ T cell subsets that can increase disease severity [35, 36]. For example, the most pathogenic CD4+ T cells in models of type 1 diabetes mellitus, arthritis, and multiple sclerosis are those that secrete both IL-17 and IFN-γ, i.e. exhibit a dual Th1/Th17 phenotype [37]. Thus, the development of autoimmune disease may represent a breakdown in normal DNA methylation patterns in a manner that increases the differentiation of pathogenic effector/memory CD4+ T cells. Demonstrating that a toxicant such as TCE can disrupt the methylome is important for understanding how toxicants promote autoimmunity.
We reported previously that a 12-week exposure to TCE in drinking water altered global methylation in effector/memory CD4+ T cells from MRL+/+ mice [38]. A subsequent study, using targeted bisulfite next generation sequencing of amplicons generated on a Fluidigm Access Array, examined TCE- and time-dependent changes in DNA methylation associated with 16 functionally important genes in CD4+ T cells. TCE was found to increase gene-specific methylation variance in effector/memory CD4+ cells [6], and to induce a time-dependent cumulative increase in DNA methylation in the CpG sites of the promoter of the Ifng gene [39].
The methylome is even larger than the transcriptome. Thus, although a gene targeted evaluation of DNA methylation may provide important information about TCE-induced epigenetic alterations of specific genes, it may not recognize more global alterations in the methylome induced by TCE exposure. Consequently, the current study used reduced representation bisulfite sequencing (RRBS) [40], for a more comprehensive look at the impact of TCE exposure on CpG sites across the entire genome. A concurrent transcriptomic analysis was conducted so that TCE-induced alterations in DNA methylation could be compared with associated changes in gene expression.
Materials and Methods
Ethics Statement
All work was approved by the Animal Care and Use Committee at the University of Arkansas for Medical Sciences, and conformed to the USDA Animal Welfare Act and Regulations.
Mouse Treatment
Female MRL+/+ mice were selected for this study. Autoimmune disease in humans is known to involve an ill-defined genetic predisposition, and is most often found in women. Young adult female MRL+/+ mice, with a propensity for autoimmunity but absence of overt disease, can be used to mimic these requirements, and are used to test for the ability of different toxicants to trigger or augment autoimmunity as previously described [38]. Eight week-old female MRL+/+ mice (Jackson Laboratories; Bar Harbor, ME, USA) were housed in polycarbonate ventilated cages and provided with lab chow (Harlan 7027) and drinking water ad libitum. TCE (purity 99+%; Aldrich Chemical Co. Inc.; Milwaukee, WI, USA) was suspended in drinking water with 1% emulsifier Alkamuls EL-620 from Rhone-Poulenc (Cranbury, NJ). The mice (8–9 mice/group) received either 0 or 0.5 mg/ml TCE in their drinking water for 40 weeks. Freshly made TCE-containing drinking water was provided every 3–4 days. The mice were weighed once a month. On the basis of water intake, body weight and measured TCE degradation in the water bottles the mice were exposed to an average of 40–50 mg/kg/day TCE. This does is occupationally relevant based on the current 8-h Permissible Exposure Limit [established by the Occupational Safety and Health Administration (OSHA)] for TCE is 100ppm or ∼76 mg/kg/day.
CD4+ T Cells
Mice were sacrificed after 40 weeks, and splenic CD4+ T cells were isolated using Dynabeads FlowComp Mouse CD4 kit (Invitrogen). The CD4+ T cells were then further separated into CD62Llo or CD62Lhi CD4+ T cell populations using Dynabeads M-280 Streptavidin (Invitrogen) conjugated with biotinylated anti-CD62L antibody (eBiosciences, 13-0621-85). The resulting CD62Llo CD4+ T cells (effector/memory CD4+ T cells) were stimulated with immobilized anti-CD3 antibody and anti-CD28 antibody for 18 has described [41], and the activated CD4+ T cells were frozen for subsequent examination of DNA methylation or transcriptomics. To ensure sufficient cells for use in all the assays each sample of CD4+ T cells used in the study originated in an equal number of pooled spleen cells from 2 to 3 mice resulting in four samples/treatment group.
DNA Methylation Analysis by RRBS
DNA from the CD4+ T cells (four control samples and four TCE samples) was isolated using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific). The resulting DNA is tested on the NanoDrop 2000c (Thermo Fisher Scientific) for an A260/A280 range of 1.8–2.0. DNA quality is then confirmed using standard gel electrophoresis. The DNA was then restriction digested, end-repaired, purified, and attached with barcode adapters [42]. The RRBS libraries were generated, bisulfite converted, PCR enriched, size selected, purified, and sequenced (2 × 100 paired end) using an Illumina HiSeq sequencer (UAMS Translational Research Institute Genomics Core).
The Fasta files containing sequenced reads were first quality checked using FastQC program v0.11.5 and trimmed for adapter and low-quality sequences using trim galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with a Phred score of 20 as cut-off. Sequences were mapped using paired end mapping to mouse genome assembly mm9 using Bismark software v0.16.1 with Bowtie 2 v2.2.8 short read aligner. In the final step of bismark_methylation_extractor module, C in CpG, CHH and CHG were extracted with the parameter—no_overlap, to ensure that overlapping reads from the paired reads were not measured twice in the final analysis. Additionally, due to detection of consistently higher methylation rate at the ends of the sequencing reads in M-bias plots (data not shown), three bases from the ends of each pair of reads were discounted for methylation extraction. Bismark’s methylation_extracter output files were then read into MethylKit R package [43] for further statistical analyses.
Differential Methylation Analysis
A Phred Quality Score (Q) is used to represent the confidence level in assignment of each base call by the sequencer. It is logarithmically related to error probability and gives an estimated probability of a base call being wrong. In bisulfite sequencing a Phred score of 20 is normally used as a cut-off, and is the default value used in many open source sequencing tools [44]. Similarly, 10 reads is the minimum number of reads required for accurate determination of DNA methylation if individual CpG sites are analyzed for methylation differences [45]. Consequently, only sequence reads with a Phred score >20 and a minimum of 10 reads per CpG were accepted for downstream statistical analysis. Reads above the 99.9 percentile were also filtered out since these reads are either mapped against repeat elements or have very high PCR bias. A logistic regression model was fitted per CpG site to test for TCE effect on methylation level using a FDR cut off of 5% and a methylation difference of at least 10%.
In order to test the difference between TCE and control mice in terms of the relationship between mean methylation variance and mean percent methylation, quadratic regression models including the quadratic and linear interaction between groups and average percent methylation were fit to the data. A likelihood ratio test was used to compare the TCE and control curves by fitting and comparing a full model and a reduced model. The full model includes quadratic and linear interaction between groups (TCE vs control) and average percent methylation, while the reduced model includes common quadratic and linear terms of average percent methylation for both TCE and control.
qRT-PCR
Fluorescence-based quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was conducted using RNA isolated from effector/memory CD4+ T cells using techniques and primers as described [21]. Fold differences (log2 scale) in expression were determined using expression levels of resting (unactivated) CD4+ T cells of the appropriate subset of control mice as the control (1×) expression level. The threshold for statistical significance in fold change was set at P < 0.05. Differences between experimental groups were tested first with analysis of variance (ANOVA), and where the F test was significant, subsequent pairwise contrasts were tested using a two-sample t-test. CD4+ T cell concentration and gene expression values were right-skewed, and therefore these data were log-transformed for statistical analyzes. Adjusting for multiple comparisons, P-values from pairwise comparisons that were smaller than the Bonferroni-adjusted significance level indicated statistical significance.
Gene Arrays
This assessment was conducted by the Genomics Core at the University of Arkansas for Medical Sciences. All RNA samples extracted from CD4+ T cells had RIN (RNA integrity number) values of 8.0 or above. Total RNA (500 ng) was converted to cDNA, amplified and biotinylated by use of the Ambion Illumina TotalPrep™-96RNA Amplification Kit (Life Technologies, Carlsbad, CA, USA). Gene expression profiling was performed using the Expression BeadChip System from Illumina (Illumina Inc., San Diego, CA, USA) following the manufacturer’s instructions. Raw data were log2 transformed and normalized to the median intensity signal of 47 231 genes on the array. After normalization and filtering of low intensity spots, two-sample Student’s t-tests were performed and these data were plotted against fold-change measurements. Statistical significance was set at false discovery rate (FDR) < 0.05. Ingenuity Pathway Analysis software (Redwood City, CA, USA) was used for network identification.
Modeling
Fractional Polynomials were fit to model the percentage of total CpG sites that displayed a particular level of mean methylation (e.g. CpG sites that averaged 0–5% methylation or 40–45% methylation). This model has more flexibility to obtain a wide range of shapes of the distribution of the data than regular polynomial models. The power of mean methylation binning were chosen among {-2, −1, −0.5, 0, 0.5, 1, 2, 3} and were allowed to be repeated. The best-fitting first-degree FP model was the one with the lowest deviance among all first-degree powers. The best-fitting second-degree FP model was determined the same way after searching through all possible second-degree power combinations. The final best-fitting model was decided among the four models: null, linear, best-fitting first-degree FP, and best-fitting second-degree FP using a close testing procedure [46]. The final sets of powers selected for the best-fitting model for each chromosome in control or TCE groups were then compared. Having the same set of power suggests that the distribution is similar while a different set of powers suggests the distribution is different.
Annotation of the CpGs and Regulatory Elements was done using the University of California, Santa Cruz, Genome Browser (mouse NCBI37/mm9).
