Abstract
Purpose
Integrating genomic sequencing in clinical care requires standardization of variant interpretation practices. The Clinical Genome Resource has established expert panels to adapt the ACMG/AMP classification framework for specific genes and diseases. The Cardiomyopathy Expert Panel (CMP-EP) selected MYH7, a key contributor to inherited cardiomyopathies, as a pilot gene to develop a broadly applicable approach.
Methods
Expert revisions were tested with 60 variants using a structured double review by pairs of clinical and diagnostic laboratory experts. Final consensus rules were established via iterative discussions.
Results
Adjustments represented disease/gene-informed specifications (12) or strength adjustments of existing rules (5). Nine rules were deemed not applicable. Key specifications included quantitative frameworks for minor allele frequency thresholds, the use of segregation data, and a semi-quantitative approach to counting multiple independent variant occurrences where fully controlled case-control studies are lacking.
Initial inter-expert classification concordance was 93%. Internal data from participating diagnostic laboratories changed the classification of 20% of the variants (n=12), highlighting the critical importance of data sharing.
Conclusions
These adapted rules provide increased specificity for use in MYH7-associated disorders in combination with expert review and clinical judgment and serve as a stepping stone for genes and disorders with similar genetic and clinical characteristics.
Keywords: myosin heavy chain 7, cardiomyopathy, variant interpretation, HCM, ClinGen
INTRODUCTION
Determining the clinical significance of sequence variants is a complex process that involves gathering and assigning relative weights to a multitude of data gathered from a diverse set of resources. Variant classification has evolved in a de-centralized fashion leading to a multitude of approaches, most developed by molecular diagnostic laboratories for internal use.
As sequencing tests become a routine tool in managing health, it is vital to harmonize and centralize knowledge and approaches. Key advancements included the creation of NCBI’s ClinVar database (www.ncbi.nlm.nih.gov/clinvar/), which has quickly become a valuable centralized resource for clinically classified variants and the NHGRI-funded Clinical Genome Resource (ClinGen, www.clinicalgenome.org), which serves as a body for managing and centralizing clinically relevant genomic knowledge. The historic lack of standardization is a major contributor for interpretation differences, which have been revealed by increased data sharing through ClinVar1–3. Many of these differences represent misclassifications which can have serious consequences, especially for medically actionable variants as illustrated by Gaba et al. 20164 where ICDs were implanted based on the incorrect classification of a variant as causative for long QT syndrome.
The American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) have responded to the urgent need for an updated variant classification framework by releasing a landmark guidance document5, which has since been adopted by many US and international laboratories6. However, this framework was designed to have universal applicability, and therefore requires significant expertise to be applied correctly for specific diseases and genes.
Lack of clinical domain expert knowledge is emerging as a key contributor to incorrect classifications2,7. Besides adding disease and gene specifications for certain rules (including flagging rules that are not applicable), there is a critical need to provide more granularity surrounding frequency thresholds that drive the benign spectrum of rules, to specify thresholds for assigning increasing weight depending on the degree of segregation with disease, and to provide guidance surrounding the use of functional data7. Some studies have proposed approaches and solutions8–11; however, systematic expert-led efforts are needed to create disease- and/or gene-specific derivatives of the original ACMG/AMP framework. ClinGen has emerged as a critical facilitator in this domain and has established a rich infrastructure including disease-specific expert working groups that have been charged with accomplishing this goal3.
Here we report the work of ClinGen’s Inherited Cardiomyopathy Expert Panel (CMP-EP), which adapted the ACMG/AMP framework for use in myosin heavy chain 7 (MYH7)-associated cardiomyopathies with the aim of improving consistency for variant interpretation and expert curation of reported MYH7 variants for submission to ClinVar (3 star). These disorders include hypertrophic, dilated and restrictive cardiomyopathy (HCM, DCM, and RCM), which are collectively among the most prevalent Mendelian conditions and affect 1 in 200–500 individuals12,13. MYH7 is the second most common inherited cause of HCM and third most common inherited cause of DCM, primarily due to missense variants that are dominantly inherited, though de novo variants have been reported 14,15.
MATERIALS AND METHODS
ClinGen’s Inherited Cardiomyopathy Expert Panel (CMP-EP)
The CMP-EP operates under the umbrella of the Cardiovascular Domain Working Group. Members were selected to provide a balanced representation of expertise in clinical cardiology, clinical research, molecular diagnostics, genetic counseling and genomic medicine. Additional emphasis was placed on global representation (USA, Great Britain, Netherlands, and Australia) to lay the foundation for international harmonization.
Decision making framework
A subset of the CMP-EP formed a core task team with clinical as well as molecular diagnostic expertise and representation from three institutions (MAK: Partners HealthCare Laboratory for Molecular Medicine, CC: Stanford University, AM: Ohio State University). This task team reviewed the original ACMG/AMP framework5 and developed proposed changes to adapt them for MYH7. Proposed adaptations were discussed by the CMP-EP using conference calls, email or electronic surveys to arrive at consensus decisions (Text S1).