Results
Large-Scale Effects of TCE on DNA Methylation in Effector/Memory CD4+ T Cells
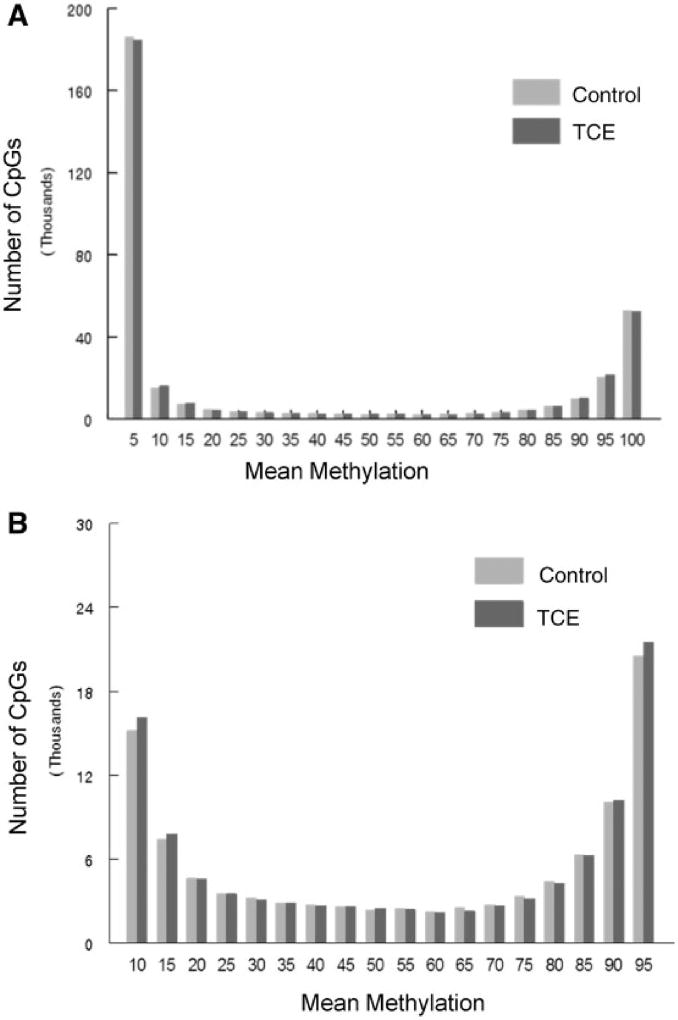
RRBS analysis of the effector/memory CD4+ T cells collected after 40 weeks of adult exposure to TCE was conducted. The analysis incorporated ∼337 770 CpGs sites that were assayed with at least 10× coverage. Bisulfite conversion efficiency in all samples was > 99%. Figure 1A presents histograms showing the average methylation of all the CpG sites examined after binning for average methylation (e.g. 0–5% methylation or 20–25% methylation). These profiles demonstrated that 54.7% and 16.5% of CpG sites from control mice were hypomethylated (0–5% methylation) or hypermethylated (95–100% methylation), respectively. These values were slightly altered in CD4+ T cells from TCE-treated mice; with 54.2% hypomethylated and 15.9% hypermethylated CpG sites respectively. Representing the binned mean methylation results without the hyper- and hypo-methylation skewing the extreme ends of the histogram (Fig. 1B) illustrated TCE-induced differences in the percentage of CpGs methylated at 90–95% (4.9% vs 6.0%; P < 0.05).
Figure 1.
Average DNA methylation levels of all CpGs interrogated. RRBS analysis of the effector/memory CD4+ T cells collected after 40 weeks of adult exposure to TCE was conducted. (A) Histograms show the average methylation of all 337 770 CpG sites examined in CD4+ T cells from either control or TCE-treated mice after binning for average methylation (e.g. 0–5% methylation or 20–25% methylation). (B) The same histograms are shown without inclusion of the CpGs that were either 0–5% or 95–100% methylated
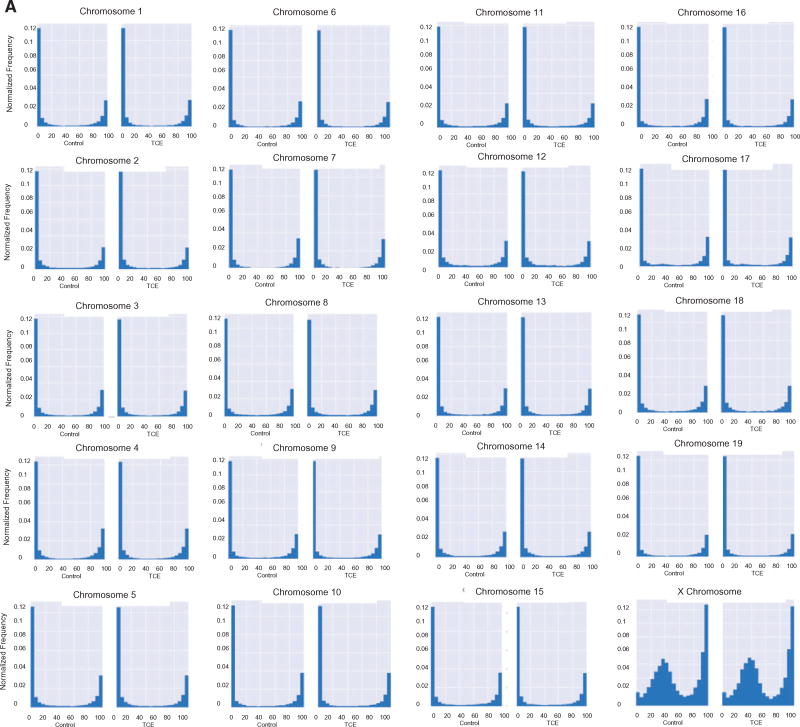
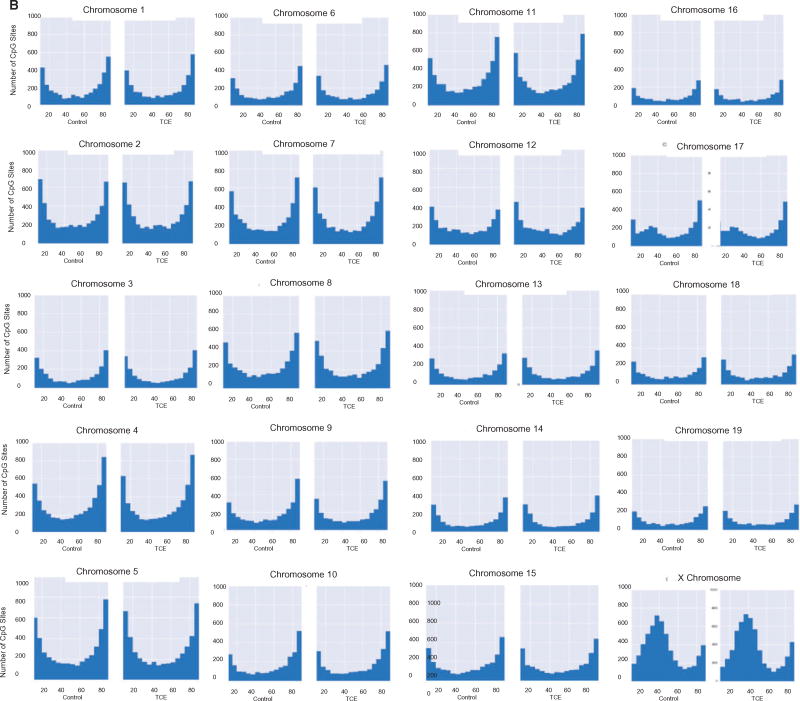
The average methylation levels of CpGs on individual chromosomes were also examined. With the exception of the X chromosome, most of the chromosomes showed very similar methylation profiles (Fig. 2A). In other words, most had similar percentages of CpGs with hypo-, hyper- and mid-range methylation levels, regardless of whether they came from CD4+ T cells from control or TCE-treated mice. Unlike the 19 autosomal chromosomes, very few (∼2.4%) of the CpGs interrogated in the X chromosome displayed methylation levels between 0 and 5%. Instead, the X chromosome had a slightly higher percentage (18.3%) of CpGs with mean methylation levels between 95 and 100%, and a considerably higher percentage (51%) of CpGs with mean methylation levels between 15 and 60%, peaking around 40% mean methylation. This effect was more visually striking after removing from the histograms all CpGs with 0–5% or 95–100% methylation (Fig. 2B). The difference in the average methylation histogram of the X chromosome was confirmed by Fractional Polynomial modeling (Supplementary Table S1 and Supplementary Fig. S1). This modeling showed that the basic shape of the methylation distribution did not differ among representative autosomal chromosomes (chromosomes 1, 3 or 12), regardless of TCE exposure, but that all are different from the X chromosome. Figure 2B also reveals other apparent chromosome-specific differences in the shape of the methylation distribution histograms. For example, chromosome 17 has increased mid-range DNA methylation levels, while several other chromosomes (e.g. 16, 18 and 19) had flattened U-shape histograms indicated less skewing toward hypo- or hyper-methylation status. The chromosome-specific differences in CpG mean methylation appeared to be inherent, i.e. were not induced by TCE exposure.
Figure 2.
Chromosome-specific mean DNA methylation levels. (A) The results from the RRBS analysis described in Fig. 1 were sorted into individual chromosomes, and presented after binning for average methylation of the CpGs. The area of each histogram was normalized to one to make it easier to compare the chromosomes. (B) The RRBS results were presented (as total number of CpG sites in the different bins) after excluding the CpG sites that were either 0–5% or 95–100% methylated
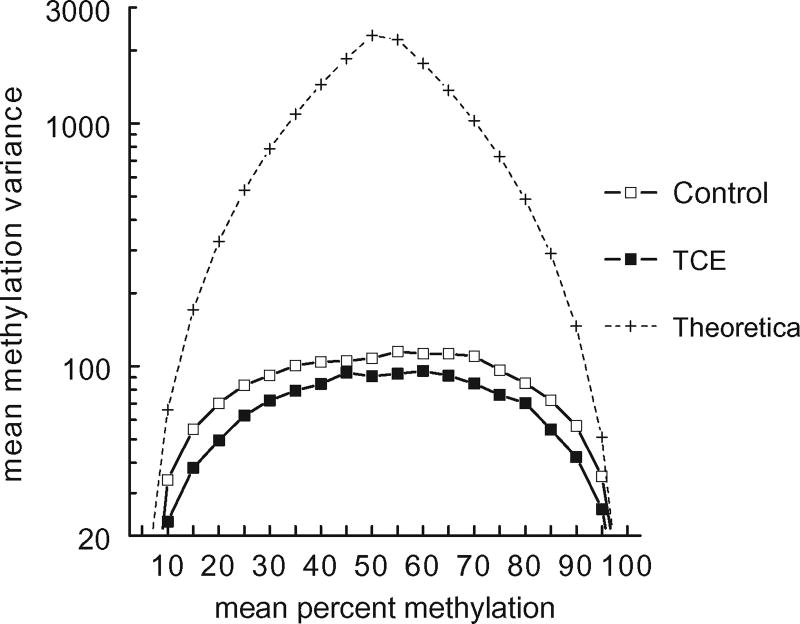
When the mean methylation of thousands of CpG sites was examined, as shown in Figs 1 and 2, no TCE-induced effect was detected. However, global effects on DNA methylation can also occur at the level of methylation variance [47]. Methylation variance reflects the group-specific inter-sample variation in the methylation of each CpG site, rather than mean methylation of each CpG site. An examination of all the CpGs interrogated showed that mean methylation variance detected in effector/memory CD4+ T cells, regardless of TCE treatment, was substantially lower than the theoretical variance that would be achieved by random distribution in a percent mean methylation bin (Fig. 3). However, as we and others have previously reported, inter-sample methylation variance at the CpG sites examined in effector/memory CD4+ T cells correlated with distance to either end of the 0–100% methylation scale [6, 48]. Thus, mean methylation variance in both control and TCE samples was highest at those CpG sites that averaged 30–80% methylation (Fig. 3). Interestingly, exposure to TCE decreased the methylation variance at CpGs at almost all levels of mean methylation. This was true for the X chromosome as well, despite its different mean methylation distribution (data not shown). A likelihood test statistic of 75.8 (P-value < 0.0001) suggested that the curves documenting methylation variance for all the CpGs interrogated (Fig. 3) are significantly different between the samples from control and TCE-treated mice. We evaluated the methylation variance for each of the CpG sites that averaged between 50 and 60% methylation for four individual control samples (Supplementary Fig. S2). This revealed that the variance in the control group could not be attributed to an outlier sample, but appeared to represent a consistent difference among all the samples. Thus, the ability of TCE to impact CpG methylation on a genome-scale, as evidenced by this decrease in methylation variance, did not appear to be artefactual.
Figure 3.
TCE exposure decreased total methylation variance. The RRBS results for the effector/memory CD4+ T cells from control or TCE-treated mice were sorted by treatment group and binned for mean methylation. The inter-sample methylation variance at all the CpG sites in the different bins was then calculated. The dotted line represents a prediction of highest possible methylation variance based on a theoretical value spread of four samples in each bin
Effects of TCE at the Individual CpG Level
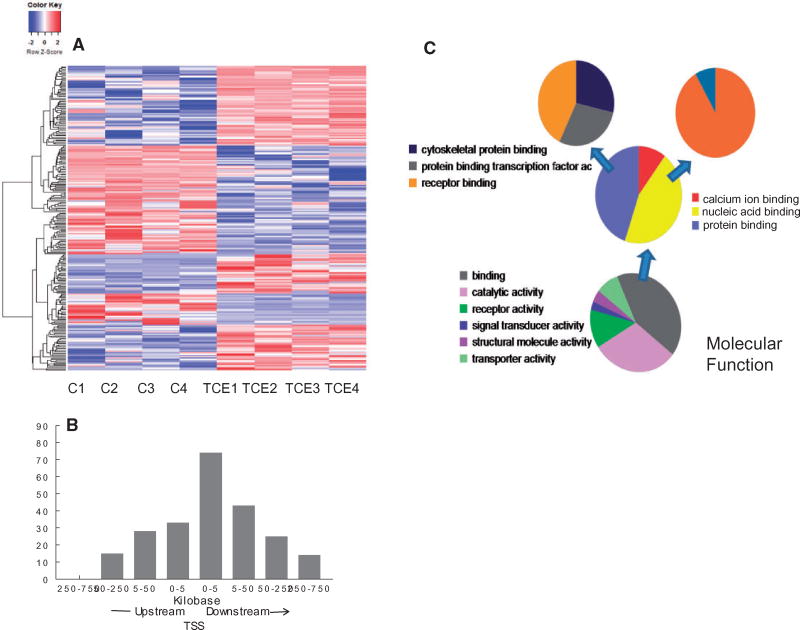
In addition to assessing the genome-scale effects of TCE on DNA methylation, individual CpG sites that were differentially methylated by TCE exposure were identified. Comparison of effector/memory CD4+ T cells from control and TCE-treated mice revealed 233 differentially methylated sites (DMS, q value < 0.005, and methylation differences ≥ 10%). Hierarchical clustering of the DMS is shown in Fig. 4A. Annotation of the DMS indicated that the 233 DMS were associated with 216 genes after taking into account those instances in which two or more CpGs were associated with the same gene. Further evaluation for potential functional significance narrowed down the list to 157 DMS that were actually located in a gene, or within 5 kb upstream of the transcription start site (TSS), and thus in a possible promoter region. As shown in Fig. 4B, more DMS were found downstream compared with upstream of the TSS. Distribution from TSS was not normally distributed (P-value of Shapiro–Wilk tests is <0.001, rejects the null hypothesis which is data are normally distributed). Examination of symmetry plot and quantile–quantile plot shows that the data are heavy-tailed. The greatest number of DMS were located within 5 kB downstream of the TSS. Thus, TCE tended to alter methylation of CpG sites in the gene body close to the TSS more often than it altered sites in a traditional promoter region.
Figure 4.
Identification of CpG sites differentially methylated by TCE exposure. (A) RRBS analysis of effector/memory CD4+ T cells from control and TCE-treated mice revealed 233 DMS (methylation difference ≥ 10%, q value <0.005). Hierarchical clustering of the gene-associated DMS is shown here. (B) The genomic location of the 233 DMS detected in the effector/memory CD4+ T cells relative to the nearest transcription start site (TSS) is shown. (C) The genes associated with the DMS identified by the RRBS were subjected to a gene list functional analysis by the Panther Gene Ontology Classification System
Pathway analysis of the CpG sites differentially methylated by TCE indicated that, in terms of molecular function, TCE primarily altered methylation of genes associated with binding (GO:0005488) (Fig. 4C). Drilling down in the binding molecular function category showed that TCE effects were focused on genes associated with nucleic acid binding (GO:003676), that were in turn enriched for genes associated with DNA binding (GO:0003677). TCE also differentially methylated genes associated with protein binding (GO:0005515), specifically transcription factor binding activity (GO:0000988). Taken together, TCE exposure altered DNA methylation in a manner that seemed primed to impact epigenetic function and gene expression.
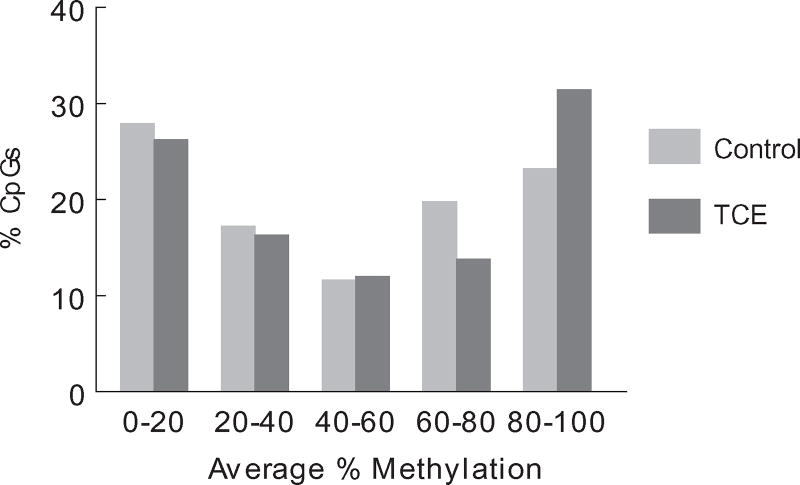
When the CpG sites that were differentially methylated between control and TCE samples were binned by average percent methylation in effector/memory CD4+ T cells from control mice the profile included many CpG sites with mid-range methylation (Fig. 5). The percentage of DMS with hypo-methylated (0–20%) and hyper-methylated (80–100%) status was much lower than those found in the evaluation of all the CpGs interrogated (as shown in Fig. 2). When the differentially methylated CpG sites were binned by average percent methylation in effector/memory CD4+ T cells from TCE-treated mice, it showed that, compared with controls, TCE decreased the number of CpG sites with 60–80% methylation, and increased the number of CpG sites that averaged 80–100% methylation. This suggested that the effect of TCE on DNA methylation was skewed toward inducing hyper- rather than hypo-methylation.
Figure 5.
Average percent methylation of CpG sites differentially methylated by TCE. RRBS analysis of effector/memory CD4+ T cells from control and TCE-treated mice revealed 233 DMS (methylation difference ≥ 10%, q value < 0.005). These DMS were sorted separately for control and TCE-treated samples and binned for mean methylation
Enrichment for Polycomb Protein Binding Sites
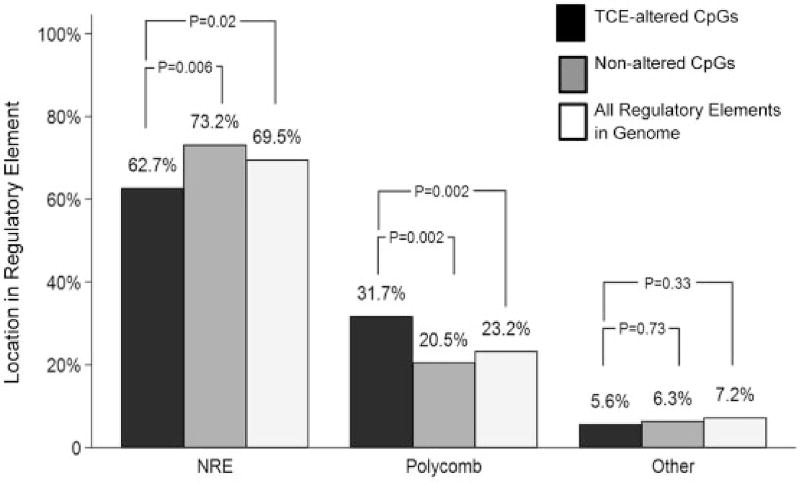
A single CpG may indicate the DNA methylation status of the surrounding region in which differential methylation of other individual sites may not reach the level of statistical significance. Thus, alterations in methylation of single CpGs in regulatory regions can have potential functional importance. The Mouse NCB137/mm9 genome in the UCSC Genome Browser was used to determine whether the DMS were located in regulatory elements that bound transcription factors, and thus might impact transcription. Of the 233 DMS, 87 (37.3%) were found in regulatory elements with annotated transcription factor binding sites. Analysis of these 87 DMS revealed that 85% were found in regions exclusively used to bind one or more of four different Polycomb group (PcG) proteins, namely Ezh2, Suz12, Mtf2 or Jarid2 (Fig. 6; Table 1). The remaining 15% DMS in regulatory regions were found in binding sites for other transcription factors (e.g. Ebf1, Gata1, Nfe212, and ATOH). This distribution was somewhat surprising since only ∼19 000 (4.5%) or the ∼399 000 unique transcription factor binding sites in the whole mouse genome are thought to bind one or more PcG protein. The TCE-induced modifications of CpG sites in the PcG protein binding regions were evenly divided between increased and decreased methylation. Of the DMS in polycomb binding sites, most (85%) flanked a TSS. In comparison, none of the DMS in binding sites for other transcription factors occurred in a region that flanked a TSS. Indeed, an evaluation of all 337 770 CpGs sites interrogated in the effector/memory CD4+ T cells revealed that only 23.2% were found in PcG protein binding regions, while only 7.2% were found in regulatory regions targeted by other transcription factors. Thus, our evaluation of effector/memory CD4+ T cells suggests that PcG protein binding regions are enriched for CpG sites. It is possible that the CpG sites in these regions of effector/memory CD4+ T cells may be particularly sensitive to TCE-induced alterations.
Figure 6.