Project design
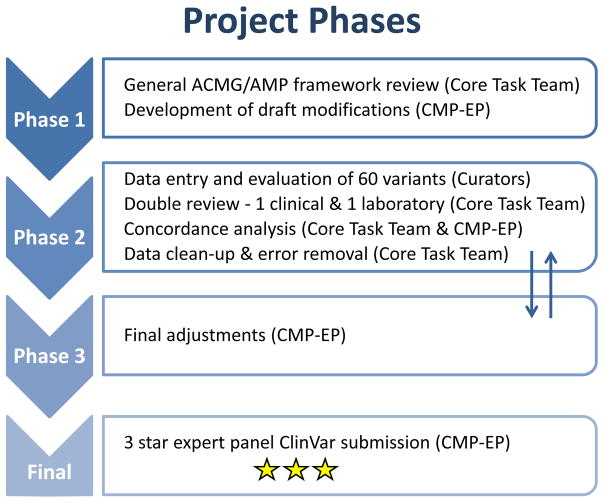
An initial classification exercise included 10 variants and served as a foundation for subsequent rule adaptations. These variants were independently scored by members of the core task team using the original ACMG/AMP framework as well as their own institutions’ variant interpretation criteria. Subsequent work was carried out as shown in Figure 1. Draft rules were applied to 60 MYH7 variants (including the initial 10), which were selected a) as a representative spectrum of variant types for MYH7, b) to test as many rules as possible, c) to cover a range of classifications and d) to include discrepant ClinVar assertions. Evidence for each variant was compiled by a set of curators (KO, KS, MT) and applicable rules were selected. Each variant was then reviewed by two task team members (one with clinical expertise and one with laboratory expertise; MAK, JB, CC, AM) and conflicts resulting from clerical errors and rule misuse were corrected. A discussion with the full CMP-EP was triggered when reviewers did not agree or raised concerns regarding the “fit” of a rule. Variants representing controversial items were used to drive additional rule adjustments by the CMP-EP. Adjusted rules were disseminated to the CMP-EP to allow for a final comment period. Additional guidance was provided by ClinGen’s Sequence Variant Interpretation working group that harmonizes framework adaptation efforts by various clinical domain working groups (https://www.clinicalgenome.org/working-groups/genomic-variant-workgroup/sub-groups/sequence-variant-interpretation-wg/). Variants and the adapted rule framework were submitted to ClinVar under a 3-star (expert panel reviewed) status.
Figure 1. Summary of ClinGen Inherited Cardiomyopathy Expert Panel (CMP-EP) involvement.
Phase 1: Disease-/gene- and other specifications made to established ACMG/AMP framework. Phase 2: Selection and review of 60 pilot variants by two independent reviewers. Classifications were then compared and discussed to resolve any conflicts. Phase 3: Additional adjustments to variant classifications. Final: Expert panel variant classifications submitted to ClinVar for public accessibility. Expert panel ratings in ClinVar are denoted with a three star rating.
Curation data sources and data collection method (publicly available data)
Variants were curated using the variant assessment process and data sources described in Duzkale et al.16 (Table S1). All databases accessed 08-09/2015; reference transcript: NM_000257.3).
Additional case level data
The number of independent observations of the 60 pilot variants, basic phenotype information and segregation with disease was available from several diagnostic or research cohorts (Partners HealthCare Laboratory for Molecular Medicine; Invitae, Inc; the Sarcomeric Human Cardiomyopathy Registry - SHaRe, https://theshareregistry.org/; the Australian Genetic Heart Disease Registry - http://www.heartregistry.org.au/; the NIHR Cardiovascular Biomedical Research Unit at Royal Brompton Hospital and Imperial College London; the National Heart Centre Singapore). Consideration was taken to account for cases that were known or suspected to be part of more than one cohort.
Statistical approaches
Multiple proband rules (PS4, PS4_Moderate, PS4_Supporting)
The current ACMG/AMP framework assigns weight to increased prevalence of a variant in cases compared to controls (PS4), but does not provide guidance for combining separate studies reporting the same variant. We created thresholds of proband occurrences that qualify for supporting, moderate, and strong weight as follows and outlined in more detail in Table S2. This approach represents a “quasi case-control” analysis. Proband cohort sizes were modeled based on the cohorts available for this study, focusing on Caucasian and African American ancestries (reflecting the main ancestries represented among cohorts available in this study). The corresponding ExAC cohorts were used as proxies for healthy controls. Odds ratios and p-values were computed for 1–15 probands carrying a variant assuming absence/extreme rarity in controls (rule PM2 is met) using the two-sided Fisher’s exact test to evaluate the null hypothesis of conditional independence. This approach has limitations (for details see S table 2) but provides a practical means for clinical variant assessment workflows.