Many CpG sites differentially methylated by TCE are found in PcG protein binding sites. RRBS analysis of effector/memory CD4+ T cells from control and TCE-treated mice revealed 233 DMS (methylation difference ≥ 10%, q value < 0.005). Annotation of these DMS described whether they were found outside of a transcription binding site (NRE: no regulatory element), or in a regulatory element that bound PcG proteins or other transcription factors. The percentage of CpG sites differentially methylated by TCE was compared with 400 randomly selected CpG sites not altered by TCE (Random CpGs), and to the total number of individual OREG sites known to bind Suz12, EZH2, Mtf2 or Jarid2 or other transcription factors (as identified in Mouse NCB137/mm9 genome in the UCSC Genome Browser) (All Regulatory Elements in Genome)
Table 1.
DMS found in genes or gene promoter regions (q value < 0.005 and differential methylation > 10%)
| Chr | Position | Difference between TCE and control methylation |
Refseq ID | Feature name | PcG | Other TF with PcG |
|---|---|---|---|---|---|---|
| chr1 | 52733087 | 19.80 | NM_133829 | Mfsd6 | None | None |
| chr1 | 72871428 | 10.11 | NM_008342 | Igfbp2 | None | None |
| chr1 | 84692366 | −12.74 | NM_L52915 | Dner | Mtf2 | |
| chr1 | 87915123 | 12.38 | NM_027029 | Spata3 | Mtf2, Suz12, EZH2, Jarid2 | |
| chr1 | 87915176 | 10.91 | NM_027029 | Spata3 | Mtf2, Suz12, EZH2, Jarid2 | |
| chr1 | 88566769 | −11.67 | NM_010933 | Nppc | Suz12, EZH2, Jarid2 | |
| chr1 | 95079010 | 21.05 | NM_001310428 | Crocc2 | None | None |
| chr10 | 57695435 | 19.10 | NM_001081954 | Dux | Mtf2 | |
| chr10 | 59348501 | 38.10 | NM_019965 | Dnajb12 | None | Ebf1 |
| chr10 | 70622501 | 12.01 | NM_031397 | Bicc1 | Mtf2, Suz12, EZH2, Jarid2 | |
| chr10 | 80297554 | 26.59 | NM_001013758 | Lingo3 | Mtf2, Suz12, EZH2 | |
| chr10 | 80306503 | 13.08 | NM_001013758 | Lingo3 | Mtf2, Suz12, EZH2 | |
| chr10 | 114239413 | −17.84 | NM_146241 | POL2 | Mtf2, Suz12, EZH2, Jarid2 | |
| chr10 | 126954849 | 24.03 | NM_001098789 | Shmt2 | None | |
| chr10 | 126962175 | −15.64 | NM_028230 | Nxph4 | Mtf2, Suz12 | |
| chr10 | 127733967 | −12.19 | NM_031252 | Il23a | None | |
| chr11 | 8496658 | 22.56 | NM_001083587 | Tns3 | None | None |
| chr11 | 50416921 | −11.01 | NM_175643 | Adamts2 | Mtf2, Suz12, EZH2, Jarid2 | |
| chr11 | 53271010 | −23.32 | NM_027917 | Schroom1 | Mtf2, Suz12 | |
| chr11 | 61267631 | 27.93 | NM_009548 | Rnf112 | None | None |
| chr11 | 92958940 | −13.88 | NM_028296 | Car 10 | Mtf2, Suz12 | |
| chr11 | 95692072 | 29.58 | NM_025659 | Abi3 | Mtf2, Suz12, EZH2 | |
| chr11 | 103222902 | 10.07 | NM_001205236 | h3d20 | None | |
| chr11 | 113664146 | −20.41 | NM_172800 | Sdk2 | None | |
| chr11 | 117829957 | −13.33 | NM_007707 | Socs3 | None | |
| chr11 | 121691565 | 22.39 | NM_029049 | Ptchd3 | Mtf2, Suz12 | |
| chr12 | 25366541 | 16.22 | NM_001004455 | Cys1 | Mtf2, EZH2 | |
| chr12 | 28026694 | 18.76 | NM_009234 | Sox11 | Mtf2, Suz12, EZH2 | |
| chr12 | 51749480 | 14.75 | NM_008858 | Prdk1 | Mtf2, Jarid2 | |
| chr12 | 51749594 | 22.12 | NM_008858 | Prdk1 | Mtf2, Jarid2 | |
| chr12 | 77506059 | 13.22 | NM_008301 | Hspa2 | Mtf2, Jarid2, EZH2, Jarid2 | |
| chr12 | 81071379 | 17.08 | NM_001252562 | Rad51b | None | Ebf1 |
| chr12 | 81216463 | −18.69 | NM_007564 | Zfp3611 | Mtf2 | |
| chr12 | 81794135 | 20.44 | NM_001177503 | Plekhd1 | Mtf2, Suz12 | |
| chr12 | 84828460 | 27.36 | NM_001267625 | Dpf3 | Mtf2, Jarid2, EZH2, Jarid2 | |
| chr12 | 84828462 | 32.76 | NM_001267625 | Dpf3 | Mtf2, Jarid2, EZH2,Suz12 | |
| chr12 | 85758595 | 20.16 | NM_025525 | Bbof1 | None | None |
| chr12 | 86630006 | −21.49 | NM_172414 | Zc2hclc | None | None |
| chr12 | 111468610 | 28.44 | NM_012023 | Ppp2r5c | None | None |
| chr13 | 49168362 | −23.74 | NM_001290313 | Wnk2 | None | None |
| chr13 | 53560005 | −23.88 | NM_013601 | Msx2 | Mtf2 | |
| chr13 | 55097204 | −16.10 | NM_001206390 | Unca5 | None | None |
| chr13 | 69137738 | 17.13 | NM_153534 | Adcy2 | EZH2, Mtf2 | |
| chr13 | 104899736 | −10.66 | NM_029447 | N1n | None | None |
| chr13 | 110249606 | −21.25 | NM_011056 | Pde4d | None | None |
| chr14 | 55195182 | −22.04 | NM_010590 | Ajuba | None | None |
| chr14 | 64478548 | −26.58 | NM_028228 | Pinx1 | None | None |
| chr15 | 7763733 | −10.39 | NM_001301333 | Gdnf | Mtf2, Jarid2, EZH2, Suz12 | |
| chr15 | 39751326 | 26.27 | NM_172814 | Lrp12 | None | None |
| chr15 | 78548459 | −21.71 | NM_183141 | Elfn2 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr15 | 78873745 | −10.38 | NM_015738 | Galr3 | Suz12, EZH2 | |
| chr15 | 79920093 | 22.09 | NM_009303 | Syngr1 | None | None |
| chr15 | 91729910 | 11.19 | NM_198927 | Mucl9 | None | None |
| chr15 | 101858889 | −20.17 | NM_010664 | Krt18 | Mtf2 | |
| chr15 | 22440834 | 24.56 | NM_023794 | Etv5 | None | None |
| chr16 | 65815534 | 17.40 | NM_028572 | Vg113 | None | ATOH1 |
| chr16 | 72682095 | 11.64 | NM_019413 | Robo1 | None | None |
| chr16 | 84989074 | 13.29 | NM_001198823 | App | None | None |
| chr17 | 11806358 | 26.52 | NM_016694 | Park2 | None | None |
| chr17 | 26204880 | −17.06 | NM_001162868 | Rab11fip3 | Mtf1 | |
| chr17 | 27787774 | 30.42 | NM_001286743 | Pascin1 | None | None |
| chr17 | 32326330 | 14.78 | NM_001033163 | Ephx3 | Mtf2 | |
| chr17 | 35033560 | 20.63 | NM_001286575 | Zbtb12 | None | None |
| chr17 | 47809520 | 26.89 | NM_198421 | Usp49 | None | None |
| chr17 | 80112667 | 10.22 | NM_009994 | Cyb1b1 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr17 | 86014548 | 20.07 | NM_198421 | SIX30S1 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr18 | 7170227 | −15.42 | NM_001081393 | Armc4 | None | None |
| chr18 | 11997673 | 11.79 | NM_001146287 | Cables1 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr18 | 37926514 | 26.30 | NM_033595 | Pcdhga12 | Suz12 | |
| chr18 | 46372273 | −16.68 | NM_178872 | Trim 36 | Suz12, Mtf2 | |
| chr18 | 74603497 | −26.02 | NM_201600 | Myo5b | EZH2, Mtf2 | |
| chr18 | 75980831 | −36.51 | NM_L45356 | Zbtb7c | Mtf2, Jarid2, EZH2, Suz12 | |
| chr19 | 5332047 | −28.01 | NM_L39301 | Catsper1 | None | None |
| chr19 | 47388615 | 22.47 | NM_008018 | Sh3pxd2a | None | None |
| chr19 | 47388661 | 24.44 | NM_008018 | Sh3pxd2a | None | None |
| chr2 | 5636284 | 11.52 | NM_177343 | Camkid | None | None |
| chr2 | 37649110 | 20.86 | NM_001163566 | Crb2 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr2 | 37649121 | 20.12 | NM_001163566 | Crb2 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr2 | 76176253 | 15.65 | NM_001081033 | Pdella | Mtf2 | |
| chr2 | 91316809 | −23.84 | NM_172668 | Lrp4 | None | None |
| chr2 | 91475664 | 16.00 | NM_010168 | F2 | None | None |
| chr2 | 126379285 | −22.21 | NM_011978 | Slc27a2 | None | None |
| chr2 | 147874612 | −14.49 | NM_010446 | Foxa2 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr2 | 165242677 | 16.72 | NM_054055 | Slc13a3 | None | None |
| chr3 | 69120629 | −12.04 | NM_178726 | Ppm11 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr3 | 89229249 | 12.43 | NM_001113331 | Shc1 | None | None |
| chr3 | 128906125 | 15.67 | NM_011098 | Pitx2 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr3 | 151928358 | −26.03 | NM_199465 | Nexn | Mtf2, Jarid2, EZH2, Suz12 | |
| chr4 | 46728412 | −11.11 | NM_001081141 | Gabbr2 | None | None |
| chr4 | 65065259 | −33.06 | NM_019514 | Astn2 | None | None |
| chr4 | 80557862 | 10.89 | NM_026821 | Lurap11 | None | None |
| chr4 | 117928909 | 15.15 | NM_011213 | Ptprf | None | None |
| chr4 | 124682634 | −26.36 | NM_138683 | Rspo1 | None | None |
| chr4 | 133430503 | −18.16 | NM_001285506 | Rps6ka1 | None | None |
| chr4 | 134711341 | 26.82 | NM_019732 | Runx3 | Suz12 | |
| chr4 | 134711350 | 23.31 | NM_019732 | Runx3 | Suz12 | |
| chr4 | 139379960 | −22.47 | NM_011039 | Pax7 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr4 | 141640595 | −21.15 | NM_145402 | GM10565 | None | None |
| chr4 | 148423617 | 29.44 | NM_019781 | Pexl4 | None | None |
| chr4 | 149745860 | 17.35 | NM_001085492 | Rere | None | None |
| chr4 | 151089350 | 26.29 | NM_001081557 | Camta1 | None | None |
| chr4 | 152834168 | 18.58 | NM_001099299 | Ajap1 | None | None |
| chr5 | 34066416 | 11.04 | NM_001163217 | Fgfr3 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr5 | 37187480 | 10.53 | NM_026242 | Mrfap1 | None | None |
| chr5 | 114723943 | −11.11 | NM_148935 | Foxn4 | None | None |
| chr5 | 122278672 | 22.15 | NM_001306126 | Sh2b3 | Mtf2 | |
| chr5 | 131782939 | −23.02 | NM_145218 | Wbscr17 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr5 | 140447989 | 17.64 | NMJL75522 | Rlfn1 | None | None |
| chr5 | 148138165 | −13.33 | NM_001039678 | Urad | None | None |
| chr6 | 23210525 | −18.28 | NM_028462 | Cadps2 | Mtf2 | |
| chr6 | 63207478 | 17.79 | NM_008167 | Grid2 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr6 | 83135914 | −26.38 | NM_007835 | Dctn1 | None | None |