Segregation thresholds
Since cardiomyopathies are characterized by variable age at onset and reduced penetrance, LOD scores were estimated by counting the number of informative meioses separating affected variant carriers across all families with this variant (i.e. without considering unaffected individuals). Affected non-carriers indicated non-segregation. Under these conditions the calculation of LOD (logarithm of odds) score simplifies to: LOD = log10(2n) where n is the number of informative meiosis observed.
RESULTS AND DISCUSSION
MYH7 is a major contributor to several cardiomyopathies (HCM, DCM, RCM). Due to their high combined prevalence and severe health outcomes, MYH7 is one of the most frequently tested genes in a clinical setting. As a first step towards modifying the ACMG/AMP framework, the CMP-EP conducted a preliminary exercise comparing inter-expert concordance for 10 representative MYH7 variants classified using the ACMG/AMP framework alongside the experts’ respective institutional criteria. As in similar previously reported studies7, this revealed low inter-expert concordance (20%) for the ACMG/AMP framework compared to high concordance (90%) when institutional criteria were used (data not shown). The subsequent adaptation of the ACMG/AMP framework for MYH7 was carried out as shown in Figure 1.
Summary of specifications
Two characteristics of the ACMG/AMP framework had a major influence on the specifications made. Due to its design to have general applicability across all Mendelian disorders, some rules are overly conservative in the setting of a specific disorder. This is best illustrated by BA1, the allele frequency threshold above which a variant is considered benign. For many Mendelian conditions, the default threshold of 5% is orders of magnitude higher than it needs to be. In addition, the framework contains several areas of vagueness (such as the absence of quantitative guidance for increasing the weight depending on the extent of “segregation with disease”. Table 1 provides a summary of the adapted ACMG/AMP framework for use in MYH7-associated disease along with instructions on how to combine the rules. Of the original 28 ACMG/AMP rules, nine were deemed not applicable and another 12 required disease- and/or gene-specific adjustments. Five rules were given modified strength criteria. A full description of rules with additional detail is provided in Text S2. The following sections highlight approaches and key specifications.
Table 1.
Summary of the adapted ACMG/AMP pathogenic and benign criteria.
| Pathogenic Criteria | Combination of Criteria Needed to Meet Classification | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rule | Specification Type | Rule Description | Pathogenic | Likely Pathogenic | |||||||
| VS | PVS1 | Removed | Null variant in gene with established LOF as disease mechanism | ||||||||
| STRONG | PS1 | No change | Different nucleotide change (same amino acid) as a previously established pathogenic variant | 2 | 1 | 1 | 1 | 1 | |||
| PS2 | Disease/gene | De novo (paternity confirmed) in a patient with disease and no family history | |||||||||
| PS3 | Disease/gene | Functional studies of mammalian knock-in models supportive of a damaging effect on the gene or gene product | |||||||||
| PS4 | Disease/gene | Prevalence of the variant in affected individuals is significantly increased compared to the prevalence in controls -OR- Variant identified in ≥15 probands with consistent phenotypes | |||||||||
| PP1_Strong | Modif. Strength | Variant segregates with ≥7 meioses | |||||||||
| MODERATE | PM1 | Disease/gene | Hotspot/est. functional domain (amino acids 181–937) without benign variation | 3 | 2 | 1 | 1 | 3 | 2 | ||
| PM2 | Disease/gene | Absent/extremely rare (<0.004%) from large population studies | |||||||||
| PM3 | Removed | Detected in trans with a pathogenic variant (recessive) | |||||||||
| PM4 | No change | Protein length changes due to in-frame deletions/insertions of any size in a non-repeat region or stop-loss variants | |||||||||
| PM5 | No change | Missense change at an amino acid residue where a different missense change previously established as pathogenic | |||||||||
| PM6 | Disease/gene | Confirmed de novo without confirmation of paternity | |||||||||
| PVS1_Moderate | Modif. Strength | Null variant in gene with evidence supporting LOF as disease mechanism | |||||||||