| chr6 | 85324297 | −10.98 | NM_001003955 | Rab11fip5 | None | None |
| chr6 | 98926364 | 30.44 | NM_001197322 | Foxp1 | None | None |
| chr6 | 113342775 | 33.59 | NM_133923 | Tt113 | None | None |
| chr6 | 125262312 | −21.28 | NM_010736 | Tltpr | Mtf2 | |
| chr7 | 13628786 | −15.20 | NM_145819 | Mzf1 | Suz12, Mtf2 | |
| chr7 | 16726237 | −11.83 | NM_148946 | Slc8a2 | None | None |
| chr7 | 24096775 | −16.75 | NM_001004194 | Nirpfe | None | None |
| chr7 | 29606786 | −24.74 | NM_016772 | Hnrnp1 | None | Ebf1 |
| chr7 | 35015176 | 13.38 | NM_008155 | Gpi1 | None | None |
| chr7 | 52940127 | 18.03 | NM_001289693 | Sec1/Ntn5 | None | None |
| chr7 | 63520836 | −24.05 | NM_021879 | Oca2 | None | None |
| chr7 | 87481393 | −12.53 | NM_L33952 | Unc45a | None | None |
| chr7 | 93301399 | 28.53 | NM_001102578 | Vmn2r75 | None | None |
| chr7 | 104376316 | 21.11 | NM_001177412 | Gab2 | None | None |
| chr7 | 133957534 | 27.42 | NM_026884 | Fam57b | None | ATOH1 |
| chr7 | 138078229 | −22.20 | NM_019564 | Htra1 | Mtf2, EZH2, Suz12 | |
| chr8 | 12430585 | −10.48 | NM_009233 | GM5607 | Mtf2 | |
| chr8 | 72406588 | 18.95 | NM_026818 | Cilp2 | Mtf2, Jarid2, EZH2, Suz12 | |
| chr8 | 72898699 | 14.06 | NM_016685 | Comp | Mtf2, EZH2, Suz12 | |
| chr8 | 73296263 | −11.49 | NM_008841 | Pik3r2 | None | None |
| chr8 | 83263159 | −24.64 | NM_053124 | Smarca5 | None | None |
| chr8 | 94882246 | −12.94 | NM_018826 | Irx5 | Mtf2, EZH2, Suz12 | |
| chr8 | 107880886 | −12.28 | NM_001081332 | Slc9a5 | None | None |
| chr8 | 116729816 | 13.57 | NM_L73016 | Vat11 | Mtf2, Suz12 | |
| chr8 | 121971793 | −19.02 | NM_054095 | Necab2 | Jarid2, EZH2, Suz12 | |
| chr8 | 125088900 | 26.17 | NM_026014 | Cdt1 | None | Ebf1 |
| chr8 | 125389116 | −36.05 | NM_007662 | Cadherin 15 | None | Ebf1 |
| chr8 | 125389194 | −30.65 | NM_007662 | Cadherin 15 | None | Ebf1 |
| chr8 | 125389244 | −29.38 | NM_007662 | Cadherin 15 | None | Ebf1 |
| chr8 | 125389246 | −28.44 | NM_007662 | Cadherin 15 | None | Ebf1 |
| chr9 | 21549579 | 12.84 | NM_026282 | Ldlr | None | None |
| chr9 | 31720357 | −14.51 | NM_013800 | Barx2 | Jarid2, EZH2, Suz12 | |
| chr9 | 56994574 | −14.04 | NM_028347 | Nei11 | None | None |
| chr9 | 107611227 | −21.43 | NM_011349 | Sema3f | Mtf2, EZH2, Suz12 | |
| chrX | 11655662 | −14.96 | NM_029510 | Bcor | Mtf2, Jarid2, EZH2, Suz12 | |
| chrX | 11658411 | −23.91 | NM_175046 | Bcor | Mtf2, Jarid2, EZH2, Suz12 | |
| chrX | 34415040 | 22.79 | NM_019668 | Ube2a | None | None |
| chrX | 34415077 | 21.67 | NM_019668 | Ube2a | None | None |
| chrX | 56387270 | 27.18 | NM_010200 | Fgf13 | Mtf2, EZH2 | |
| chrX | 68917537 | −24.71 | NM_010340 | Gpr50 | Mtf2, Jarid2, EZH2, Suz12 | |
| chrX | 97454071 | 21.48 | NM_001177943 | Eda | None | None |
| chrX | 110412278 | 23.36 | NM_033605 | Dach2 | None | None |
| chrX | 130221716 | 30.33 | NM_001105245 | Pcdh19 | Mtf2, Jarid2, Suz12 | |
| chrX | 158346841 | −25.22 | NM_198409 | Nhs | None | None |
| chrX | 160347121 | 27.82 | NM_001290379 | Apls2 | None | None |
TCE Alters CD4+ T-Cell Gene Expression
The functional effects of TCE exposure were further assessed at the gene expression level. This was accomplished using a microarray assessment of the same cells as those profiled in the RRBS analysis: i.e. effector/memory (CD62Llo) CD4+ T cells collected from control mice or from mice exposed to TCE for 40 weeks. Gene expression was examined 20h after activation of the CD4+ T cells in vitro. At a cutoff of FDR < 0.05 and a fold-change > 1.25, the expression of ∼560 genes was found to be significantly altered in the activated effector/memory CD4+ T cells of mice exposed to TCE compared with similarly activated effector/memory CD4+ T cells from control mice (data not shown). Of these differentially expressed genes, those associated with immune function are listed in Table 2. A network evaluation suggested that pathways with the most number of genes altered by TCE after 40 weeks were those with decreases in gene expression that centered on Ifng and Tnf (Supplementary Fig. S3A). qRT-PCR analysis confirmed the TCE-induced decrease in the expression of Ifng and Tnf in the effector/memory CD4+ T cells (Supplementary Fig. S3B). Of the genes altered by TCE in the effector/memory CD4+ T cells, none contained the previously described DMS.
Table 2.
TCE vs Control annotated immune gene expression
| Symbol | REFSEQ_ID | FC | P value | adj. P value | q-Value | PcG | Other TF with PcG |
|---|---|---|---|---|---|---|---|
| Cytokines | |||||||
| Ifit2 | NM_008332.2 | −2.91422 | 6.49E–06 | 0.008901 | 0.007645 | None | Cdx1, Myod1 |
| Tnf | NM_013693.1 | −2.43709 | 3.44E–05 | 0.015926 | 0.01368 | None | None |
| Ifitm3 | NM_025378.2 | −2.04188 | 0.000198 | 0.031075 | 0.026692 | None | Stat5a |
| Amica1 | NM_001005421.3 | −2.01204 | 0.000128 | 0.025726 | 0.022097 | None | None |
| Ifi202b | NM_008327.1 | −2.00414 | 1.99E–06 | 0.005447 | 0.004679 | None | None |
| Il4 | NM_021283.1 | −1.98215 | 0.00038 | 0.041971 | 0.03605 | None | Nfatc2 |
| Ifitm1 | NM_026820.2 | −1.86847 | 0.00039 | 0.04252 | 0.036523 | None | None |
| Irf7 | NM_016850.2 | −1.84292 | 0.000657 | 0.054756 | 0.047032 | None | None |
| Isg20 | NM_020583.4 | −1.75296 | 1.48E–05 | 0.011839 | 0.010169 | None | Foxa2 |
| Il17a | NM_010552.3 | −1.68794 | 0.000793 | 0.060267 | 0.051766 | None | Bhlhe40 |
| Lif | NM_001039537.1 | −1.50454 | 1.05E–05 | 0.01061 | 0.009113 | None | Pax6 |
| Il16 | NM_010551 | 1.305869 | 6.96E–07 | 0.00394 | 0.003384 | None | Foxa2, ATOH1 |
| Tnfrsf26 | NM_175649.5 | 1.346837 | 0.00032 | 0.038826 | 0.03335 | None | Cdx1 |
| Ing4 | NM_133345.2 | 1.366442 | 0.000634 | 0.054238 | 0.046587 | None | Rxra, Sox3 |
| Traf1 | NM_009421.3 | 1.401221 | 0.000837 | 0.062156 | 0.053389 | None | Bhlhe40, Myod1 |
| Ncf4 | NM_008677.1 | 1.405497 | 8.09E–05 | 0.021965 | 0.018867 | None | Ebf1, Myod1, Bhlhe40 |
| Il1r2 | NM_010555.4 | 1.459921 | 0.00052 | 0.049157 | 0.042223 | None | Foxa2 |
| Chemokines | |||||||
| Cxcl10 | NM_021274.1 | −1.76465 | 0.000219 | 0.032733 | 0.028116 | None | None |
| Ccrl2 | NM_017466.4 | −1.66005 | 1.25E–05 | 0.010725 | 0.009212 | None | None |
| Cxcr4 | NM_009911.2 | 1.634843 | 0.000334 | 0.020502 | 0.016849 | Suz12,Jarid2, Mtf2 | ATOH1 |
| Transcription factors & enzymes | |||||||
| Mycbp2 | NM_207215.2 | −1.64382 | 0.000189 | 0.030347 | 0.026066 | Mtf2 | Meis1, Bhlhe40, Ebf1 |
| Trafd1 | NM_172275.1 | −1.58643 | 7.10E–05 | 0.021467 | 0.018439 | None | Foxa2, Nkx2-5 |
| Sp100 | NM_013673.2 | −1.31215 | 0.000377 | 0.041921 | 0.036008 | None | Bhlhe40, Cdx1 |
| Cxxc1 | NM_028868.3 | 1.321214 | 7.64E–05 | 0.021965 | 0.018867 | None | Bhlhe40 |
| Csk | NM_007783.2 | 1.344270 | 0.0003704 | 0.041861 | 0.035956 | None | Ebf1, ATOH1 |
| Mapk11 | NM_011161.4 | 1.341871 | 0.000238 | 0.034005 | 0.029208 | None | Egr2, Sox3 |
| Rap1gap | NM_001081155.1 | 1.344891 | 0.000792 | 0.060267 | 0.051766 | Mtf2 | Bhlhe40 |
| Elk3 | NM_205536.1 | 1.3791 | 1.47E–05 | 0.011839 | 0.010169 | None | Ebf1, Stat5a |
| Nfkbib | NM_010908.3 | 1.410298 | 0.000119 | 0.025478 | 0.021884 | None | Foxa2 |
| Rag1ap1 | NM_009057.2 | 1.42048 | 0.000158 | 0.027165 | 0.023333 | None | Ebf1 |
| Mt1 | NM_013602.2 | 2.12539 | 7.29E–06 | 0.009447 | 0.008114 | None | None |
| Cell cycle | |||||||
| Gadd45g | NM_011817.1 | −1.30109 | 7.79E–05 | 0.021965 | 0.018867 | Suz12, Mtf2 | ATOH1 |
| Cdc23 | NM_178347 | 1.340161 | 5.13E–06 | 0.0083 | 0.007129 | None | Foxa2 |
| Apoptosis | |||||||
| Daxx | NM_007829.3 | −1.79108 | 3.82E–05 | 0.016024 | 0.013764 | None | None |
| Fas1 | NM_010177.3 | −1.45356 | 0.000115 | 0.025294 | 0.021726 | None | Cdx1 |
| Bcl11b | NM_021399.2 | 1.311508 | 0.000262 | 0.035376 | 0.030386 | EZH2, Suz12, Jarid2, Mtf2 | |
| Pdcd2 | NM_008799.2 | 1.356754 | 3.22E–06 | 0.006902 | 0.005928 | None | Bhlhe40, Meis1 |
| Pdcd4 | NM_011050.3 | 1.407455 | 0.000546 | 0.05012 | 0.04305 | None | Foxa2, Myod1 |
| Integrins | |||||||
| Sdc3 | NM_011520.3 | −1.84389 | 0.000208 | 0.031629 | 0.027168 | None | Hoxc9, Sox3 |
| Ly6c1 | NM_010741.2 | −1.50191 | 0.000146 | 0.026013 | 0.022343 | None | Cdx1, Bhlhe40 |
| Cd247 | NM_031162.1 | 1.300788 | 0.000203 | 0.031216 | 0.026812 | None | Foxa2, Creg1, Tal1 |
| Leng9 | NM_175529.3 | 1.303316 | 0.000444 | 0.045017 | 0.038667 | Mtf2 | None |
| Mic2l1 | NM_138309.1 | 1.33617 | 9.20E–05 | 0.022878 | 0.019651 | None | Stat5a |
| Hist1h2ai | NM_178182.1 | 1.419197 | 0.000898 | 0.063308 | 0.054378 | None | None |
| Hist1h1c | NM_015786.1 | 1.592885 | 0.000622 | 0.054158 | 0.046519 | None | Cdx1, Bhlhe40 |
| Ctla4 | NM_009843.3 | 1.472701 | 0.000221 | 0.03284 | 0.028208 | None | None |
| Miscellaneous | |||||||
| Birc2 | NM_007465.1 | 1.372856 | 1.75E–05 | 0.012794 | 0.010989 | None | Rxra, Cdx1 |
| Ddb2 | NM_028119.4 | 1.37404 | 1.22E–05 | 0.010725 | 0.009212 | None | Cdx1, Rxra, Tal1 |
| Rfx1 | NM_009055.2 | 1.519909 | 0.000101 | 0.023946 | 0.020568 | None | Myod1 |
Although TCE-induced gene changes and DMS did not coincide they did share some common features. Evaluation of the first intron and 5 kb region upstream of the TSS for the immune genes altered by TCE (Table 2) showed that 10.2% had binding sites for PcG proteins, while an additional 69.4% had binding sites for other transcription factors that were in turn regulated by PcG proteins at their own TSS. This suggests that at least 80% of the genes altered by TCE in the effector/memory CD4+ T cells had the potential for PcG protein regulation.
Discussion
Our RRBS evaluation of CpG methylation distributions in autosomal chromosomes in effector/memory CD4+ T cells conformed to the general trend: hypomethylation > hypermethylation > mid-range methylation. On a genome-scale TCE appeared to decrease methylation variance of CpG sites that averaged <95%, or >5% methylation. Increased methylation variance at CpG sites that average between 30 and 60% methylation is thought to be important for maintaining flexibility in the methylation and associated expression of functionally important genes [47]. Such regions may be protected from suppressed, fully methylated states or permissive, unmethylated states. When we examined all the CpGs interrogated by RRBS in the effector/memory CD4+ T cells from control mice, the variance was highest for CpGs with intermediate methylation levels. This has been described previously [6] and is related to the fact that percentage scales tend to restrict variability near the edges of the scale. In the current study, we found that TCE exposure decreased variance in CpG sites with intermediate methylation levels. The ability of TCE to impact intermediate methylation may have more functional significance than effects at the ends of the methylation scale; increasing DNA methylation from 5 to 20%, or decreasing methylation from 95 to 80% is less likely to alter gene expression.
Using a methylation difference of 10% between the two groups as the cutoff, only 233 CpGs (216 genes) of the 337 770 CpGs interrogated in effector/memory CD4+ T cells were differentially methylated by TCE exposure. The relatively small TCE affect (0.07% of the CpGs examined) compares to the 0.83% of total CpGs that were found to be differentially methylated when naive CD4+ T cells were contrasted to memory CD4+ T cells in humans [24]. It is perhaps not surprising that two populations of effector/memory CD4+ R cells that differed only in relatively low level adult exposure to a toxicant would demonstrate less epigenetic modifications than that which accompanies CD4+ T cell differentiation. A transcriptomic analysis of the same effector/memory CD4+ T cells used to generate the RRBS data identified a number of immune-associated changes following TCE exposure. However, none of the differentially expressed genes overlapped with DMS in the CD4+ T cells from the TCE-treated mice despite the fact that many of the DMS were found in gene bodies within 5 kb downstream of the TSS. Although cytosine methylation of promoters is negatively correlated with gene expression, the question of whether methylation of a particular cytosine impacts expression is still unclear. Others have also seen a lack of correlation between gene expression and treatment-related changes in methylation status [49–53]. In one study of over 230 000 cytosines, only 16.6% demonstrated a significant association between methylation and expression of a closely located TSS [54]. The association between gene body methylation and gene expression appears to be complex and context dependent [55, 56]. Even in cancerous cells with their often more robust changes in methylation, associations between gene expression and methylation are surprisingly small, and include both positive and negative correlations [57]. This may be attributed to the time-dependent sensitivity of gene expression. Alternatively, the epigenetic impact of exogenous factors such as TCE on gene expression may be indirect; via methylation-induced changes in the expression or function of some upstream regulator, and thus not obviously correlative. Alterations in methylation may only play a permissive, rather than direct, role in regulating gene expression.
Compared with the 19 autosomal chromosomes, the X chromosome had a very different profile of mean methylation levels, regardless of TCE exposure. In dramatic contrast to the autosomal chromosomes the X chromosome had very few hypomethylated CpG sites, and a much larger percentage of CpG sites with mid-range DNA methylation. Differences in X chromosome DNA methylation profiles are not surprising due to the epigenetically regulated X chromosome silencing that occurs in the blastocyst. This silencing is accomplished by a combination of epigenetic modifications involving histone deacetylation, RNA methylation, and DNA methylation [58, 59]. Comparing the methylation status of individual CpGs in the X chromosome from peripheral blood leukocytes of males and females indicated both had a set of highly methylated CpGs, while CpGs that were hypomethylated in males (under 11%) tended to be methylated in the 30–40% range in females [60]. This is largely in agreement with our analysis of the X chromosome from effector/memory CD4+ T cells of female mice. The mean methylation distribution observed in the X chromosome does not reflect averaging of two chromosomes, one of which was completely hypermethylated. Instead, the profile indicates the presence of a more complex methylation pattern. It remains to be determined whether this pattern is due to the DNA methylation on the inactivated X chromosome, the active X chromosome, or a combination of both.
One surprising result was the apparent connection between TCE-altered CpGs and PcG protein binding regions. PcG proteins were first identified as regulators of embryonic development and stem cell pluripotency [61]. PcG proteins form two complexes in mammals; polycomb-repressive complex 1 (PRC1) and PRC2. PRC2 mediates H3K27me3, which is thought to inhibit transcription by a mechanism involving H2A ubiquitination and/or chromatin compaction [62, 63]. The initial work on PcG proteins conducted in embryonic stem cells identified a complex interaction between PRC2 binding and DNA methylation. Although the majority of CpG islands do not normally recruit PcG proteins, there is an anomalous conservation of CpG sites at PRC2-binding domains [64, 65]. This connection seems to involve a certain level of reciprocal regulation. For example, PRC2 binding represses DNA methylation at the PRC2 target regions in embryonic stem cells [66]. Similarly, DNA methylation appears to regulate PRC2 binding. A high density of unmethylated CpG sites reportedly promotes PcG protein binding [67]. Removal of DNA methylation promotes the accumulation of the PRC2 complex in inappropriate genomic loci, indicating that DNA methylation is capable of attenuating PRC2 binding [68]. However, there is evidence in somatic cells and cancer cells that DNA methylation and PRC2 binding may not be mutually exclusive, and may in fact work together to suppress specific gene expression [65].
The role of PcG proteins in the regulation of immune function specifically is still being defined. Late stages of human B cell differentiation showed methylation gain at PcG-repressed areas, thus suggesting a need for DNA methylation to block PcG protein binding in non-transformed lymphocytes [69]. In terms of T cells, PcG proteins have been shown to form a complex with the Ikaros transcription factor to regulate thymocyte development [70]. They can also regulate the function of mature peripheral T cells [71]. For example, EZH2, a component of PRC2, has recently been shown by others to be highly expressed in CD4+ T cells [72], where it reportedly associates with Foxp3 to mediate gene repression and suppressive function [73]. Loss of EZH2 in vivo caused increase immune pathology, including colitis, in part due to a lack of functional Treg cells [73, 74]. EZH2 also controls differentiation and plasticity of CD4+ Th1 and Th2 cells by binding and controlling expression of Tbx21 and Gata3 [75]. Deletion of EZH2 leads to increased generation of effector/ memory CD4+ T cells with an increased production of effector cytokines including IFN-γ [73]. Cell differentiation is accompanied by losses and gains of H3K27me3 at many promoters at many stages of the process, while DNA methylation is altered at only a relatively small number of promoters during differentiation. This suggests that PcG protein binding represents a more robust suppression than DNA methylation.