| PS4_Moderate | Modif. Strength | Variant identified in ≥6 probands with consistent phenotypes | |||||||||
| PP1_Moderate | Modif. Strength | Variant segregates in ≥5 meioses | |||||||||
| SUPPORTING | PP1 | Disease/gene | Variant segregates in ≥3 meioses | 2 | 4 | 2 | 2 | ||||
| PP2 | Removed | Missense variant in a gene that has a low rate of benign missense variation and where missense variants are a common mechanism of disease | |||||||||
| PP3 | No change | Multiple lines of computational evidence support a deleterious effect on the gene or gene product | |||||||||
| PP4 | Removed | Phenotype specific for disease with single genetic etiology | |||||||||
| PP5 | Removed | Reputable source reports as pathogenic | |||||||||
| PS4_Supporting | Modif. strength | Variant identified in ≥2 probands with consistent phenotypes | |||||||||
| Benign Criteria | Combination of Criteria Needed to Meet Classification | ||||||
|---|---|---|---|---|---|---|---|
| Rule | Specification Type | Rule Description | Benign | Likely Benign | |||
| SA | BA1 | Disease/gene | Allele frequency is ≥0.1% based on the filtering allele frequency (FAF) in ExAC | 1 | |||
| STRONG | BS1 | Disease/gene | Allele frequency is ≥0.02% based on the filtering allele frequency (FAF) in ExAC provided there is no conflicting information | 2 | 1 | ||
| BS2 | Removed | Observed in healthy adult with full penetrance expected at an early age | |||||
| BS3 | No change | Functional studies of mammalian knock-in models supportive of no damaging effect on protein function or splicing | |||||
| BS4 | Disease/gene | Non-segregation in affected members of a family | |||||
| SUPPORTING | BP1 | Removed | Missense variant in gene where only LOF causes disease | 2 | |||
| BP2 | Disease/gene | Observed as comp het (in trans) or double het in genes with overlapping function (e.g. sarcomere genes) without increased disease severity -OR- Observed in cis with a pathogenic variant in any inheritance pattern | |||||
| BP3 | Removed | In-frame deletions/insertions in a repetitive region without a known function | |||||
| BP4 | No change | Multiple lines of computational evidence suggest no impact on gene or gene product | |||||
| BP5 | Disease/gene | Variant found in a case with an alternate molecular basis for disease | |||||
| BP6 | Removed | Reputable source reports as benign | |||||
| BP7 | No change | A silent variant for which splicing prediction algorithms predict no impact to the splice consensus sequence nor the creation of a new splice site -AND- the nucleotide is not highly conserved | |||||
VS = Very Strong. SA = Stand alone. Removed - not applicable to MYH7-associated disease. Modif. strength: Modified rule strength. Numbers under each classification refer to the number of rules with that strength required to classify the variant as its header category. Example: A likely pathogenic classification may be made with 1 piece of strong and 2 pieces of supporting evidence.
Disease- and gene- specific adaptations
Minor allele frequency (MAF) driven rules (BA1, BS1, and PM2)
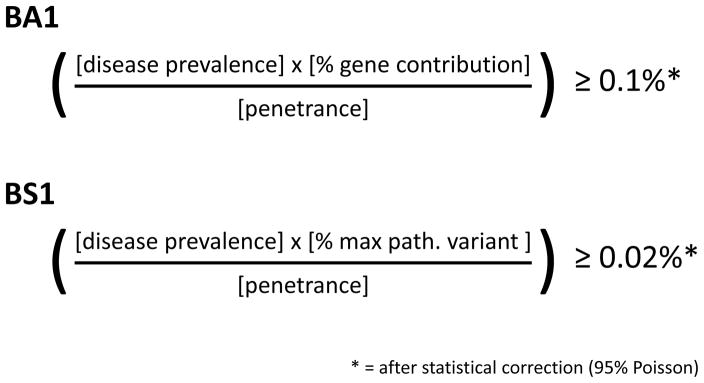
The CMP-EP modified BA1 using extremely conservative values for disease prevalence, gene contribution, and estimated penetrance of MYH7 variants (Figure 2, for additional detail see Text S3). Across MYH7-associated diseases, prevalence values were compiled from the literature and the most conservative one was selected to derive a threshold that is applicable to all (1/200). Penetrance was set deliberately low at 30%. To control for uncertainty in estimated MAFs of very rare variants in smaller cohorts, a statistical correction (95% Poisson distribution) was added11. This correction (termed “filtering allele frequency”; FAF) is now available for each variant for all ExAC cohorts, http://exac.broadinstitute.org/). The final BA1 threshold was set at ≥0.1%. A variant that is observed at this MAF in the general population could theoretically be pathogenic under the assumption that it is the ONLY pathogenic variant. Because of the extremely conservative approach, this threshold is likely two orders of magnitude higher than the true threshold and can be used safely in a diagnostic setting.
Figure 2. Derivation of allele frequency thresholds for rules BA1 and BS1.
Disease prevalence = 1/200 individuals (1/400 chromosomes). Penetrance = 30%. % gene contribution = 10.6%. % maximum pathogenic variant contribution (max path. variant) = 2%.
The threshold for BS1 (allele frequency too high for disorder) was derived using the same approach except that allelic heterogeneity was now considered (i.e. gene contribution was replaced by the maximum credible variant contribution, which encompasses both gene and variant contribution, Figure 2). This gives rise to a more aggressive threshold with less room for error, which is acceptable given the less definitive classification. The prevalence of the most common pathogenic cardiomyopathy variant was used to define the maximum theoretical population frequency for a pathogenic allele (Whiffin et al.11 and Text S3). The final BS1 threshold was set at a FAF of ≥0.02%. A variant observed at this MAF can be assumed to be likely benign provided that there is no substantial contradictory evidence supporting pathogenicity. Allowing a variant to reach a likely benign classification based on BS1 alone represents a revision of the original ACMG/AMP framework by ClinGen’s Sequence Variant Interpretation Working Group 17. The CMP-EP added the following safeguard to BS1: Because our current knowledge of the genetic architecture of HCM is largely derived from predominantly Caucasian proband cohorts, the threshold should only be applied to populations where sufficient numbers of probands have been deeply analyzed, leaving the possibility open that more common pathogenic variants may exist in less well-characterized populations. Both, BA1 and BS1 were tested in a large diagnostic cohort (LMM internal data) and no currently known likely pathogenic or pathogenic MYH7 variant would be misclassified as Likely Benign or Benign (data not shown).
The CMP-EP recommends activating rule PM2 (absent in population databases) when the filtering allele frequency is <0.004%. The PM2 threshold used more realistic prevalence and penetrance values (1/500, 50%), which typically represents a very small/negligible number of alleles in the ExAC database.
Segregation with disease (PP1, PP1_Moderate, PP1_Strong)
The ACMG/AMP framework assigns supporting evidence to co-segregation (PP1) and states that higher weight can be assigned with an increasing degree of segregation, but does not define thresholds. The CMP-EP specified three levels of evidence using autosomal dominant likelihood ratios of 10 (3 meioses, LOD 0.9), 30 (5 meioses, LOD 1.5) and 100 (7 meioses, LOD 2.1) to count as supporting, moderate and strong evidence provided that PM2 (absent or rare in large population cohorts) is met. Finally, the CMP-EP waived the ACMG/AMP recommendation for demonstrating segregation in >1 family given that MYH7 is a well-established disease.
Increased prevalence of variant in probands versus controls (PS4, PS4_Moderate, PS4_Supporting)
Rule PS4 is designed for variants that are significantly enriched in probands. While traditional case-control studies using phenotyped case and control cohorts are typically not available for rare, Mendelian variants, it is not uncommon that multiple separate studies report the same variant in cohorts of modest size. To be able to utilize combined proband counts across different studies, the CMP-EP created a framework using the ExAC cohort as a proxy for healthy controls. As with segregation, evidence levels were assigned based on likelihood ratios with ideal target thresholds being 10 (supporting), 30 (moderate) and 100 (strong). To simplify use in current molecular diagnostic practice, where statistical tools are typically not embedded in routine workflows, conservative universal thresholds were set to proband counts of ≥2 (supporting), ≥6 (moderate) and ≥15 (strong). Moderate (but acceptable) deviations from the targeted odds ratios (ORs) for the two main racial cohorts used (Non Finnish European: NFE and African American: AFR) were deemed acceptable in return for ease of use (supporting: OR [AFR|NFE]=[10.4|13.4], moderate: OR [AFR|NFE]=[31.3|40.1], and strong; OR [AFR|NFE]=[79.1|100]). To apply these rules, rule PM2 must be met. These proband counts are extremely conservative to balance the limitations of the underlying statistical approach and the risk of double-counting probands that is inherent when working with published data.
De novo occurrence
Rule PS2 (de novo in a patient with the disease and no family history, paternity and maternity confirmed): The CMP-EP removed the requirement to prove maternity as the likelihood of undisclosed non-maternity (e.g. due to surrogacy) was considered rare. In addition, the following specifications were added: (a) “no family history” is defined as the absence of diagnosed disease or suspicious findings in a three-generation pedigree and (b) both parents must be genotype and phenotype negative after a thorough clinical evaluation that ideally includes a combination of ECG and echo or CMR for maximum sensitivity (Text S2). When paternity has not been established, de novo occurrence receives moderate weight (PM6) but the CMP-EP allowed upgrading to “strong” (PS2) when ≥3 de novo occurrences have been documented.
Dealing with ambiguous phenotypes when counting multiple or segregation with disease
Left ventricular non-compaction (LVNC)
It is currently debated whether LVNC is a distinct cardiomyopathy, a morphological trait shared by different cardiomyopathies or an entirely benign structural variant 18–20. The CMP-EP therefore recommended that individuals with isolated LVNC (no additional cardiomyopathy such as HCM or DCM present) should NOT be added to proband or segregation counts in the context of HCM and DCM. End-stage HCM: Due to the challenge in distinguishing between end-stage HCM and DCM, a conservative approach was taken to not include DCM cases in proband or segregation counts for HCM variants, unless earlier clinical evidence supported the HCM phenotype.