There were some limitations associated with the current study. RRBS analysis does not distinguish between 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). The ten-eleven translocation (TET) family of proteins can oxidize 5mC to 5hmC, a mark not effectively maintained by Dnmt1, thus leading to demethylation as cells divide. Polarization of CD4+ T cells toward Th1 and Th2 lineages is accompanied by changes in 5hmc-mediated DNA demethylation of key genes [76], and defects in DNA hydroxymethylation have been demonstrated in both thymocytes and peripheral CD4+ T cells from patients with autoimmune diseases [77, 78]. A direct correlation between levels of 5hmc and H3K27me has been described in a variety of somatic tissues [79]. It will be important to distinguish whether the enrichment of PcG protein binding sites in the current study are associated with TCE-induced alterations DNA methylation or DNA hydroxymethylation.
Despite its limitations, the current study has demonstrated effects of TCE on genome-wide and gene-specific DNA methylation. This included a TCE-induced decrease in methylation variance, and the observation that TCE-induced changes in CpG methylation tended to occur in regulatory elements that bound suppressive PcG proteins. These effects may be mechanistically important since many autoimmune diseases are driven by effector/memory CD4+ T cells which are resistant to several mechanisms designed to guard against the expansion of autoreactive CD4+ T cells. Thus, any epigenetic mechanism that targeted effector/memory CD4+ T cells could have important functional consequences. Activation and subsequent gene expression in CD4+ T cells is a complex process. Aside from epigenetic mechanisms such as DNA methylation and histone acetylation, this process is also regulated by the levels and/or phosphorylation state of transcription factors and other signaling molecules. Understanding the contribution of all these factors toward CD4+ T cell activation is going to require complex modeling. The epigenetic alteration of polycomb protein binding may be another component in this process. The possibility that TCE alters DNA methylation in PcG protein binding sites, suggests that an associated alteration in PRC2 binding, and downstream upregulation of proinflammatory Th1 cytokines could play a role in the ability of TCE to promote autoimmunity.
Supplementary Material
Acknowledgments
We thank Dr Damir Herman for initial coaching on the RRBS method, Dr Kartik Shankar for recommendations on RRBS sample preparation, and Dr Stewart Macleod for helpful advice on next generation sequencing, and for performing the Illumina sequencing.
Funding
This work was supported by grants from the Arkansas Biosciences Institute, the National Institutes of Health (R01ES021484), and the UAMS Translational Research Institute (National Institutes of Health UL1RR029884).
Footnotes
Data Availability
Data are available in the Supplementary Material.
Supplementary Data
Supplementary data is available at EnvEpig online.
Conflict of interest statement. None declared.
References
- 1.Kawakami N, Odoardi F, Ziemssen T, Bradl M, Ritter T, Neuhaus O, Lassmann H, Wekerle H, Flugel A. Autoimmune CD4+ T cell memory: lifelong persistence of encephalitogenic T cell clones in healthy immune repertoires. J Immunol. 2005;175:69–81. doi: 10.4049/jimmunol.175.1.69. [DOI] [PubMed] [Google Scholar]
- 2.Oling V, Reijonen H, Simell O, Knip M, Ilonen J. Autoantigen-specific memory CD4+ T cells are prevalent early in progression to Type 1 diabetes. Cell Immunol. 2012;273:133–9. doi: 10.1016/j.cellimm.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Elyaman W, Kivisakk P, Reddy J, Chitnis T, Raddassi K, Imitola J, Bradshaw E, Kuchroo VK, Yagita H, Sayegh MH, et al. Distinct functions of autoreactive memory and effector CD4+ T cells in experimental autoimmune encephalomyelitis. Am J Pathol. 2008;173:411–22. doi: 10.2353/ajpath.2008.080142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams JL, Kithcart AP, Smith KM, Shawler T, Cox GM, Whitacre CC. Memory cells specific for myelin oligodendrocyte glycoprotein (MOG) govern the transfer of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2011;234:84–92. doi: 10.1016/j.jneuroim.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ATSDR: Toxicological Profile for Trichloroethylene. US Department of Health and Human Services, Agency for Toxic Substances and Disease Registry; 2014. [PubMed] [Google Scholar]
- 6.Gilbert KM, Blossom SJ, Erickson SW, Reisfeld B, Zurlinden TJ, Broadfoot B, West K, Bai S, Cooney CA. Chronic exposure to water pollutant trichloroethylene increased epigenetic drift in CD4+ T cells. Epigenomics. 2016;8:633–49. doi: 10.2217/epi-2015-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byers VS, Levin AS, Ozonoff DM, Baldwin RW. Association between clinical symptoms and lymphocyte abnormalities in a population with chronic domestic exposure to industrial solvent-contaminated domestic water supply and a high incidence of leukemia. Cancer Immunol Immunother. 1988;27:77–82. doi: 10.1007/BF00205762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanez DS, Moran M, Unamuno P, Armijo M. Silica and trichloroethylene-induced progressive systemic sclerosis. Dermatol. 1992;184:98–102. doi: 10.1159/000247513. [DOI] [PubMed] [Google Scholar]
- 9.Hansen BL, Isager H. A scleroderma-resembling disease-exposure to trichloroethylene and trichloroethane, is there a causal connection? Ugeskr Laeger. 1988;150:805–8. [PubMed] [Google Scholar]
- 10.Saihan EM, Burton JL, Heaton KW. A new syndrome with pigmentation, scleroderma, gynaecomastia, Raynaud’s phenomenon and peripheral neuropathy. Br J Dermatol. 1978;99:437–40. doi: 10.1111/j.1365-2133.1978.tb06184.x. [DOI] [PubMed] [Google Scholar]
- 11.Flindt-Hansen H, Isager H. Scleroderma after occupational exposure to trichloroethylene and trichlorethane. Toxicol Lett. 1987;95:173–81. [PubMed] [Google Scholar]
- 12.Czirjak L, Pocs E, Szegedi G. Localized scleroderma after exposure to organic solvents. Dermatology (Basel) 1994;189:399–401. doi: 10.1159/000246888. [DOI] [PubMed] [Google Scholar]
- 13.Lockey JE, Kelly CR, Cannon GW, Colby TV, Aldrich V, Livingston GK. Progressive systemic sclerosis associated with exposure to trichloroethylene. J Occup Med. 1997;29:493–6. [PubMed] [Google Scholar]
- 14.Dubrow R, Gute DM. Cause-specific mortality among Rhode Island jewelry workers. Am J Ind Med. 1987;12:579–93. doi: 10.1002/ajim.4700120511. [DOI] [PubMed] [Google Scholar]
- 15.Gist GL, Burg JR. Trichloroethylene – a review of the literature from a health effects perspective. Toxicol Ind Health. 1995;11:253–307. doi: 10.1177/074823379501100301. [DOI] [PubMed] [Google Scholar]
- 16.Kilburn KH, Washaw RW. Prevalence of symptoms of systemic lupus erythematosus (SLE) and of fluorescent antinuclear antibodies associated with chronic exposure to trichloroethylene and other chemicals in well water. Environ Res. 1992;57:1–9. doi: 10.1016/s0013-9351(05)80014-3. [DOI] [PubMed] [Google Scholar]
- 17.Clark LC, Giulano A, Walsh B, Guernsey de Zaplen J, Alberts DS, Meister J, Manson TS. The Santa Cruz County Community Health Survey. Phoenix, AZ: Arizona Department of Health Services; 1994. [Google Scholar]
- 18.Nietert PJ, Sutherland SE, Silver RM, Pandey JP, Knapp RG, Hoel DG, Dosemeci M. Is occupational organic solvent exposure a risk factor for scleroderma? Arthritis Rheum. 1998;41:1111–9. doi: 10.1002/1529-0131(199806)41:6<1111::AID-ART19>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 19.Iavicoli I, Marinaccio A, Carelli G. Effects of occupational trichloroethylene exposure on cytokine levels in workers. J Occup Environ Med. 2005;47:453–7. doi: 10.1097/01.jom.0000161728.23285.66. [DOI] [PubMed] [Google Scholar]
- 20.Yi J, Teng YX, Zang D, Zhou W, Dong HY, Niu Y, Bin P, Huang XQ, Zheng YX, Dai YF. Analysis of subgroups of lymphocyte in peripheral blood among dermatitis medicamentosa-like of trichloroethylene patients and healthy exposed workers. Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45:1017–21. [PubMed] [Google Scholar]
- 21.Gilbert KM, Przybyla B, Pumford NR, Han T, Fuscoe J, Schnackenberg LK, Holland RD, Doss JC, MacMillan-Crow LA, Blossom SJ. Delineating liver events in trichloroethylene-induced autoimmune hepatitis. Chem Res Toxicol. 2009;22:626–32. doi: 10.1021/tx800409r. [DOI] [PubMed] [Google Scholar]
- 22.Griffin JM, Blossom SJ, Jackson SK, Gilbert KM, Pumford NR. Trichloroethylene accelerates an autoimmune response in association with Th1 T cell activation in MRL+/+ mice. Immunopharmacology. 2000;46:123–37. doi: 10.1016/s0162-3109(99)00164-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Wang J, Ma H, Ansari GA, Khan MF. N-Acetylcysteine protects against trichloroethene-mediated autoimmunity by attenuating oxidative stress. Toxicol Appl Pharmacol. 2013;273:189–95. doi: 10.1016/j.taap.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori HK, Hart T, LaMere SA, Chew PV, Salomon DR. Defining CD4 T cell memory by the epigenetic landscape of CpG DNA methylation. J Immunol. 2015;194:1565–79. doi: 10.4049/jimmunol.1401162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto S, Ogoshi K, Sasaki A, Abe J, Qu W, Nakatani Y, Ahsan B, Oshima K, Shand FH, Ametani A, et al. Coordinated changes in DNA methylation in antigen-specific memory CD4 T cells. J Immunol. 2013;190:4076–91. doi: 10.4049/jimmunol.1202267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lleo A, Zhang W, Zhao M, Tan Y, Bernuzzi F, Zhu B, Liu Q, Tan Q, Malinverno F, Valenti L, et al. DNA methylation profiling of the X chromosome reveals an aberrant demethylation on CXCR3 promoter in primary biliary cirrhosis. Clin Epigenetics. 2015;7:61–0098. doi: 10.1186/s13148-015-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altorok N, Coit P, Hughes T, Koelsch KA, Stone DU, Rasmussen A, Radfar L, Scofield RH, Sivils KL, Farris AD, et al. Genome-wide DNA methylation patterns in naive CD4+ T cells from patients with primary Sjogren’s syndrome. Arthritis Rheumatol. 2014;66:731–9. doi: 10.1002/art.38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer B, Chavez RA, Munro JE, Chiaroni-Clarke RC, Akikusa JD, Allen RC, Craig JM, Ponsonby AL, Saffery R, Ellis JA. DNA methylation at IL32 in juvenile idiopathic arthritis. Sci Rep. 2015;5:11063. doi: 10.1038/srep11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Shu Y, Xiao Y, Wang Q, Kanekura T, Li Y, Wang J, Zhao M, Lu Q, Xiao R. Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin Epigenetics. 2014;6:25–6. doi: 10.1186/1868-7083-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Z, Mei X, Zhao D, Sun Y, Song J, Pan W, Shi W. DNA methylation modulates HERV-E expression in CD4+ T cells from systemic lupus erythematosus patients. J Dermatol Sci. 2015;77:110–6. doi: 10.1016/j.jdermsci.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Coit P, Renauer P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, Sawalha AH. Renal involvement in lupus is characterized by unique DNA methylation changes in naive CD4+ T cells. J Autoimmun. 2015;61:29–35. doi: 10.1016/j.jaut.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J Autoimmun. 2013;41:92–9. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, Richardson BC. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupuslike disease in syngeneic mice. J Clin Invest. 1993;92:38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–35. [PubMed] [Google Scholar]