Functional data
The ACMG/AMP framework assigns strong weight to well-established in vitro or in vivo functional studies that are supportive of a damaging effect on the gene or protein (PS3). The normal function of the protein encoded by MYH7 is to convert energy from ATP hydrolysis into mechanical force to allow for muscle contractility21. After reviewing the assays used to measure this function for the 60 pilot variants, the CMP-EP determined that “strong functional evidence” can only be provided by a mammalian variant-specific knock in model (Table S3). Other in vivo models that alter dosage of the normal protein (transgenic or knockout mice, zebrafish knock-downs) are not acceptable as they do not provide clues about the importance of a particular variant. Typically performed MYH7 in vitro assays were generally deemed to have relatively low positive predictive value (either due to low accuracy or low correlation between an observed effect and the ability to cause disease) and are therefore currently not considered strong evidence. However, the CMP-EP recognized that in the event that in vitro models that accurately predict the effect in vivo become available, their weight can be reconsidered.
Incorporating protein domain information
The ACMG/AMP framework assigns supporting evidence of pathogenicity to missense variants in a gene that has a low rate of missense variation, provided that missense variants are a common mechanism of disease (PP2) and moderate evidence for variants located in a hotspot and/or critical domain without benign variation (PM1). It is well-established that missense variants in MYH7 are the predominant class of pathogenic alleles. The ExAC database (http://exac.broadinstitute.org/22) provides a metric to express the deviation of variant counts from the expected number (constraint score). Positive scores indicate intolerance to variation, which is the case for MYH7 (z=6.54). However, recent studies suggest that this is driven by statistically significant clustering of pathogenic variants in the head region (p<3×10−15, amino acids 181 – 937, NM_000257)23,24. The CMP-EP concluded that this evidence was most appropriately weighted as moderate through application of the critical domain rule (PM1). PP2 was deemed no longer applicable because it does not apply to variants outside the head domain and to avoid double counting the same evidence twice for variants in the head domain.
Other modifications
Rules deemed not applicable
Four rules of the pathogenic framework (PVS1, PM3, PP2, PP4) and three rules of the benign framework (BS2, BP1, BP3) were deemed non-applicable either entirely or in the original strength level suggested and two additional rules were removed for other reasons. Select rules are discussed here and a full list of not applicable and removed rules, along with a summary of the rationale, are provided in Text S2). PVS1 (null variant in a gene where loss of function (LOF) is a known disease mechanism): MYH7 LOF variants are very rare and their contribution to inherited cardiomyopathy is incompletely understood. While there is currently no evidence for a disease-causing role in the heterozygous state, compound heterozygosity of LOF variants along with missense variants can lead to extremely severe presentations, mimicking recessive inheritance25,26. The CMP-EP assigned moderate weight to a LOF variant (PVS1_Moderate, Table 1), which yields a classification of VUS in the absence of case level data supporting pathogenicity. PM3 (variant detected in trans with a pathogenic variant): While compound heterozygosity leading to a more severe phenotype has been documented, this rule was designed for traditional recessive inheritance.
Removed rules
(PP5/BP6: reputable source reports variant as pathogenic/benign, but evidence is not accessible): The CMP-EP decided expert curations should only be used if accompanied by the evidence used. Platforms such as ClinVar enable laboratories to share the evidence on which an interpretation is based, and the CMP-EP encourages this practice.
Performance of the new MYH7 specific rules
Sixty pilot variants were selected to cover a broad spectrum of scenarios, while focusing on the types of data most commonly encountered (Figure S1, Table S4). The majority of the rules were applied at least once (Figure S2). The rules supporting benign evidence were used the least, reflecting the intentional bias towards the pathogenic spectrum. After application of the modified ACMG/AMP framework by two independent expert reviewers, 8/60 variants were discordantly classified. Factors underlying discordance revealed two causes: (a) 4 data errors (differences in the data used, such as arriving at different proband or segregation counts), and (b) 4 rule applications that deviated from the intended use (Figure S3). After correcting for data errors, concordance was 93%. Rules requiring a high level of expert knowledge were more vulnerable to error (e.g. those relying on segregation counts), exposing a significant overhead associated with training curators. All discrepancies were resolved upon review by the task team and did not require full CMP-EP review. This represents a significant improvement compared to the initial 10 variant pilot. The pilot variants (31 associated with HCM and 6 with DCM) have been submitted to ClinVar with a “3-star” (expert panel approved) label. ClinVar IDs are listed in Table S4.
Laboratory internal data has a high impact and shows a critical need to enable sharing of case-level information
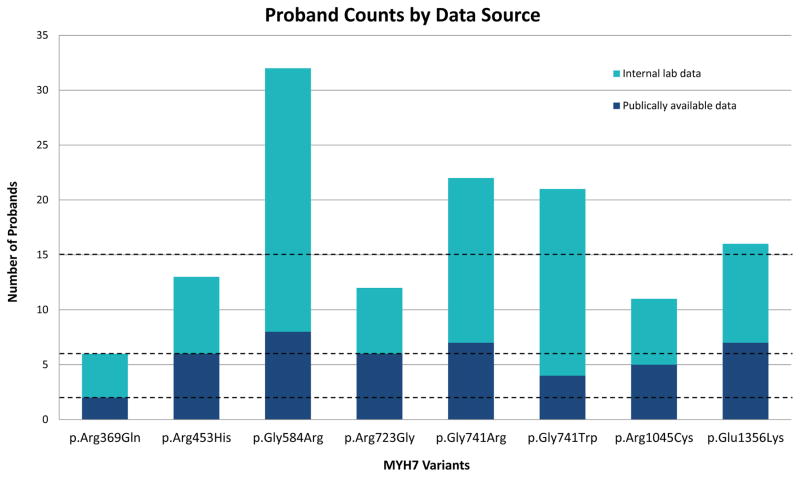
When available, the CMP-EP also reviewed internal laboratory data from several of its members that was unpublished at the time of curation (see materials and methods). For 25 pilot variants, this impacted the application or strength for at least one of the multiple proband, segregation, or de novo rules and impacted the final classification of 12 variants (48%). Seven variants were upgraded from “likely pathogenic” to “pathogenic” and another five increased from “uncertain significance” to “likely pathogenic” (Figure 3, Table S5). This is a powerful demonstration of the impact of historically de-centralized, private data and illustrates an urgent need to incentivize data sharing as well as to establish infrastructure and standards to share and aggregate such case-level data.
Figure 3. Impact of data sharing on proband counts.
Increased proband counts obtained from internal lab data changed the variant classification for eight variants. Hashed lines correspond to the thresholds for supporting (≥2, PS4_Supporting), moderate (≥6, PS4_Moderate), and strong (≥15, PS4). Publically available data was collected from PubMed, Google, HGMD Professional, ClinVar, and relevant locus-specific variant databases. Internal laboratory data was collected from the Partners HealthCare Laboratory for Molecular Medicine, Invitae, Inc, the Sarcomeric Human Cardiomyopathy Registry – SHaRe (https://theshareregistry.org/), the Australian Genetic Heart Disease Registry (http://www.heartregistry.org.au/), the NIHR Cardiovascular Biomedical Research Unit at Royal Brompton Hospital and Imperial College London, and the National Heart Centre Singapore.
Clinical judgment
Although the ACMG/AMP framework represents a major step forward in our ability to classify variants in the context of Mendelian disease, it will need continued improvement and refinement as our understanding of these diseases develops. It is premature to expect that even this improved, rule-based framework will function without additional clinical judgment, which is a hallmark of medical practice. Clinical judgment represents the capability of experts to consider additional, as of yet un-quantified factors, and adjust the weighting of specific evidence elements intuitively based on deep experience and exposure to many cases over time. Prior studies have begun to quantitate this phenomenon in the context of variant classification7. In our pilot, judgment led to overriding the classification for two MYH7 variants where the CMP-EP upgraded the rule-based classification of VUS to Likely Pathogenic: p.Arg1420Trp (PM2: absent/rare in controls; PS4_Moderate: present in 11 probands; PP3: computational predictions favor pathogenic) and p.Arg1909Pro (PM2: absent/rare in controls; PM6: de novo occurrence without confirmed paternity; PP3: computational predictions favor pathogenic). Considering the extremely conservative approaches used for several rules, the CMP-EP felt that the evidence for both variants was sufficiently borderline (just one additional supporting criterion required to meet the criteria for likely pathogenic) and the available evidence provided additional specificity not accounted for by the rules to warrant this upgrade (p.Arg1420Trp: additional probands not counted due to conservative nature of the approach; p.Arg1909Pro: phenotype included DCM and myopathy, and additional segregations).
LIMITATIONS
Proband cohorts used to derive data on multiple occurrences of variants largely represent diagnostic cases from broad referral populations where clinical diagnoses were based on information provided by ordering healthcare providers.
Allele frequency thresholds were developed assuming autosomal dominant inheritance and were deliberately designed as overly stringent to minimize the risk for false positive interpretations, which can cause harm to the patients and their families.
In accordance with the ACMG/AMP parent framework, these rules assume a single variant disease paradigm.
CONCLUSIONS AND FUTURE DIRECTIONS
The CMP-EP created modified ACMG/AMP variant classification rules that can be used for all MYH7-associated cardiomyopathies. It is expected that our framework will undergo iterative refinements catalyzed by its use, as well as continuous improvements made to the parent framework by the community and the CMP-EP will provide updated versions as needed. Future work will extend the MYH7 framework to other cardiomyopathy genes, which will likely require only minimal additional specifications.
Finally, the CMP-EP will establish a sustained curation process to apply the MYH7 rules to all variants presently in the public domain with the goal to submit 3-star (expert panel endorsed) variant classifications along with the associated evidence into ClinVar.