- 35.Ivanova EA, Orekhov AN. T helper lymphocyte subsets and plasticity in autoimmunity and cancer: an overview. Biomed Res Int. 2015;2015:327470. doi: 10.1155/2015/327470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirahara K, Nakayama T. CD4+ T-cell subsets in inflammatory diseases: beyond the Th1/Th2 paradigm. Int Immunol. 2016;28:163–71. doi: 10.1093/intimm/dxw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cosmi L, Maggi L, Santarlasci V, Liotta F, Annunziato F. T helper cells plasticity in inflammation. Cytometry A. 2014;85:36–42. doi: 10.1002/cyto.a.22348. [DOI] [PubMed] [Google Scholar]
- 38.Gilbert KM, Nelson AR, Cooney CA, Reisfeld B, Blossom SJ. Epigenetic alterations may regulate temporary reversal of CD4(+) T cell activation caused by trichloroethylene exposure. Toxicol Sci. 2012;127:169–78. doi: 10.1093/toxsci/kfs093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert KM, Blossom SJ, Erickson SW, Broadfoot B, West K, Bai S, Li J, Cooney CA. Chronic exposure to trichloroethylene increases DNA methylation of the Ifng promoter in CD4+ T cells. Toxicol Lett. 2016;260:1–7. doi: 10.1016/j.toxlet.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–81. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 41.Gilbert KM, Pumford NR, Blossom SJ. Environmental contaminant trichloroethylene promotes autoimmune disease and inhibits T-cell apoptosis in MRL+/+ mice. J Immunotox. 2006;3:263–7. doi: 10.1080/15476910601023578. [DOI] [PubMed] [Google Scholar]
- 42.Boyle P, Clement K, Gu H, Smith ZD, Ziller M, Fostel JL, Holmes L, Meldrim J, Kelley F, Gnirke A, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13:R92–13. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87–13. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JH, Park SJ, Kenta N. An integrative approach for efficient analysis of whole genome bisulfite sequencing data. BMC Genomics. 2015;16(Suppl. 12):S14. doi: 10.1186/1471-2164-16-S12-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziller MJ, Stamenova EK, Gu H, Gnirke A, Meissner A. Targeted bisulfite sequencing of the dynamic DNA methylome. Epigenetics Chromatin. 2016;9:55. doi: 10.1186/s13072-016-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambler G, Royston P. Fractional polynomial model selection procedures: investigation of Type I error rate. J Stat Simul Comput. 2001;69:89–108. [Google Scholar]
- 47.Elliott G, Hong C, Xing X, Zhou X, Li D, Coarfa C, Bell RJ, Maire CL, Ligon KL, Sigaroudinia M, et al. Intermediate DNA methylation is a conserved signature of genome regulation. Nat Commun. 2015;6:6363. doi: 10.1038/ncomms7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacoby M, Gohrbandt S, Clausse V, Brons NH, Muller CP. Interindividual variability and co-regulation of DNA methylation differ among blood cell populations. Epigenetics. 2012;7:1421–34. doi: 10.4161/epi.22845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaneko KJ, Rein T, Guo ZS, Latham K, DePamphilis ML. DNA methylation may restrict but does not determine differential gene expression at the Sgy/Tead2 locus during mouse development. Mol Cell Biol. 2004;24:1968–82. doi: 10.1128/MCB.24.5.1968-1982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moarii M, Boeva V, Vert JP, Reyal F. Changes in correlation between promoter methylation and gene expression in cancer. BMC Genomics. 2015;16:873–1994. doi: 10.1186/s12864-015-1994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bendall ML, Luong K, Wetmore KM, Blow M, Korlach J, Deutschbauer A, Malmstrom RR. Exploring the roles of DNA methylation in the metal-reducing bacterium Shewanella oneidensis MR-1. J Bacteriol. 2013;195:4966–74. doi: 10.1128/JB.00935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willems E, Guerrero-Bosagna C, Decuypere E, Janssens S, Buyse J, Buys N, Jensen P, Everaert N. Differential expression of genes and DNA methylation associated with prenatal protein undernutrition by albumen removal in an avian model. Sci Rep. 2016;6:20837. doi: 10.1038/srep20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bysani M, Perfilyev A, de Mello VD, Ronn T, Nilsson E, Pihlajamaki J, Ling C. Epigenetic alterations in blood mirror age-associated DNA methylation and gene expression changes in human liver. Epigenomics. 2017;9:105–22. doi: 10.2217/epi-2016-0087. [DOI] [PubMed] [Google Scholar]
- 54.Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan MS, Kawaji H, Lassmann T, Harbers M, Forrest AR, Bajic VB. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics. 2014;15:119–5. doi: 10.1186/1471-2164-15-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–67. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. 2013;5:553–68. doi: 10.2217/epi.13.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long MD, Smiraglia DJ, Campbell MJ. The genomic impact of DNA CpG methylation on gene expression; relationships in prostate cancer. Biomolecules. 2017;7:E15. doi: 10.3390/biom7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinheiro I, Heard E. X chromosome inactivation: new players in the initiation of gene silencing. F1000Res. 2017;6:1–10. doi: 10.12688/f1000research.10707.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balaton BP, Cotton AM, Brown CJ. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol Sex Differ. 2015;6:35. doi: 10.1186/s13293-015-0053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet. 2015;24:1528–39. doi: 10.1093/hmg/ddu564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan AA, Lee AJ, Roh TY. Polycomb group protein-mediated histone modifications during cell differentiation. Epigenomics. 2015;7:75–84. doi: 10.2217/epi.14.61. [DOI] [PubMed] [Google Scholar]
- 62.Eskeland R, Freyer E, Leeb M, Wutz A, Bickmore WA. Histone acetylation and the maintenance of chromatin compaction by Polycomb repressive complexes. Cold Spring Harb Symp Quant Biol. 2010;75:71–8. doi: 10.1101/sqb.2010.75.053. [DOI] [PubMed] [Google Scholar]
- 63.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–35. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 64.Tanay A, O’Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A. 2007;104:5521–6. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCabe MT, Lee EK, Vertino PM. A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. Cancer Res. 2009;69:282–91. doi: 10.1158/0008-5472.CAN-08-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thornton SR, Butty VL, Levine SS, Boyer LA. Polycomb Repressive Complex 2 regulates lineage fidelity during embryonic stem cell differentiation. PLoS ONE. 2014;9:e110498. doi: 10.1371/journal.pone.0110498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lynch MD, Smith AJ, De GM, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–29. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–38. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kulis M, Merkel A, Heath S, Queiros AC, Schuyler RP, Castellano G, Beekman R, Raineri E, Esteve A, Clot G, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–56. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oravecz A, Apostolov A, Polak K, Jost B, Le GS, Chan S, Kastner P. Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat Commun. 2015;6:8823. doi: 10.1038/ncomms9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Kinkel S, Maksimovic J, Bandala-Sanchez E, Tanzer MC, Naselli G, Zhang JG, Zhan Y, Lew AM, Silke J, et al. The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood. 2014;124:737–49. doi: 10.1182/blood-2013-12-544106. [DOI] [PubMed] [Google Scholar]
- 72.Onodera A, Tumes DJ, Watanabe Y, Hirahara K, Kaneda A, Sugiyama F, Suzuki Y, Nakayama T. Spatial interplay between polycomb and trithorax complexes controls transcriptional activity in T lymphocytes. Mol Cell Biol. 2015;35:3841–53. doi: 10.1128/MCB.00677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang XP, Jiang K, Hirahara K, Vahedi G, Afzali B, Sciume G, Bonelli M, Sun HW, Jankovic D, Kanno Y, et al. EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci Rep. 2015;5:10643. doi: 10.1038/srep10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sarmento O, Xiong Y, Sun Z, Svingen P, Bamidele A, Smyrk T, Nair A, Baheti S, McGovern D, Friton J, et al. O-015 YI alterations in the FOXP3-EZH2 pathway associates with increased susceptibility to colitis in both mice and human. Inflamm Bowel Dis. 2016;22(Suppl. 1):S5–6. [Google Scholar]
- 75.Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, Hosokawa H, Koseki H, Tokoyoda K, Suzuki Y, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39:819–32. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 76.Nestor CE, Lentini A, Hagg NC, Gawel DR, Gustafsson M, Mattson L, Wang H, Rundquist O, Meehan RR, Klocke B, et al. 5-Hydroxymethylcytosine remodeling precedes lineage specification during differentiation of human CD4(+) T cells. Cell Rep. 2016;16:559–70. doi: 10.1016/j.celrep.2016.05.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao M, Wang J, Liao W, Li D, Li M, Wu H, Zhang Y, Gershwin Me, Lu Q. Increased 5-hydroxymethylcytosine in CD4(+) T cells in systemic lupus erythematosus. J Autoimmum. 2016;69:64–73. doi: 10.1016/j.jaut.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 78.Liu T, Sun J, Wang Z, Yang W, Zhang H, Fan C, Shan Z, Teng W. Changes in the DNA methylation and hydroxymethylation status of the intercellular adhesion molecule 1 gene promoter in thyrocytes from autoimmune thyroiditis patients. Thyroid. 2017;27:838–45. doi: 10.1089/thy.2016.0576. [DOI] [PubMed] [Google Scholar]
- 79.Haffner MC, Pellakuru LG, Ghosh S, Lotan TL, Nelson WG, De Marzo AM, Yegnasubramanian S. Tight correlation of 5-hydroxymethylcytosine and Polycomb marks in health and disease. Cell Cycle. 2013;12:1835–41. doi: 10.4161/cc.25010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.