Supplementary Material
Acknowledgments
The CMP-EP thanks Lisa Kurtz for her help revising and submitting the manuscript. Research reported in this publication was supported by the NHGRI and funding from the NICHD and NCI (awards U41HG006834 (HR) and U01HG007437 (JB)). JI was supported by a National Heart Foundation of Australia Future Leader Fellowship (#100833). CS was supported by a National Health and Medical Research Council (NHMRC) Practitioner Fellowship (#1059156). JW was supported by the Wellcome Trust [107469/Z/15/Z]; British Heart Foundation; Medical Research Council (UK); and the NIHR Royal Brompton Biomedical Research Unit. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
A complete list of secondary authors from the ClinGen Cardiovascular Clinical Domain Working Group is provided in the Supplementary Appendix online.
Supplementary information is available at the Genetics in Medicine website.
COI statement:
The authors declare no conflict of interest.
References
- 1.Balmana J, Digiovanni L, Gaddam P, et al. Conflicting Interpretation of Genetic Variants and Cancer Risk by Commercial Laboratories as Assessed by the Prospective Registry of Multiplex Testing. J Clin Oncol. 2016;34(34):4071–4078. doi: 10.1200/JCO.2016.68.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pepin MG, Murray ML, Bailey S, Leistritz-Kessler D, Schwarze U, Byers PH. The challenge of comprehensive and consistent sequence variant interpretation between clinical laboratories. Genet Med. 2016;18(1):20–24. doi: 10.1038/gim.2015.31. [DOI] [PubMed] [Google Scholar]
- 3.Rehm HL, Berg JS, Brooks LD, et al. ClinGen--the Clinical Genome Resource. N Engl J Med. 2015;372(23):2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaba P, Bos JM, Cannon BC, et al. Implantable cardioverter-defibrillator explantation for overdiagnosed or overtreated congenital long QT syndrome. Heart Rhythm. 2016;13(4):879–885. doi: 10.1016/j.hrthm.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consensus statment on adoption of the American College of Medical Genetics and Genomics (ACMG) guidelines for sequence variant classification and interpretation. 2016 Nov 11; [press release]. http://www.acgs.uk.com/media/1032817/acgs_consensus_statement_on_adoption_of_acmg_guidelines__1_.pdf.
- 7.Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98(6):1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song W, Gardner SA, Hovhannisyan H, et al. Exploring the landscape of pathogenic genetic variation in the ExAC population database: insights of relevance to variant classification. Genet Med. 2016;18(8):850–854. doi: 10.1038/gim.2015.180. [DOI] [PubMed] [Google Scholar]
- 9.Jarvik GP, Browning BL. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am J Hum Genet. 2016;98(6):1077–1081. doi: 10.1016/j.ajhg.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiffin N, Minikel E, Walsh R, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med. 2017 doi: 10.1038/gim.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65(12):1249–1254. doi: 10.1016/j.jacc.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10(9):531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 14.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17(11):880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 15.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16(8):601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 16.Duzkale H, Shen J, McLaughlin H, et al. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84(5):453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavtigian SV, Greenblatt MS, Harrison SM, et al. Modeling the ACMG/AMP Variant Classification Guidelines as a Bayesian Classification Framework. 2017 doi: 10.1038/gim.2017.210. Submitted Genetics in Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weir-McCall JR, Yeap PM, Papagiorcopulo C, et al. Left Ventricular Noncompaction: Anatomical Phenotype or Distinct Cardiomyopathy? J Am Coll Cardiol. 2016;68(20):2157–2165. doi: 10.1016/j.jacc.2016.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towbin JA, Lorts A, Jefferies JL. Left ventricular non-compaction cardiomyopathy. Lancet. 2015;386(9995):813–825. doi: 10.1016/S0140-6736(14)61282-4. [DOI] [PubMed] [Google Scholar]
- 20.Sen-Chowdhry S, McKenna WJ. Left ventricular noncompaction and cardiomyopathy: cause, contributor, or epiphenomenon? Curr Opin Cardiol. 2008;23(3):171–175. doi: 10.1097/HCO.0b013e3282fdc939. [DOI] [PubMed] [Google Scholar]
- 21.Sivaramakrishnan S, Ashley E, Leinwand L, Spudich JA. Insights into human beta-cardiac myosin function from single molecule and single cell studies. J Cardiovasc Transl Res. 2009;2(4):426–440. doi: 10.1007/s12265-009-9129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19(2):192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homburger JR, Green EM, Caleshu C, et al. Multidimensional structure-function relationships in human beta-cardiac myosin from population-scale genetic variation. Proc Natl Acad Sci U S A. 2016;113(24):6701–6706. doi: 10.1073/pnas.1606950113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hougs L, Havndrup O, Bundgaard H, et al. One third of Danish hypertrophic cardiomyopathy patients with MYH7 mutations have mutations [corrected] in MYH7 rod region. Eur J Hum Genet. 2005;13(2):161–165. doi: 10.1038/sj.ejhg.5201310. [DOI] [PubMed] [Google Scholar]
- 26.Girolami F, Ho CY, Semsarian C, et al. Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. J Am Coll Cardiol. 2010;55(14):1444–1453. doi: 10.1016/j.jacc.2009.11.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.